Abstract
The tetraheme cytochrome c554 (cyt c554) from Nitrosomonas europaea is believed to function as an electron-transfer protein from hydroxylamine oxidoreductase (HAO). We show here that cyt c554 also has significant NO reductase activity. The protein contains one high-spin and three low-spin c-type hemes. HAO catalyzed reduction of the cyt c554, ligand binding, intermolecular electron transfer, and kinetics of NO reduction by cyt c554 have been investigated. We detect the formation of a NO-bound ferrous heme species in cyt c554 by EPR and Mössbauer spectroscopies during the HAO catalyzed oxidation of hydroxylamine, indicating that N-oxide intermediates produced from HAO readily bind to cyt c554. In the half-reduced state of cyt c554, we detect a spin interaction between the [FeNO]7 state of heme 2 and the low-spin ferric state of heme 4. We find that ferrous cyt c554 will reduce NO at a rate greater than 16 s−1, which is comparable to rates of other known NO reductases. Carbon monoxide or nitrite are shown not to bind to the reduced protein, and previous results indicate the reactions with O2 are slow and that a variety of ligands will not bind in the oxidized state. Thus, the enzymatic site is highly selective for NO. The NO reductase activity of cyt c554 may be important during ammonia oxidation in N. europaea at low oxygen concentrations to detoxify NO produced by reduction of nitrite or incomplete oxidation of hydroxylamine.
Introduction
The chemoautotrophic soil bacterium, Nitrosomonas europaea, derives energy for growth from the oxidation of ammonia to nitrite. NO is produced by Nitrosomonas by a periplasmic copper-containing nitrite reductase, using nitrite as a terminal electron acceptor under limiting oxygen conditions.1,2 However, the production of NO (and then N2O) during oxidation of ammonia to nitrite under low [O2] in N. europaea has been found to occur in the absence of a functional NirK gene,3 indicating that enzyme(s) other than nitrite reductase also produce NO. The presence of excess NO induces denitrification in nitrifying bacteria.4-8 Recent studies have shown that free NO acts as a signaling molecule in regulating the growth pattern and metabolic activity of N. europaea.9 The presence of excess nitrite10 and NO11 has been found to be toxic to Nitrosomonas. A gene, NorB coding for a membrane cytochrome cbb3 NO-reductase is found in the genome of Nitrosomonas.12 Genetic removal of the NorB gene has been shown to strongly diminish the conversion of nitrite to NO and then N2O in cells of Nitrosomonas indicating an important role for the membrane cytochrome cbb3 NO-reductase.13 There are, however, indications that some other catalyst for reduction of NO to N2O might exist; N2O production and resistance to NO toxicity continues in the NorB mutant.13,14 The NO reductase activity of cytochrome c554, shown here, may be a candidate for providing at least a low flux of NO reduction to counter NO toxicity.
Cyt c554 is a monomeric protein (26 kDa) found in the periplasm with an R-helical structure containing four c-type hemes covalently bound via two cysteine thioether linkages in the –C–X–Y–C–H– sequence.15 The crystal structure of cyt c554 shows that hemes 1, 3, and 4 have bis-His axial coordination. Heme 2 is five coordinate with one axial His ligand.16,17 The four hemes in cyt c554 are present in two diheme pairs (hemes 2/4, hemes 1/3). Similar heme packing motifs have been observed in a family multiheme proteins, which includes hydroxylamine oxidoreductase (HAO),18 cytochrome c nitrite reductase (NiR),19,20 fumarate reductase,21-23 NapB,24 diheme cytochrome c,25 and the split-Soret cytochrome.26 Only HAO and NiR have significant conservation of primary sequence.27
Cyt c554 can act as the physiological electron acceptor of the enzyme HAO.28-30 Electrons from cyt c554 are thought to pass through a membrane-anchored tetraheme cytochrome cM552 to ubiquinone and then to ammonia monooxygenase (AMO) and the respiratory pathway leading to ATP synthesis.31 In keeping with key roles in the energy-yielding reactions of Nitrosomonas, the cellular concentrations of AMO, HAO, cyt c554, and cytochrome aa3 are within the same order of magnitude.30
The structural arrangement of heme 2 and heme 4 in cyt c554 aligns with that of the active site heme pairs in the enzymes HAO and NiR (each containing one 5-coordinate heme). However, the structure of oxidized and reduced cyt c554 show that, unlike HAO and NiR, the sixth coordination site of the five coordinate heme 2 in cyt c554 is tightly shielded by three hydrophobic amino acid residues (T154, P155, F156). Consistent with this aspect of the structure, cyt c554 does not bind the common heme ligands CN− and CO.32 Therefore, while a possible enzymatic role for cyt c554 in ammonia metabolism has been speculated, until now, there was no evidence in support.
We demonstrate here the formation and characterization of a NO bound complex with heme 2 of cyt c554. The presence of NO is shown to result in fast oxidation of the fully ferrous protein, and in multiple turnover reactions, the NO oxidation of the protein is observed to be catalytic. The kinetics of NO reduction by cyt c554, and intermolecular electron transfer in cyt c554 have been measured. Thus, Cyt c554 may account for the some of the unidentified NO reductase activity in Nitrosomonas.
Materials and Methods
Purification of the Protein
Growth and 57Fe enrichment of Nitrosomonas europaea and purification of cyt c554 and HAO were as described previously.32,33 All experiments were carried out in 50 mM potassium phosphate buffer pH 7.0, unless otherwise noted. The concentration of cyt c554 was determined spectrophotometrically with ∊554 = 24.6 mM−1 cm−1 per heme.34 All chemicals were reagent grade or better. Doubly distilled or Millipore super Q water was used throughout.
Preparation of the Reduced Samples
HAO-mediated reduction of cyt c554 by hydroxylamine (hydroxylamine/HAO reduced cyt c554) or hydrazine (hydrazine/HAO reduced cyt c554) was accomplished with 5-fold excess of substrate (Sigma) in the presence of a catalytic amount (5% molar ratio) of HAO. The protein solutions were incubated for 10 min at room temperature before freezing in liquid nitrogen. The electrochemically half-reduced samples (−50 mV poised) were prepared as reported previously.32 Reduction with sodium dithionite (Sigma) was done with thoroughly degassed protein samples under argon by adding the required amount of sodium dithionite. The sodium dithionite solution was standardized with methyl viologen (∊570 = 9000 M−1 cm−1). Photoreduced samples were prepared under argon with 10% riboflavin (vs protein concentration) and 5-fold excess of EDTA by illuminating the sample for 2 h with a projector lamp.35 During illumination, the sample holder was in a chilled water bath in a Pyrex beaker.
Preparation of NO Treated Samples
NO gas (Matheson) was passed through water, followed by a solid KOH column to remove trace amounts of acidic N-oxide impurities. The samples were degassed thoroughly in a Schlenk line under argon in septa sealed vials. Measured volumes of NO gas were then bubbled through the degassed protein solutions, using a gastight syringe (Hamilton), followed by inversion of the vial a few times to mix. The protein sample was then transferred, with a gastight syringe, to an EPR tube or Mössbauer cup under argon, and frozen in liquid nitrogen within five minutes of NO addition. For optical experiments, the protein samples (50 mM KPi, pH 7.0) were degassed in septa sealed cuvettes using a Schlenk line under argon, and reduced with sodium dithionite. A measured volume of NO gas was then bubbled through the cuvette using a gastight syringe, and spectra were recorded immediately.
