Abstract
Faithful transmission of genetic material to daughter cells involves a characteristic temporal order of DNA replication, which may play a significant role in the inheritance of epigenetic states. We developed a genome-scale approach—Repli Seq—to map temporally ordered replicating DNA using massively parallel sequencing and applied it to study regional variation in human DNA replication time across multiple human cell types. The method requires as few as 8,000 cytometry-fractionated cells for a single analysis, and provides high-resolution DNA replication patterns with respect to both cell-cycle time and genomic position. We find that different cell types exhibit characteristic replication signatures that reveal striking plasticity in regional replication time patterns covering at least 50% of the human genome. We also identified autosomal regions with marked biphasic replication timing that include known regions of monoallelic expression as well as many previously uncharacterized domains. Comparison with high-resolution genome-wide profiles of DNaseI sensitivity revealed that DNA replication typically initiates within foci of accessible chromatin comprising clustered DNaseI hypersensitive sites, and that replication time is better correlated with chromatin accessibility than with gene expression. The data collectively provide a unique, genome-wide picture of the epigenetic compartmentalization of the human genome and suggest that cell-lineage specification involves extensive reprogramming of replication timing patterns.
Keywords: chromatin structure, gene expression, tissue specificity
The eukaryotic cell cycle consists of an orderly process in which cells grow, replicate their chromosomes, and segregate the duplicated genome to their daughter cells. DNA replication is central to this process, and occurs by a complex series of events involving the activation of thousands of replication initiation zones, usually in a defined sequential order (1). The molecular details and the mechanisms establishing and maintaining the replication program are far from clear. In particular, it is not known to what degree the human replication program varies between different cell types, nor what the relationship is between such variation and the structure and accessibility of human chromatin.
Beginning with the experiments of Taylor et al. using tritiated thymidine for visualizing DNA replication (2), nucleotide incorporation studies have established that replication in animal cells is organized into discrete zones of similar replication timing consisting of multiple replication origins and their associated replicons that vary in size from 30 to 450 kb (1, 3). Multireplicon zones of similar replication timing are interspersed in the genome and can be visualized in metaphase chromosomes as heritable “replication bands” that generally correspond to classical cytogenetic Giemsa bands (light early and dark late) (4). Replication time zones can also be visualized in interphase cells as stable structures with defined subnuclear positioning that is highly heritable in daughter cells (5). Like classical banding, replication banding appears to be quite similar between different cell types (4), suggesting a largely invariant replication program. Some large-scale molecular studies of different human cell types appear to confirm the basic similarity of the replication program across developmental lineages (6–9).
In contrast to these stereotypical patterns, localized plasticity in the human replication program has been described in the context of several cell-type- or developmental stage-specific genes, where earlier replication is associated with expression and later replication with repression (1, 10–13). More recently, genome-wide studies in both mouse and Drosophila have observed replication time plasticity over about 20% of the genome (14, 15). The regions differing in replication time between cell types include those with a subset of genes showing the expected pattern of early replication in expressed cells and later replication in repressed cells, as well as those lacking expression differences.
DNA replication has also been linked to other epigenetic features, including chromatin modification and DNA methylation. In both mouse and Drosophila, early replication is more closely associated with histone acetylation than with gene expression per se (14, 15), although replication origin efficiency has been suggested to be more dependent on transcription than on open chromatin (16). The DNA methylation-replication time connection comes from observations of demethylation-induced advancement of replication commonly seen at a number of loci, including the inactive X chromosome (12); advanced replication time is also associated with abnormal hypomethylation seen in ICF syndrome cells deficient in the DNMT3B methyltransferase (13, 17).
Although it is clear that replication timing, DNA methylation, and chromatin modification are important epigenetic factors with respect to transcription, their relative importance and interdependence have yet to be fully elucidated. For example, the fully methylated HPRT gene on the inactive X can undergo a spontaneous advance in replication that is not associated with transcription, although demethylation-induced reactivation occurs at a much higher frequency in the advanced state (12). Similarly, escape from X inactivation in ICF syndrome cells for the abnormally unmethylated G6PD gene only occurs if replication is advanced toward an active-X-like pattern (13).
It is apparent from these and other studies that replication timing is a critical epigenetic component of cellular function in addition to its role in the faithful replication of DNA sequence information (18). To ensure the transmission of cellular identity and function, it is evident that the replication-associated inheritance of epigenetic templates must be faithfully copied as well as the sequence. The presence at the replication fork of chromatin assembly factors, histone-modifying enzymes, and DNA methyltransferases is consistent with this function (18). Additionally, the fidelity of DNA replication itself appears to be influenced by time of replication, because late domains exhibit increased mutation frequency (19).
