Abstract
Despite improvements in the treatment of gastrointestinal cancers, 5-year survival rates for advanced-stage patients remain disappointing. Therefore, the need exists to develop innovative new therapies while optimizing the current ones. Pharmacogenetics can be helpful in this context. Metabolism of cancer drugs varies according to age, gender, diet, concurrent use of other drugs, and existing comorbidities, including impaired liver and renal function. In addition, metabolizing enzymes, drug-transport proteins, metabolites, and drug receptors are genetically determined. It has also been demonstrated that genetic mutations within a tumor can be a determining factor with regard to response to treatment. The most common agents used in the treatment of digestive system tumors — 5-fluorouracil, oxaliplatin, cisplatin, irinotecan, gemcitabine, and newly developed biologic agents bevacizumab, cetuximab, panitumumab, and erlotinib—will be reviewed from a pharmacogenetic perspective. The US Food and Drug Administration has approved the UDP-glucuronosyltransferase 1A1 test in patients treated with irinotecan, and additional approval of newer tests is anticipated. Increasing availability of these sophisticated assays is expected to facilitate the delivery of more effective, less toxic chemotherapy regimens in the management of relatively resistant tumors of the gastrointestinal tract.
Tumors of the digestive system are the leading group of cancers in terms of frequency and cause of cancer deaths.1 These malignancies are generally diagnosed at an advanced stage, metastatic disease is rarely curable, and the rate of recurrence is quite high in earlier-stage resectable gastrointestinal cancers. The objective of adjuvant therapy is to decrease postoperative recurrence rate and prolong disease-free and overall survival. At certain stages of colon, rectum, gastric, and pancreas cancers, the benefit of adjuvant chemotherapy alone or in combination with radiotherapy has been demonstrated. As for cancers of the anal canal, a high rate of cure can be achieved with neoadjuvant chemoradiotherapy. On the other hand, the role of adjuvant therapy in esophagus, liver, and biliary tract cancers has not been established clearly.
Despite recent advances in medical treatment of gastrointestinal cancers, 5-year survival rates remain disappointing. Effective new treatments are urgently needed, and existing therapies need to be refined and improved.2,3 Moreover, the ability to distinguish which patients are or are not likely to respond to a given therapy, as well as to identify patients at risk of developing severe toxicity, will enable clinicians to optimize cancer treatment, increase the chances of therapeutic success, and improve patient outcomes.4 In this respect, pharmacogenetics represents, perhaps, the most promising method of offering patients such individualized therapy.5
Doses for anticancer agents are generally close to maximum tolerated levels, as determined in phase I trials; however, optimal therapeutic doses have not been established for many cancer drugs. Thus, a certain incidence of grade 3/4 side effects might be expected in some patient populations. Metabolism of chemotherapy agents varies depending on patient age, gender, diet, concomitant drug use, comorbidities, and hepatic and renal functions. However, enzymes that metabolize the drug, proteins that transport the drug and its metabolites, and drug receptors are determined based on a patient’s genetic profile. Pharmacogenetics focuses on investigating the influence of genetic structure on cancer treatment. In this report, primary drugs used in the treatment of gastrointestinal cancers — 5-fluorouracil (5-FU), oxaliplatin, cisplatin, irinotecan, gemcitabine, and the novel biologic agents, bevacizumab, cetuximab, panitumumab, and erlotinib—will be reviewed from a pharmacogenomic perspective. Table 1 shows mechanisms of action, metabolisms, and excretions of these drugs.
Table 1.
Mechanisms of action, metabolism, and excretion of drugs used in patients with gastrointestinal cancers.
