Abstract
Substantial evidence has accumulated indicating a significant role for oligomerization in the function of E3 ubiquitin ligases. Among the many characterized E3 ligases, the yeast U-box protein Ufd2 and its mammalian homolog E4B appear to be unique in functioning as monomers. An E4B U-box domain construct (E4BU) has been sub-cloned, over-expressed in E. Coli and purified, which enabled determination of a high resolution NMR solution structure and detailed biophysical analysis. E4BU is a stable monomeric protein that folds into the same structure observed for other structurally characterized U-box domains, all of which are homodimers. Multiple sequence alignment combined with comparative structural analysis reveals substitutions in the sequence that inhibit dimerization. The interaction between E4BU and the E2 conjugating enzyme UbcH5c has been mapped using NMR and this data has been used to generate a structural model for the complex. The E2 binding site is found to be similar to that observed for dimeric U-box and RING domain E3 ligases. Despite the inability to dimerize, E4BU was found to be active in a standard autoubiquitination assay. The structure of E4BU and its ability to function as a monomer are discussed in light of the ubiquitous observation of U-box and RING domain oligomerization.
Ubiquitination is a critical post-translational modification shown to target proteins in a variety of cellular signaling contexts, ranging from transcriptional activation, endocytosis, DNA repair, and proteasomal degradation [1–4]. The molecular mechanism underlying the transfer of ubiquitin (Ub) to a substrate consists of three key enzymatic steps. First, ubiquitin itself is adenylated at its C-terminal glycine residue by an activating enzyme (E1). Second, the adenylated Ub forms a covalent linkage to a conjugating enzyme (E2). Finally, a ligating enzyme (E3) recruits both the Ub-charged E2 species and the target substrate protein. Co-localization of the charged E2 and the substrate catalyzes the transfer of Ub to an e-amine group on lysine residues of the target protein [5].
There are three classes of E3 enzymes: HECT, RING, and U-box, which are distinguished on the basis of their E2-recruiting domains. Whereas HECT and RING domain E3 ligases have been studied extensively, much less is know about the U-box proteins. The U-box and RING classes of E3 ligases act as scaffolding molecules that recruit and co-localize both a Ub-charged E2 and the substrate concomitantly. The recruitment of substrate in these proteins involves common protein interaction modules such as a WD-40 repeat, TPR and armadillo repeat domain [6–8].
In addition to a common organization, the architecture of U-box and RING domains are similar. Both contain a central alpha helix flanked by two surface exposed loops arranged in a cross-brace formation. The structure of RING domains is built around two zinc binding sites that are critical to its stability. In contrast, U-boxes do not bind zinc, but have evolved instead networks of hydrogen bonds and salt bridges in corresponding locations in the structure [9]. Other similarities between these two domains include an antiparallel β-sheet type arrangement involving the first surface exposed loop and the central alpha helix. The β-sheet is stabilized by highly conserved hydrophobic residues responsible for the core packing and stability of the molecule. It has been reported that the L1 – α1 – L2 motif of U-box and RING domains are critical elements for E2 interactions [10–12]. Most U-box and RING domain structures also contain an elongated C-terminal helix. The physical basis and physiological rationale for evolving distinct U-box and RING E3 ligases are not yet known.
U-box proteins constitute a unique class of E3 ubiquitin ligases that appear to function in association with heat shock proteins in protein folding, refolding or complex formation. Ufd2 was first recognized as a U-box containing protein by Koegl and colleagues who designated the protein as an E4 ubiquitin ligase because it was shown to catalyze the elongation of previously ubiquitinated substrates [13]. Ufd2 plays a role in the Cdc48 chaparone pathway. Ufd2 ubiquitinates Cdc48 clients targeting them for proteasome mediated degradation. Interactions have been reported between Ufd2 and Cdc48, and this interaction is stimulated by Cdc48 binding with its cofactors Npl4 and Ufd1 [14]. Additionally, the proteasome shuttling protein, Rad23 binds Ufd2 through its UBL domain, implicating Ufd2 in proteasome shuttling [15]. It has yet to be determined if these interactions are conserved for the mammalian homolog E4B.
In addition to the ability to interact with E2 enzymes, it was also found that Ufd2 could interact with other E3 proteins, including another member of the Ufd family, Ufd4. Ufd4 is a HECT E3 ligase that was shown to only monoubiquitinate the UNC-45 substrate. However, together with Ufd2, Ufd4 stimulates rapid and efficient substrate poly-ubiquitination. Ufd2, and its mammalian homologue, E4B, have also been reported to interact with another U-box containing protein, CHIP [16, 17]. It appears that this interaction also promotes an increased rate of substrate poly-ubiquitination.
