Abstract
We report a rapid gene mutation screening method by making use of the mechanical properties of single-strand DNA (ssDNA) tethered to a solid surface. With proper temperature control, ssDNA in solution undergoes intra-base pairing and forms a specific complex 3D structure. By tethering such ssDNA strands to a solid surface, a DNA film can be formed. The mechanical properties of such DNA films, probed by devices such as a quartz crystal resonator, are directly related to the specific structure of the ssDNA which is characteristic of its base sequence, and thus can be used as the basis for mutation screening. Using this approach, we have detected a single base mutation among 545 bases in P53 gene. This result suggests promising potential of employing the mechanical properties of DNA strands for rapid screening of mutations.
INTRODUCTION
The complete sequencing of the human genome has created a tremendous interest in large scale DNA characterization and sequencing.1,2 With the rapid increase in the number of mutations identified, detection of genetic variations in patients would bring about early diagnosis and even prevention of certain diseases like cancer. The current gold standard for identifying gene mutation is based on DNA sequencing technologies such as the capillary array electrophoresis. However, precisely sequencing a large number of samples is currently time-consuming and expensive. Screening of gene mutations prior to precise mutation identification would significantly reduce the number of samples that need to be sequenced and, therefore, development of rapid, sensitive, and inexpensive technologies for DNA mutation screening is of great interest. Some of the current mutation screening technologies include allele specific oligonucleotides, protein truncation test, DNA microarray technology and electrophoretic mobility shift assays.1,3–8 Although these screening methods have all been explored to a reasonable degree of success,9 they are limited by the length of the DNA that can be effectively screened per operation, which is apparently an important factor that determines the cost of mutation screening. Among them, the electrophoretic mobility shift assays, including single strand conformational polymorphism (SSCP) and heteroduplex analysis (HA), are able to screen longer DNA strands than other methods and have gain clinical popularity.1 However, the maximum length of the DNA that can be effectively screened by SSCP and HA is still limited to be less than 300 base pairs (bps) and the optimization of these electrophoretic mobility shift assays is highly empirical.8
In this paper, we demonstrate another effective approach for rapid gene mutation screening by making use of the mechanical properties of single-strand DNA (ssDNA). It is based on the folding of ssDNA strands in aqueous solution and the immobilization of such strands to a quartz crystal resonator. We show that appreciable differences in the shear modulus can be detected between ssDNA films consisting of wild type and mutant type DNA strands of P53 gene, due to a single base mutation in a total of 545 bases (see Supporting Materials for their sequences). We choose to use the P53 gene and its mutant as an example to demonstrate the capability of this technology, because P53 gene has been used as examples for other mutation screening technologies1,3 and its importance has been extensively studied as its mutation leads to cancers such as breast cancer, bone cancer and bladder cancer.10
Experimental Details
DNA sample preparation
The gene samples used were P53 wild type gene and P53 R type mutant, both containing 1182 bps. A fragment of the genes containing 545 bases was amplified by PCR. By modifying one primer with a biotin group at the 5’ end, biotin dsDNA strands were obtained. (See Supplementary Materials for the sequences of the DNA samples and primers). The amplification product was purified to remove the reaction buffer and the primers. The purified dsDNA was denatured at 95°C and cooled suddenly at −20°C to form ssDNA, with one strand containing biotin at the 5’ end and the other strand containing the complementary base sequence to the biotinylated strand. 100 µl of the purified ssDNA was diluted in 1.5 mL of phosphate buffered saline (PBS, pH 7.4) and this formed the sample that was introduced to the quartz crystal resonator.
Crystal preparation
The gold-coated quartz crystals having a fundamental resonant frequency of 5 MHz were first cleaned in an ultra violet ozone chamber for 10 minutes and then cleaned in a 5:1:1 mixture of deionized water, ammonium hydroxide, and sodium peroxide at 74°C for 5–8minutes. They were then washed with deionized water and dried with nitrogen gas. It was again placed in the UV ozone chamber for 10 minutes to oxidize any organic impurities present on the crystal surface before the surface was modified with the biotin-thiol molecule.