Preparation of the CO Treated Samples
CO gas (Sigma) was passed through degassed water and then bubbled through the protein solutions for 20 min. To establish that the solution contained CO, a myoglobin (Sigma) solution was prepared under similar conditions and checked optically for binding of CO.
Kinetic Measurement of NO Reduction
The kinetic parameters for single turnover were determined by monitoring the rate of oxidation of a fully reduced cyt c554 sample in the presence of NO. A 1.85 μM sample of cyt c554 in an optical cell was titrated with dithionite to the fully reduced state with minimal dithionite in solution. The reaction was initiated with by injection of 100 μL of NO saturated solution (~2 mM at 20 °C)36 to give a final concentration of approximately 125 μM NO. All solutions were degassed thoroughly under argon.
EPR and Mössbauer Spectroscopy
X-band EPR spectra were recorded on a Bruker 300 spectrometer equipped with an Oxford ESR-910 liquid helium cryostat and a Bruker bimodal cavity. The quantification of all signals was relative to a CuEDTA spin standard. The spectra were obtained with a field modulation of 1 mTpp at 100 kHz. The magnetic field was calibrated with an NMR gaussmeter and the microwave frequency was measured with a counter. The Q-band spectra were recorded on a Bruker 200 spectrometer with a home-built microwave probe and cryostat.37 Mössbauer spectra were obtained on a constant acceleration instrument and isomer shifts are reported with respect to an iron metal standard. All displayed spectra were recorded on 57Fe enriched protein.
The EPR simulation software was written by the authors. The program diagonalizes the spin Hamiltonian
| (1) |
where J is the isotropic exchange coupling between two heme centers, D′ is a symmetric traceless tensor, d is a polar vector of the antisymmetric contribution, and g1, g2 are the intrinsic g-tensors of two interacting hemes. The simulations are generated with consideration of all intensity factors, which allows computation of simulated spectra for a specified sample concentration. The simulations therefore allow a quantitative determination of protein signal intensities. The Windows software package (SpinCount) is available for general application to any mono- or dinuclear metal complex by contacting Prof. Hendrich.
Results
Mössbauer Spectroscopy
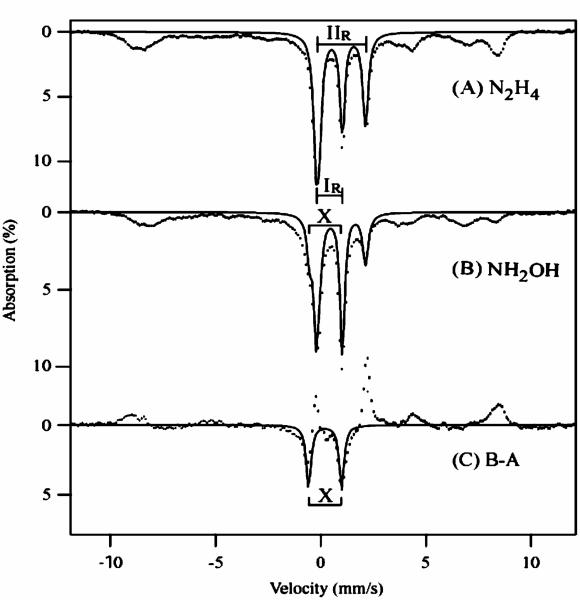
We have shown previously that two of the hemes (heme 1 and 2) of cyt c554 can be reduced with the substrates of HAO (hydrazine or hydroxylamine) in the presence of catalytic amounts (5%) of HAO.32 Cyt c554 does not react directly with these substrates. As mentioned briefly in our previous work, the spectroscopic properties of cyt c554 reduced by HAO in the presence of NH2OH as opposed to N2H4 are quite different, and are not attributable to an altered redox state. Low temperature (4.2 K) Mössbauer spectra, in the absence of an applied magnetic field, of the hydrazine/HAO and hydroxylamine/HAO reduced samples are shown in Figure 1A and B, respectively. The hydrazine reduced spectrum shows paramagnetic hyperfine features from the two low-spin (S = 1/2) ferric hemes (hemes 3 and 4), and two diamagnetic doublets from high-spin (S = 2) ferrous heme 2 (IIR) and low-spin (S = 0) ferrous heme 1 (IR). The solid line overlaid on the spectrum is a fit to the diamagnetic ferrous doublets of heme 1 (IR) and heme 2 (IIR) with δ = 0.42 mm/s, ΔEQ = 1.3 mm/s and δ = 0.98 mm/s, ΔEQ = 2.2 mm/s, respectively, each accounting for 25% of the total iron. The hydroxylamine/HAO reduced spectrum (Figure 1B) shows paramagnetic hyperfine features similar to the hydrazine/HAO spectrum, however, the intensity of the diamagnetic doublets has changed. The intensity of the high-spin ferrous doublet (IIR) has dropped to 50% of the hydrazine/HAO reduced sample, with the appearance of a new diamagnetic doublet (X). The new doublet X is apparent in the difference spectrum hydroxylamine/HAO minus hydrazine/HAO (Figure 1C). The theoretical curve overlaid on the difference spectrum is a fit with δ = 0.23 mm/s, ΔEQ = 1.53 mm/s, accounting for 12% of the total iron. The positive peaks in the subtraction are due to the decrease in the intensity of the doublet associated with the high-spin ferrous heme. The solid line overlaid in Figure 1B is a theoretical curve which shows the effect of loss of 50% of the high-spin heme and the addition of species X.
Figure 1.
Mössbauer spectra of (A) hydrazine/HAO reduced and (B) hydroxylamine/HAO reduced 1.7 mM cyt c554, temperature 4.2 K, in absence of an applied magnetic field. The brackets labeled IR and IIR mark the doublets arising from ferrous low-spin heme 1 and ferrous high-spin heme 2. The spectrum in (C) is the difference B–A. The solid lines are fits (see text for parameters), all using line widths of 0.32 mm/s.
The Mossbauer parameters of species X are close to those previously reported for ferrous heme-NO [FeNO]7 protein species,38-41 and differ significantly from those of known ferric heme-NO [FeNO]6 protein species.42 Heme [FeNO]7 complexes have spin S = 1/2 and display paramagnetic Mössbauer spectra, but species X we observe displays a quadrupole doublet. As we shall demonstrate from the EPR data below, the (heme 2)-NO [FeNO]7 complex is spin-coupled to low-spin (S = 1/2) ferric heme 4. The (heme 2)NO complex is S = 1/2, but the spin-coupling to heme 4 results in fast spin relaxation and low spin expectation, and thus the spectrum shows a quadrupole doublet.
To test for binding of NO or CO to the high-spin heme 2 center, Mössbauer samples of fully reduced cyt c554 were treated with excess of NO or CO gas. The high temperature (150 K) Mössbauer spectrum (not shown) of reduced cyt c554 prior to addition was the same as reported previously.34 Upon addition of NO, the spectrum showed a minor (10%) drop in the intensity of the high-spin ferrous doublet (IIR) without a significant change in the other doublets. Upon addition of CO, the spectrum showed no change in any of the doublets. Thus, NO and CO did not appear to bind in significant amount to the fully reduced protein. However, as we will show below, NO does specifically react with the high-spin ferrous heme 2 center in the fully reduced state of the protein, forming an transient NO bound species that is detectable under limiting reductant conditions.