An X-ray crystal structure of Ufd2 has been determined [18]. The structure reveals three domains: an N-terminal variable domain, a conserved central armadillo-like repeat, and a C-terminal U-box domain. The U-box and armadillo-like repeat domains likely serve for recruiting E2 and substrate, respectively. The N-terminal domain probably serves in regulation of function. Sequence alignment of the yeast Ufd2 sequence with the mammalian E4B homologues shows a very high degree of conservation within the armadillo-like repeat domain and U-box region; however, the N-terminal region is not conserved, and in fact, the yeast protein is 212 residues shorter than the mammalian protein. Differences in the putative regulatory domain is consistent with the more complex regulatory mechanisms in higher eukaryotes.
Our group has undertaken a comprehensive investigation of the structure and function of the U-box domain of mammalian Ufd2, E4B. We began by examining the state of oligomerization of the E4B U-box and show that it exists primarily as a monomer in solution under a range of concentrations. The structure was determined in solution by NMR and chemical shift perturbation experiments were used to map the interaction with the E2 enzyme UbcH5c. This data was then used to generate a model of the complex. We also show the U-box alone is fully active in an in vitro autoubiquitination assay.
MATERIALS AND METHODS
Expression Plasmids
Mouse E4B DNA was amplified with overhanging 5′ BamH1 and 3′ Xho1 restriction sites for subcloning into in-house pBG vectors (L. Mizoue; Center for Structural Biology, Vanderbilt University). Residues 1072-1173, 1082-1173, and 1092-1173 were subcloned into the pBG102 vector, which produces an N-terminal 6xHis-SUMO tag fusion protein. Human UbcH5c was produced as an untagged construct from pET28 as described previously [19].
Protein Purification
The E4B U-box constructs as well as UbcH5c were overexpressed in BL21(DE3) Escherichia coli strains. The E4B proteins were purified by Nickel affinity chromatography and the His-SUMO tag was cleaved by H3C precision protease. A second purification step with the same column removed the affinity tag from the sample. The protein was further purified by anion-exchange chromatography using a SourceQ column and Superose6 gel filtration chromatography. UbcH5c was purified by cation exchange chromatography using SP resin, followed by size-exclusion chromatography with an S75 column. The proteins were dialyzed against 20 mM TRIS pH 7.0, 50 mM NaCl and concentrated for further studies. Protein concentrations were determined by using a calculated extinction coefficient for each protein. Protein samples for NMR experiments were expressed in minimal media with NH4Cl and glucose as the sole nitrogen and carbon sources, respectively. 15NH4Cl and [13C6]glucose (Cambridge Isotope Laboratories) were used as needed to prepare the requisite isotopically enriched proteins.
Electrospray Mass Spectrometry
Experiments were conducted with an ESI-oaTOF (electrospray ionization orthogonal accelerating time-of-flight) mass spectrometer (micrOTOF, Bruker Daltonics, Inc., Billercia, MA), which has been modified for enhanced collisional cooling in the source for the analysis of noncovalent protein complexes. This modification and methods have been previously described [20]. E4B residues 1092-1173 was prepared in 25mM sodium acetate at pH 7.0 at concentrations of 250 μM, 500 μM, and 1 mM.
Analytical Ultracentrifugation
The state of oligomerization of E4B was analyzed by sedimentation velocity analysis. Samples to be analyzed by AUC were first dialysed extensively against 20 mM TRIS buffer (pH 7.0) containing 50 mM NaCl, and dialysis buffer was saved to be used in the reference sector of each sample cell. AUC analysis was performed at 20 °C in a Beckman Optima XL-I analytical ultracentrifuge. Sample concentrations of 100 μM, 50 μM, and 25 μM were loaded into standard 12 mm 2-sector Epon-charcoal cells, and housed in an An-60 Ti rotor. Sedimentation behavior resulting from a rotor speed of 50,000 rpm was observed using the optical absorbance of each sample at a wavelength of 280 nm. The initial 200 scans obtained from each cell were fitted according to the continuous c(S) Lamm equation model in the SEDFIT software package (version 9.4) in order to obtain the relative prevalence of each oligomeric species [21].
NMR Spectroscopy
NMR experiments were conducted using Bruker DRX 600 and 800 MHz spectrometers equipped with z-axis gradient TXI cryoprobes. The 15N-1H HSQC experiments to compare different E4B U-box constructs were acquired at 25 °C with 750 μM protein in 25 mM NaH2PO4 at pH 6.5, 25 mM NaCl, and 5% D2O. To search for oligomerization-dependent changes in linewidth, 15N-1H HSQC experiments were acquired at concentrations of 100 μM and 1 mM 15N-E4B(1092-1173) under identical conditions.
To obtain backbone resonance assignments, 750 μM 13C,15N-E4B(1092-1173) was prepared in 25 mM NaH2PO4 at pH 6.5, 25 mM NaCl, and 5% D2O. 2D 15N-1H HSQC and 3D HNCACB, CBCA(CO)NH, HNCA, and HNCO experiments were conducted. To obtain the side chain resonance assignments of E4B (1092-1173), the same sample was used to acquire 2D 13C-1H HSQC and 3D HCCH-TOCSY and HCCH-COSY experiments. The final resonance assignments were deposited at the Biological Magnetic Resonance Data Bank under entry XXXX. 2D 1H-NOESY and 3D 15N-NOESY-HSQC and 13C-NOESY-HSQC experiments were recorded to assign intramolecular NOEs. All NMR data were processed using NMRPipe [22] and analyzed with Sparky [23].