| Drugs | Indication | Mechanism of action | Metabolism | Renal excretion | Biliary excretion | Dose reduction |
|---|---|---|---|---|---|---|
| 5-Fluorouracil | Colorectal cancer, gastric cancer | Antimetabolite, thymidylate synthase blocking | Thymidylate synthase, thymidylate phosphorylase, dihydropyrimidine dehydrogenase | Inactive metabolites in urine | Inactive metabolites in bile | Enzymatic catabolism by DPD in the liver (>90%); Biluribin > 5 mg/dL: omit use |
| Oxaliplatin | Colorectal cancer | Alkylator of DNA | – | 50% | – | Reduce dose for renal dysfunction |
| Cisplatin | Anal cancer | Alkylator of DNA | – | 90% | <10% | Creatinine clearance 10–50 mL/min: 25% reduction |
| Irinotecan | Colorectal cancer, gastric cancer | Topoisomerase I inhibition | Carboxylesterase, CYP3A4, UGT1A1, topoisomerase I | >25% | >50% hepatic metabolism and biliary excretion | Metabolized in the liver. Consider in the presence of hepatic dysfunction |
| Gemcitabine | Pancreatic carcinoma | Metabolized intracellularly to the active diphosphate and triphosphate | Cytidine deaminase, deoxycytidylate deaminase | Nearly entirely excreted in urine | – | Metabolized in the liver. Consider in the presence of hepatic dysfunction |
| Bevacizumab | Colorectal cancer | Monoclonal antibody for VEGF-A | – | – | – | |
| Cetuximab | Colorectal cancer | Monoclonal antibody to EGFR | – | – | No adjustment needed for renal or hepatic impairment | |
| Panitumumab | Colorectal cancer | Monoclonal antibody to EGFR | ||||
| Erlotinib | Pancreas cancer | Inhibits intracellular phosphorylation of EGFR tyrosine kinase | 8% renal elimination | Hepatic metabolism through CYP 3A4; 83% hepatic elimination | Consider in severe liver impairment |
Abbreviations: VEGF = vascular endothelial growth factor; EGFR = epidermal growth factor receptor
5-FLUOROURACIL
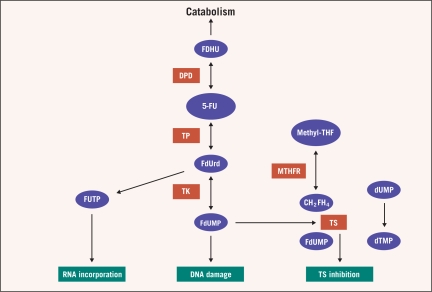
5-FU has been used in the treatment of gastrointestinal cancers for over 50 years.6 5-FU itself is a prodrug that is converted to 5-fluoro-2′-deoxyuridine-5′-monophosphate (5-FdUMP) within the cell to inhibit thymidylate synthase (TS). Thus, synthesis of pyrimidine and subsequently DNA is suppressed. Eighty-five percent of 5-FU is catabolized to its inactive metabolites via dihydropyrimidine dehydrogenase (DPD) (Figure 1).7
Figure 1.
5-Fluorouracil metabolism
Abbreviations: 5-FU = 5-fluorouracil; DPD = dihydropyrimidine dehydrogenase; dTMP = thymidine monophosphate; dUMP = deoxyuridine monophosphate; FDHU = 5-fluorodihydrouracil; FdU = fluorodeoxyuridine; FdUMP = 5-fluoro-2-deoxyuridine monophosphate; FdUrd = 5-fluorodeoxyuridine; FUTP = 5-fluorouridine triphosphate; MTHFR = methylenetetrahydrofolate reductase; THF = tetrahydrofolate; TK = thymidine kinase; TP = thymidilate phosphorylase; TS = thymidylate synthase
Dihydropyrimidine Dehydrogenase
In cases of DPD enzyme deficiency, blood levels of 5-FU and its active metabolites increase. DPD enzyme deficiency can result in fatal myelotoxicity, mucositis, and cerebellar toxicity.7–15 Determination of DPD enzyme activity in mononuclear cells may be of benefit; however, it is technically very difficult.12 The gene encoding DPD enzyme is located at 1p22 and consists of 23-exons.13 Conditions resulting in DPD deficiency include base substitutions, splicing deficits, and frameshift mutations. More than 40 different polymorphisms related to this enzyme have been reported.15 Severe 5-FU toxicity is associated with 17 of these mutations. In the general population, homozygote and heterozygote DPD dysfunction is estimated to be 0.1% and 3% to 5%, respectively. DPYD*2A is themost common DPD polymorphism associated with 5-FU toxicity. In this case, exon 14 is skipped as a result of the G→A translocation at intron 14, and inactive enzyme is formed. With this polymorphism, the heterozygote form is characterized by severe toxicity; the homozygote form is characterized by mental deficiency.
Dihydropyrimidine dehydrogenase enzyme deficiency was demonstrated in 61% of patients exhibiting severe 5-FU toxicity. DPYD*2A polymorphism was identified in 50% of patients with grade 4 neutropenia. However, DPD enzyme activity is normal in most patients with severe 5-FU toxicity. Therefore, multiple factors and genes are thought to be involved in 5-FU toxicity.14,15 Genetic variations related to other enzymes involved in 5-FU metabolism are also important, particularly TS and thymidylate phosphorylase (TP).