To define comprehensively the DNA replication program in human cells and explore its importance for genome organization and higher-order transcriptional regulation, we developed a high-throughput approach for quantifying DNA replication time as a function of genome position using massively parallel sequencing (Repli-Seq) and applied it to define the “replicomes” of multiple human cell lineages including undifferentiated embryonic stem cells (hESCs). Our analysis shows that a large fraction of the human genome-at least 50%-exhibits tissue-specific variation in DNA replication time. Additionally, we find several genomic regions with biphasic patterns that are compatible with allelic variation in replication timing. We also examined the correlation between replication time and either tissue-specific gene expression, and chromatin accessibility, revealing strong association with the latter.
Results and Discussion
Whole-Genome Profiling of DNA Replication Timing by Repli-Seq.
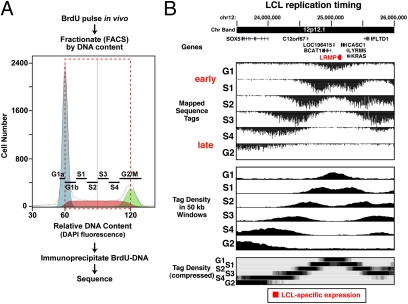
To explore the human replication timing program in different cell lineages, we developed an approach-Repli-Seq-that couples next-generation sequencing with a well-validated strategy originally developed for use with sequence-tagged site (STS) markers (20, 21) (Fig. 1). We labeled newly synthesized DNA strands in vivo with a pulse of 5-bromo-2-deoxyuridine (BrdU), which is incorporated by DNA polymerase into newly replicating DNA in place of thymidine. We then sorted labeled cells into six fractions that span all of the DNA synthesis phase of cell division (G1b, S1, S2, S3, S4, and G2; Fig. 1A). We isolated the BrdU-labeled nascent strands (BrdU-DNA) using an anti-BrdU monoclonal antibody, and assembled the isolated BrdU-DNA fractions into separate Illumina sequencing libraries. Following 27-cycle sequencing of about 2000 cell equivalents per replication time fraction, we obtained about 3 million high-quality reads that mapped uniquely to the human genome reference sequence. To produce a visual profile of the progress of DNA replication along the genome, we computed the normalized density of mapped tags for each cell-cycle fraction (sequence tag summaries are given in Table S1). Fig. 1B illustrates the resulting data from a 3 Mb region of chromosome 12 profiled in a B-lymphoblastoid cell line (LCL; GM06990). Replication in this region initiates very early in the cell cycle (G1b fraction) in the vicinity of the lymphoid-specific gene LRMP, and then progresses bidirectionally into late-replicating flanking regions that contain repressed genes (SOX5 and IFLTD1).
Fig. 1.
Repli-Seq approach. (A) Cell-cycle fractionation of newly synthesized DNA. Exponentially growing cells are pulse-labeled with BrdU, stained with DAPI, and sorted into different fractions of the cell cycle according to DNA content as shown for this normal lymphoblastoid cell line (LCL). Fractionation is continuous across the cell cycle (see Materials and Methods). Antibody-purified BrdU-labeled DNA is made into sequencing libraries and sequenced on the Illumina platform, and the sequence tags are mapped to the hg18 reference genome. (B) Visualization of replication patterns as exemplified at the LRMP locus in the GM06990 LCL. Mapped sequence tags in this region of chromosome 12 reveal a very early peak of replication at the lymphoid-specific LRMP gene. Sequence tag densities for each cell-cycle fraction (below) were calculated over 50-kb windows, normalized according to their genome-wide sequence tag counts, and further normalized at each genomic position by calculating the percentage of total replication (to avoid biases due to nucleotide composition or mapability).
Validation of Repli-Seq with PCR-Based Replication Timing.