Formation of the biotin-thiol, streptavidin and ssDNA layers and data collection
The crystals were soaked overnight in a solution of biotin-terminated tri(ethylene glycol)hexadecanethiol (concentration of 1 mM in anhydrous ethanol). They were then washed three times with anhydrous ethanol and deionized water and dried with nitrogen gas. A monolayer of the biotin-thiol was deposited onto the surface via a covalent bond between the thiol group and the gold. The biotin end of the molecule was directed away from the surface forming a biotin surface on the crystal (Fig. 1a). The resonant frequency and energy dissipation data were collected and processed by a commercial module (Q-sense E1). All measurements were made in PBS (pH 7.4) at a constant temperature of 23°C. The crystals were checked for the resonant frequency for odd harmonic resonances (1 to 13) in the aqueous phase before any measurements were made. The flowrate of PBS was kept at 200 µl/min. After obtaining a stable frequency baseline in PBS, streptavidin (1 µM in PBS, purchased from Sigma Aldrich) was flowed over the crystal (Fig. 1b). The fast irreversible binding of streptavidin and biotin was confirmed by a sharp decrease in the resonate frequency of the crystals. The biotinylated ssDNA sample was then flowed over the streptavidin layer to which the biotin end of the ssDNA binds (Fig. 1c and Fig. 1d).
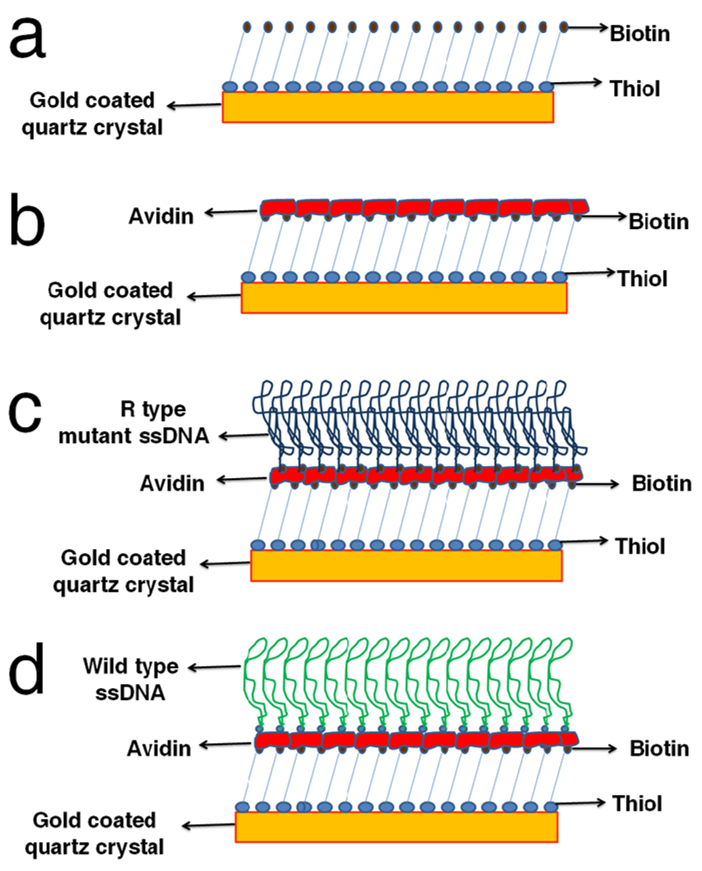
FIGURE 1.
Schematic process for immobilizing ssDNA strands with a complex 3-D structure on a quartz crystal. (a) A monolayer of biotin-thiol is immobilized onto the Au film by covalently linking the thiol group to Au. (b) A layer of streptavidin binds to the biotin layer. (c) Biotinylated mutant type ssDNA with a complex 3-D structure is immobilized through binding with the streptavidin layer. (d) Biotinylated wild type ssDNA (with a different 3-D structure from the mutant type) is immobilized onto the crystal by the same method.