Thymidylate Synthase
Thymidylate synthase is a target of 5-FU. It plays a significant role in folate metabolism. TS enables conversion of deoxyuridylate to deoxythymidylate.16,17 In tumors, increased TS enzyme expression is associated with resistance to 5-FU and capecitabine. 18 In particular, intratumoral TS levels in metastatic lesions are indicative of 5-FU resistance.18,19 This is a result of differences between TS expressions of primary and metastatic lesions.20
In adjuvant treatment of stage III colon cancer, response of patients with TSER3 polymorphism is poor. TS 5′-untranslated region (5′-UTR) polymorphism is useful for determining response to 5-FU. While the response rate is 50% in those with 2R/2R, it is 8% in those with 3R/3R.20 Apart from response, TS polymorphism also affects survival. Median survival in cases of 2R/2R is 16 months vs. 12 months in cases of 3R/3R.21 TS polymorphism is also relevant in predicting response to neoadjuvant 5-FU treatment in rectal cancer. Cases of 3R/3R are associated with a poor response.22
Thymidylate Phosphorylase
Thymidylate phosphorylase mRNA levels in patients not responding to 5-FU were 2.6-fold higher than in responding patients in pretreatment biopsies of patients with colorectal cancer.23 Survival was significantly increased in patients with both TS and TP under nonresponse cutoff values, and low intratumoral expression of TS and TP was associated with a response to 5-FU and improved survival.21,23,24
GEMCITABINE
Gemcitabine is a nucleoside analog of deoxycytidine used in treatment of localized and advanced-stage pancreatic cancer.25 Gemcitabine undergoes metabolic activation by kinases to form a cytotoxic trinucleotide in the cell.26 Metabolic inactivation of gemcitabine by deamination is catalyzed by cytidine deaminase (CDA) or — after phosphorylation — by deoxycytidylate deaminase (DCTD).27 In a study of patients with pancreatic carcinoma treated with gemcitabine, intratumoral SLC29A1 expression (as measured by immunohistochemistry) was related to prolonged survival.28 This is because functional nucleoside transporters are necessary for gemcitabine cytoxicity, and nucleoside-transporter–deficient cells are highly resistant to gemcitabine. 29 SLC29A1 is the most abundant of the nucleoside transporters.30 Analysis of SLC29A1 mRNA expression also showed a significant correlation between increased mRNA expression and longer survival in patients with pancreatic cancer treated with gemcitabine.31
Deoxycytidine kinase (DCK) deficiency was previously the most common form of acquired resistance to gemcitabine in vitro.32 Studies have shown a correlation between higher levels of DCK activity and increased gemcitabine sensitivity in patients with advanced pancreatic cancer treated with gemcitabine, whereas low tumoral DCK immunohistochemical expression was associated with a worse overall survival and progression-free survival.30
Ribonucleotide reductase (RR) is a target enzyme for gemcitabine.33 The pharmacology and pharmacogenetics of ribonucleotide reductase subunit M1 (RRM1) is of particular interest due to its potential role in gemcitabine chemosensitivity and synergy with other chemotherapeutic agents, particularly cisplatin.33–35 In a study of genetically modified lung cancer cell lines, RRM1 expression correlated inversely with gemcitabine sensitivity.36
Deactivating enzymes of gemcitabine include 5′, 3′-nucleotidase, cytosolic (NT5C), deoxycytidylate deaminase (DCTD), and cytidine deaminase (CDA).26,33 Upregulation of CDA may play a role in gemcitabine resistance, while impaired activity may result in increased toxicities.33,34 Germline genotyping revealed homozygosity for CDA 208A in a recent report of a Japanese patient with pancreatic cancer treated with gemcitabine and cisplatinum who developed severe hematologic and nonhematologic toxicity.35
IRINOTECAN (CPT-11)
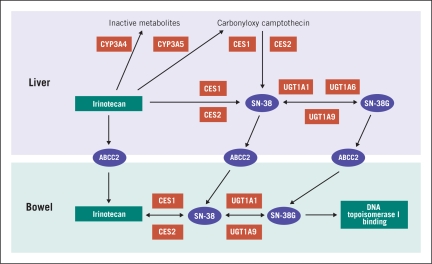
Irinotecan is a topoisomerase I inhibitor.37 Among gastrointestinal malignancies, it is used in the treatment of colorectal cancer and gastric cancer. Activation, transportation, and deactivation of irinotecan is complex, and many enzymes, including carboxylesterase (CE), CYP3A4, uridine diphosphate glucuronosyltransferase (UGT1A1), and topoisomerase I are involved in the metabolism of this drug.37–39 Irinotecan is converted to its active metabolite, SN38, by CE present in the gastrointestinal tract (Figure 2).38,39 This enzyme has many allelic variations and genotypes and is involved in irinotecan toxicity.38
Figure 2.
Irinotecan metabolism
UGT1A1, present in the liver, is the enzyme that inactivates SN38 via glucuronidation. 40,41 The association of irinotecan toxicity and UGT1A1 polymorphism is currently under investigation. UGT1A1 inactivates SN38 via phase II reaction.40,41 The UGT1A1*28 polymorphism is associated with reduced UGT1A1 expression and, as a result, decreased glucuronidation of SN38. This, in turn, increases blood levels of active metabolites resulting in increased toxicity.39–42 The pharmacokinetics of irinotecan is poorly associated with body surface area.