EPR Spectroscopy of Substrate/HAO Reduced cyt c554
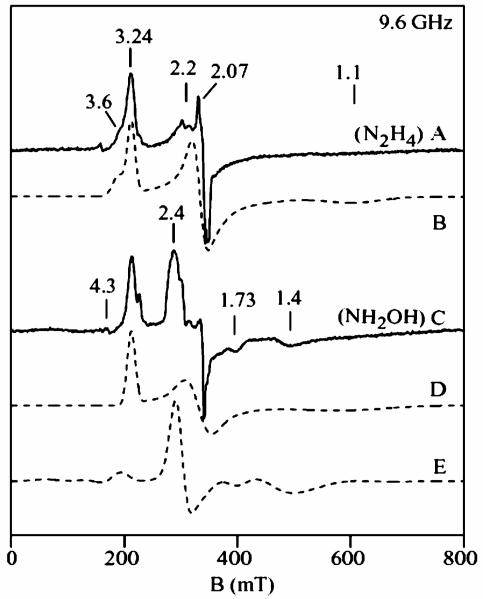
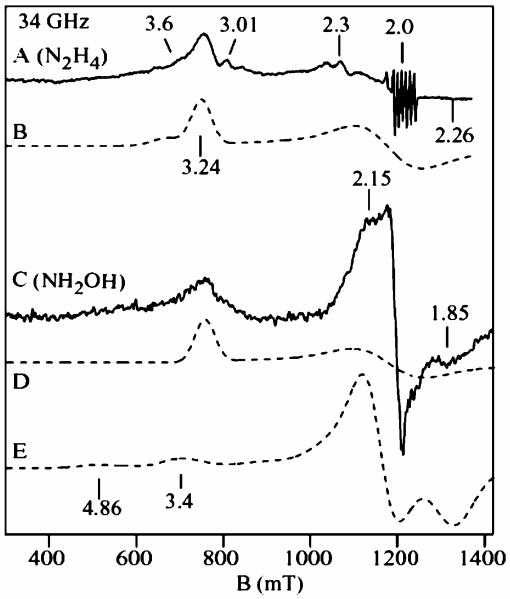
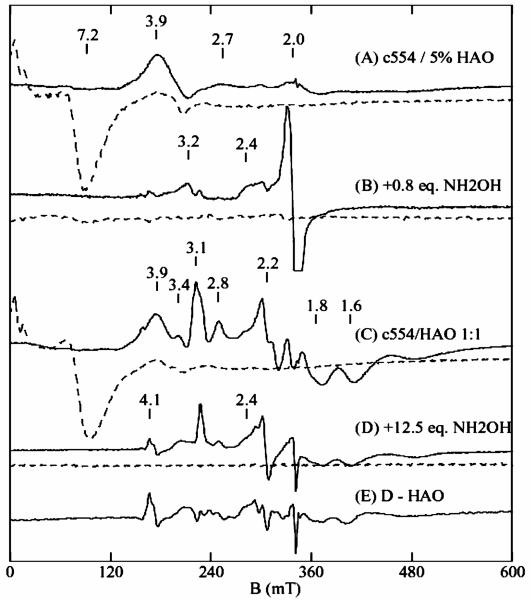
X- and Q-band EPR spectra of substrate/HAO reduced cyt c554 samples are shown in Figure 2 and Figure 3. For both figures, the experimental spectra (solid lines) of the hydrazine/HAO reduced samples are shown in A, and hydroxylamine/HAO reduced samples in C, and the dashed lines are quantitative simulations. The sharp signal near g = 2.07 (Figure 2A) is from a Cu2+ impurity (<3%). The small signals near g = 3.01 and 2.3 are from the catalytic amount of HAO in the samples. The sharp six line signal near g = 2.0 (Figure 3A) is from a Mn2+ impurity (<1%). The hydrazine/HAO reduced samples show signals at g = 3.24, 2.06, and 1.10 from heme 3, and a broad shoulder at gmax = 3.6 from heme 4. Figure 2B and Figure 3B show quantitative simulations for these signals using the same g-values. The hydroxylamine/HAO reduced samples (Figure 2C and Figure 3C) show the signal from heme 3 at g = 3.24, however, the signal from heme 4 at g = 3.6 is absent, and new signals appears at g = 2.4, 1.73, 1.46 (X-band) and g = 2.15, 1.85 (Q-band). The sharp signal at g = 2.0 is a five coordinate [FeNO]7 species accounting for less than 5% of a heme. The addition of more hydroxylamine to the samples did not change the spectra. The hydrazine/HAO and hydroxylamine/HAO reduced samples did not show signals in parallel mode. Samples of hydroxylamine/HAO reduced cyt c554 prepared under anaerobic conditions gave the same signals, thus the appearance of the new signals do not depend on oxygen.
Figure 2.
X-band EPR spectra of (A) hydrazine/HAO reduced 1.5 mM cyt c554, and (C) hydroxylamine/HAO reduced cyt c554. Experimental conditions: temperature 15 K, frequency 9.62 GHz, power 0.2 mW. (B) Quantitative simulation for two noninteracting low-spin (S = 1/2) hemes: heme 3 (g1 = 1.1, 2.06, 3.24), heme 4 (g2 = 0.6, 2.04, 3.6). (D) Quantitative simulation for low-spin heme 3. (E) Quantitative simulation for two exchange-coupled low-spin hemes (S1 = S2 = 1/2). Parameters: g1 = (2.11, 2.07, 1.92), g2 = (0.65, 2.1, 3.6), J = (0.49, 0.43, 0.36) cm−1, σJ = 0.03 cm−1, coordinate rotation (α = 45°, β = 180°, γ = 135°).
Figure 3.
Q-band EPR spectra of the samples of Figure 2. (A) hydrazine/HAO and (C) hydroxylamine/HAO reduced cyt c554. Experimental conditions: temperature 10 K; frequency 34.0 GHz, power 5 μW. The dashed lines are quantitative simulations using the same parameters as that of Figure 2.
The dashed lines in Figure 2D and Figure 3D are quantitative simulations of the heme 3 signals, which match the corresponding signals in the hydrazine/HAO reduced samples, indicating heme 1 is reduced and heme 3 is oxidized in both the samples, and consequently, no spin interaction between heme 1 (S = 0) and heme 3 (S = 1/2). This is consistent with the above result from Mössbauer spectroscopy. The new signals at g = 2.4, 1.73, 1.46 (X-band) and 2.15, 1.85 (Q-band) from the hydroxylamine/ HAO reduced samples show frequency dependent shifts, which suggest a spin-coupled system.
The Mössbauer data on the hydroxylamine/HAO reduced sample indicates the presence of a new quadrupole doublet (X) with parameters similar to those reported for ferrous-NO heme complexes [FeNO]7, with relative abundance equivalent to 50% of a heme in the protein. An [FeNO]7 heme species is paramagnetic with one unpaired electron (S = 1/2). Consistent with these observations, the dashed lines in Figure 2E and Figure 3E are quantitative simulations obtained for the two interacting low-spin hemes (S1 = 1/2, S2 = 1/2), for X- and Q-band frequencies, using the same set of parameters and for 0.5 equiv. of a heme (per protein). The two g-tensors of the simulations are g1 = (2.12, 2.07, 1.92) and g2 = (3.59, 2.1, 0.61), with an antiferromagnetic coupling tensor between the two spin centers of J = (0.49, 0.45, 0.37) cm−1. The two g-tensors are rotated with respect to each other by a set of Euler angles (α, β, γ) = (45, 180, 135). The z-axes of the two g-tensors are antiparallel, and the x-axis of g1 is along the y-axis of g2 and visa-versa for the other axes. This choice of angles is loosely based on the crystal structure, since the orientation of the g-tensor with respect to the heme-NO coordinates is not known. The three g-values in the g1 tensor are similar to those reported for a NO bound low-spin ferrous hemes, [Fe–NO]7 species.39,41,43 Thus, the EPR data of the hydroxylamine/HAO reduced sample also indicates NO binding to 50% of the high-spin ferrous heme 2 in the half-reduced state of the protein. This is the first report of a low-spin ferrous heme-nitrosyl [Fe–NO7] species interacting with a low-spin ferric heme.
