SUMMARY
Seventy-five percent of lung adenocarcinomas with epidermal growth factor receptor (EGFR) mutations respond to treatment with the tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib; however, drug-resistant tumors eventually emerge. In 60% of cases, resistant tumors carry a secondary mutation in EGFR (T790M), amplification of MET, or both. Here, we describe the establishment of erlotinib resistance in lung tumors, which were induced by mutant EGFR, in transgenic mice after multiple cycles of drug treatment; we detect the T790M mutation in five out of 24 tumors or Met amplification in one out of 11 tumors in these mice. This preclinical mouse model, therefore, recapitulates the molecular changes responsible for resistance to TKIs in human tumors and holds promise for the discovery of additional mechanisms of drug resistance in lung cancer.
INTRODUCTION
Lung adenocarcinomas with mutations in exons encoding the tyrosine kinase domain of the epidermal growth factor receptor (EGFR) gene are associated with sensitivity to tyrosine kinase inhibitors (TKIs) (Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004). Nevertheless, tumors that initially respond to TKI treatment invariably progress on therapy. In approximately 50% of drug-resistant tumors, resistance is associated with the presence of a secondary mutation in EGFR that substitutes methionine for threonine at position 790 (T790M) in the kinase domain of the protein (Kobayashi et al., 2005; Pao et al., 2005a). MET amplification has also been documented in 20% of TKI-resistant lung cancers, irrespective of the presence of a T790M mutation (Bean et al., 2007; Engelman et al., 2007). The molecular basis for drug resistance in the remaining 30–40% of tumors remains elusive, and effective therapies to prevent and overcome resistance to the currently used TKIs in lung cancer are not known.
Previously, we developed tetracycline-inducible mouse models of EGFR-dependent lung cancer. In these animal models, pneumocyte-specific expression of a human transgene containing either the EGFRL858R point mutant or an exon 19 deletion mutant (EGFRΔL747-S752), two common EGFR mutants that are observed in human lung adenocarcinoma (Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004), gives rise to lung adenocarcinomas with bronchioloalveolar features that are dependent on the continued presence and activity of the mutant receptor for survival (Ji et al., 2006; Politi et al., 2006). Importantly, treatment of lung tumor-bearing mice with the TKI erlotinib causes tumor regression. These similarities between the animal models and the human disease prompted us to test whether long-term erlotinib treatment of mutant EGFR-driven lung tumors in mice would lead to the emergence of drug-resistant tumors that could then provide further insight into the molecular basis of TKI resistance in human patients.
RESULTS
Generation of erlotinib-resistant tumors in transgenic mice
In an initial pilot experiment, we treated two lung tumor-bearing mice with 25 mg/kg/day of erlotinib, 5 days per week for 5 months, and observed sustained and complete tumor regression. This suggested that continuous erlotinib treatment at this dose was excellent therapy but an inefficient means of generating drug-resistant tumors. We then reasoned that intermittent treatment might allow the remaining cells to expand during drug-free intervals and acquire additional mutations; if a mutation conferred drug resistance, the mutant clone would continue to grow. We further predicted that mice with a large tumor burden [as measured by magnetic resonance imaging (MRI)] would be more likely to have genetically complex tumors that would exhibit either primary or secondary resistance to erlotinib.
To test this, we used 11 mice with lung tumors that were induced by mutant EGFR and treated them with erlotinib using an intermittent dosing protocol (Fig. 1A; Table 1). The presence of lung tumors at the beginning of treatment was determined by MRI in ten of the 11 mice. When possible, mice with clearly discernible tumor nodules and/or consolidations involving a whole lung lobe were selected for treatment. Animals received 25 mg/kg/day of erlotinib, 5 days per week for 4 weeks, and then erlotinib was discontinued for 4 weeks while maintaining the mice on a diet containing doxycycline to ensure continued expression of the transgene (Fig. 1A). This ‘on drug/off drug cycle’ was repeated one to three times. Tumor growth and regression were monitored by MRI at the end of each treatment period and at the end of each drug-free month. We used these images to measure tumor volume in mice in which dense consolidations and/or tumor nodules could easily be demarcated and distinguished from the heart (supplementary material Table S1). Although multiple tumor nodules were generally observed in each mouse at the time of necropsy, only the largest one or two were amenable to tumor volume measurements using MRI. Nine of eleven mice that underwent multiple rounds of erlotinib treatment had tumors at the time of sacrifice (Table 1).
