Abstract
Cofilin is a major regulator of actin dynamics involved in the regulation of cell spreading and migration through its actin depolymerizing and severing activities. V-Src is an activated Src tyrosine kinase and a potent oncogene known to phosphorylate a variety of cellular proteins in cell transformation process including altered cell adhesion, spreading and migration. Recently, it has been suggested that cofilin is a potential substrate of v-Src (Rush et al., 2005). Here, we show direct tyrosine phosphorylation of cofilin by v-Src and identify Y68 as the major phosphorylation site. Cofilin phosphorylation at Y68 did not change its activity per se, but induced increased ubiquitination of cofilin and its degradation through the proteosome pathway. Furthermore, the negative effect of cofilin on cellular F-actin contents was inhibited by co-expression of v-Src, whereas that of cofilin mutant Y68F (Y68 mutated to F) was not affected, suggesting that v-Src-mediated cofilin phosphorylation at Y68 is required for degradation of cofilin in vivo. Lastly, inhibition of cell spreading by v-Src was rescued partially by co-expression of cofilin, and to a greater extent by the Y68F mutant which is not subjected to v-Src induced degradation through phosphorylation, suggesting that v-Src mediated changes in cell spreading is, at least in part, through inhibiting the function of cofilin via phosphorylating it at Y68. Together, these results suggest a novel mechanism by which cofilin is regulated by v-Src through tyrosine phosphorylation at Y68 that triggers degradation of cofilin via ubiquitination-proteosome pathway and consequently inhibits cofilin activity in reducing cellular F-actin contents and cell spreading.
Keywords: cofilin, protein phosphorylation, protein degradation, cell spreading
Introduction
Cell migration is an essential cellular activity for various biological and disease processes such as embryonic development, wound healing, angiogenesis and cancer metastasis (Caswell & Norman, 2006; Christopher & Guan, 2000; Lauffenburger & Horwitz, 1996). It is a multi-step process initiated by forward extension of cell membrane, called lamellipodia, which is also considered to be similar to the membrane protrusion in cell spreading (Wakatsuki et al., 2003). Lamellipodia plays a major role in cell migration by forming new attachments to the substrate and pulling the cell body forward (Condeelis, 2001; Zigmond, 2004). The force for membrane protrusion is provided by localized actin filament elongation and branching at the leading edge of lamellipodia, which requires coordinated functions of both actin-polymerizing and -depolymerizing factors (Pollard & Borisy, 2003; Stossel et al., 2001).
Cofilin is a major regulator of actin dynamics in cell spreading and migration (Bamburg, 1999; Bamburg et al., 1999; Carlier et al., 1999; Huang et al., 2006; Ichetovkin et al., 2002). It plays a key role in enhancing cell membrane protrusion at the leading edge of migrating cells by disassembling F-actin from the rear part of actin network to recycle G-actins to the front for further rounds of polymerization. In addition, F-actin severing activity of cofilin also promotes actin filament elongation by creating filament ends that are used as new nucleation sites for filament growth (Bailly & Jones, 2003; Carlier et al., 1997; Dawe et al., 2003). The activity of cofilin is tightly regulated due to its crucial function in the regulation of actin dynamics (Bailly & Jones, 2003; Bamburg & Wiggan, 2002). Phosphorylation of the conserved serine 3 (S3) residue of cofilin has been identified as a critical regulatory mechanism for cofilin (Agnew et al., 1995; Moriyama et al., 1996). LIM kinase (LIMK) and the related testicular kinase (TESK) have been shown to phosphorylate cofilin at this site specifically, which inhibits its actin binding activity and mediates various signals to remodel actin cytoskeleton (Gungabissoon & Bamburg, 2003; Meberg, 2000). Conversely, Chronophin and Slingshot (SSH1L) act as specific phosphatases for cofilin at S3 and mediate dephosphorylation of cofilin by several stimuli (Niwa et al., 2002; Samstag et al., 1996). A recent report using proteomic analysis of v-Src transformed NIH3T3 cells has identified phosphorylation of Y140 of cofilin in these cells, although the functional consequence of Y140 phosphorylation was not further investigated (Rush et al., 2005). It is also unclear whether cofilin could be regulated by other post-translational modifications and whether these may play a role in the regulation of cofilin activity in actin dynamics during cell spreading and migration.