Since SN38 undergoes glucuronidation to a lesser extent in patients with Gilbert and Crigler Najjar syndromes, irinotecan toxicity increases in these patients, because of reduced or deficient expression levels of UGT1A1. Gilbert syndrome results from the UGT1A1*28 homozygote transition of promoter polymorphism caused by seven TA repetitions. In UGT1A1*28 polymorphism, transcription is decreased by 70% and toxicity is increased. Patients with the 7/7 genotype (homozygous for seven TA repetitions) exhibit a 9.3-fold increase in risk of grade 4 neutropenia, and irinotecan is associated with severe side effects in this population.40–43
In a meta-analysis of 10 studies assessing the correlation between irinotecan-induced hematologic toxicities in UGT1A1*28 patients, irinotecan dose, and overall toxicity, risk of experiencing irinotecaninduced hematologic toxicity for homozygous UGT1A1*28 patients was found to be a function of the dose of irinotecan administered, and genotyping was recommended at high doses (> 200 mg/m2) of irinotecan. 42 Genotyping was of modest benefit at intermediate doses, such as 180mg/m2 used in the FOLFIRI (folinic acid/5-FU/irinotecan) regimen. Unless administered concomitantly with another myelotoxic agent, UGT1A1*28 testing was not recommended at doses < 150 mg/m2.
PLATINUM COMPOUNDS (Cisplatin and Oxaliplatin)
Cisplatin and oxaliplatin are commonly used in gastrointestinal cancers.44,45 Platinum analogs block DNA replication by forming different DNA adducts. DNA repair enzymes ERCC1 and ERCC2—also known as XPD and glutathione S transferase (GSTP) enzymes—are involved in the activity of these agents.46 High expression of the genes that code for these enzymes is inversely correlated with therapeutic response in colorectal and gastric cancer.47
In single nucleotide polymorphisms involving transformation of lysine forming on ERCC2 gene, codon 751, to glutamine, the response rate is 24% in lysine, the inactive form of this enzyme, and 10% in other forms.48 This trial was performed in patients who had previously received chemotherapy. These results have not been confirmed in patients receiving first-line therapy.
COMBINATION CHEMOTHERAPY
The N9741 trial is a phase III, randomized trial designed to compare the efficacy of FOLFOX (folinic acid/5-FU/oxaliplatin), IROX (irinotecan/oxaliplatin), and IFL (irinotecan/bolus 5-FU/folinic acid) in patients with metastatic colorectal cancer.49 The pharmacogenetic evaluation performed in this trial revealed that both the objective response rate and incidence of grade 3/4 side effects, particularly diarrhea, were lower in black patients. The low rate of response was particularly marked in the FOLFOX arm. Overall, the rate of response was 41% and 30% in white and black patients, respectively (P = .015). The rate of severe toxicity was 48% in whites and 34% in black patients in the FOLFOX arm (P = .047). Although no significant median survival difference was observed between these two patient groups in the FOLFOX arm, median survival was lower in black patients in both the IFL and IROX groups.
In all arms, black patients experienced less toxicity, particularly diarrhea, than white patients did. In this trial, UGT1A1 7/7 involved in irinotecan metabolism was identified at a rate of 21% and 9% in black and white patients, respectively. However, the determinant role of UGT1A1 gene polymorphism with respect to response and toxicity could not be demonstrated. Significant differences were also detected between white and black patients in prevalence of other pharmacogenetic variances such as CYP3A, MDR (multidrug resistance), ERCC1, ERCC2, and GSTP. These genes are important in the metabolism and detoxification of irinotecan and oxaliplatin.50 In the same trial (Intergroup N9741), GSTP1 polymorphism was demonstrated to be associated with early development of oxaliplatin neuropathy in patients receiving FOLFOX.
A small study showed that ERCC codon 118 polymorphism predicted response to oxaliplatin/5-FU chemotherapy in patients with advanced colorectal cancer. In this study, response rate was 61.9%, 42.3%, and 21.4% in T/T, C/T, and C/C groups, respectively (P: 0.018).51
BIOLOGIC AGENTS
Biologic agents used in gastrointestinal cancers include bevacizumab, cetuximab, panitumumab, and erlotinib. Increased VEGF expression is involved in tumoral angiogenesis and associated with poor prognosis. The therapeutic benefit of bevacizumab has been shown in the treatment of patients with advanced-stage colorectal cancer. Thus far, however, adequate pharmacogenetic data have not been produced to predict toxicity, response, or resistance.