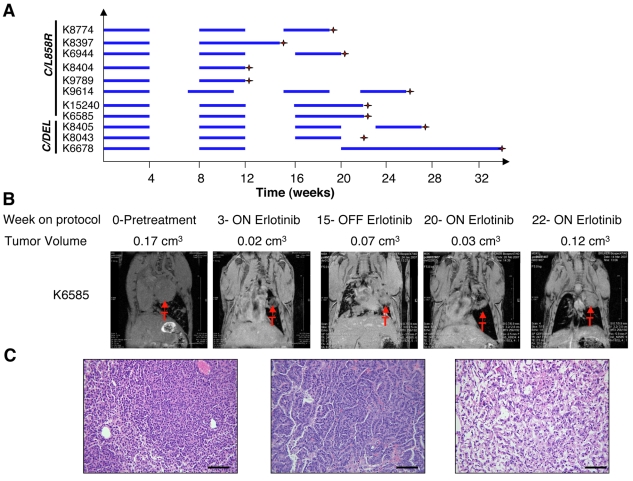
Fig. 1.
Erlotinib-resistant lung adenocarcinomas emerge after long-term intermittent drug treatment. (A) Line chart depicting the schedule used to treat individual mice with erlotinib. Mice were treated 5 days per week for 4 weeks (blue horizontal bars indicate treatment with 25 mg/kg/day of erlotinib) after which treatment was interrupted for 4 weeks (no bars). Erlotinib was administered sooner to animals that became cachectic during this period (see, for example, K9614). The treatment cycle was repeated up to three times. Doxycycline administration was initiated shortly after weaning and subsequently kept constant throughout the life of the animal. Red crosses indicate the time point at which the mouse was sacrificed. K15240, K6585 and K8043 were heterozygous for p53, and K6678 was heterozygous for Ink4a/Arf (also known as Cdkn2a). C=CC10-rtTA, L858R=TetO-EGFRL858R, DEL=TetO-EGFRΔL747-S752. (B) Coronal magnetic resonance images of lungs from a C/L858R/p53+/− (K6585) mouse subjected to the erlotinib treatment protocol. At the end of the final treatment cycle, erlotinib treatment was continued and the tumor volume increased despite the presence of the drug. Tumor volume measurements are shown. (C) Hematoxylin and eosin images of lung adenocarcinomas in mice after multiple cycles of erlotinib treatment (left and center panels). Areas of scarring where tumors had apparently regressed were also observed in the lungs of the mice (right panel). Bars, 200 μm.
Table 1.
Summary of sequencing and quantitative PCR data
During the first round of treatment, six of seven tumors for which tumor volume measurements were available showed a decrease in tumor volume of greater than 40%. Four of these tumors shrank by greater than 90%. In one of the mice (K6944) (see supplementary material Table S1), one tumor nodule responded to drug treatment (90% decrease in tumor volume) whereas a second nodule did not (6% decrease in tumor volume). In humans, decreases in tumor size of greater than 30% are considered to be partial responses according to RECIST (response evaluation criteria in solid tumours) criteria, and increases in tumor size must be greater than 20% to qualify as progressive disease. Following these guidelines, we can conclude that six of these tumors responded to erlotinib treatment and one tumor showed evidence of stable disease. Three animals with symptoms of respiratory distress that did not improve during the second round of treatment were sacrificed. The remaining animals underwent a total of three or four rounds of erlotinib treatment and were sacrificed when MRI indicated that recurrent or persistent lung tumors were present despite prolonged treatment, or when the mice displayed signs of illness. Tumor volume measurements obtained during the final round of treatment indicated that five of six tumors (in five mice) had become smaller. However, the decreases in tumor volume were consistently less than the decreases observed during the first round of treatment (supplementary material Table S1). The diminished response during the final round of treatment suggests that the drug-free intervals permitted the outgrowth of both drug-resistant and drug-sensitive tumor cells. Notably, in two cases in which mice with measurable tumor nodules were maintained on erlotinib for more than 4 weeks during the final round of treatment, tumor volume increased significantly during the additional treatment time (by 43% for K6678 and by 275% for K6585) (Fig. 1B); this implies that tumor cells had acquired the ability to grow despite the continued administration of the drug.