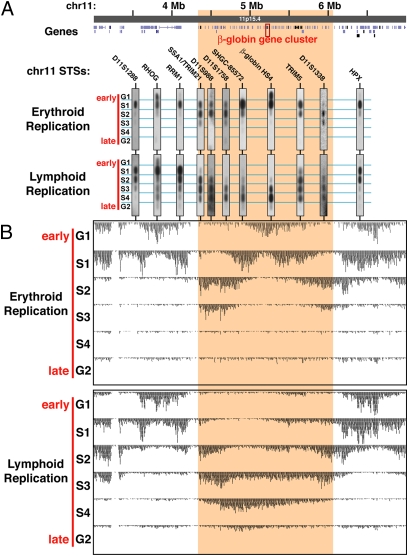
To confirm that Repli-Seq provides replication patterns that parallel those obtained with a conventional STS marker and PCR-based assay, we applied both approaches to examine the replication timing of the human β-globin locus on chromosome 11 in B-lymphoblastoid cells (in which the β-globin locus is silent) and K562 erythroid cells (in which the locus is active) (Fig. 2). This analysis revealed a tight correspondence between STS-based patterns and those from Repli-Seq. STS markers in the β-globin locus control region (LCR) and a surrounding region of about 2 Mb replicate during mid- to late-S phase in lymphoblastoid cells, whereas markers in the far up- and downstream flanking regions replicate early (Fig. 2A). Repli-Seq tag densities paralleled the STS patterns and provided compatible replication timing patterns for the inter-STS regions (Fig. 2B). In K562 cells the β-globin locus replicates in the very early cell-cycle fraction, G1b, consistent with previous BrdU-based replication studies (22). Not previously appreciated, however, is that most of the surrounding region extending ±1 Mb that replicates late in non-globin-expressing cells is early-replicating in K562 cells (Fig. 2, shaded region). K562 and LCLs display similar patterns of early replication in the far-flanking regions. Interestingly, the peak of early replication at the globin gene cluster in K562 corresponds to the preferred replication origin locations previously identified near the HBB globin gene (1).
Fig. 2.
Validation of Repli-Seq with STS-based replication timing. (A) Repli-Seq replication timing measurements compared to a standard PCR-based assay at specific genomic locations in the distal portion of 11p15.4 for erythroid and lymphoblastoid cells. Using standard STS-based replication timing, 12 markers define a 2-Mb domain (shaded) containing the β-globin gene cluster that is early-replicating in erythroid cells that express globin genes (K562) and late-replicating in lymphoid cells where globin genes are silent (GM08729). (B) Sequence-based replication timing. The Repli-Seq whole-genome sequence-based data closely match the replication patterns found for specific STSs, but provide more comprehensive coverage of this 3-Mb region.
Whole-Genome Replication Time Profiles Across Multiple Cell Lineages.
To identify systematically regions that differ in replication timing between distinct human cell types genome-wide, we produced additional whole-genome Repli-Seq profiles from normal dermal fibroblasts (BJ), hESCs (BG02), and a third LCL (TL010). We also confirmed the high reproducibility of Repli-Seq by reapplying it to fibroblasts from a new frozen stock (r = 0.98; Fig. S1).
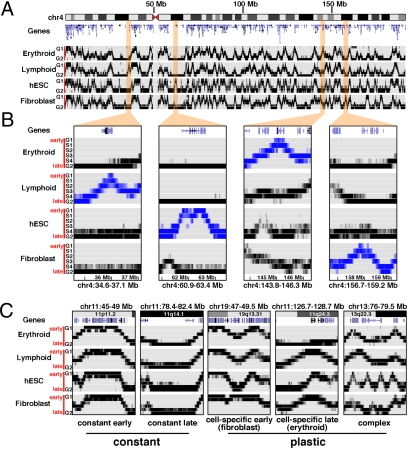
We observed each cell type to display a unique whole-genome replication timing profile composed of regions with fixed replication behavior across all cell types (“constant” domains) in combination with domains manifesting both quantitative and qualitative cell-specific patterns (“plastic” domains; Fig. 3; Fig. S2). We also identified several stereotypical patterns of regional replication timing. Prominent among these are a large number of “inverted-V” regions that exhibit cell-type-specific early replication flanked by symmetrical early-to-late transitions from G1b/S1 to S4/G2 (Fig. 3 A and B). We also observed analogous “V” regions displaying cell-specific late replication, as well as more complex patterns (Fig. 3C).
Fig. 3.
Cell-lineage-specific replication timing. (A) Comparison of replication timing profiles from four cell types across chromosome 4, illustrating unique lineage patterns. (B) Lineage-specific early-replication patterns. Cell-lineage-specific early patterns are highlighted in expanded chromosome 4 subregions. The lymphoid-specific CENTD1 gene in the 34.6–37.1 Mb region is at the apex of an early-replication peak in the GM06990 LCL, whereas this region is uniformly late-replicating in the other three lineages. Similar patterns are seen in the other expanded regions: LPHN3 in the 60.9–63.4 Mb region (hESC-specific), GYPA-GYPB-GYPE in the 143.8–146.3 Mb region (erythroid-specific), and PDGFC in the 156.7–159.2 region (fibroblast-specific). (C) Stereotypical replication timing patterns. Shown are major patterns of DNA replication timing observed across the genome, including (i) regions of constant early replication across cell lineages; (ii) regions of constant late replication; (iii) regions with cell-specific early replication; (iv) regions with lineage-specific late replication (i.e., one cell type late, all others early); and (v) complex patterns that vary considerably between lineages.