The Src family tyrosine kinases are implicated in a variety of signaling pathways, leading to the stimulation of DNA synthesis, cell proliferation and cytoskeletal reorganization (Bjorge et al., 2000; Brown & Cooper, 1996; Thomas & Brugge, 1997). A highly activated viral counterpart of Src, v-Src, is responsible for the transforming properties of Rous Sarcoma virus (Yeatman, 2004). A number of cellular substrates have been identified for v-Src, whose phosphorylation contributes to the altered cell adhesion, migration and invasion of the transformed cells that likely play a role in cancer metastasis in vivo (Brown & Cooper, 1996; Frame, 2002; Lin et al., 2006). These substrates include FAK, Cas and paxillin, proteins that are localized in focal adhesions and important for the regulation of cell adhesion, spreading, migration and invasion (Brown & Turner, 2004; Defilippi et al., 2006; Mitra & Schlaepfer, 2006; Parsons, 2003). They also include proteins that regulate actin dynamics directly such as cortactin, whose activity for actin filament cross-linking is inhibited by Src-mediated tyrosine phosphorylation (Huang et al., 1997; Wu & Parsons, 1993). In addition, the activity of Ste20-like kinase (SLK) to induce actin stress fiber disassembly was shown to be inhibited by v-Src kinase activity (Chaar et al., 2006; Sabourin et al., 2000).
While these results have clearly shown multiple potential mechanisms by which v-Src may influence actin dynamics in cell spreading and migration, it is not clear whether v-Src could target other and potentially more direct major regulators of actin assembly or disassembly. Given the key functions of cofilin in the disassembly of F-actin in cell migration, we have examined potential regulation of cofilin by v-Src through phosphorylation. We identified Y68 of cofilin as a major phosphorylation site for v-Src and showed that this phosphorylation induced degradation of cofilin through the ubiquitination-proteosome pathway. Consistent with this, we found that co-expression of v-Src with cofilin, but not the Y68F mutant significantly inhibited the function of cofilin to reduce cellular F-actin contents and that the inhibitory effect of v-Src on cell spreading was partially rescued by co-expression of cofilin, but not Y68F mutant. These results establish cofilin as a key substrate of v-Src in the regulation of actin dynamics and cell adhesion.
Results
Phosphorylation of cofilin by v-Src at Y68
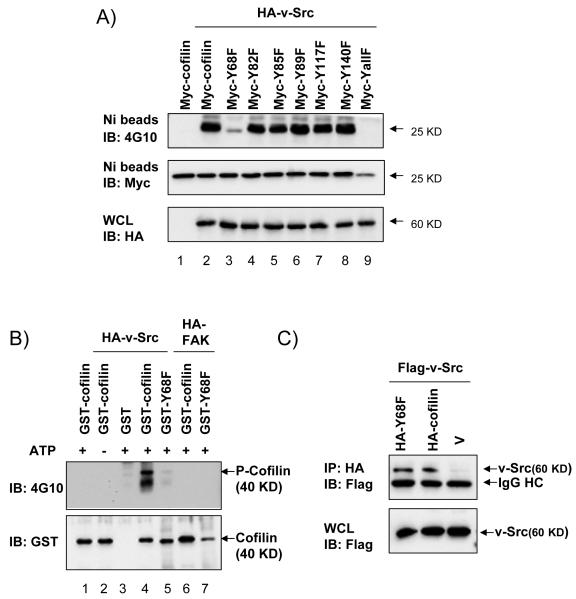
To study the potential regulatory mechanism of cofilin by v-Src, we first determined whether v-Src can mediate tyrosine phosphorylation of cofilin. 293T cells were transfected with plasmids encoding HA-tagged v-Src and His-Myc-tagged cofilin. The overexpressed cofilins were then pulldown with Ni-beads, followed by immunoblotting with anti-phosphotyrosine antibody, 4G10. Fig. 1A shows that cofilin is specifically phosphorylated on tyrosine in cells co-transfected with v-Src (compare lane 2 with lane 1). To identify the site of phosphorylation by v-Src, each of the 6 tyrosine residues in cofilin was mutated to phenylalanine and the mutants were examined for their phosphorylation by v-Src. As shown in Fig. 1A, the Y68F mutant exhibited the greatest reduction in phosphorylation by v-Src (compare lane 3 with lanes 2 and 4–8), suggesting that Y68 is the major phosphorylation site of v-Src. Mutation of all six sites completely abolished cofilin phosphorylation by v-Src, as expected (lane 9). Co-transfection of plasmids encoding HA-FAK and Myc-cofilin or its mutants in similar experiments did not result in tyrosine phosphorylation of cofilin (data not shown), providing further support for the specificity of cofilin phosphorylation by v-Src. We then examined whether v-Src could directly phosphorylate cofilin at Y68. In vitro phosphorylation assay was performed using purified GST fusion proteins GST-cofilin or GST-Y68F, and recombinant HA-v-Src or HA-FAK immobilized on agarose beads by immunoprecipitation of lysates from 293T cells expressing the kinases, as described in the Materials and Methods. Fig. 1B shows that GST-cofilin was phosphorylated by v-Src in an ATP-dependent manner in vitro (lanes 1–4). Interestingly, mutation of Y68 to F significantly decreased cofilin phosphorylation by v-Src in vitro (compare lane 5 and lane 4), suggesting that Y68 is a major phosphorylation site by v-Src. The weak signal in lane 5 could be due to phosphorylation of GST-Y68F mutant at other sites by v-Src. We also observed a lower band in both lanes 4 and 5, which is probably a degradation product of GST-cofilin with the GST part degraded at least partially as this lower band was not recognized by anti-GST (lower panel). Consistent with the co-transfection studies, recombinant FAK did not phosphorylate either GST-cofilin or GST-Y68F in vitro (lanes 6 and 7). Lastly, we examined whether v-Src could associate with its substrate cofilin by co-immunoprecipitation of lysates from 293T cells that had been transfected with plasmids encoding Flag-tagged v-Src and HA-tagged cofilin, Y68F mutant, or vector alone. Fig. 1C shows that v-Src associated with both its substrate cofilin and the Y68F mutant, suggesting that the lack of phosphorylation of the Y68F mutant was not caused by a disruption of v-Src association with the mutant. Taken together, these results demonstrate that v-Src binds to cofilin and phosphorylates it at Y68.