Upon sacrifice, all tumors that were present on the lungs of the mice were collected for molecular analysis (and, when large enough, histopathological evaluation) and included in our study of drug resistance, regardless of whether we had determined that they were growing using MRI. We did this because it proved difficult to identify growing tumors reliably using MRI, especially in animals with multiple tumors after multiple rounds of therapy. Since most tumors showed significantly reduced responses to erlotinib after several rounds of treatment, we assumed that any residual tumors probably harbored drug-resistant tumor cells; hence they were harvested and analyzed to avoid missing any drug-resistant tumors. Thus, in contrast to the definition of resistance in human disease, our criteria for including tumors in the study of resistance did not include progressive disease, as defined by RECIST (i.e. a 20% increase in tumor size), during drug treatment. Formally, it is possible that some of the tumors collected were persistent rather than bona fide drug-resistant tumors, leading to an underestimate of the frequency of any observed resistance-conferring mutation.
Tumors identified in mice that had undergone multiple rounds of treatment with erlotinib were solid and/or papillary lung adenocarcinomas (Fig. 1C, left and center panels), mostly surrounded by normal lung. Occasional focal areas of atypical adenomatous hyperplasia (AAH) and bronchioloalveolar carcinoma (BAC), and areas indicative of tumor regression (Fig. 1C, right panel), were also observed in the lung epithelium of these mice. These tumors were histologically indistinguishable from adenocarcinomas arising in untreated mice (data not shown).
Secondary mutations in the EGFR transgene and Met amplification in erlotinib-resistant tumors
We first asked whether secondary mutations in the EGFR transgene could account for the drug-resistant tumors. We generated cDNA from RNA that was extracted from individual tumor nodules and from matched normal lung, and sequenced part of the human EGFR transgene cDNA spanning the kinase domain in these samples (nucleotides 2150–2600). We detected a secondary cytosine (C)-to-thymine (T) point mutation at position 2369, causing the T790M amino acid substitution, in five of 24 (21%) tumors harvested from mice that were subjected to multiple cycles of therapy (Fig. 2A; Table 1), but the mutation was not detected in matched normal lung from the same animals or in 17 tumors from untreated animals. As expected, the EGFR mutation that was associated with drug sensitivity and present in the original transgene (either L858R or ΔL747-S752) was detected in all of the above samples (Fig. 2B).
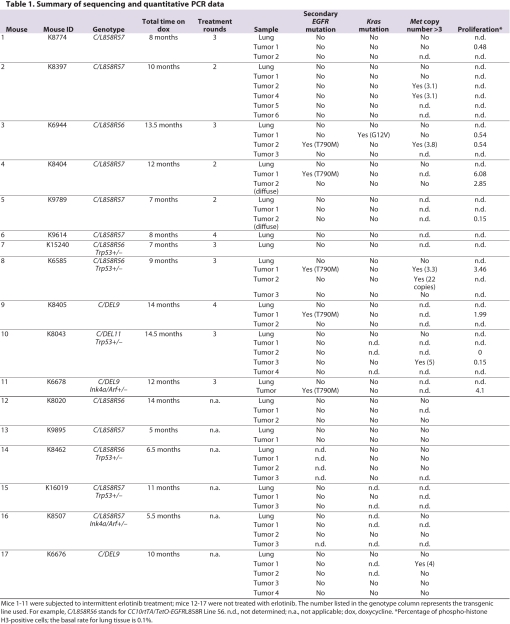
Fig. 2.
Secondary mutations in EGFR and Met amplification in drug-resistant tumors. (A,B) DNA sequencing chromatograms reveal the presence of the T790M mutation in a lung tumor from an L858R-expressing mouse after multiple rounds of erlotinib treatment. (A) The exon 20 T790M mutation is detected only in the lung tumor and not in matched normal lung. The mutant peak is red in the forward direction and green in the reverse direction. Both the wild-type and mutant sequence are detected in the resistant tumor. (B) The exon 21 L858R mutation is detected in the lung tumor and adjacent normal lung. (C,D) Met amplification and overexpression in a lung tumor from a C/L858R/p53+/− mouse harvested after three cycles of erlotinib treatment. (C) Met copy number assayed by quantitative PCR. The copy number for each tumor sample was calculated relative to the adjacent lung from the same animal, under the assumption that the Met copy number in the adjacent lung is two. (D) Met expression assayed by quantitative reverse transcriptase PCR. Met expression in the tumor sample relative to the adjacent lung from each individual mouse is plotted.