The NMR chemical shift perturbation assays employed either 15N-E4BU and unlabeled UbcH5c or 15N-UbcH5c and unlabeled E4BU. The NMR samples contained 100 μM 15N-protein in 25 mM NaH2PO4 at pH 7.0, 150 mM NaCl, and 5% D2O. Unlabeled protein in the same buffer was added to the labeled sample until the molar ratio reached 1:2 (15N labeled:unlabeled). The normalized chemical shift change was calculated using the equation Δσ = √[(ΔH)2 + (ΔN/5)2], where ΔH and ΔN are chemical shift changes in proton and nitrogen, respectively [24].
Structure Calculations
Upper bounds for distance restraints were generated from using the CALIBA module in CYANA [25]. The reference volume determined by CALIBA was increased 5 times before conversion in order to loosen the distance restraints. Backbone torsion angle restraints for E4B (1092-1173) were estimated from Cα and Cβ chemical shifts using TALOS [26].
The structural analysis involved multiple iterations of CYANA calculations and inspection of the restraints and the spectra. NOEs were identified through a combination of manual and automatic assignments using the CALIBA module of CYANA. At each iteration, 100 structures were generated using CYANA. In the later stages, the 50 structures with lowest target function were selected for further refinement by restrained molecular dynamics (rMD) calculations in AMBER 9 with implicit treatment of the water solvent [27]. After 1 ps of energy minimization to regularize CYANA structures in the AMBER force field, the temperature of the system was rapidly increased to 1200 K over 5 ps, then was slowly cooled to 0 K over 15 ps. NMR restraints were slowly turned on during the first 3 ps and kept for the rest of the simulation. Force constants for distance and torsion angle constraints were set to 32 and 50 kcal/mol, respectively. To select the final representative ensemble of conformers, the structures were placed in order of increasing restraint violation energy and the top 20 were selected. These structures were inspected for any local regions of high restraint violation. The quality of the final ensemble was assessed using the Procheck-NMR and MOLPROBITY programs [28, 29]. The final ensemble of 20 conformers was deposited at the Protein Data Bank under code XXXX.
Free Molecular Dynamics Simulation of UbcH5c
Using chain A of PDB ID 1X23, the first N-terminal 6 residues from the cloning vector were removed so as to not interfere with modeling. Initially, 500 steps of minimization over a period of 2 fs were run to allow relaxation of the initial structure. A 1000 fs molecular dynamic simulation was run in which coordinates were extracted every 50 fs, generating an ensemble of 20 structures. The molecular dynamics simulations were run in AMBER9 with the ff03 force field at a constant temperature of 300 K. The structure was solvated in a cubic box of TIP3P water using a minimum distance of 14 Å between the protein and solvent edges.
E4BU-UbcH5c docking using HADDOCK
A model for the E4BU-UbcH5c complex was generated using HADDOCK2 [30]. The four unstructured N-terminal residues of the E4BU construct were removed from the coordinate file so as to not interfere with docking of UbcH5c. The average solvent accessibilities per residue in the ensemble of E4BU and UbcH5c were calculated using NACCESS [31]. Residues with a solvent accessible surface higher than 50% and chemical shift perturbations more than 1 S.D. above the mean were designated as active for HADDOCK calculations. The adjacent residues with solvent accessibility greater than 50% were designated as passive residues. In the first iteration of the calculation, an initial ensemble of 1000 rigid body docking models was generated. The 200 lowest energy models were selected for a second iteration with semi-flexible simulated annealing and these were further refined in explicit water. The ensemble of structures was generated using the final ensemble of 20 E4BU structures and 20 UbcH5c structures generated by a room temperature MD simulation in AMBER starting from the crystal structure of UbcH5c, as described above. The 20 lowest energy models were selected from the most populated cluster with the lowest HADDOCK score and used for analysis.
In vitro autoubiquitination assay
Ubiquitination experiments were carried out at a final volume of 20 μl including: E1 (BostonBiochem) at 52 nM, bacterially expressed His6-E2-UbcH5a at 0.6 μM, ubiquitin (BostonBiochem) at 50 μM, E4B 1072-1173 at 1 μM. The assay was performed in buffer containing 100 mM NaCl, 1 mM DTT, 5 mM MgCl2 and 25 mM Tris-Cl at pH 7.5. The reactions were activated with 5 mM ATP and incubated at 30 °C for the different times indicated. To stop the ubiquitination reaction, the samples were incubated for 15 minutes at 90 °C after the addition of 5 μl SDS-loading buffer. Reactions were resolved on a NuPAGE 4–12% Bis-Tris gradient gel (Invitrogen) and detected by a SimplyBlue SafeStain (Invitrogen).