In the BOND-2 trial, metastatic colorectal cancer patients progressing after irinotecan-based chemotherapy were randomized to receive irinotecan plus bevacizumab plus cetuximab (CBI) or bevacizumab and cetuximab (CB).52 In this trial, germline polymorphisms involved in angiogenesis (vascular endothelial growth factor [VEGF], interleukin-8 [IL-8], transforming growth factor [TGF]-β), the epidermal growth factor receptor (EGFR) pathway (EGFR, cyclooxygenase-2, E-cadherin), DNA repair (ERCC1, ERCC2, XRCC1, xeroderma pigmentosum group D [XPD]), and drug metabolism pathway (GSTP1, UGT1A1) were investigated.52,53 Results showed a correlation between TGF-β polymorphism and response; UGT1A1, cyclin D1, and time to disease progression (TTP); and TGF-β polymorphism and tumor response in the CBI arm. In addition, a correlation between EGFR 497 and overall survival was found. As for the CB arm, the investigators found a trend in association between polymorphisms of Fc fragment of IgG–3A and tumor response; a significant correlation between ERCC2, TGF-β and TTP; and ERCC2 and overall survival were correlated.52–54
A correlation between RAS mutation and resistance to the EGFR antibodies cetuximab and panitumumab has been demonstrated. KRAS mutations account for approximately 30% to 40% of patients who are not responsive to treatment with these agents.55–57 Patients with RAS mutation in tumor tissue have lower rates of response to cetuximab and panitumumab and shorter progression-free survival time. Recently, a BRAF V600E mutation was detected in 11 of 79 patients who had wild-type KRAS.58 This BRAF mutation is associated with no response to cetuximab and panitumumab with significantly shorter progression-free and overall survival compared to wild-type patients.58,59
Erlotinib is a small-molecule tyrosine kinase inhibitor that targets HER1/EGFR.60,61 In addition, it is a potent inhibitor of CYP1A1, and a moderate inhibitor of CYP3A4 and CYP2C8, as well as a strong inhibitor of glucuronidation by UGT1A1 in vitro.62 Potent inducers of CYP3A4 may reduce the efficacy of erlotinib, whereas potent inhibitors of CYP3A4 may lead to increased toxicity.60–62 Concomitant treatment with these types of agents should be avoided. The inhibition of glucuronidation may cause interactions with substrates of UGT1A1 exclusively cleared by this pathway. Patients with low expression of UGT1A1 or genetic glucuronidation disorders (eg, Gilbert’s disease) may exhibit increased serum concentrations of bilirubin and must be treated with caution.62
In a recent study including 569 patients with advanced pancreatic cancer, erlotinib in combination with gemcitabine showed statistically superior overall survival compared with gemcitabine alone (6.4 months vs. 5.9 months, respectively).63 In this study, patients responded equally well to treatment with erlotinib regardless of whether their tumors expressed abnormal levels of EGFR. Survival was longer in patients with wild-type KRAS in comparison to KRAS-mutated patients (7.5 months vs. 3.7 months, respectively) in erlotinib-treated patients. In the wild-type group, median survival was 3.4 months in the erlotinib arm and 7 months in the placebo arm. Survival was also longer in EGFR wild-type patients and in patients with high EGFR copy numbers as demonstrated by fluorescence in situ hybridization.64
In the AViTA study, patients with advanced-stage pancreatic cancer were treated with gemcitabine plus erlotinib with or without bevacizumab. In this study, survival was positively correlated with severity of rash, but no statistically significant difference was detected in terms of overall survival with the addition of bevacizumab.65 Therefore, reassessment of erlotinib treatment is recommended in patients who do not develop rash within the first 4 to 8 weeks of treatment.
DISCUSSION
Based on the results of pharmacogenetic studies, the US Food and Drug Administration has approved the UGT1A1 test for patients receiving irinotecan. Additionally, RAS mutation assay is required to determine whether patients are candidates for treatment with cetuximab and panitumumab. New tests are on the horizon and, based on data from many of the trials reviewed herein, are expected to be approved. Increasing availability of these sophisticated assays is expected to facilitate the delivery of more effective, less toxic chemotherapy regimens by individualizing treatments for patients with relatively resistant tumors of the gastrointestinal tract.
Footnotes
Disclosures of Potential Conflicts of Interest
Dr. Yalçin indicated no potential conflicts of interest.
REFERENCES