To characterize and compare replication patterns across cell types genome-wide, we simplified the data for each cell type by computing the ratio of early- (G1+S1) to late- (S4+G2) replicating DNA as a function of genomic position. We then analyzed all pairwise combinations between cell types to ascertain domains of significant replication time variability, excluding regions with very low sequence coverage (see Methods). In contrast to the striking concordance of replication timing in the three different LCLs studied (r ≥ 0.88 for whole-genome comparisons), we found a surprisingly large fraction of the human genome—49%—to exhibit replication timing plasticity when four different cell lineages were compared (Tables S2 and S3). Moreover, it is likely that this is a lower-limit estimate, as inclusion of additional cell types could substantially increase the spectrum of observed variability.
Replication Time and Genomic Features.
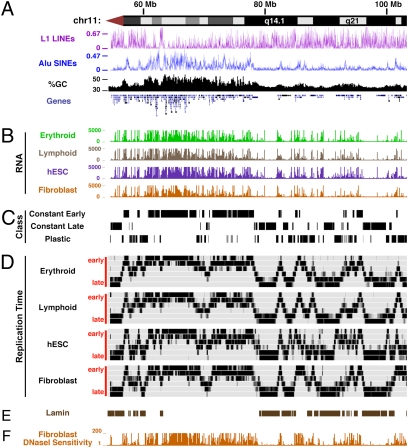
We next compared replication timing patterns with major genomic annotations (Fig. 4; Table S3). Regions of constant early replication are significantly (P < 0.05) associated with high gene density, gene expression, Alu density, GC content, CpG density, and vertebrate nonexonic conservation. Constant early regions are also significantly depleted of L1 LINEs and satellite sequences, whereas constant late regions are enriched in these features (Fig. 4; Table S3).
Fig. 4.
Replication timing patterns versus annotated genomic features. (A) Major annotated genomic features across a 50 Mb portion of the long arm of chromosome 11 including the densities of L1 LINEs and Alu SINEs; G+C content; and gene density. (B) Total RNA output measured by Affymetrix exon arrays across four cell types. (C) Constant early, constant late, and plastic replication time regions. (D) Repli-Seq data from four cell types (see Fig. 1 for description). (E) Regions of nuclear lamina association recently reported for human fibroblasts (23) correlate well with the regions of late replication we found in BJ fibroblasts. (F) Chromatin accessibility and density of DNaseI-hypersensitive sites in BJ fibroblasts. Note the extremely close correspondence with early-replicating regions in fibroblasts (D above, lower track), and the inverse relationship with lamin-associated late-replicating regions.
Many of these genomic features are variable in regions exhibiting replication plasticity; thus, their patterns of overall enrichment or depletion are generally more modest than those in the replication constant categories. The gene content in plastic regions, for example, is depleted by 30% compared to the rest of the genome, whereas constant early regions have an enrichment of 135% and constant late have a depletion of 83% (Fig. 4; Table S3). Overall, the plastic region enrichment patterns resemble constant late domains to some extent by having lower levels of genes, CpG islands, and Alus, yet they also resemble constant early domains by being depleted for satellite sequences and L1 LINEs.
Replication Time, Gene Expression, and Chromatin Structure.
To compare replication timing with gene expression, we measured RNA levels in each cell type using Affymetrix tiling arrays targeting 1.5 million known and predicted exons (see Methods). We found only a moderate correlation between early replication and transcriptional output (r = 0.40). We next examined transcriptional output specifically in regions of replication time plasticity and found that they follow the expected classic pattern of late replication when repressed and early replication when expressed (Fig. S3). On a global level, however, these results suggest that gene expression is largely a surrogate marker for replication time and that other factors such as chromatin structure may be more closely or indeed causally related to replication timing patterns. We therefore examined replication time patterns in relation to chromatin and nuclear structure.
Association with the nuclear lamina appears to be a physical marker of repressed genomic domains, and regions displaying this association have recently been defined in human fibroblasts (23). We compared these regions with fibroblast replication timing patterns and observed a striking correlation between sites of nuclear lamina association and regions displaying late replication (Fig. 4). This observation suggests that late-replicating regions generically define not only a repressed but also a physically segregated nuclear compartment.