RESULTS
To produce a stable E4B U-box domain, three C-terminal bacterial expression constructs were designed based on analysis of predicted secondary structure and comparison to the x-ray crystal structure of Ufd2: Ile1072-His1173, Val1082-His1173, and Ala1092-His1173. To determine the optimal construct for structural and biophysical analysis, all three constructs were expressed and purified with uniform 15N enrichment then characterized by 15N-1H-HSQC NMR. The constructs exhibited similar spectra with the same group of well-dispersed signals, but with a progressively larger number of signals in the central region of the spectrum in proportion to the length of the construct (Figure S1). This observation indicated all constructs contained a common globular domain but that the region Ile1072-Ala1092 lacks stable structure. Moreover, the two longer constructs degraded over time, as evident from the appearance of heterogeneity in HSQC spectra acquired one week after the samples were first prepared. Consequently, all further NMR experiments were performed on the shortest construct, Ala1092-His1173, which will be referred to as E4BU.
E4BU is a monomer in solution
All U-box E3 ligases characterized to date are single chain proteins that form homooligomers, except for the yeast homolog of E4B, Ufd2 [12, 18, 32–34]. During the purification of E4BU, the protein was found to elute as a dimer with an apparent molecular weight of approximately 21 kDa according to size-exclusion chromatography (SEC) experiments. However, since molecular weight is notoriously difficult to quantify by SEC, additional experiments were performed to directly determine the oligomerization state of E4BU. The approaches included SEC-MALS, which indicated an ~10 kDa species in the primary peak (data not shown) and native mass spectrometry using electrospray ionization, which revealed a 10:1 ratio of monomer to dimer (Figure 1A). The predominance of monomer over dimer was further confirmed by analytical ultracentrifugation sedimentation velocity experiments, which also showed a 10:1 ratio of monomer over dimer (Figure 1B). Concentration-dependent NMR chemical shifts provide an additional sensitive indicator of self-association. 15N-1H HSQC spectra were acquired for 0.10 mM and 1.0 mM samples of 15N-E4BU and no difference was observed between the two spectra (Figure S3), indicating no significant population of dimer or higher aggregates even at the higher protein concentration.
Figure 1.
Characterization of self-association of E4BU. (A) Electrospray ionization mass spectrometry of E4BU. All ions detected correspond to monomeric species, except for a single additional (+9) charge state corresponding to a dimer (insert). All mass-to-charge (m/z) species correlate to a monomeric protein. (B) Analytical ultracentrifugation of a 100 μM solution of E4BU. The results of the sedimentation velocity experiment yielded ~87% of monomer and < 9% of dimer. The remaining protein appears to form higher order aggregates. (C) Multiple sequence alignment of U-box domains for which structures are deposited in the PDB. The arrows indicate residues corresponding to two key interacting regions in the CHIP and Prp19 U-box dimer interface, which are not conserved in mammalian E4BU or yeast Ufd2U.
NMR Structure Determination of E4BU
A high resolution solution structure of E4BU was determined by multidimensional heteronuclear NMR spectroscopy using isotopically enriched protein samples. A total of 1565 NOE-derived distance constraints and 92 dihedral angle constraints were used as input for structure calculations. The input constraint list was refined by using iterative cycles of CYANA. The final ensemble of 50 CYANA starting structures was further refined by restrained molecular dynamics in AMBER. The final representative ensemble consists of the 20 conformers selected on the basis of lowest restraint violation energy (Figure 2A). The structured region of the ensemble (residues 1096-1170) has a final rmsd versus the mean of 0.45 Å for the backbone and 1.13 Å for all atoms. The high quality of the structure is reflected in the results from PROCHECK analysis. For example, 99% of all backbone torsion angles occupy the most favored and favored regions of the Ramachandran plot. Further details, and structural statistics are reported in Table 1.
Figure 2.
Solution NMR structure of E4BU. (A) Stereo view of the final ensemble of 20 conformers representing the structure. (B) Ribbon diagram of the single representative structure. (C) Comparison of E4BU with the U-box domain of Ufd2 extracted from the crystal of the intact protein. Best-fit superposition of the two structures gives an rmsd over all backbone atoms of 1.150 Å.
TABLE 1.