RESULTS AND DISCUSSION
The scheme for immobilizing ssDNA stands, amplified from either the wild type or mutant type, on the surface of the resonator is presented in Figure 1. The quartz crystal resonator is coated with a thin gold (Au) film. A monolayer of biotin modified with a thiol functional group (biotin-thiol) is first immobilized onto the Au film by covalently linking the thiol group to Au (Fig. 1a). A layer of streptavidin then binds to the biotin layer (Fig. 1b). Afterwards, ssDNA with a biotin group at the 5’ end (biotinylated ssDNA) is immobilized through binding with the streptavidin layer (Fig. 1c and Fig. 1d). The biotinylated ssDNA is produced by amplifying a fragment of the P53 gene with a biotin-modified primer, followed by denaturing the amplified dsDNA at 95°C and suddenly cooling at −20°C. Upon the sudden temperature quench, each ssDNA strand undergoes intra-base pairing and forms a particular complex 3-D structure which is characteristic of its base sequence. Because of the presence of the mutant base, ssDNA strands amplified from wild type and mutant type genes form different 3D conformations before they are tethered to the surface of the resonator, which would eventually result in films of different mechanical properties.
The quartz crystal resonator is driven at its fundamental resonant frequency of 5 MHz in PBS buffer. The odd harmonic resonances with resolution down to 1 Hz and the energy dissipation of the resonator are recorded as a function of time. The energy dissipation is given in terms of the dissipation factor D which is inversely proportional to the quality factor (Q) and defined in terms of energy dissipated versus energy stored in the oscillator as:
| (1) |
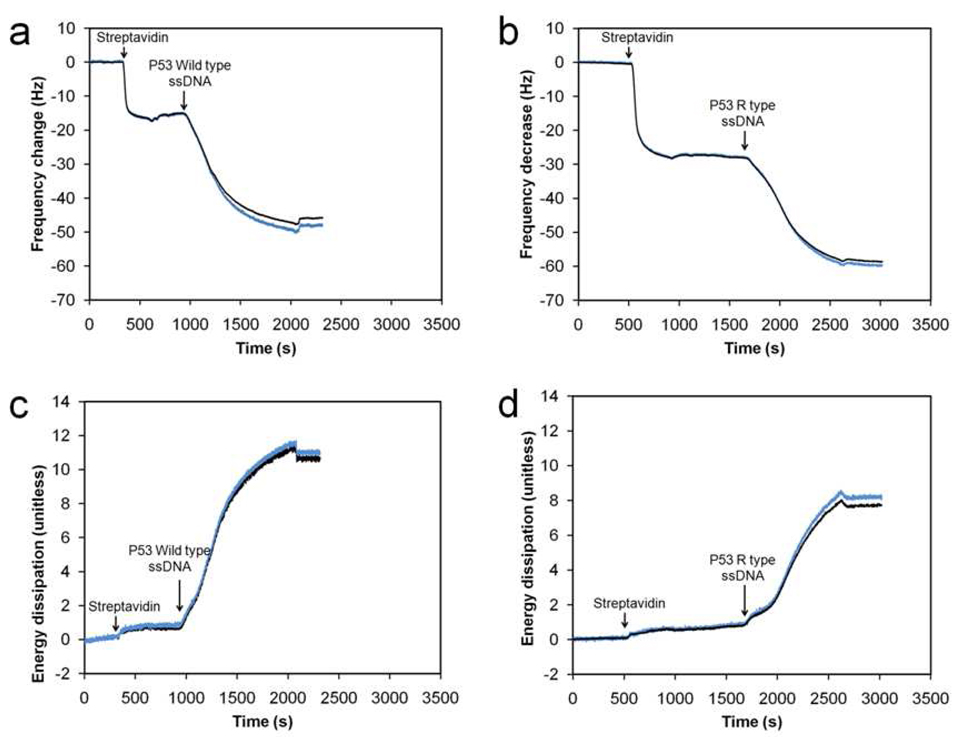
Figure 2 shows a typical response of the fifth harmonic frequency (Fig. 2a and Fig. 2b) and the energy dissipation (Fig. 2c and Fig. 2d) of the resonator recorded during the immobilization of wild type and mutant type ssDNA strands respectively. The first sharp decrease in the resonant frequency and the slight increase in energy dissipation indicate a strong binding of streptavidin to the biotin layer on the crystal. The biotin on the crystal binds to two of the four binding sites of each streptavidin molecule, thus forming a reasonably rigid layer.12,13 Upon flowing the ssDNA sample over the crystal, the biotin end of the ssDNA quickly binds to the streptavidin present on the crystal surface. This is seen as a large decrease in frequency and a more apparent increase in energy dissipation. For both the wild and mutant type ssDNA, the frequency decrease due to binding of the ssDNA (Fig. 2a and Fig. 2b) is about the same, indicating that the mass adsorbed is about the same. This is expected as the number of bases for the wild type and the mutant type ssDNA are both 545 bases. However, interestingly, appreciable differences in the energy dissipation due to binding of the wild type (Fig. 2c) and the mutant type (Fig. 2d) are observed, indicating that the viscoelastic properties of the films consisting of the two types of ssDNA strands are different.