To examine the relationship between replication timing and chromatin accessibility, we analyzed genome-wide DNaseI sensitivity measurements obtained from the same BJ fibroblast and K562 cell cultures produced under the ENCODE Project (Fig. 4). This analysis revealed that early replication is significantly more closely associated with chromatin accessibility and the density of DNaseI hypersensitive sites than with transcriptional output per se (raw data correlation r = 0.64 versus 0.40 over chromosome 11, for example). Indeed, we identified numerous plastic replication regions without any observed lineage-specific differences in RNA levels, although with clearly compatible alterations in chromatin accessibility (Fig. S4).
Very Early Replication and Replication Initiation Zones.
A distinctive feature of the replication time profiles is the presence throughout the genome of hundreds of early-replicating regions flanked by symmetrical, monotonic early-to-late transitions (inverted-Vs in Figs. 2–4). These regions are often specific to one cell type (Fig. 3), and apparently represent replication initiation zones together with associated bidirectional replication forks that travel the entire length of S phase (14, 24, 25). Because the inverted-V apex regions and other strong G1 replication zones should be enriched in constitutive and cell-type-specific replication origins, we cataloged these segments for hESCs, LCLs, and fibroblasts using conservative criteria (see Methods). We identified 1131 such G1 regions in H0287 LCLs, 1199 in GM06990 LCLs, 1809 in BG02 hESCs, and 1547 in BJ fibroblasts (Table S4).
Few human replication origins have been fully described with high confidence, but a recent HeLa cell study identified 266 replication origins in ENCODE regions (16); 190 of these (71%) overlap with our combined early-replicating G1 segments, suggesting that the latter represent bona fide initiation zones. Pairwise comparisons between the different cell lines indicated that only about half of the G1 initiation zones we identified overlap between any two cell types, whereas 85% are shared between two LCLs. Such lineage-specific variability is consistent with the replication time plasticity analysis reported above and suggests that early-replication origin firing is highly variable with respect to cell type.
Biphasic Replication Timing.
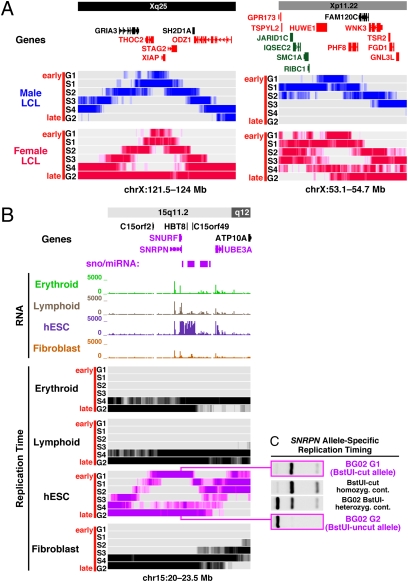
Among the replication timing regions discovered to have plastic replication is a class of biphasic replication domains. Many such regions are seen along the X chromosome in the female GM06990 LCLs (Fig. S2). To systematically identify biphasic regions in all cell types, we devised an automatic search method based on finding low signals in one cell-cycle fraction that are interposed between strong signals of both earlier and later replication (see Methods). Using this approach, we identified many regions exhibiting biphasic replication (Table S5), although the regions flagged account for only a small portion of the measured genome (0.32%; Tables S3 and S5). Included among these is the imprinted domain associated with Prader-Willi syndrome at the SNRPN-SNURF locus on chromosome 15, as well as many loci on the X chromosome in the female LCL (GM06990) that would be expected to derive from the replication differences between the active and inactive Xs. Interestingly, the majority of biphasic domains appear to have genomic features similar to the pan-lineage/constant early regions (Table S3).
Biphasic replication zones on the X chromosome in female cells (Fig. 5A) are perhaps the easiest to understand because they follow the classic pattern of allelic asynchrony due to X chromosome inactivation (12, 26). Certain imprinted domains would also be expected to exhibit replication asynchrony. The extreme biphasic replication found for the 2 Mb Prader-Willi region on chromosome 15 (G1 and G2) was specific to hESCs (Fig. 5B), although the region exhibits imprinted gene expression in most cell types (27), including LCLs and fibroblasts, where replication asynchrony at SNRPN is known to be moderate, requiring allele-specific methods to ascertain (28). The extreme asynchrony in hESCs is apparently related to hESC-specific high expression within the region that includes SNRPN-SNURF transcription and transcription of more than 70 genes encoding C/D box snoRNAs (Fig. 5B).
Fig. 5.