- 1.Parkin MD, Bray F, Ferlay J, et al. Global Cancer Statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Yalcin S, Oksuzoglu B, Tekuzman G, et al. Biweekly irinotecan (CPT-11) plus bolus 5-fluourouracl (5-FU) and folinic acid in patients with advanced stage colorectal cancer (ACRC) Jpn J Clin Oncol. 2003;33:580–583. doi: 10.1093/jjco/hyg111. [DOI] [PubMed] [Google Scholar]
- 3.Aksoy S, Karaca B, Dincer M, et al. Common etiology of capecitabine and fluorouracil-induced coronary vasospasm in a colon cancer patient. vasospasm in a colon cancer patient. Ann Pharmacother. 2005;39:573–574. doi: 10.1345/aph.1E252. [DOI] [PubMed] [Google Scholar]
- 4.Yong WP, Innocenti F, Ratain MJ. The role of pharmacogenetics in cancer therapeutics. Br J Clin Pharmacol. 2006;62:35–46. doi: 10.1111/j.1365-2125.2006.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vesell ES. Pharmacogenetic perspectives gained from twin and family studies. Pharmacol Ther. 1989;41:535–552. doi: 10.1016/0163-7258(89)90130-7. [DOI] [PubMed] [Google Scholar]
- 6.Toxicity of fluorouracil in patients with advanced colorectal cancer: effect of administration schedule and prognostic factors. Meta-Analysis Group in Cancer. J Clin Oncol. 1998;16:3537–3541. doi: 10.1200/JCO.1998.16.11.3537. [DOI] [PubMed] [Google Scholar]
- 7.Daher GC, Harris BE, Diasio RB. Metabolism of pyrimidine analogues and their nucleosides. Pharmacol Ther. 1990;48:189–222. doi: 10.1016/0163-7258(90)90080-l. [DOI] [PubMed] [Google Scholar]
- 8.Diasio RB. Clinical implications of dihydropyrimidine dehydrogenase on 5-FU pharmacology. Oncology. 2001;15:21–26. [PubMed] [Google Scholar]
- 9.Fleming RA, Milano GA, Gaspard MH, et al. Dihydropyrimidine dehydrogenase activity in cancer patients. Eur J Cancer. 1993;29A:740–744. doi: 10.1016/s0959-8049(05)80358-2. [DOI] [PubMed] [Google Scholar]
- 10.Milano G, Etienne MC, Pierrefite V, et al. Dihydropyrimidine dehydrogenase deficiency and fluorouracil-related toxicity. Br J Cancer. 1999;79:627–630. doi: 10.1038/sj.bjc.6690098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uetake H, Ichikawa W, Takechi T, et al. Relationship between intratumoral dihydropyrimidine dehydrogenase activity and gene expression in human colorectal cancer. Clin Cancer Res. 1999;5:2836–2839. [PubMed] [Google Scholar]
- 12.Lu Z, Zhang R, Diasio RB. Dihydropyrimidine dehydrogenase activity in human peripheral blood mononuclear cells and liver: population characteristics newly identified deficient patients and clinical implications in 5-fluorouracil in cancer patients. Cancer Res. 1993;53:5433–5438. [PubMed] [Google Scholar]
- 13.Wei X, Elizondo G, Sapone A, et al. Characterization of the human dihydropyrimidine dehydrogenase gene. Genomics. 1998;51:391–400. doi: 10.1006/geno.1998.5379. [DOI] [PubMed] [Google Scholar]
- 14.Mattison L, Soong R, Diasio R. Implications of dihydropyrimidine dehydrogenase on 5FU pharmacogenetics and pharmacogenomics. Pharmacogenomics. 2002;3:485–491. doi: 10.1517/14622416.3.4.485. [DOI] [PubMed] [Google Scholar]
- 15.Ridge S, Sludden J, Wei X, et al. Dihydropyrimidine dehydrogenase pharmacogenetics in patients with colorectal cancer. Br J Cancer. 1998;77:497. doi: 10.1038/bjc.1998.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller CR, McLeod HL. Pharmacogenomics of cancer chemotherapy-induced toxicity. Support Oncol. 2007;5:9–14. [PubMed] [Google Scholar]
- 17.Johnston PG, Lenz HJ, Leichman CG, et al. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res. 1995;55(7):1407–1412. [PubMed] [Google Scholar]
- 18.Kidd EA, Yu J, Li X, et al. Variance in the expression of 5-fluorouracil pathway genes in colorectal cancer. Clin Cancer Res. 2005;11:2612–2619. doi: 10.1158/1078-0432.CCR-04-1258. [DOI] [PubMed] [Google Scholar]
- 19.Pullarkat ST, Stoehlmacher J, Ghaderi V, et al. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenomics J. 2001;1:65–70. doi: 10.1038/sj.tpj.6500012. [DOI] [PubMed] [Google Scholar]
- 20.Marsh S, McKay JA, Cassidy J, et al. Polymorphism in the thymidylate synthase promoter enhancer region in colorectal cancer. Int J Oncol. 2001;19:383–386. doi: 10.3892/ijo.19.2.383. [DOI] [PubMed] [Google Scholar]
- 21.Salonga D, Danenberg KD, Johnson M, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyridine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 22.Villafranca E, Okruzhnov Y, Dominguez MA, et al. Polymorphisms of the repeated sequences in the enhancer region of the thymidylate synthase gene promoter may predict downstaging after preoperative chemoradiation in rectal cancer. J Clin Oncol. 2001;19:1779–1786. doi: 10.1200/JCO.2001.19.6.1779. [DOI] [PubMed] [Google Scholar]
- 23.Metzger R, Danenberg K, Leichman CG, et al. High basal level gene expression of thymidine phosphorylase (platelet-derived endothelial cell growth factor) in colorectal tumors is associated with nonresponse to 5-fluorouracil. Clin Cancer Res. 1998;4:2371–2376. [PubMed] [Google Scholar]