NO Addition to Half-Reduced cyt c554
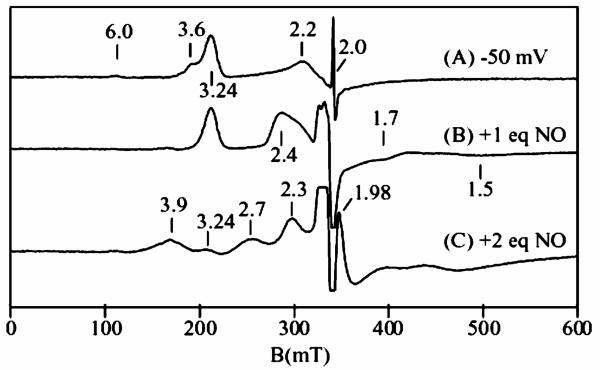
A sample of cyt c554 was electrochemically poised at −50 mV to reduce the two hemes with midpoint potential of +47 mV, and then titrated with NO gas. Figure 4A shows the perpendicular mode X-band EPR spectrum of the −50 mV reduced sample. The sample shows no parallel mode signal in this state. The spectrum of Figure 4A is similar to that of the hydrazine/HAO reduced samples, with signals at g = 3.24 and 2.2 from heme 3, and a broad gmax = 3.6 signal from heme 4. The sharp signal at g = 2.0 is from the mediator used for the electrochemical reduction. The weak signal at g = 6 is from a ferric high-spin minority species (<5%) presumably from denatured protein. Upon addition of approximately 1 equiv. of NO (vs cyt c554) (Figure 4B), the gmax = 3.6 signal from heme 4 of the protein disappears, and a new signals appears at g = 2.4, 1.7, and 1.5, like those observed in the hydroxylamine/HAO reduced spectrum. Thus, NO addition produces the same state of the protein in which heme 3 and heme 4 are oxidized, heme 1 and heme 2 are reduced, and NO is bound to heme 2. This confirms that the g = 2.4 signal observed in the hydroxylamine/HAO reduced cyt c554 spectrum originates from binding of NO to heme 2.
Figure 4.
X-band EPR spectra of half-reduced 2 mM cyt c554 and with NO. (A) Electrochemically poised at −50 mV. (B) After addition of approximately one equivalent of NO to the sample of (A). (C) After addition of another equivalent of NO to the sample of (B). Experimental conditions: temperature 15 K, frequency 9.62 GHz, power 0.2 mW.
In the absence of excess NO, this state of the protein is stable against further oxidation. The addition of another equivalent of NO to the above sample causes more than 90% loss of the g = 3.24 signal from heme 3, with the appearance of signals at g = 2.7 and 3.9 (Figure 4C). The signals at g = 2.7 and 3.9 are both from the fully oxidized state of cyt c554; the g = 2.7 signal is from the heme 1/3 pair, and the g = 3.9 signal is from the heme 2/4 pair.32 The intensities of these signals in comparison to oxidized cyt c554 indicates that heme 1 is now nearly fully oxidized (>90%), whereas only a small amount of heme 2 is oxidized (<10%). Concomitant with the oxidation of heme 1 (and loss of the g = 3.24 signal), the g = 2.4 signal shifts to g = 2.3. We do not yet understand the origin of the shift, but it appears to be associated with a protein conformational change when heme 1 is oxidized. Such a shift could be attributed to a rotation of one of the axial His ligands on heme 2 or 4. The addition of NO to a hydroxylamine/HAO reduced sample also was found to show a signal similar to that Figure 4C. The sharp signal at g = 1.98 is from excess NO in solution. The stronger signal at g = 2 is from a minority (<5%) species of 5-coordinate heme [FeNO]7, from binding of NO to the reduced high-spin impurity. Thus, in the presence of excess NO and limiting reductant, cyt c554 equilibrates to a state in which hemes 1, 3, and 4 are oxidized, and heme 2 is bound with NO as [FeNO].7
Hydroxylamine Titration
HAO catalyzed oxidation of hydroxylamine to nitrite is known to produce NO or HNO as a byproduct.3,44 Clearly, the FeNO species observed in cyt c554 in the presence of HAO and hydroxylamine is due to this byproduct. Presumably, the production of NO or HNO is due to a rate-limited flow of electrons out of HAO. To investigate the transfer and reduction of NO, two samples were titrated with hydroxylamine containing protein ratios [HAO]/[cyt c554] equal to 0.05 and 1.0, under argon atmosphere.
Figure 5A shows the perpendicular (solid line) and parallel mode (dashed line) EPR spectra recorded on a 0.5 mM cyt c554 sample in the presence of 25 μM (5%) HAO, before addition of hydroxylamine. This sample was titrated with hydroxylamine up to 1.2 equivalents per cyt c554. The full titration data set is shown in Supporting Information Figure S1. Figure 5B shows the spectrum recorded after addition of 0.8 equivalents of hydroxylamine. Less than 10% of the fully oxidized protein is present, based on the signals at g = 3.9 and 7.2. Generation of the NO bound signal at g = 2.4 is observable in samples containing 0.6 equiv. and higher of hydroxylamine. The g = 2 signal is from a minority 5-coordinate heme [FeNO]7 species (<10%), similar to that observed in Figure 4, which is sample preparation dependent.
Figure 5.
Selected perpendicular (solid) and parallel (dashed) mode X-band EPR spectra of hydroxylamine titrations of cyt c554 with HAO. (A) 0.5 mM cyt c554 containing 25 μM HAO. (B) Sample A plus 0.4 mM hydroxylamine. (C) 0.8 mM cyt c554 containing 0.8 mM HAO. (D) Sample C plus 10 mM hydroxylamine. (E) spectrum D minus a spectrum of hydroxylamine reduced HAO. All spectra displayed on the same concentration scale, except B and E are magnified by 2. Experimental conditions: temperature 15 K (B1⊥B), 2K (B1∥B), frequency 9.62 GHz (B1⊥ B), 9.24 GHz (B1∥B), power 0.2 mW.