Biphasic replication timing. (A) Biphasic replication timing (simultaneous early and late replication of the same genomic region) associated with X chromosome inactivation. Shown are two examples of biphasic replication timing associated with X chromosome inactivation in females. In the chrX:121.5–124 Mb region, male LCLs (H0287) have an early peak associated with several genes known to be subject to X inactivation in females (38) (depicted in red; X inactivation status of genes in black is unknown). The region is biphasic in female LCLs (GM06990) with the early component resembling the male pattern (active X) and the very late component (G2) thus representing the inactive X. In the chrX:53.1–54.7 Mb region there is a transition from a more synchronous region containing many genes that escape X inactivation (in green) to a biphasic region that is subject to X inactivation (in red). (B) Biphasic replication in the imprinted Prader-Willi/Angelman syndrome region on chromosome 15. Replication is markedly biphasic in hESCs over a 2–3 Mb region associated with Prader-Willi syndrome (27), including the imprinted SNRPN gene. The early component is specific to hESCs and correlates with the much higher level of RNA expression in this region relative to other cell types. (C) Biphasic patterns indicate allele-specific replication programs. Early and late hESC fractions were examined for the presence or absence of an informative restriction site polymorphism in the SNRPN gene (BstUI) to confirm that biphasic patterns are allelic. Early-replicating DNA is exclusively the cleaved allele that is expressed (paternal), whereas the late-replicating DNA is exclusively the uncleaved allele that is repressed (maternal). Controls shown include BG02 DNA (for an equal allelic contribution) and a homozygous-cleaved DNA control for digestion.
Because hESCs are known to exhibit imprinted SNRPN expression (29), the G1-replicating portion should correspond to the expressed paternal allele whereas the G2 portion should correspond to the repressed maternal allele, similar to the order of allele-specific replication timing in LCLs (28). This allelic asynchrony was confirmed using an STS-based replication assay that interrogates a polymorphic BstUI restriction site (28). The early-replicating DNA component was exclusively the expressed BstUI-cut allele, whereas the late-replicating DNA component was exclusively the silent BstUI-uncut allele (Fig. 5C).
Several other biphasic replication regions we identified were previously recognized as asynchronous, or were candidates for imprinting in human cells. These loci include the FRA3B region (30) and a region on chromosome 2 containing the LRRTM1 gene that appears to be imprinted with a variable pattern of maternal down-regulation (31). Additionally, 4 of 18 regions previously identified to replicate asynchronously in LCLs and fibroblasts (32, 33) are within 1 Mb of a biphasic replication locus (Table S5). One such region is near IL12B and contains the EBF1 gene that was recently identified as exhibiting monoallelic expression in cloned LCLs (34).
Aside from such examples, however, we observed a poor correspondence between biphasically replicating regions and reported loci with monoallelic expression. Of 371 genes exhibiting random monoallelic expression in LCL studies (33, 34), only three—PIP3E, TIRAP, and XYLT1—overlap a biphasic replication zone identified in H0287 and GM06990 LCLs. These findings ostensibly suggest that random monoallelic expression need not significantly perturb replication time patterns and, conversely, that allelic variation in replication timing does not necessitate allelic variation in gene expression.
Our analysis of biphasic regions only flagged 0.3% of the measured genome, whereas previous whole-genome studies of replication time in HeLa cells (35, 36) and mouse leukemia cells (25) have suggested that 9–20% of these genomes replicate asynchronously. The greater stringency of our definition of asynchrony may partially explain this discrepancy, but relaxation of our criteria is unlikely to significantly close the gap based on our preliminary analyses and visual inspection of the whole-genome replication patterns. Differences between the cell lines used in these studies and between mouse and human may also explain some of this discordance. Additionally, the previous studies use lower-resolution cell-cycle fractionation, and cell synchrony also appears to be lower than attained by our cytometry-based method and this could result in the erroneous appearance of asynchrony for some loci at the extremes of the replication program.
Perspective.
In summary, we developed an approach for quantifying human DNA replication time at high-resolution genome-wide and applied it across multiple human cell types. Even when performed on a limited range of cell types, the resulting data reveal a surprising plasticity in human replication programs. Repli-Seq is readily extensible to additional cell types as well as to diverse eukaryotic organisms. Because of its low cell-input requirements (conservatively about 5000 cell equivalents per replication time point, ≈1000-fold lower than for a typical histone modification), the method is uniquely suited to studying rare cell populations such as finely differentiated or developing cell types.
Direct sequencing of cell-cycle-ordered replication fractions has several important advantages over array-based approaches that typically ascertain only early and late portions of the cell cycle (9, 14, 15). Whereas array-based approaches are confounded by repetitive regions, massively parallel sequencing with 27–36 bp read lengths has the potential to interrogate up to 90% of the raw human genome sequence. Profiling of multiple temporally distinct replicating fractions coupled with a sequencing-based approach enables ascertainment of biphasic replication timing that is difficult or impossible to attain with any methods employing early- versus late-replication ratios. Continuing improvements in the throughput of DNA sequencing, combined with the ability to multiplex samples using barcoding, provide Repli-Seq with considerable cost advantages over other approaches.