In four of the five cases in which we detected a T790M mutation, the size of the peak representing a thymine at position 2369 of human EGFR was equal to, or smaller than, the size of the peak representing a cytosine (Fig. 2A). This is consistent with the original mRNA encoded by the transgene being more abundant than, or equal to, that of the mRNA with the secondary mutation, similar to the situation observed in human lung tumors that are resistant to TKIs (Pao et al., 2005a; Engelman et al., 2006). Since many of the TKI-resistant human tumors exhibit EGFR amplification, it is likely that only a fraction of the copies of the EGFR gene have the T790M mutation (Engelman et al., 2006). An analogous scenario is possible in which multiple copies of the EGFR transgene are integrated in the mouse genome but only some of the copies contain the secondary mutation. There may be other explanations for the low abundance of the T790M mutation: it could be that only a fraction of the cancer cells in the drug-resistant tumors have the T790M mutation, or the tumors may contain normal lung epithelial cells.
We also sequenced endogenous Kras from cDNA generated from 21 tumors that were harvested from mice that underwent multiple rounds of erlotinib treatment, to test whether mutations in this gene were associated with drug resistance. We detected a guanine (G)-to-thymine (T) transversion leading to a G12V amino acid substitution in one of these tumors (4.7%; K6944, tumor 1) (Table 1). Lung tumors bearing Kras mutations can occur spontaneously in aging mice with variable frequencies depending on the mouse strain (Dragani et al., 1995). This tumor did not respond to erlotinib treatment (it appeared to decrease by 6% in the first round of treatment and to increase by 2% in the final round of treatment) (see supplementary material Table 1), strongly indicating that it was a Kras-driven tumor, unrelated to the expression of mutant EGFR. Of note, KRAS mutations have been implicated as a mechanism of primary resistance but not acquired resistance to TKIs in humans (Pao et al., 2005a; Pao et al., 2005b). No Kras mutations were detected in cDNA derived from matched normal lung or from ten tumors from untreated mice (Table 1).
Since the T790M mutation was found in only about 20% of tumors tested for resistance mutations, we asked whether Met amplification, also reported as a mechanism of erlotinib resistance, could be detected in any of the tumors. Using quantitative PCR, we found that one of 11 tumors harvested from mice that were subjected to multiple rounds of erlotinib had 22 copies of the Met proto-oncogene (Fig. 2C; Table 1). An additional five tumors showed an increased Met copy number (four tumors with a copy number of three to four, and one tumor with a copy number of five) (Table 1). Of nine untreated tumors that were analyzed, only one showed a copy number increase (to four copies) (Fig. 2C; Table 1). To establish whether Met amplification was associated with increased production of Met RNA, we analyzed 15 untreated tumors and 20 tumors that were presumed to be resistant by quantitative reverse transcriptase PCR (results for a subset of tumors are shown in Fig. 2D). We observed that the tumor sample displaying high-level amplification of the Met gene also showed an approximately 20-fold increase in expression of Met RNA compared with the adjacent lung (Fig. 2D). However, we did not find increased levels of Met RNA in the tumor samples from the untreated and treated mice that showed Met copy numbers of three to five (data not shown).
MET amplification has been shown to produce resistance to TKIs by activating the ERBB3-PI3K (phosphoinositide 3-kinase) signaling pathway (Engelman et al., 2007). Moreover, mutations in PIK3CA and loss of PTEN have been observed in lung tumors harboring EGFR mutations, indicating that potentiation of the PI3K signaling pathway may play a role in mediating tumorigenesis induced by mutant EGFR (Endoh et al., 2006; Kawano et al., 2006). To investigate whether Erbb3 was amplified in erlotinib-resistant tumors, we examined Erbb3 copy number and expression using quantitative PCR, but did not observe any differences between tumors from treated animals and untreated samples (data not shown). To further explore whether perturbations in the PI3K pathway could constitute a mechanism of erlotinib resistance, we sequenced Pik3ca and Pten mutation hotspots (exons 9 and 20 in Pik3ca, and exons 2–8 in Pten; these also correspond to mutation hotspots in the human genes) in cDNA generated from RNA that was extracted from 20 and ten tumors, respectively, from mice that received multiple cycles of drug treatment; however, we did not find mutations in these regions of either gene.
In an effort to identify novel mechanisms of drug resistance using these mouse models, we have begun to collect the expression data derived from drug-resistant lung adenocarcinomas. To investigate whether upregulation of drug-efflux transporters could contribute to drug resistance, we examined the expression of Abcb1a, Abcb1b and Abcg2 in six tumors harvested from mice that underwent multiple rounds of treatment compared with matched normal tissue. These genes were modestly downregulated in the tumors (Abcb1a, twofold; Abcb1b, 1.88-fold; Abcg2, 1.52-fold), regardless of whether the tumor harbored a T790M mutation (n=1), a Kras mutation (n=1) or no known mutation (n=4), indicating that upregulation of drug transporters is unlikely to account for drug resistance.