An EPR spectrum for the titration of a 1:1 mixture of (both 0.8 mM) cyt c554 and HAO prior to the addition of hydroxylamine is shown in Figure 5C. The spectrum show signals from both oxidized cyt c554 (g = 3.9, 7.2) and HAO (other signals).45 The parallel mode EPR signal from HAO is significantly weaker and hidden under the cyt c554 parallel mode signal. This sample was titrated with hydroxylamine up to 12.5 equivalents per cyt c554. The full titration data set is shown in Supporting Information Figure S2. Figure 5D shows the spectrum recorded after addition of 12.5 equivalents of hydroxylamine. The oxidized signals from cyt c554 at g = 3.9 (solid line) and 7.2 (dashed line) are absent for this sample, indicating reduction of cyt c554 hemes 1 and 2. For HAO, and consistent with previous results, hydroxylamine causes reduction of the +285 mV heme (sharpening of the g = 3.1 feature), and 50% loss of the 0 mV heme signals (g = 3.4, 2.8, 1.84, 1.67).45,46 Figure 5E shows a difference spectrum of D minus a spectrum of hydroxylamine reduced HAO (see Figure S2). The difference spectrum shows the NO bound signal at g = 2.4. Unlike the previous titration with 5% HAO where the cyt c554 NO species is observed with 0.6 equivalents of hydroxylamine, for the 1:1 titration, a cyt c554 NO species is only observed with more than 10 equivalents of hydroxylamine. The difference spectra for all the titration data (see Figure S3) show that the cyt c554 NO species is not present in the samples with lesser amounts of hydroxylamine. The signal at g = 4.1 is from an [FeNO]7 impurity species (<3%).
Optical Spectroscopy of Ligand Binding to Reduced cyt c554
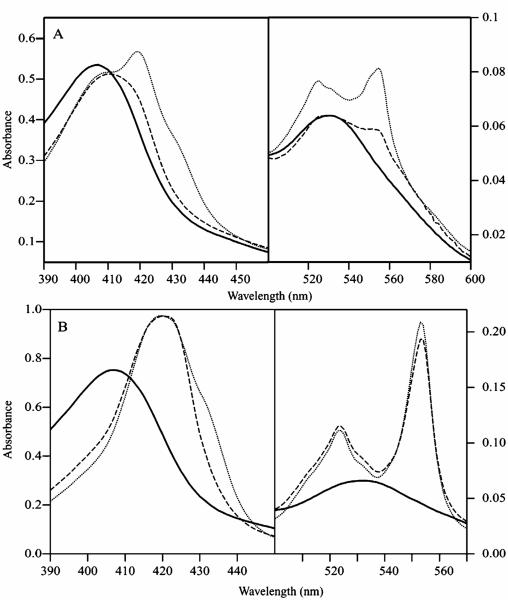
Optical spectra showing the NO addition to the half-reduced and fully reduced cyt c554 are given in Figure 6A and B, respectively. The spectrum of fully oxidized cyt c554 (solid line) shows a broad Soret band from the high-spin and low-spin hemes at 407 nm, and a band at 530 nm from the low-spin hemes.34 Upon half-reduction of the protein with hydrazine in the presence of 5% of HAO (dotted line, Figure 6A), the intensity of the oxidized heme signal at 407 nm drops, and new peaks appear at 420, 524, 554 nm, with a broad shoulder near 430 nm from high-spin ferrous heme 2. The addition of excess NO to the half-reduced protein (dashed line, Figure 6A) caused immediate loss of intensity of the ferrous low-spin heme bands at 420, 554, and 524 nm, indicating oxidation of the low-spin heme 1 center. The high-spin ferrous band at 430 nm disappears, and the Soret maximum shifts to 412 nm, indicating formation of a six coordinate ferrous heme-nitrosyl complex.47-51 The optical spectrum of the half-reduced cyt c554 with NO is therefore consistent with the other spectroscopic data, indicating binding of NO to ferrous high-spin heme 2. The addition of excess NO to the fully oxidized protein (not shown) resulted in no change in the optical spectrum, indicating that cyt c554 does not bind NO in the oxidized state. The addition of excess CO or NO2− under argon atmosphere to the half-reduced protein also showed no change (not shown), indicating that cyt c554 does not bind CO or NO2−.
Figure 6.
Optical spectra showing the effect of NO addition to cyt c554. (A) oxidized (solid line), half-reduced (dotted line), half-reduced + NO (dashed line); (B) oxidized (solid line), fully reduced (dotted line), fully reduced + NO (dashed line).
Figure 6B shows optical spectra for NO addition to the fully reduced cyt c554. The broad shoulder near 430 nm in the reduced spectrum is from high-spin ferrous heme 2, and is similar to that observed in the half-reduced sample. Treatment of the fully reduced protein with NO gas again shows a drop in the intensity of the ferrous high-spin heme band near 430 nm, indicating NO binding to the high-spin heme. The NO bound state of the fully reduced protein is unstable and regenerates the fully reduced state of the protein in the presence of excess reductant in the solution. The addition of excess NO oxidizes the protein to the same state as observed for the half-reduced protein plus NO in Figure 6A. The reoxidation process is dependent upon the amount of excess reductant (dithionite) and NO concentration in the reaction mixture. The addition of excess CO or NO2− to the reduced sample resulted in no significant change, indicating that cyt c554 does not bind CO or NO2− in the fully reduced state of the protein.
Kinetics of NO Reduction
The rate of NO reduction by fully reduced cyt c554 was measured under single turnover conditions. A 1.5-mL portion of 1.85 μM cyt c554 sample was titrated with dithionite to ensure complete reduction without a large excess of reductant in the solution. To the resulting solution, 100 μL of a saturated NO solution (~2 mM at 20 °C) was added to give a final concentration of approximately 125 μM NO in the cuvette. The change in the intensity of absorption band at the 554 nm band were followed at 0.5 s intervals (see Figure S4). The time trace of the 554 nm band minus the absorption at 750 nm (baseline) is shown in the inset. From the optical data of Figure 6 and previous studies,52 the extinction of the band at 554 nm is mainly dependent on the oxidation state of the three low-spin hemes, thus the band represent 3 equivalents of electrons. The time trace appears to be composed of a fast and a slow oxidation process. We suspect that the fast process is the oxidation of the two low potential hemes (heme 3 and 4), and the slower process is oxidation of heme 1. We assume that the NO concentration used in our experiments is sufficiently high to attain a maximum reaction rate and calculate Vmax from the initial slope using an average extinction of 23 mM−1 cm−1 per heme for the oxidation of two lowest potential hemes.52 This gives a turnover number for NO reduction of 16 s−1. This rate represents a lower bound, since its detection is limited by the minimum cycle time of the spectrophotometer (0.5 s) and the mixing time of NO in solution.
We have also attempted to measure the rate of multiple turnover reduction of NO by catalytic amounts of cyt c554 monitoring the loss of the reducing substrate, dithionite, at 315 nm. This band decreased significantly faster in the presence of cyt c554, indicating catalysis of NO reduction by the protein. However, this measurement was complicated by the direct reaction between dithionite and NO. We have not yet been able to develop a better assay because of the reactivity of possible reductants with NO. In attempts to find a suitable reductant, cyt c554 could not be reduced with NADH, malic, or glutamic acids, all of which have reduction potentials lower than the two highest potential hemes (+47 mV) of cyt c554. Another possibility would be to use hydrazine/HAO as the electron source, however, HAO also has a significant interaction with NO.42,53
Measurement of Intermolecular Electron Transfer in cyt c554
The intermolecular electron-transfer rates of cyt c554 was studied by mixing 200 μL of the photoreduced sample with 100 μL of oxidized sample cyt c554 at the same concentration. This mixing ratio was chosen to generate a final protein mixture with 2 reducing equivalents. The samples were frozen at various times and the changes in the EPR signal intensities were monitored after scaling for protein concentration (not shown). The reduction potential of photoactivated riboflavin is −230 mV,54 and will reduce 3 of the hemes (two +47 mV, −147 mV) of cyt c554. The EPR spectra of a photoreduced sample showed only a broad signal at g = 3.6 from the oxidized heme 4 of cyt c554. Thus, hemes 1, 2, and 3 are reduced in this sample. The spectra of the mixed sample showed signals different from the simple sum of the two samples. The EPR spectrum recorded after one minute incubation of the mixed sample at room temperature under argon shows significant reduction in the oxidized signals and appearance of new signal from oxidized heme 3 (g = 3.24). After another one minute of incubation at room temperature under argon, the oxidized signal intensities reduce to less than 10% of the original oxidized sample and the EPR signals typical of the half-reduced state of the protein becomes prominent. Additional incubation at room-temperature did not show any signal change. The results indicate that intermolecular electron transfer occurs on a time scale of one minute, or an approximate rate of 0.01 s−1.