Our studies of replication timing and gene expression in multiple lineages and DNaseI sensitivity in erythroid cells and fibroblasts suggest that replication time reflects an important component of the epigenetic compartmentalization of the genome. These data serve as a basis for further exploration of cell-type-specific replication programs, their evolutionary conservation, their variation in different genetic backgrounds, and their potential alteration in disease and disease susceptibility.
Materials and Methods
Expanded experimental procedures are provided in SI Methods.
Cell Culture and RNA Analysis, and the Repli-Seq Procedure.
All cells were grown in exponential phase for replication timing and gene expression studies using standard culture conditions. Affymetrix Human Exon 1.0 ST arrays were used to examine RNA levels in all cultures (Affymetrix, Santa Clara, CA) and the data are available as released from the ENCODE Project through the UCSC Genome Browser at http://genome.ucsc.edu (see SI Methods).
Repli-Seq is based on our STS replication assay (20, 21) and has been modified for sequence analysis of newly replicated DNA (Fig. 1). The procedure is similar for attached and unattached cell types.
The density of BrdU-DNA-derived sequence tags along the genome was calculated for each cell-cycle fraction using 50 kb sliding windows at 1 kb intervals and was normalized to a global density of 4 million tags per genome for each fraction (see SI Methods). To avoid potential variability in signal and background related to tag mapability variation, sequence bias, or copy-number differences, we further normalized the 50 kb densities of each cell line to a percentage of total replication for that line at each genomic coordinate.
Analysis of Lineage-Specific Differences in Replication Timing and Biphasic Replication.
To simplify the computational search for variations in replication timing in different cell lines, we combined the percent-normalized sequence tag density value (PNDV) for G1 and S1 for each 1 kb window of the genome to yield a cumulative “early” replication signal for each cell type. Similarly, a “late” signal was calculated by adding the PNDV for S4 and G2. We identified regions having significant differences in the early:late ratio between any two cell lines and classified these as “plastic” replication domains (Table S2).
To determine regions where replication time is biphasic, cell-cycle PNDV signals were added in pairs to generate expanded replication time categories (G1+S1, S1+S2, S2+S3, S3+S4, S4+G2). A biphasic region was defined as a 1 kb bin that contained at least 40% of the total replication signal in each of two nonadjacent expanded replication time categories (Table S5).
DNaseI Sensitivity and Genomic Feature Analysis.
To compare replication timing with chromatin sensitivity to DNaseI digestion, we digested BJ fibroblast and K562 erythroid nuclei according to previously described procedures (37), DNaseI-derived fragments were sequenced and mapped, and 150 bp densities were determined (see SI Methods for data accession).
Enrichments for particular genomic features were calculated for the different replication classes by comparing their frequencies within the regions of interest to the genome as a whole.
Supplementary Material
Acknowledgments
We thank Ping Luo (STS-based replication timing), Jeff Goldy (cell culture), and Andrew Haydock (RNA isolation) for technical assistance. Carol Ware and Angel Nelson cultured the BG02 hESCs. This work was supported by National Institutes of Health grants R01HD16659, P30HD02274, and U54HG004592.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA levels determined by Affymetrix Exon array analysis are available from the NCBI GEO database under accession numbers GSM472898, GSM472903, GSM472910, GSM472944, and GSM472945. The digital DNaseI chromatin accessibility data are available as released from the ENCODE Project through the UCSC Genome Browser at http://genome.ucsc.edu (subId = 295 and subId = 106, narrow peak data). Newly replicated DNA sequencing data are deposited to the NCBI Short Read Archive under the Study accession number SRP001393.1.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912402107/DCSupplemental.