Variable signaling pathway activation in erlotinib-resistant tumors
We previously showed that mutant EGFR-induced tumors exhibited activation of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) and PI3K signaling pathways (Politi et al., 2006). Presumably, one or both of these signaling pathways must be activated for erlotinib-resistant tumors to emerge. To gain insight into the signaling pathways that remain active in erlotinib-resistant tumors, and that possibly contribute to resistance itself, we stained tumor sections with an antibody that recognizes phosphorylated EGFR (p-Tyr 992, this tyrosine residue is in the cytoplasmic tail of EGFR and is phosphorylated when the kinase is active). Seven out of eight tumors arising in mice after multiple cycles of erlotinib treatment stained positively for phosphorylated EGFR (the exception being the tumor that harbored the KrasG12V mutation) (Fig. 3A). This is consistent with the notion that resistance requires restoration of the kinase activity of EGFR, or activation of some other kinase that is capable of phosphorylating EGFR, despite continued administration of erlotinib. Furthermore, the lungs of mice that had undergone multiple rounds of erlotinib treatment also contained tumor nodules that were too small to be screened for secondary mutations, and cells in these nodules also stained positive for phosphorylated EGFR.
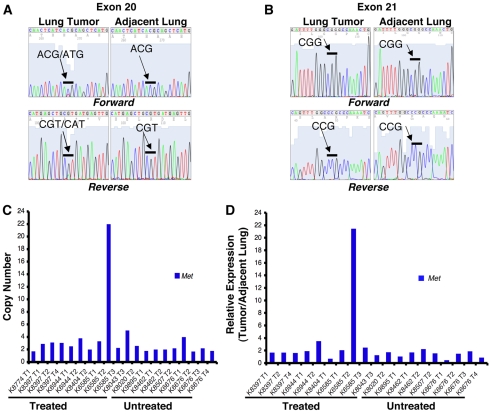
Fig. 3.
Signaling pathway activation and proliferation of erlotinib-resistant tumors. (A) Representative immunohistochemical staining of lung adenocarcinomas, harvested from mice that underwent multiple cycles of erlotinib treatment or that were left untreated, for phospho-EGFR (top panels), phospho-Erk (middle panels) and phospho-Akt (bottom panels). Phospho-EGFR and phospho-Akt staining were observed in all samples with the exception of the tumor harboring a Kras mutation. (Note that there is macrophage staining for phospho-Akt in this sample, but not tumor cell staining.) Phospho-Erk staining was more variable, but was strong in tumors without the T790M mutation. (B) Phospho-histone H3 staining indicating that tumors harvested from mice subjected to multiple rounds of erlotinib treatment are proliferating. The presence of known secondary mutations in the samples is indicated. Bars, 200μm.
We next stained sections with antibodies to phosphorylated Erk (p-Thr 202, p-Tyr 204) and Akt (p-Ser 473) to examine the components of signaling pathways downstream of EGFR. When we compared phospho-Akt and phospho-Erk staining in tumors from untreated mice with those that had received multiple cycles of erlotinib, in most cases Akt was consistently phosphorylated in both sets of tumors (excluding the tumor harboring KrasG12V) (Fig. 3A). By contrast, strong uniform staining for phospho-Erk was observed in two tumors that did not have the T790M mutation (Fig. 3A) and in the tumor with the KrasG12V mutation, but not in tumors with the T790M mutation (n=4) (Fig. 3A). The tumors with T790M mutations were either negative for phospho-Erk (n=1) or exhibited non-uniform patches of positively stained cells (n=3), similar to patterns observed in untreated tumors (Fig. 3A). This pattern of phospho-Erk staining is consistent with staining that we have observed in lung tumors from transgenic mice expressing the EGFRL858R+T790M mutation (K.P. and W. Pao, unpublished).
Taken together, these results indicate that phosphorylation of EGFR and activation of the PI3K-Akt signaling pathway, but not the Mapk/Erk pathway, appear to be consistently associated with EGFR-induced tumorigenesis, regardless of whether the tumors are untreated or drug resistant. The strong phospho-Erk staining observed in the tumor with a Kras mutation indicates that the Mapk/Erk pathway is associated with oncogenic Ras signaling and suggests that any drug-resistant tumors that show strong phospho-Erk staining should be examined further for Ras pathway activation.