- 24.Meropol NJ, Gold PJ, Diasio RB, et al. Thymidine phosphorylase expression is associated with response to capecitabine plus irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24(25):4069–4077. doi: 10.1200/JCO.2005.05.2084. [DOI] [PubMed] [Google Scholar]
- 25.Burris HA, 3rd, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15(6):2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 26.Plunkett W, Huang P, Xu YZ, et al. Gemcitabine: metabolism, mechanisms of action, and self-potentiation. Semin Oncol. 1995;22:3–10. [PubMed] [Google Scholar]
- 27.Gandhi V, Plunkett W. Modulatory activity of 2′, 2′-difluorodeoxycytidine on the phosphorylation and cytotoxicity of arabinosyl nucleosides. Cancer Res. 1990;50:3675–3680. [PubMed] [Google Scholar]
- 28.Spratlin J, Sangha R, Glubrecht D, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res. 2004;10:6956–6961. doi: 10.1158/1078-0432.CCR-04-0224. [DOI] [PubMed] [Google Scholar]
- 29.Mackey JR, Mani RS, Selner M, et al. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res. 1998;58:4349–4357. [PubMed] [Google Scholar]
- 30.Sebastiani V, Ricci F, Rubio-Viqueira B, et al. Immunohistochemical and genetic evaluation of deoxycytidine kinase in pancreatic cancer: relationship to molecular mechanisms of gemcitabine resistance and survival. Clin Cancer Res. 2006;12:2492–2497. doi: 10.1158/1078-0432.CCR-05-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giovannetti E, Mey V, Nannizzi S, et al. Pharmacogenetics of anticancer drug sensitivity in pancreatic cancer. Mol Cancer Ther. 2006;5:1387–1395. doi: 10.1158/1535-7163.MCT-06-0004. [DOI] [PubMed] [Google Scholar]
- 32.Bergman AM, Pinedo HM, Peters GJ. Determinants of resistance to 2′, 2′-difluorodeoxycytidine (gemcitabine) Drug Resist Update. 2002;5:19–33. doi: 10.1016/s1368-7646(02)00002-x. [DOI] [PubMed] [Google Scholar]
- 33.Mini E, Nobili S, Caciagli B, et al. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17(suppl 5):v7–v12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- 34.Ueno H, Kiyosawa K, Kaniwa N. Pharmacogenomics of gemcitabine: can genetic studies lead to tailor-made therapy? Br J Cancer. 2007;97:145–151. doi: 10.1038/sj.bjc.6603860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yonemori K, Ueno H, Okusaka T, et al. Severe drug toxicity associated with a single-nucleotide polymorphism of the cytidine deaminase gene in a Japanese cancer patient treated with gemcitabine plus cisplatin. Clin Cancer Res. 2005;11:2620–2624. doi: 10.1158/1078-0432.CCR-04-1497. [DOI] [PubMed] [Google Scholar]
- 36.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24:4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 37.Mathijssen RH, van Alphen RJ, Verweij J, et al. Clinical pharmacokinetics and metabolism of irinotecan (CPT-11) Clin Cancer Res. 2001;7:2182–2194. [PubMed] [Google Scholar]
- 38.Charasson V, Bellott R, Meynard D, et al. Pharmacogenetics of human carboxylesterase 2, an enzyme involved in the activation of irinotecan into SN-38. Clin Pharmacol Ther. 2004;76:528–535. doi: 10.1016/j.clpt.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Khanna R, Morton C, Danks MK, et al. Proficient metabolism of irinotecan by a human intestinal carboylesterase. Cancer Res. 2000;60:4725–4728. [PubMed] [Google Scholar]
- 40.Innocenti F, Undevia SD, Iyer L, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22:1382. doi: 10.1200/JCO.2004.07.173. [DOI] [PubMed] [Google Scholar]
- 41.Iyer L, Das S, Janisch L, et al. UGT1A1*28 polimorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43–47. doi: 10.1038/sj.tpj.6500072. [DOI] [PubMed] [Google Scholar]
- 42.Hoskins J, Goldberg R, Qu P, et al. UGT1A1*28 genotype and irinotecan induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99:1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- 43.McLeod HL, Sargent DJ, Marsh S, et al. Pharmacogenetic analysis of systemic toxicity and response after 5-fluorouracil (5FU)/CPT-11, 5FU/oxaliplatin (oxal), or CPT-11/oxal therapy for advanced colorectal cancer (CRC): Results from an intergroup trial. Proc Am Soc Clin Oncol. 2003;22 (abstr 1013) [Google Scholar]
- 44.Vermorken JB, van der Vijgh WJF, Klein I, et al. Pharmacokinetics of free and total platinum species after short infusion of cisplatin. Cancer Treat Rep. 1984;68:505–513. [PubMed] [Google Scholar]
- 45.Raymond E, Faivre S, Woynarowski JM, et al. Oxaliplatin: mechanism of action and antineo-plastic activity. Semin Oncol. 1998;25(2 suppl 5):4–12. [PubMed] [Google Scholar]
- 46.Levi F, Metzger G, Massari C, et al. Oxaliplatin: pharmacokinetics and chronopharmacological aspects. Clin Pharmacokinet. 2000;5:1–21. doi: 10.2165/00003088-200038010-00001. [DOI] [PubMed] [Google Scholar]