FIGURE 2.
Plot of raw (blue curve) and fitted (black curve) data for a typical frequency response and energy dissipation of the resonator at its fifth harmonic resonance as a function of time during the immobilization of the streptavidin layer and the ssDNA film. (a) Frequency response plot for the wild type sample. (b) Frequency response plot for the mutant type sample. (c) Energy dissipation plot for the wild type sample. (d) Energy dissipation plot for the mutant type sample.
To extract the viscoelastic properties of the ssDNA films from the frequency response and energy dissipation of the resonator, the film is modeled using the Voight model of viscoelasticity. The Voight model is represented as a spring and a dashpot in parallel, the spring constituting the pure elastic response and the dashpot, the viscous response. The calculation of shear modulus of polymer films from the electrical response of the quartz resonator using transmission line analysis has been well investigated earlier.14–16 A more direct method is the continuum mechanics approach, in which the materials physical properties such as the film shear modulus, film viscosity, and film thickness are directly related to the experimentally measured resonant frequency change and the energy dissipation response.17 Within the framework of continuum mechanics, the shear modulus used in the Voight model is a complex shear modulus (μ*) which can be written as:
| (2) |
where μ is the elastic shear modulus and η is the shear viscosity of the film. For viscoelastic materials, as the strain lags the stress by some amount, describing this behavior by a complex shear modulus makes it convenient to separate out the in-phase and out-of-phase stress components.14 When a shear stress σxy is applied to the film, the stress strain relationship on the Voight element is given as follows:17
| (3) |
where ux is the displacement in the × direction and vx is the velocity in the × direction. μ, η, and the thickness of the film can be obtained by solving the wave equation for the propagation of bulk shear waves in a viscoelastic medium using appropriate boundary conditions17,18 at each layer on the crystal surface and fitting the frequency response and energy dissipation into the model. The fitted frequency response and energy dissipation of the resonator according to this model are plotted and compared with the raw data in Figure 2, which shows reasonably good agreement.
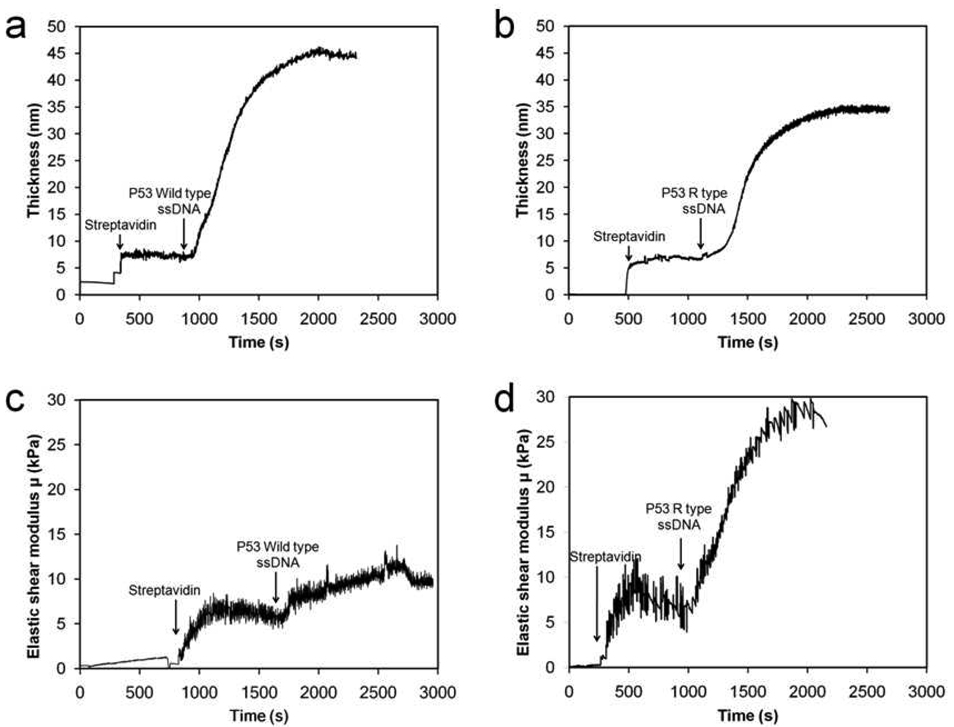
Figure 3 plots the thickness and μ of the films obtained from fitting the Voight model to the raw data. It is obvious that an appreciable difference in the plots of μ can be observed between the two types of samples, although the difference in the plot of film thickness is insignificant. The most apparent difference in the plot of μ is that after the DNA strands are immobilized, μ is about 11 kPa for the wild type sample but is about 28 kPa for the mutant type one. To confirm the result, the experiments for both types of samples are each repeated four times, and the result is presented in Table 1. It is found that the thickness of the biotin-streptavidin layer is about 10.0 nm with a standard deviation of 1.6 nm, the total thickness of the film after immobilizing the wild type ssDNA is about 30.0 nm with a standard deviation of 5.6 nm, and the total film thickness for the mutant type ssDNA is about 36 nm with a standard deviation of 3.7 nm. This indicates that no significant difference in the film thickness can be observed between the two types of samples, and the two types of DNA strands are immobilized on the surface with a similar aerial mass density consistently from experiment to experiment. However, μ of the films with immobilized wild type samples is about 13,715 Pa with a standard deviation of 1,713 Pa, while μ of the films with immobilized mutant type samples is about 24,399 Pa with standard deviation of 1,808 Pa.