To determine whether the staining of phosphoproteins implicated in cell signaling correlated with proliferation of cells in drug-resistant lung adenocarcinomas, we stained tumors with a phospho-histone H3 antibody that detects cells undergoing mitosis. Ten of 11 tumors harvested from mice that had undergone multiple rounds of erlotinib treatment had a percentage of phospho-histone H3-positive cells that was slightly or markedly above the basal rate for normal lung (0.1%) (Table 1). These results indicate that, in most cases, the tumor cells are proliferating despite treatment with erlotinib (Fig. 3B). Within this limited subset of tumors it also appears that tumors with the T790M mutation are undergoing cell division more rapidly than tumors without this mutation. However, future studies of additional tumors will conclusively determine whether the specific secondary mutation influences the proliferation rate of individual drug-resistant tumors. When lung tissue samples from these same mice were stained for phospho-histone H3, cells in the small foci of AAH or BAC, mentioned earlier, did not appear to be proliferating above the basal rate for lung tissue and may represent dormant tumor cells that persisted despite drug treatment.
DISCUSSION
Genetically engineered mouse models of cancer have potential as preclinical models for testing new treatment regimens and for studying mechanisms of acquired resistance to conventional chemotherapies or targeted therapies (Rottenberg and Jonkers, 2008). Acquired resistance to anti-vascular endothelial growth factor receptor 2 (VEGFR2) treatment in a pancreatic islet tumor model has been attributed to upregulation of Fgf ligands that promote angiogenesis (Casanovas et al., 2005), and upregulation of the drug transporters Mdr1a and Mdr1b (also known as Abcb1a and Abcb1b, respectively) was observed in doxorubicin- and docetaxel-resistant tumors arising in Brca1−/−; p53−/− mammary glands (Rottenberg et al., 2007). Here, we describe the identification of resistance-conferring point mutations and gene amplification in genetically engineered mouse models after treatment with targeted therapy. Importantly, these genetic changes are identical to those observed in TKI-resistant human lung cancer, strongly supporting the use of these preclinical models for the discovery of additional mechanisms of drug resistance and for the testing of novel therapeutics.
The most common cause of resistance to TKIs in human lung cancer is a secondary mutation in EGFR that leads to substitution of a methionine for a threonine at position 790 in the protein (T790M); this mutation is found in 50% of cases. We identified this mutation in 20% of candidate drug-resistant tumors in our mouse model. The relatively low frequency of the T790M mutation observed here may reflect the fact that, in addition to collecting and analyzing tumors that were growing in the presence of the drug, we collected and analyzed tumors that showed a diminished response to erlotinib after multiple rounds of treatment: in humans, only the former have been studied.
In drug-resistant human tumors, the T790M mutation is frequently found in only a minority of mutant EGFR sequences (Pao et al., 2005a). It is therefore possible that tumors in which we did not detect the T790M mutation harbor the mutation in a fraction of transgene copies and that it is undetectable using direct sequencing. We are currently undertaking high-throughput DNA sequencing studies to determine the exact relationship between transgene copy number and the abundance of the T790M mutation.
One drug-resistant tumor harbored a mutation in Kras. Mutations in KRAS account for primary resistance to TKIs in human tumors (Pao et al., 2005b), but are not observed in cases of acquired resistance (Pao et al., 2005a). Aging mice spontaneously develop lung tumors with Kras mutations (Dragani et al., 1995), providing the most likely explanation for the presence of this mutation in the tumor in our model. The immunohistochemical staining pattern observed in this tumor, showing intense phospho-Erk signaling and the absence of phospho-EGFR and phospho-Akt staining (Fig. 3A), further supports the possibility that this is a Kras-driven tumor.
Our success in generating erlotinib-resistant tumors is probably due, in part, to the use of animals with a large tumor burden: these tumors are histologically advanced heterogenous adenocarcinomas and are likely to harbor cells with additional genetic hits that may contribute to erlotinib resistance. In addition, our use of an intermittent treatment schedule, which allows persistent tumor cells to expand during the breaks from treatment, probably provides an opportunity for the emergence of additional mutations that could lead to erlotinib-resistant tumor growth. Because secondary EGFR mutations and MET amplification account for only about 60% of TKI-resistant lung adenocarcinomas in humans, future studies of mouse tumors with unexplained drug resistance may identify additional mechanisms of TKI resistance that are also found in human lung adenocarcinomas. These studies include using novel ‘deep’ sequencing techniques to investigate whether additional genes are mutated in erlotinib-resistant tumors, expression profiling, and comparative genomic hybridization, coupled to analysis of the signaling pathways that are active in individual drug-resistant tumors.