- 47.Ruzzo A, Graziano F, Loupakis F, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol. 2007;25:1247–1254. doi: 10.1200/JCO.2006.08.1844. [DOI] [PubMed] [Google Scholar]
- 48.Stoehlmacher J, Park DJ, Zhang W, et al. A multivariate analysis of genomic polymorphisms: Prediction of clinical outcome to 5-fluorouracil/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91:344–354. doi: 10.1038/sj.bjc.6601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldberg RM, McLeod HL, Sargent DJ, et al. Genetic polymorphisms, toxicity, and response rate in African Americans (AA) with metastatic colorectal cancer (MCRC) compared to Caucasians (C) when treated with IFL, FOLFOX or IROX in Intergroup N9741. J Clin Oncol. 2006;24(18S June 20 suppl) (abstr 3503) [Google Scholar]
- 50.Grothey A, McLeod HL, Green EM, et al. Glutathione S-transferase P1 I105V (GSTP1 I105V) polymorphism is associated with early onset of oxaliplatin-induced neurotoxicity. J Clin Oncol. 2005;23(16S June 1 suppl) (abstr 3509) [Google Scholar]
- 51.Viguier J, Boige V, Miquel C, et al. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaliplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res. 2005;11:6212–6217. doi: 10.1158/1078-0432.CCR-04-2216. [DOI] [PubMed] [Google Scholar]
- 52.Lenz H, Zhang W, Yang D, et al. Pharmacogenomic analysis of a randomized phase II trial (BOND 2) of cetuximab/bevacizumab/irinotecan (CBI) versus cetuximab/bevacizumab (CB) in irinotecan-refractory colorectal cancer. Gastrointestinal Cancers Symposium. 2007. (abstr 401)
- 53.Zhang W, Vallböhmer D, Yang D, et al. Genomic profile associated with clinical outcome of EGFR-expressing metastatic colorectal cancer patients treated with epidermal growth factor receptor (EGFR) inhibitor cetuximab. J Clin Oncol. 2005;23(16S June 1 suppl) (abstr 3557) [Google Scholar]
- 54.Zhang W, Yang D, Capanu M, et al. Pharmacogenomic analysis of a randomized phase II trial (BOND 2) of cetuximab/bevacizumab/irinotecan (CBI) versus cetuximab/bevacizumab (CB) in irinotecan-refractory colorectal cancer. J Clin Oncol. 2007;25(18S June 20 suppl) doi: 10.1200/JCO.2007.12.0949. (abstr 4128) [DOI] [PubMed] [Google Scholar]
- 55.Van Cutsem E, Lang I, D’haens G, et al. KRAS status and efficacy in the first-line treatment of patients with metastatic colorectal cancer (mCRC) treated with FOLFIRI with or without cetuximab: The CRYSTAL experience. J Clin Oncol. 2008;26(15S May 20 suppl):5s. (abstr 2) [Google Scholar]
- 56.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 57.Bokemeyer C, Bondarenko I, Hartmann J, et al. KRAS status and efficacy of first-line treatment of patients with metastatic colorectal cancer (mCRC) with FOLFOX with or without cetuximab: The OPUS experience. J Clin Oncol. 2008;26(15S May 20 suppl):178s. (abstr 4000) [Google Scholar]
- 58.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 59.Di Fiore F, Van Cutsem E, Laurent-Puig P, et al. Role of KRAS mutation in predicting response, progression-free survival, and overall survival in irinotecan-refractory patients treated with cetuximab plus irinotecan for a metastatic colorectal cancer: analysis of 281 individual data from published series. J Clin Oncol. 2008;26(15S May 20 suppl) (abstr 4035) [Google Scholar]
- 60.Hidalgo M, Siu LL, Nemunaitis J, et al. Phase I and pharmacologic study of OSI-774, an epidermal growth factor receptor tyrosine kinase inhibitor, in patients with advanced solid malignancies. J Clin Oncol. 2001;19:3267–3279. doi: 10.1200/JCO.2001.19.13.3267. [DOI] [PubMed] [Google Scholar]
- 61.Li J, Zhao M, He P, et al. Differential metabolism of gefitinib and erlotinib by human cytochrome P450 enzymes. Clin Cancer Res. 2007;13:3731–3737. doi: 10.1158/1078-0432.CCR-07-0088. [DOI] [PubMed] [Google Scholar]
- 62.Rudin CM, Liu W, Desai A, et al. Pharmacogenomic and pharmacokinetic determinants of erlotinib toxicity. J Clin Oncol. 2008;26:1119–1127. doi: 10.1200/JCO.2007.13.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 64.Moore MJ, da Cunha Santos G, Kamel-Reid S. The relationship of K-rasmutations and EGFR gene copy number to outcome in patients treated with erlotinib on National Cancer Institute of Canada Clinical Trials Group trial study PA.3. J Clin Oncol. 2007;25(18S June 20 suppl) (abstr 4521) [Google Scholar]
- 65.Verslype C, Vervenne W, Bennouna J, et al. Rash as a marker for the efficacy of gemcitabine plus erlotinib-based therapy in pancreatic cancer: Results from the AViTA Study. Rash as a marker for the efficacy of gemcitabine plus erlotinib-based therapy in pancreatic cancer: Results from the AViTA Study. J Clin Oncol. 2009;27(15S suppl) (abstr 4532) [Google Scholar]