Discussion
Cyt c554 is kinetically competent to accept electrons from HAO during the oxidation of hydroxylamine in keeping with its proposed function in electron transfer for the oxidation of ammonia to nitrite.29 Although one heme is 5-coordinate, no evidence previously indicated that this heme would bind ligands, except for the slow reaction of ferrous cyt c554 with O2 and H2O2. In our previous work, we concluded that cyt c554 would not bind the ligands CN−, CO, NO, and F−.32 Considering the possibility that cyt c554 may bind NO in the reduced state, in this previous work, we reduced samples with an excess of dithionite and then added NO gas under anaerobic conditions. The EPR spectra of those samples before and after NO addition showed no significant changes. We now understand that in the presence of excess reductant, the NO was consumed during the preparation of those samples, resulting in the reduced protein state without NO bound, and thus no change to the observed spectrum.
The results presented here under limiting reductant conditions indicate that cyt c554 binds NO to heme 2 only in the ferrous state and that the binding is highly selective for NO. The common heme binding ligands CN− and NO do not bind ferric cyt c554 in the oxidized state, and CO and nitrite will not bind in the ferrous state. Previous studies have shown that dioxygen will react with reduced cyt c554.55 Importantly, this selection of ligands indicates a substrate specificity favoring a bent mode of ligand coordination to the iron. In general, the ligands of Fe(II)CO, Fe(III)CN, and Fe(III)NO heme complexes bind in a linear conformation, whereas the ligands of Fe(II)NO and Fe(II)O2 complexes adopt a bent conformation.56-58 A partial understanding of the specificity comes from the crystal structure that shows, for both the oxidized and reduced protein, the vacant sixth coordination site of heme 2 is shielded by three hydrophobic residues. Apparently, the steric constraints imposed by these residues do not allow binding of larger molecules, including the substrates of HAO, hydroxylamine or hydrazine, which do not react with cyt c554. However, it is not yet clear which structural aspects impose the more strict specificity observed for the smaller molecules.
We show here the first evidence that cyt c554 has NO reductase activity. In the presence of excess NO, ferrous cyt c554 is oxidized to a state in which the three 6-coordinate hemes are ferric, and the enzymatic site (heme 2) remains ferrous with NO bound. The single turnover kinetic measurements indicate that the rate of the reduction of NO by cyt c554 (>16 s−1) is of the same order of magnitude reported for the nitric oxide reductases (NOR) isolated from Paracoccus denitrificans (10–50 s−1), E. coli (15 s−1), and Pseudomonas stutzeri (<2 s−1).59-62 In multiple turnover reactions, cyt c554 catalytically reduces NO. The spectroscopic data indicate that this reaction proceeds until the depletion of either the reductant or NO. The rate of reduction of NO is nearly 100-fold faster in comparison to O2,55 indicating specificity toward the reaction with NO. These results suggests a possible new function for cyt c554 as an NO reductase.
Reduction of NO by half-reduced cyt c554 is observed here in the presence of excess NO. Samples of the half-reduced state in the presence of no greater than approximately stoichiometric amounts of NO are stable; i.e., in the absence of an excess of NO, heme 2 binds NO without oxidation of heme 1. This suggests a mechanism of NO reduction involving a bimolecular reaction between the NO bound intermediate species and free NO in the solution resulting in the production of N2O. In cyt P450nor, the spontaneous decomposition of the intermediate Fe(II)-NO state has been reported to be extremely slow (0.027 s−1), compare to the fast turnover (1200 s−1) of the catalytic reaction in the presence of excess of free NO in the solution, providing precedent for a bimolecular mechanism of NO reduction.63
Physiological Significance of NO Binding and Reduction
NO is produced in Nitrosomonas by a periplasmic copper-containing nitrite reductase (NiR), using nitrite as a terminal electron acceptor under limiting oxygen conditions. Since the diversion of electrons to nitrite- and nitric oxide reductases from the electrochemical gradient-producing cytochrome aa3 oxidase decreases energy yield, NO is not produced aerobically. It is possible that NO is also produced by premature release from HAO when the limiting oxygen concentration decreases the steady-state flux of electron removal from HAO and therefore poises the hemes of the enzyme at a lower potential. A recent report suggested that cellular concentration of hydroxylamine in N. europaea is approximately 0.8 M.64 If so, NO production observed in the present work with excess of hydroxylamine may possibly be the case in vivo. Under anaerobic conditions, cells must deal with the potential toxicity of NO. The formation of the heme 2 ferrous-NO complex of cyt c554 may prevent the flux of electron transfer from HAO, and thus ultimately prevent NO production by putting the cell into a dormant state. Alternatively, 2 of the 4 electrons derived from the oxidation of a molecule of hydroxylamine to nitrite, are used for the oxidation of ammonia to hydroxylamine and the remaining two electron equivalents can reduce nitrite to NO (by nitrite reductase) and NO to N2O (by NO reductase and/or cyt c554). Such an NOR reductase may serve to the protect the bacteria from the toxic effects of NO. A similar detoxifying function has been reported for pentaheme periplasmic cyt c nitrite reductase from E. coli, type II cyt c′, and cyt p450nor.63,65-67 Recent studies suggest that NO regulates the growth pattern and metabolic pathways of N. europaea, possibly interacting with an as yet unknown target protein in the cell, and altering metabolic pathways.9 Binding of NO to cyt c554, as observed in the present work, may influence the electron-transfer process from HAO to membrane bound cyt cM552,31,68 which in turn can affect ammonia metabolism in N. europaea.
In our previous work, we noted that the hydroxylamine/HAO reduction of cyt c554 results in a different state of cyt c554 than for hydrazine/HAO reduction.32 It is now clear that the differences are due to intermediate products in the oxidation of hydroxylamine by HAO. In the titration of cyt c554 with hydroxylamine in the presence of catalytic amounts of HAO (HAO/cyt c554 = 0.05), a significant amount of NO is detected at heme 2 of cyt c554 for samples containing 0.6 equivalents or greater of hydroxylamine. Whereas for an HAO/cyt c554 ratio of 1, 12 equivalents of hydroxylamine were required to detect the NO bound species. The differences in these two titrations can be rationalized by the limitations in the rates of electron transfer relative to the rate of substrate oxidation by HAO. For low ratios of HAO/cyt c554, the transfer of electrons to cyt c554 is limited by the availability of binding sites to HAO, and the rate of intermolecular electron transfer between cyt c554. The measurements of the intermolecular electron transfer between cyt c554 indicate a slow transfer rate (0.01 s−1) relative to the catalytic turnover rate of HAO (30 s−1). The inefficient donation of electrons out of HAO results in increased amounts of intermediate product formation as detected by binding of NO to cyt c554. For the 1:1 ratio of HAO/cyt c554, the transfer of electrons to cyt c554 is efficient. The NO intermediates generated by HAO are now more efficiently consumed by cyt c554, thus NO detectable species are only observable with higher amount of substrate.