METHODS
Animal husbandry and genotyping
All animals were kept in specific pathogen-free housing with abundant food and water under guidelines approved by the MSKCC Institutional Animal Care and Use Committee and Research Animal Resource Center. TetO-EGFRL858R, TetO-EGFRΔL747-S752, CCSP-rtTA, TetO-KrasG12D, p53-null and Ink4A/Arf-deficient mice have been described previously (Jacks et al., 1994; Serrano et al., 1996; Tichelaar et al., 2000; Fisher et al., 2001; Politi et al., 2006). Tail DNA was isolated using the Qiaprep Tail DNeasy isolation kit (Qiagen), according to the manufacturer’s protocol. Mice were genotyped according to the protocols described in the original papers. Doxycycline was administered by feeding mice with doxycycline-impregnated food pellets (625 ppm) (Harlan-Teklad). Erlotinib (provided by Genentech) was suspended in 0.5% (w/v) methylcellulose and injected intraperitoneally at the dose and times indicated in the experiments.
Histology and immunohistochemistry
Animals were sacrificed with a lethal dose of CO2 per institutional guidelines. The lungs were excised; areas of normal lung and tumor nodules were macrodissected and flash-frozen in liquid nitrogen for molecular analyses. When the tumor nodules were large enough, half of each was fixed with 4% paraformaldehyde in PBS. The remaining lung tissue was also fixed. Tissues were fixed in 4% paraformaldehyde overnight at room temperature, placed in 70% ethanol, and sent for paraffin embedding and sectioning (Histoserv). Slides were reviewed by a board-certified pathologist (M.F.Z.).
The primary antibodies used for immunohistochemistry were anti-phospho-histone H3 (Ser10) (used at a 1:200 dilution; Cell Signaling Technology), anti-EGFRL858R (used at a 1:400 dilution) (Politi et al., 2006), anti-phospho-EGFR (p-Tyr-992) (used at a 1:200 dilution; Cell Signaling Technology), anti-phospho-Erk (used at a 1:100 dilution; Cell Signaling Technology) and anti-phospho-Akt (used at a 1:100 dilution; Cell Signaling Technology).
Sequencing
Flash-frozen tumor samples and adjacent normal tissue were crushed and used for RNA extraction using Trizol reagent (Invitrogen). 3 μg of RNA was treated with DNase I and 1.5 μg was used for first-strand cDNA synthesis (Superscript III First-Strand Synthesis kit, Invitrogen). The cDNA was used as a template to amplify regions of the EGFR transgene, endogenous Kras, Pik3ca and Pten. PCR products were analyzed using direct dideoxynucleotide sequencing.
Quantitative PCR
Analysis of genomic DNA: murine Met levels were evaluated in SYBR Green assays using the following primers: Met-sense, 5′-GCCGCTCATTCAACTACC-3′ and Met-antisense, 5-TTCC-′ CAGTGATAACCAGTGTGTAG-3′. 20 ng of genomic DNA was amplified for 40 cycles (15 seconds at 95°C, 30 seconds at 60°C) in an IQ5 iCycler (Bio-Rad) using the SYBR Green Supermix (Bio-Rad) and 400 nm of primers. Triplicate CT values were averaged, and the amounts of target were interpolated from standard curves and normalized to Gapdh (Taqman assay Mm99999915_g1, AB).
Analysis of mRNA: 20 ng of cDNA was used in a quantitative PCR reaction using an iCycler and the pre-designed TaqMan ABI gene expression assay for Met (Mm00434924_m1). Primers were chosen based on their ability to span the most 3′ exon-exon junction. Amplification was carried out for 40 cycles (15 seconds at 95°C, 1 minute at 60°C). Triplicate CT values were averaged, and the amounts of target were interpolated from standard curves and normalized to Hprt1 (Taqman assay Mm00446968_m1).
Magnetic resonance imaging (MRI)
Mice were anesthetized with 2% isofluorane oxygen gas. Respiratory-gated lung magnetic resonance images were acquired on a Bruker 4.7T Biospec scanner (Bruker Biospin, MA) in the Small Animal Imaging MRI Core Facility at MSKCC, as described previously (Politi et al., 2006). Tumor volume was quantified by calculating the area of visible lung opacities present in each image sequence per mouse using ParaVision 3.0.2 imaging software, and then multiplying the total sum of the areas by 0.09 cm (the distance between each MRI sequence).