Structural Statistics for E4BU
| Restraints for Calculation | ||
| Total NOE Restraints | 1565 | |
| Short Range | 780 | |
| Medium Range | 370 | |
| Long Range | 415 | |
| Dihedral Angle Restraint | 92 | |
| Constraint Violations, mean ± S.D. | ||
| Distance Violations | ||
| 0.1Å < d < 0.2 Å | 0.20±.41 | |
| D > 0.2Å | 0 | |
| Average maximum distance violations (Å) | 0.08±.03 | |
| Torsion angle violations > 5.0° | 0 | |
| Average maximum torsion angle violations (degree) | 0 | |
| AMBER energies, mean ± S.D. (kcal mol−1) | ||
| Restraint | 1.08±0.14 | |
| van der Waals | −651.24±8.71 | |
| Total molecular | −3222.35±7.44 | |
| Precision, root mean square deviation from the mean (Å) | ||
| Ordered | All residues | |
| Backbone | 0.46±0.09 | 1.11±0.08 |
| All heavy atoms | 1.53±0.33 | 1.87±0.26 |
| Ramachandran statistics (%) | ||
| Most favored | 87.7 | |
| Additionally allowed | 10.1 | |
| Generously allowed | 1.2 | |
| Disallowed | 1.0 | |
The structure of E4BU consists of the ββαβα-fold typical of U-box and RING domains. The central α-helix is flanked by two prominent surface exposed loop regions, spanning residues Arg1104 to Thr1110 (L1) and Thr1138 to Gln1144 (L2) (Figure 2B). The characteristic network of hydrogen bonds within each loop stabilizes the overall structure. Critical polar contacts exist throughout L1. For example, the side chain of Arg1104 hydrogen bonds to the backbone carboxyl group of Asp1109. The side chain carboxyl group of Asp1105 is distinctive, forming a hydrogen bond to the backbone amide of Leu1107 in 16 out of 20 structures of the ensemble. Of these 16, 10 show a bifurcated hydrogen bond to the amides of both Leu1107 and Met1108. L2 contributes to a network of interactions similar to that involving L1. Important stabilizing hydrogen bonds include two from the carboxyl side chain of Asp1139: one oxygen atom hydrogen bonds to the backbone amides of Phe1141 and Asn1142 and the other interacts with Gln1144. Both of these interactions are observed in all 20 structures of the ensemble. Asp1139 appears to play a central role in the stabilization network as its backbone amide also is involved in a hydrogen bond with the side chain of Arg1143. This stabilizing bond is also seen in all 20 structures of the ensemble.
Comparison of E4BU to other U-box and RING domains
Overall, the structure of E4BU has a high degree of structural similarity to other U-box and RING domains. An overlay of the backbone atoms of E4BU with the U-box domain of Ufd2 extracted from the crystal structure of the full protein (2QIZ) is shown in Figure 2C. The two structures superimpose with a backbone rmsd of 1.2 Å. Overlays with CHIP U-box, Prp19 U-box, and BRCA1 RING domains give rmsds of 1.3 Å, 1.4 Å, and 1.3 Å, respectively.
In order to obtain insights into the differences between monomeric E4B and the dimeric U-box and RING domains, the sequence was analyzed by multiple sequence alignment. Two residues in the E4B U-box domain sequence were found to diverge significantly from the other U-boxes: Arg1117 and Glu1152, which are hydrophobic residues in all other U-boxes (Figure 3A, black arrows). In scPrp19 and mmCHIP, the residues corresponding to Arg1117 (Leu15 and Ile246, respectively) make crucial hydrophobic contacts across the dimer interface [12, 33]. The critical importance of this residue was shown for Prp19; mutation of Leu15 abrogated dimerization and abolished cell viability [33]. The residues corresponding to Glu1152 in E4B are also hydrophobic in Prp19 and CHIP, and have been previously described as critical elements in the hydrophobic core and across the dimer interface. Analysis of electrostatic surface potential of U-box domains reveals that E4BU is significantly more acidic than the Prp19 and CHIP U-boxes, especially in the region corresponding to the dimer interface (Figure 3B–D). In particular, E4BU Glu1152 is responsible for a highly negative patch at the core of what would be the dimer interface. Electrostatic repulsion at the dimer interface due to substitution of hydrophobic residues with charged side chains appears to significantly destabilize the dimeric state of E4BU.
Figure 3.
E4BU has a more negative surface potential than other U-box domains. (A) Multiple sequence alignment of E4BU with Ufd2, CHIP, and Prp19. Alignments made using the CLUSTALW server at EBI [38]. The resulting alignment file was then processed through ESPript [39]. (B) Electrostatic surface representation of the archetypal U-box dimer interface of E4BU. Blue coloring corresponds to positive charge and red corresponds to negative charge. (C) Electrostatic surface representation of the Prp19 U-box dimer interface. (D) Electrostatic surface representation of the CHIP U-box dimer interface. The electrostatic fields were calculated using the Adaptive Poisson-Boltzmann Solver algorithm for electrostatics [40].