In recent work, Pacheco and co-workers have detected an one-electron oxidation event of a c-heme in HAO within 10 ms of the reaction between partially reduced HAO and NO.53 This event was NO dependent and thus associated with the reduction of [FeNO]6 at the active site heme of HAO. The fate of the product [FeNO]7 species at longer times is unclear. The byproduct of the HAO hydroxylamine reaction may be HNO/NO− or NO. We detect an [FeNO]7 species in cyt c554 which could be formed from either HNO/NO− or from NO with electron transfer through HAO. Since the time scales of our reactions are significantly longer than 10 ms, a direct comparison of results is not possible.
Conclusion
We have shown that cyt c554 binds NO and reduces it at a rate comparable to other known NO reductase. This is the first evidence that cyt c554 can function as an enzyme. This process may have an important role in regulating the free NO concentration in N. europaea. Alternatively, since cyt c554 is an important electron transfer protein in the oxidation pathway, binding of NO to this protein may significantly affect the respiratory electron transport chain in the bacteria.
Supplementary Material
Footnotes
Supporting Information Available: The complete titration sets of EPR spectra and time dependent optical spectra (all PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Abeliovich A, Vonshak A. Arch. Microbiol. 1992;158:267–270. [Google Scholar]
- 2.Schmidt I, Bock E. Arch. Microbiol. 1997;167:106–111. [PubMed] [Google Scholar]
- 3.Beaumont HJE, Hommes NG, Sayavedra-Soto LA, Arp DJ, Arciero DM, Hooper AB, Westerhoff HV, Van Spanning RJM. J. Bacteriol. 2002;184:2557–2560. doi: 10.1128/JB.184.9.2557-2560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt I, Bock E. Antonie van Leeuwenhoek. 1998;73:271–278. doi: 10.1023/a:1001572121053. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt I, Bock E, Jetten MSM. Microbiology. 2001;147:2247–2253. doi: 10.1099/00221287-147-8-2247. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt I, Hermelink C, Van de Pas-Schoonen K, Strous M, op den Camp HJ, Kuenen JG, Jetten MSM. Appl. Environ. Microbiol. 2002;68:5351–5357. doi: 10.1128/AEM.68.11.5351-5357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt I, Zart D, Bock E. Antonie van Leeuwenhoek. 2001;79:39–47. doi: 10.1023/a:1010269331350. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt I, Zart D, Bock E. Antonie Van Leeuwenhoek. 2001;79:311–318. doi: 10.1023/a:1012038314206. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt I, Steenbakkers PJM, op den Camp HJM, Schmidt K, Jetten MSM. J. Bacteriol. 2004;186:2781–2788. doi: 10.1128/JB.186.9.2781-2788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein LY, Arp DJ. Appl. Environ. Microbiol. 1998;64:4098–4102. doi: 10.1128/aem.64.10.4098-4102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu KE, Valentine AM, Wang D, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ. NATO ASI Series, Series 2: Environment. 1997;26:273–286. [Google Scholar]
- 12.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, Sayavedra-Soto L, Arciero D, Hommes N, Whittaker M, Arp D. J. Bacteriol. 2003;185:2759–2773. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt I, van Spanning RJ, Jetten MS. Microbiology. 2004;150:4107–4114. doi: 10.1099/mic.0.27382-0. [DOI] [PubMed] [Google Scholar]
- 14.Beaumont HJ, van Schooten B, Lens SI, Westerhoff HV, van Spanning RJ. J. Bacteriol. 2004;186:4417–4421. doi: 10.1128/JB.186.13.4417-4421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iverson TM, Hendrich MP, Arciero DM, Hooper AB, Rees DC. In: Handbook of Metalloproteins. Messerschmidt A, Huber R, Poulos TL, Wieghardt K, editors. John Wiley & Sons; Chichester: 2001. pp. 136–146. [Google Scholar]
- 16.Iverson TM, Arciero DM, Hsu BT, Logan MSP, Hooper AB, Rees DC. Nat. Struct. Biol. 1998;5:1005–1012. doi: 10.1038/2975. [DOI] [PubMed] [Google Scholar]
- 17.Iverson TM, Arciero DM, Hooper AB, Rees DC. J. Biol. Inorg. Chem. 2001;6:390–397. doi: 10.1007/s007750100213. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi N, Moriyama H, Fujiwara T, Fukumori Y, Tanaka N. Nat. Struct. Biol. 1997;4:276–284. doi: 10.1038/nsb0497-276. [DOI] [PubMed] [Google Scholar]
- 19.Einsle O, Messerschmidt A, Stach P, Bourenkov GP, Bartunik HD, Huber R, Kroneck PMH. Nature. 1999;400:476–480. doi: 10.1038/22802. [DOI] [PubMed] [Google Scholar]
- 20.Bamford VA, Angove HC, Seward HE, Thomson AJ, Cole JA, Butt JN, Hemmings AM, Richardson DJ. Biochemistry. 2002;41:2921–2931. doi: 10.1021/bi015765d. [DOI] [PubMed] [Google Scholar]
- 21.Bamford V, Dobbin PS, Richardson DJ, Hemmings AM. Nat. Struct. Biol. 1999;6:1104–1107. doi: 10.1038/70039. [DOI] [PubMed] [Google Scholar]
- 22.Taylor P, Pealing SL, Reid GA, Chapman SK, Walkinshaw MD. Nat. Struct. Biol. 1999;6:1108–1112. doi: 10.1038/70045. [DOI] [PubMed] [Google Scholar]
- 23.Leys D, Tsapin AS, Nealson KH, Meyer TE, Cusanovich MA, Van Beeumen JJ. Nat. Struct. Biol. 1999;6:1113–1117. doi: 10.1038/70051. [DOI] [PubMed] [Google Scholar]
- 24.Brige A, Leys D, Meyer TE, Cusanovich MA, Van Beeumen JJ. Biochemistry. 2002;41:4827–4836. doi: 10.1021/bi012144b. [DOI] [PubMed] [Google Scholar]
- 25.Heitmann D, Einsle O. Biochemistry. 2005;44:12411–12419. doi: 10.1021/bi0509999. [DOI] [PubMed] [Google Scholar]
- 26.Matias PM, Morais J, Coelho AV, Meijers R, Gonzalez A, Thompson AW, Sieker L, Legall J, Carrondo MA. J. Biol. Inorg. Chem. 1997;2:507–514. [Google Scholar]
- 27.Bergmann DJ, Hooper AB, Klotz MG. Appl. Environ. Microbiol. 2005;71:5371–5382. doi: 10.1128/AEM.71.9.5371-5382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamanaka T, Shinra M. J. Biochem. (Tokyo) 1974;75:1265–1273. doi: 10.1093/oxfordjournals.jbchem.a130510. [DOI] [PubMed] [Google Scholar]