FIGURE 3.
Plot of thickness and elastic shear modulus (μ) of ssDNA films obtained from the data in Fig. 2. (a) Film thickness as a function of time for the wild type ssDNA sample. (b) Film thickness as a function of time for the mutant type sample. (c) μ as a function of time for the wild type sample. (d) μ as a function of time for the mutant type sample.
Table 1.
Film thickness and shear modulus measured during the immobilization of P53 Wild type ssDNA and the P53 R type mutant ssDNA.
| Sample | Film thickness: (nm) |
Film Shear Modulus (Pa) |
|
|---|---|---|---|
|
P53 Wild type ssDNA |
Expt 1 | 29.5 | 15,561 |
| Expt 2 | 37.6 | 14,553 | |
| Expt 3 | 21.9 | 10,953 | |
| Expt 4 | 31.0 | 13,793 | |
|
P53 R type mutant ssDNA |
Expt 1 | 35.9 | 25,542 |
| Expt 2 | 34.3 | 23,258 | |
| Expt 3 | 42.1 | 26,687 | |
| Expt 4 | 32.1 | 22,110 | |
The large difference in μ indicates that there is an appreciable difference in the viscoelasticity of the films formed by the wild type and mutant type DNA. This difference is likely because the two types of ssDNA strands form different 3-D conformations in aqueous solution which is the direct result of one base mutation in their sequences. Upon quenching the temperature rapidly, ssDNA strands form a complex 3-D structure to minimize its free energy and hybridize with itself via intra-base pairing. During this process, even a single base mutation may significantly alter the 3-D structure of the ssDNA in solution, which leads to the appreciable differences in the mechanical properties of the DNA films formed by tethering such DNA strands to a solid surface. The resulted difference in the mechanical properties of the DNA films may be detected by a number of techniques. We believe that the example we show here by probing the elastic shear modulus of the DNA films using a quartz crystal resonator is only one approach among many others.
CONCLUSIONS
Although resonator-based sensors have been used for mutation detection,19–24 they are all based on mass change on the surface of the resonator which serves as a mass balance. Such mass change is typically caused by capturing DNA strands through hybridization with a DNA probe or use of a bioreceptor. To the best of our knowledge, no literature has reported mutation screening based on the mechanical properties of the ssDNA films. Coupling DNA films with a resonator provides an excellent means to probe the mechanical properties of DNA films. Combining it with the capability of DNA molecules to form unique 3-D structures enables us to screen the mutant from wild type DNA quickly and reliably. We have shown the capability of this technique to detect a single base mutation among a fragment of 545 bases in P53 gene, which already exceeds the reported maximum length of the DNA that can be effectively screened by other methods such as SSCP/HA and its variants. This technique can be further improved by selecting different fragments of the gene containing the mutation to optimize the screening. Similar to all the technologies based on the formation of DNA 3-D complex structures, much study still needs to be carried out to understand and predict the 3-D structure of the long DNA strand in solution as well as on a solid surface, so that better experiments could be designed to maximize the difference in the mechanical properties of the 3-D structures between the wild and mutant type DNA.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by the National Science Foundation (CBET 0747164) and the National Human Genome Research Institute of National Institute of Health.