E4BU – UbcH5 interaction
U-box and RING domains serve as E2-recruiting modules in E3 ubiquitin ligases. To determine how E4BU interacts with E2 enzymes, the binding interface between the U-box domain of E4B and a cognate E2 enzyme UbcH5c was analyzed using NMR chemical shift perturbation experiments. UbcH5c was used for these experiments since the backbone NMR assignments and the crystal structure were already available (P. Brzovic and R. Klevit, unpublished; PDB ID 1X23). Titration of UbcH5c into 15N-enriched E4BU was monitored in 15N-1H HSQC spectra acquired at molar ratios of 1:0.125, 1:0.25, 1:0.5, 1:1, and 1:2. Distinct chemical shift perturbations were observed for specific peaks even at the lowest titration point (Figure 4A). Over the course of the titration a significant number of peaks broadened beyond detection as a consequence of formation of the 27 kDa complex, and/or effects associated with intermediate exchange on the NMR timescale. Overall, the resonances of 10 residues were significantly perturbed (mean + 1 standard deviation): Leu1107, Met1108, Asp1109, Leu1118, Thr1122, Ile1129, Leu1130, Arg1131, Asn1135, and Asn1142 (Figure S2). Using the backbone assignments for E4BU, the perturbed residues were then mapped onto the structure (Figure 4B). It has previously been reported that Ile235 in CHIP and the corresponding residue in BRCA1, Ile26, are crucial for interaction with E2 [11]; the corresponding residue in E4BU, Leu1107, has one of the largest chemical shift perturbations, upon binding of UbcH5c. Overall, the perturbed residues are found to be located in and around the two surface exposed loops, including residues in the central α-helix, α1, consistent with the location of E2 binding sites in other RING and U-box domains.
Figure 4.
NMR chemical shift perturbations indicating interaction of E4BU and UbcH5c. (A) 15N-1H HSQC spectra of E4BU obtained in the absence (black) and presence (red) of 0.125 molar equivalent of UbcH5c. (B) Surface representation of E4B with residues that have significant chemical shift perturbations in panel (a) highlighted in red. (C) 15N-1H HSQC spectra of UbcH5c in the absence (black) and presence (red) of 0.125 molar equivalent of E4BU. (D) Surface representation of UbcH5c with residues that have significant chemical shift perturbations in panel (c) highlighted in red.
Reciprocal titrations were also performed using unlabeled E4BU and 15N-UbcH5c with spectra acquired at ratios of 1:0.061, 1:0.125, 1:0.25, 1:0.5, 1:1, and 1:2. In all, 14 residues were significantly perturbed including: Arg5, Ile6, Asn7, Lys8, and Ser11 in α1; Thr58, Asp59, and Phe62 in L4; Ser94, Ala96, Leu97, and Ile99, in L7; and Ile88 and Leu103 in helical regions that flank L7. When mapped out on the structure of UbcH5c, these residues form a contiguous surface on one face of the protein (Supplemental Fig. 2B, Figure 5C,D). These results are consistent with previous reports of E2 interactions with CHIP [13, 14, 32].
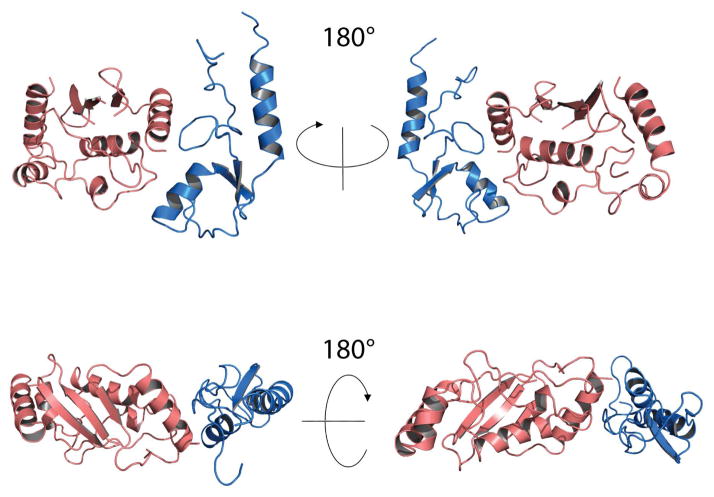
In order to gain a more thorough understanding of the interface between E4BU and UbcH5c, the chemical shift perturbation data was used to generate a molecular model with HADDOCK. It has been previously demonstrated that using crystal structures in HADDOCK docking may not offer sufficient initial flexibility for proper docking [35]. A molecular dynamics simulation was therefore carried out to generate an ensemble of 20 UbcH5c structures. Active residues for the HADDOCK ambiguous interaction restraints (AIRs) were assigned to residues exhibiting significant chemical shift perturbations (mean+1SD) or broadening and backbone and side chain regions that occupied greater than or equal to 50% solvent accessibility (as calculated by NACCESS). Overall, 7 active and 4 passive residues were assigned for the E4BU ensemble, while 6 active and 7 passive residues were assigned for UbcH5c. Cluster analysis of the 200 models generated by Haddock generated 9 groups of 4 molecules or more. However, of these, two similar clusters stood out as being highly populated (Cluster 1- 54 conformers, Cluster 2- 16 conformers) and having the most favorable HADDOCK energies. The 20 lowest energy conformers of the most highly populated cluster were selected to represent the model of the complex. This model contains interactions between L1 of E4BU and α1 of UbcH5c, α1 of E4BU and L4 of UbcH5c, and L2 of E4BU and L7 of UbcH5c (Figure 5a). Remarkably, this model aligns extremely well with the co-crystal structure of UbcH5a and the CHIP dimer, superimposing to an rmsd of only 0.81Å. Thus, this experimentally–guided model suggests monomeric E4BU binds E2 conjugating enzymes in the same manner as U-box and RING domain dimers.