- 29.Arciero DM, Balny C, Hooper AB. Biochemistry. 1991;30:11466–11472. doi: 10.1021/bi00112a014. [DOI] [PubMed] [Google Scholar]
- 30.DiSpirito AA, Taaffe LR, Hooper AB. Biochim. Biophys. Acta. 1985;806:320–330. [Google Scholar]
- 31.Whittaker M, Bergmann D, Arciero D, Hooper AB. Biochim. Biophys. Acta. 2000;1459:346–355. doi: 10.1016/s0005-2728(00)00171-7. [DOI] [PubMed] [Google Scholar]
- 32.Upadhyay AK, Petasis DT, Arciero DM, Hooper AB, Hendrich MP. J. Am. Chem. Soc. 2003;125:1738–1747. doi: 10.1021/ja020922x. [DOI] [PubMed] [Google Scholar]
- 33.Andersson KK, Kent TA, Lipscomb JD, Hooper AB, Munck E. J. Biol. Chem. 1984;259:6833–6840. [PubMed] [Google Scholar]
- 34.Andersson KK, Lipscomb JD, Valentine M, Munck E, Hooper AB. J. Biol. Chem. 1986;261:1126–1138. [PubMed] [Google Scholar]
- 35.Massey V, Stankovich M, Hemmerich P. Biochemistry. 1978;17:1–8. doi: 10.1021/bi00594a001. [DOI] [PubMed] [Google Scholar]
- 36.Wilhelm E, Battino R, Wilcock RJ. Chem. Rev. 1977;77:219–262. [Google Scholar]
- 37.Petasis DT, Hendrich MP. J. Magn. Reson. 1999;136:200–206. doi: 10.1006/jmre.1998.1657. [DOI] [PubMed] [Google Scholar]
- 38.Oosterhuis WT, Lang G. J. Chem. Phys. 1969;50:4381–4387. [Google Scholar]
- 39.Liu MC, Huynh Boi H, Payne WJ, Peck HD, Jr., Dervartanian DV, Legall J. Eur. J. Biochem. 1987;169:253–258. doi: 10.1111/j.1432-1033.1987.tb13605.x. [DOI] [PubMed] [Google Scholar]
- 40.Debrunner PG. Physical Bioinorganic Chemistry Series. 1989;4:137–234. [Google Scholar]
- 41.Costa C, Moura JJG, Moura I, Liu MY, Peck HD, Jr., LeGall J, Wang Y, Huynh Boi H. J. Biol. Chem. 1990;265:14382–14288. [PubMed] [Google Scholar]
- 42.Hendrich MP, Upadhyay AK, Riga J, Arciero DM, Hooper AB. Biochemistry. 2002;41:4603–4611. doi: 10.1021/bi011332z. [DOI] [PubMed] [Google Scholar]
- 43.Christner JA, Munck E, Janick PA, Siegel LM. J. Biol. Chem. 1983;258:11147–11156. [PubMed] [Google Scholar]
- 44.Hooper AB, Terry KR. Biochim. Biophys. Acta. 1979;571:12–20. doi: 10.1016/0005-2744(79)90220-1. [DOI] [PubMed] [Google Scholar]
- 45.Hendrich MP, Petasis D, Arciero DM, Hooper AB. J. Am. Chem. Soc. 2001;123:2997–3005. doi: 10.1021/ja002982d. [DOI] [PubMed] [Google Scholar]
- 46.Arciero DM, Golombek A, Hendrich MP, Hooper AB. Biochemistry. 1998;37:523–529. doi: 10.1021/bi972187l. [DOI] [PubMed] [Google Scholar]
- 47.Meyer TE, Kamen MD. Adv. Protein Chem. 1982;35:105–212. doi: 10.1016/s0065-3233(08)60469-6. [DOI] [PubMed] [Google Scholar]
- 48.Andrew CR, Green EL, Lawson DM, Eady RR. Biochemistry. 2001;40:4115–4122. doi: 10.1021/bi0023652. [DOI] [PubMed] [Google Scholar]
- 49.Andrew CR, George SJ, Lawson DM, Eady RR. Biochemistry. 2002;41:2353–2360. doi: 10.1021/bi011419k. [DOI] [PubMed] [Google Scholar]
- 50.Reynolds MF, Parks RB, Burstyn JN, Shelver D, Thorsteinsson MV, Kerby RL, Roberts GP, Vogel KM, Spiro TG. Biochemistry. 2000;39:388–396. doi: 10.1021/bi991378g. [DOI] [PubMed] [Google Scholar]
- 51.Ballou DP, Zhao Y, Brandish PE, Marletta MA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12097–12101. doi: 10.1073/pnas.192209799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arciero DM, Collins MJ, Haladjian J, Bianco P, Hooper AB. Biochemistry. 1991;30:11459–11465. doi: 10.1021/bi00112a013. [DOI] [PubMed] [Google Scholar]
- 53.Cabail MZ, Kostera J, Pacheco AA. Inorg. Chem. 2005;44:225–231. doi: 10.1021/ic048822a. [DOI] [PubMed] [Google Scholar]
- 54.Tollin G. J Bioenerg Biomembr. 1995;27:303–309. doi: 10.1007/BF02110100. [DOI] [PubMed] [Google Scholar]
- 55.DiSpirito AA, Balny C, Hooper AB. Eur. J. Biochem. 1987;162:299–304. doi: 10.1111/j.1432-1033.1987.tb10600.x. [DOI] [PubMed] [Google Scholar]
- 56.Reynolds MF, Burstyn JN. Nitric Oxide. 2000:381–399. [Google Scholar]
- 57.Yang J, Kloek AP, Goldberg DE, Mathews FS. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4224–4228. doi: 10.1073/pnas.92.10.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaanan B. J. Mol. Biol. 1983;171:31–59. doi: 10.1016/s0022-2836(83)80313-1. [DOI] [PubMed] [Google Scholar]
- 59.Zumft WG. Microbiol. Mol. Biol. Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hendriks J, Warne A, Gohlke U, Haltia T, Ludovici C, Luebben M, Saraste M. Biochemistry. 1998;37:13102–13109. doi: 10.1021/bi980943x. [DOI] [PubMed] [Google Scholar]
- 61.Gomes CM, Giuffre A, Forte E, Vicente JB, Saraiva LM, Brunori M, Teixeira M. J. Biol. Chem. 2002;277:25273–25276. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- 62.Forte E, Urbani A, Saraste M, Sarti P, Brunori M, Giuffre A. Eur. J. Biochem. 2001;268:6486–6490. doi: 10.1046/j.0014-2956.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- 63.Shiro Y, Fujii M, Iizuka T, Adachi S, Tsukamoto K, Nakahara K, Shoun H. J. Biol. Chem. 1995;270:1617–23. doi: 10.1074/jbc.270.4.1617. [DOI] [PubMed] [Google Scholar]
- 64.Schmidt I, Look C, Bock E, Jetten MS. Microbiology. 2004;150:1405–1412. doi: 10.1099/mic.0.26719-0. [DOI] [PubMed] [Google Scholar]
- 65.Cross R, Aish J, Paston SJ, Poole RK, Moir JW. J. Bacteriol. 2000;182:1442–1447. doi: 10.1128/jb.182.5.1442-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poock SR, Leach ER, Moir JWB, Cole JA, Richardson DJ. J. Biol. Chem. 2002;277:23664–23669. doi: 10.1074/jbc.M200731200. [DOI] [PubMed] [Google Scholar]
- 67.Yoshimura T, Iwasaki H, Shidara S, Suzuki S, Nakahara A, Matsubara T. J. Biochem. (Tokyo) 1988;103:1016–1019. doi: 10.1093/oxfordjournals.jbchem.a122372. [DOI] [PubMed] [Google Scholar]
- 68.Arp DJ, Stein LY. Crit. Rev. Biochem. Mol. Biol. 2003;38:471–495. doi: 10.1080/10409230390267446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.