REFERENCES
- 1.Hestekin CN, Barron AE. Electrophoresis. 2006;27:3805–3815. doi: 10.1002/elps.200600421. Top of Form. [DOI] [PubMed] [Google Scholar]
- 2.Kwok PY. Annual Review of Genomics and Human Genetics. 2001;2:235–258. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- 3.Cotton RG. Mutation research. 1993;285:125–144. doi: 10.1016/0027-5107(93)90060-s. [DOI] [PubMed] [Google Scholar]
- 4.Orita M, Suzuki Y, Sekiya T, Hayashi K. Genomics. 1989;5:874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi K, Yandell DW. Human Mutation. 1993;2:338–346. doi: 10.1002/humu.1380020503. [DOI] [PubMed] [Google Scholar]
- 6.Balogh K, Patocs A, Majnik J, Racz K, Hunyady L. Molecular Genetics and Metabolism. 2004;83:74–81. doi: 10.1016/j.ymgme.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Muyzer G, Smalla K. Antonie van Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 8.Larsen LA, Christiansen M, Vuust J, Andersen PS. Human Mutation. 1999;13:318–327. doi: 10.1002/(SICI)1098-1004(1999)13:4<318::AID-HUMU9>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Tabone T. Human mutation. 2008;29:886–890. doi: 10.1002/humu.20781. [DOI] [PubMed] [Google Scholar]
- 10.Hollstein M, Sidransky D, Vogelstein B, Harris CC. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 11.Rodahl M, Kasemo B. Sensors and Actuators A: Physical. 1996;54:448–456. [Google Scholar]
- 12.Su X, Wu Y-J, Robelek R, Knoll W. Langmuir. 2005;21:348–353. doi: 10.1021/la047997u. [DOI] [PubMed] [Google Scholar]
- 13.Aung K, Ho X, Su X. Sensors and Actuators B: Chemical. 2008;131:371–378. [Google Scholar]
- 14.Lucklum R, Hauptmann P. Frequency Control Symposium and PDA Proceedings of the 2001 IEEE International; 2001. pp. 408–418. [Google Scholar]
- 15.Behling C, Lucklum R, Hauptmann P. Ultrasonics, Ferroelectrics and Frequency Control, IEEE Transactions on; 1999. pp. 1431–1438. [DOI] [PubMed] [Google Scholar]
- 16.Lucklum R, Behling C, Cernosek RW, Martin SJ. Journal of Physics D: Applied Physics. 1997;30:346–356. [Google Scholar]
- 17.Voinova MV, Rodahl M, Jonson M, Kasemo B. Physica Scripta. 1999;59:391–396. [Google Scholar]
- 18.Reed CE, Kanazawa KK, Kaufman JH. Journal of Applied Physics. 1990;68:1993–2001. [Google Scholar]
- 19.Larsson C, Rodahl M, Hook F. Analytical Chemistry. 2003;75:5080–5087. doi: 10.1021/ac034269n. [DOI] [PubMed] [Google Scholar]
- 20.Su X, Wu Y, Knoll W. Biosensors and Bioelectronics. 2005;21:719–726. doi: 10.1016/j.bios.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Lucarelli F, Tombelli S, Minunni M, Marrazza G, Mascini M. Analytica Chimica Acta. 2008;609:139–159. doi: 10.1016/j.aca.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Hook F, Ray A, Norden B, Kasemo B. Langmuir. 2001;17:8305–8312. [Google Scholar]
- 23.Su X, Robelek R, Wu Y, Wang G, Knoll W. Analytical Chemistry. 2004;76:489–494. doi: 10.1021/ac035175g. [DOI] [PubMed] [Google Scholar]
- 24.Cho YK, Kim S, Kim YA, Lim HK, Lee K, Yoon D, Lim G, Pak YE, Ha TH, Kim K. Journal of Colloid and Interface Science. 2004;278:44–52. doi: 10.1016/j.jcis.2004.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.