References
- 1.Aladjem MI. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- 2.Taylor JH, Woods PS, Hughes WL. The organization and duplication of chromosomes as revealed by autoradiographic studies using tritium-labeled thymidinee. Proc Natl Acad Sci USA. 1957;43:122–128. doi: 10.1073/pnas.43.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- 4.Drouin R, Holmquist GP, Richer CL. High-resolution replication bands compared with morphologic G- and R-bands. Adv Hum Genet. 1994;22:47–115. doi: 10.1007/978-1-4757-9062-7_2. [DOI] [PubMed] [Google Scholar]
- 5.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe Y, et al. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: Disease-related genes in timing-switch regions. Hum Mol Genet. 2002;11:13–21. doi: 10.1093/hmg/11.1.13. [DOI] [PubMed] [Google Scholar]
- 7.White EJ, et al. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc Natl Acad Sci USA. 2004;101:17771–17776. doi: 10.1073/pnas.0408170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodfine K, et al. Replication timing of the human genome. Hum Mol Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- 9.Donaldson AD. Shaping time: Chromatin structure and the DNA replication programme. Trends Genet. 2005;21:444–449. doi: 10.1016/j.tig.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Goldman MA, Holmquist GP, Gray MC, Caston LA, Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- 11.Hatton KS, et al. Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol. 1988;8:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen RS, Canfield TK, Fjeld AD, Gartler SM. Role of late replication timing in the silencing of X-linked genes. Hum Mol Genet. 1996;5:1345–1353. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- 13.Hansen RS, et al. Escape from gene silencing in ICF syndrome: Evidence for advanced replication time as a major determinant. Hum Mol Genet. 2000;9:2575–2587. doi: 10.1093/hmg/9.18.2575. [DOI] [PubMed] [Google Scholar]
- 14.Hiratani I, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwaiger M, et al. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadoret JC, et al. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci USA. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Bonis ML, et al. Maintenance of X- and Y-inactivation of the pseudoautosomal (PAR2) gene SPRY3 is independent from DNA methylation and associated to multiple layers of epigenetic modifications. Hum Mol Genet. 2006;15:1123–1132. doi: 10.1093/hmg/ddl027. [DOI] [PubMed] [Google Scholar]
- 18.McNairn AJ, Gilbert DM. Epigenomic replication: Linking epigenetics to DNA replication. Bioessays. 2003;25:647–656. doi: 10.1002/bies.10305. [DOI] [PubMed] [Google Scholar]
- 19.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen RS, et al. A variable domain of delayed replication in FRAXA fragile X chromosomes: X inactivation-like spread of late replication. Proc Natl Acad Sci USA. 1997;94:4587–4592. doi: 10.1073/pnas.94.9.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen RS, Canfield TK, Lamb MM, Gartler SM, Laird CD. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 22.Epner E, Forrester WC, Groudine M. Asynchronous DNA replication within the human β-globin gene locus. Proc Natl Acad Sci USA. 1988;85:8081–8085. doi: 10.1073/pnas.85.21.8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 24.Ermakova OV, et al. Evidence that a single replication fork proceeds from early to late replicating domains in the IgH locus in a non-B cell line. Mol Cell. 1999;3:321–330. doi: 10.1016/s1097-2765(00)80459-1. [DOI] [PubMed] [Google Scholar]
- 25.Farkash-Amar S, et al. Global organization of replication time zones of the mouse genome. Genome Res. 2008;18:1562–1570. doi: 10.1101/gr.079566.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heard E, Disteche CM. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 27.Horsthemke B, Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am J Med Genet A. 2008;146A:2041–2052. doi: 10.1002/ajmg.a.32364. [DOI] [PubMed] [Google Scholar]
- 28.Kawame H, Gartler SM, Hansen RS. Allele-specific replication timing in imprinted domains: Absence of asynchrony at several loci. Hum Mol Genet. 1995;4:2287–2293. doi: 10.1093/hmg/4.12.2287. [DOI] [PubMed] [Google Scholar]
- 29.Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum Mol Genet. 2007;16(Spec no. 2):R243–R251. doi: 10.1093/hmg/ddm245. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, et al. Allele-specific late replication and fragility of the most active common fragile site, FRA3B. Hum Mol Genet. 1999;8:431–437. doi: 10.1093/hmg/8.3.431. [DOI] [PubMed] [Google Scholar]
- 31.Francks C, et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol Psychiatry. 2007;12:1129–1139. doi: 10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gimelbrant AA, Ensminger AW, Qi P, Zucker J, Chess A. Monoallelic expression and asynchronous replication of p120 catenin in mouse and human cells. J Biol Chem. 2005;280:1354–1359. doi: 10.1074/jbc.M411283200. [DOI] [PubMed] [Google Scholar]
- 33.Ensminger AW, Chess A. Coordinated replication timing of monoallelically expressed genes along human autosomes. Hum Mol Genet. 2004;13:651–658. doi: 10.1093/hmg/ddh062. [DOI] [PubMed] [Google Scholar]
- 34.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 35.Jeon Y, et al. Temporal profile of replication of human chromosomes. Proc Natl Acad Sci USA. 2005;102:6419–6424. doi: 10.1073/pnas.0405088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karnani N, Taylor C, Malhotra A, Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabo PJ, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 38.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.