Gene expression profiling
mRNA was extracted from pulverized lung samples using Trizol and then hybridized to MOE 430 2.0 chips (Affymetrix) using standard hybridization techniques.
We used the robust multichip average (RMA) method for data pre-processing, and a moderated paired t-test for comparing the gene expression levels in the tumors with the matched normal tissue samples. The moderated paired t-test is similar to a standard paired t-test except that it uses information from all of the genes to estimate variance, which is a more robust approach in the setting of microarray data analysis.
Supplementary Material
Acknowledgments
We thank Mary Ann Melnick, Jennifer Demers, Gabriela Sanchez and Andreas Giannakou for expert technical assistance; Jason Koutcher, Carl Le, Mihaela Lupu and Dov Winkelman for magnetic resonance imaging; Agnes Viale, Daoqi You and Jeffrey Zhao for quantitative PCR analysis; Genentech for providing Tarceva (erlotinib); and members of the Varmus lab and William Pao for insightful discussions and for critical reading of the manuscript. This work was funded by RO1 CA120247-01 to H.V., and by grants R24 CA83084 and P30-CA 08748, which provide partial support for the core facilities used in conducting this investigation. K.P. is currently a recipient of the Pathway to Independence Award from the NCI (K99CA131488) and was previously a recipient of the American Cancer Society-Davidson Sinai Research Foundation Postdoctoral Fellowship (PF-05-078-01-MGO). Deposited in PMC for release after 12 months. This article is freely accessible online from the date of publication.
Footnotes
COMPETING INTERESTS
The rights to a patent application on the testing of the EGFR T790M mutation have been licensed to Molecular MD by MSKCC. This applies to Katerina Politi and Harold Varmus.
AUTHOR CONTRIBUTIONS
K.P. and H.V. designed the study, analyzed the data and wrote the paper. K.P. and P.-D.F performed experiments and M.Z. analyzed the tumor histopathology. R.S. analyzed the gene expression data.
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.003681/-/DC1
REFERENCES
- Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, et al. (2007). MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA 104, 20932–20937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanovas O, Hicklin DJ, Bergers G, Hanahan D. (2005). Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer Cell 8, 299–309 [DOI] [PubMed] [Google Scholar]
- Dragani TA, Manenti G, Pierotti MA. (1995). Genetics of murine lung tumors. Adv Cancer Res. 67, 83–112 [DOI] [PubMed] [Google Scholar]
- Endoh H, Yatabe Y, Kosaka T, Kuwano H, Mitsudomi T. (2006). PTEN and PIK3CA expression is associated with prolonged survival after gefitinib treatment in EGFR-mutated lung cancer patients. J Thorac Oncol. 1, 629–634 [PubMed] [Google Scholar]
- Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, Naumov GN, Yeap BY, Jarrell E, Sun J, et al. (2006). Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 116, 2695–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. (2007). MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 [DOI] [PubMed] [Google Scholar]
- Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. (2001). Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 15, 3249–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. (1994). Tumor spectrum analysis in p53-mutant mice. Curr Biol. 4, 1–7 [DOI] [PubMed] [Google Scholar]
- Ji H, Li D, Chen L, Shimamura T, Kobayashi S, McNamara K, Mahmood U, Mitchell A, Sun Y, Al-Hashem R, et al. (2006). The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell 9, 485–495 [DOI] [PubMed] [Google Scholar]
- Kawano O, Sasaki H, Endo K, Suzuki E, Haneda H, Yukiue H, Kobayashi Y, Yano M, Fujii Y. (2006). PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer 54, 209–215 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. (2005). EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 352, 786–792 [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. (2004). Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 350, 2129–2139 [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. (2004). EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. (2004). EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 101, 13306–13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. (2005a). Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. (2005b). KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. (2006). Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 20, 1496–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S, Jonkers J. (2008). Modeling therapy resistance in genetically engineered mouse cancer models. Drug Resist Updat. 11, 51–60 [DOI] [PubMed] [Google Scholar]
- Rottenberg S, Nygren AO, Pajic M, van Leeuwen FW, van der Heijden I, van de Wetering K, Liu X, de Visser KE, Gilhuijs KG, van Tellingen O, et al. (2007). Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci USA 104, 12117–12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. (1996). Role of the INK4a locus in tumor suppression and cell mortality. Cell 85, 27–37 [DOI] [PubMed] [Google Scholar]
- Tichelaar JW, Lu W, Whitsett JA. (2000). Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J Biol Chem. 275, 11858–11864 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.