Figure 5.
Model of the complex of E4BU and UbcH5c based on NMR chemical shift perturbation. Ribbon diagram of the mean structure from the 20 lowest energy conformers of the highest populated cluster from HADDOCK calculations with E4BU (blue) and UbcH5c (salmon).
E4BU function
We next asked whether or not the monomeric E4BU is functional, as interaction with an E2 is not sufficient to prove activity. Most E3 ligases are able to modify themselves through autoubiquitination, where the ligase itself serves as the substrate, a process that serves a useful role in auto-regulation through protein turnover. Because the E4B(1092-1173) construct encodes just the U-box domain, the longer 1072-1173 construct was used for the autoubiquitination experiments. The additional 20 residues outside of the U-box domain, four of which are lysines, ensure there is an adequate substrate for ubiquitination. The E4BU in vitro autoubiquitination assay was developed based on our protocol [Y.N.D. and W.J.C., unpublished results] using recombinant bacterially expressed and purified proteins Uba1 (E1), UbcH5 (E2), E4B(1072-1173) (E3/substrate), and Ub. Analysis of the autoubiquitination reaction by SDS-PAGE reveals time-dependent formation of higher molecular weight E4B species, showing that the U-box has robust autoubiquitination activity (Figure 6A).
Figure 6.
Ubiquitination activity of the monmeric E4B U-box in an in vitro ubiquitination assay. Time course of the assay using E3B(1072-1173) and the UbcH5c E2 conjugating enzyme.
DISCUSSION
We have shown that unlike all other U-box proteins characterized to date, the U-box domain of E4B exists and functions as a monomer in solution [12, 33, 36]. It could be hypothesized that the availability of two or more E2 binding modules in oligomeric E3 ligases would increase the processivity of substrate ubiquitination through faster loading and unloading of E2 enzymes on the E3. However, this does not appear to be the case, as most oligomeric U-box and RING E3 ligases appear to possess only one active binding site. The clearest example of ‘half-of-sites’ binding is the U-box protein CHIP. The x-ray crystal structure of CHIP reveals a unique asymmetric dimer in which one of the U-box domain is blocked from interacting with E2 enzymes, ‘half-of-sites’ binding [12].
The intrinsic dimerization of isolated U-box domains appears to be weak. In the case of Prp19, analytical ultracentrifugation revealed that the Prp19 U-box (Prp19U) has only a weak propensity to dimerize. The solution NMR structure of Prp19U was determined in a monomeric state at a concentration of 1 mM, but a dimer was formed when the protein was crystallized from a solution at more than 2-fold higher concentration. A central coiled-coil domain adjacent to the U-box domain is the dominant oligomerization interface, leading to an overall tetrameric assembly [6]. The short tether to the coiled-coil domain results in a very high local concentration of pairs of U-boxes in the tetramer, which drives U-box/U-box dimerization in vivo [33]. The U-box domain of CHIP (CHIPU) exhibits similar properties. For example, isolated CHIPU crystallizes as a dimer [11]. However, like Prp19, the U-box is not the dominant oligmerization domain, but rather, it is the central coiled-coil region of the protein [12, 34]. Our studies showed E4BU has a very weak propensity to dimerize and has been resistant to crystallization. It appears that oligomerizatin of E4BU would require an additional oligomerization domain in order to dimerize. However, dimerization of E4BU seems unlikely even in the context of oligomerization of the full-length protein due to the charged residues present in the common U-box domain dimerization interface. Thus, the E4B U-box E3 ligase stands out for its structure and apparent function in a monomeric state.
Previous reports have suggested that E4B interacts with CHIP in regulation of the myosin assembly pathway in C. elegans [17]. The TPR domains of CHIP are required for interaction with E4B, but the corresponding region of E4B necessary for CHIP binding is not yet known [16]. CHIP is a dimer and it is intriguing to consider the possibility that it forms a hetero-oligomer with E4B and whether one or both E3 ligases are active in the complex. CHIP exhibits ‘half of sites’ binding, i.e. only one Ub~E2 conjugate is bound to dimer. It would also be important to determine if U-box domains from one or both E3 ligases are able to bind E2 enzymes concomitantly. If a novel U-box hetero-oligomer were formed and it allowed E2 accession to multiple U-box domains, the rates and relative processivity of substrate ubiquitination would likely increase.
Despite the information noted above, the function of E4B is not well characterized and further study is required. For example, only UbcH5 family E2 conjugating enzymes are known to function with the E4B U-box, but other E2 enzymes have not been tested. Evidence to date suggests E4B polyubiquitinates substrates leading to protein turnover [37], but more in-depth functional analysis is required to confirm this. Some clues can be obtained from characterization of the polyubiquitin chain topology of conjugates formed on E4B substrates, and such studies will be reported in due course. The initial assignment of Ufd2 as an E4 ubiquitin ligase remains an open issue. Further analysis of E4B structure and biochemical properties will help and test and refine this hypothesis and generate a clearer understanding of the cellular activities of this unique monomeric ubiquitin ligase.
Supplementary Material
Acknowledgments
We thank Markus Voehler for assistance with the NMR experiments, Brian Weiner and Benjamin Chagot for aid provided with NMR data processing and calculations, and Seth Chitayat for help with AUC experiments.
Abbreviations
- AUC
Analytical Ultracentrifugation
- BARD1
BRCA1 Associated Ring Domain protein 1
- BRCA1
Breast Cancer type 1 susceptibility protein
- CHIP
Carboxy terminus of Hsc70 Interacting Protein
- E1
ubiquitin activating enzyme
- E2
ubiquitin conjugating enzyme
- E3
ubiquitin ligase
- ESI-oaTOF
electrospray ionization orthogonal accelerating time-of-flight
- HECT
Homologous to E6 Carboxy Terminus
- HSQC
heteronuclear single quantum coherence
- MALS
Multi-Angle Light Scattering
- NMR
nuclear magnetic resonance
- NOESY
nuclear overhauser enhancement spectroscopy
- Prp19
Pre-mRNA Processing factor 19
- RING
Really Interesting New Gene
- RMSD
root mean square deviation
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEC
Size Exclusion Chromatography
- TPR
Tetratricopeptide Repeat
- TOCSY
total correlation spectroscopy
- Ufd
Ubiquitin Fusion Degradation
- Y2H
Yeast 2 Hybrid
Footnotes
This research was supported by operating grants RO1 GM075156 (to WJC), RO1 GM058008 (to RMC), and RO1 CA079963 (to REK), and support to access instrumentation facilities P30 ES0000267 (VU Center in Molecular Toxicology) and P50 CA068485 (Vanderbilt-Ingram Cancer Center) from the National Institutes of Health. SES was supported by NIH training grants T32 CA09582 and F32 GM 087050.
SUPPORTING INFORMATION AVAILABLE. Additional figures are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hochstrasser M. Ubiquitin signalling: what’s in a chain? Nat Cell Biol. 2004;6(7):571–572. doi: 10.1038/ncb0704-571. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8(6):610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Kim I, Rao H. What’s Ub chain linkage got to do with it? Sci STKE. 2006;2006(330):pe18. doi: 10.1126/stke.3302006pe18. [DOI] [PubMed] [Google Scholar]
- 4.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Ohi MD, et al. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol Cell Biol. 2005;25(1):451–460. doi: 10.1128/MCB.25.1.451-460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballinger CA, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mudgil Y, et al. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 2004;134(1):59–66. doi: 10.1104/pp.103.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohi MD, et al. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10(4):250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14(10):941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 11.Xu Z, et al. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M, et al. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20(4):525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 13.Koegl M, et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96(5):635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 14.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120(1):73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15(7):3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoppe T, et al. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell. 2004;118(3):337–349. doi: 10.1016/j.cell.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Janiesch PC, et al. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat Cell Biol. 2007;9(4):379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- 18.Tu D, et al. Inaugural Article: Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc Natl Acad Sci U S A. 2007;104(40):15599–15606. doi: 10.1073/pnas.0701369104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brzovic PS, et al. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21(6):873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Lee YT, et al. Structure of the S100A6 complex with a fragment from the C-terminal domain of Siah-1 interacting protein: a novel mode for S100 protein target recognition. Biochemistry. 2008;47(41):10921–10932. doi: 10.1021/bi801233z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuck P, et al. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J. 2002;82(2):1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2006. [Google Scholar]
- 24.Mulder FA, et al. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J Mol Biol. 1999;292(1):111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 25.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273(1):283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 26.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13(3):289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 27.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 29.Hubbard S, Thornton J. NACCESS. University of Manchester; U.K: 1992–96. [Google Scholar]
- 30.Andersen P, et al. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J Biol Chem. 2004;279(38):40053–40061. doi: 10.1074/jbc.M405057200. [DOI] [PubMed] [Google Scholar]
- 31.Vander Kooi CW, et al. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry. 2006;45(1):121–130. doi: 10.1021/bi051787e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Z, et al. Structure and interactions of the helical and U-box domains of CHIP, the C terminus of HSP70 interacting protein. Biochemistry. 2006;45(15):4749–4759. doi: 10.1021/bi0601508. [DOI] [PubMed] [Google Scholar]
- 33.Dominguez C, et al. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12(4):633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Brzovic PS, et al. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8(10):833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto M, et al. Molecular clearance of ataxin-3 is regulated by a mammalian E4. Embo J. 2004;23(3):659–669. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Gouet P, et al. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15(4):305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 38.Baker NA, et al. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.