Figure 1. v-Src phosphorylation of cofilin at Y68.
(A) 293T cells were co-transfected with His- and Myc-tagged cofilin or the mutants and HA-tagged v-Src or vector control, as indicated. Lysates were precipitated with Ni-beads, followed by western blotting with anti-phosphotyrosine, 4G10 (top) or anti-Myc (middle) antibody. Whole cell lysates (WCL) were also immunoblotted with anti-HA (bottom). (B) Recombinant GST-cofilin or GST-Y68F purified from bacteria were incubated in a kinase buffer (ATP is omitted in lane 2, as indicated by labels above the upper panel) with HA-v-Src or HA-FAK immunoprecipitated from 293T cells transfected with corresponding expression vectors, as indicated. In vitro kinase assays were performed as described in the Experimental Procedures and the reaction mixtures were subjected to immunoblotting with 4G10 antibody (upper) or anti-GST (lower). (C) 293T cells were co-transfected with Flag-tagged v-Src and HA-tagged cofilin or Y68F mutant or vector control as indicated. Lysates were immunoprecipitated with anti-HA antibody, followed by western blotting with anti-Flag antibody (upper). WCL were also immunoblotted with anti-Flag (lower). Molecular weight markers are indicated on the right.
Phosphorylation of cofilin at Y68 by v-Src regulates its ubiquitination and degradation
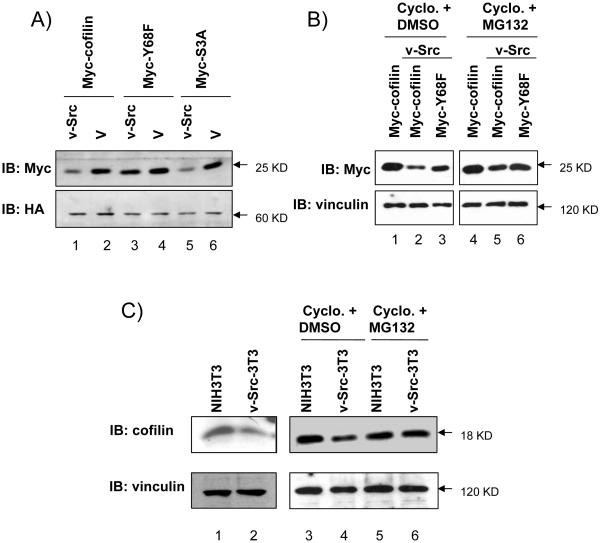
During the course of studies on cofilin phosphorylation by v-Src, we noticed that the expression level of cofilin and the S3A mutant, but not the Y68F mutant, was reduced specifically in the presence of co-expressed v-Src (Fig. 2A). Because the ectopically expressed cofilin is under the control of an exogenous promoter, these results suggest that v-Src likely affects the expression level of cofilin through posttranscriptional mechanisms. This possibility is supported by the observation that treatment of the cells with an inhibitor of protein synthesis cycloheximide did not affect the decreased protein level of cofilin induced by v-Src (Fig. 2B, lanes 1–3). We then examined the effects of v-Src on the protein levels of endogenous cofilin using a NIH 3T3 cell line stably expressing v-Src. Fig. 2C shows that expression of cofilin in these cells is also decreased compared to the control NIH 3T3 cells both in the presence and absence of cycloheximide (lanes 1–4). The expression level of endogenous cofilin is also reduced by expression of v-Src with or without treatment of cycloheximide in 293T and CHO cells (data not shown). Together, these data suggest that phosphorylation of cofilin by v-Src at Y68 may regulate the expression of cofilin by stimulation of its degradation.
Figure 2. Degradation and ubiquitination of cofilin induced by v-Src phosphorylation at Y68 residue.
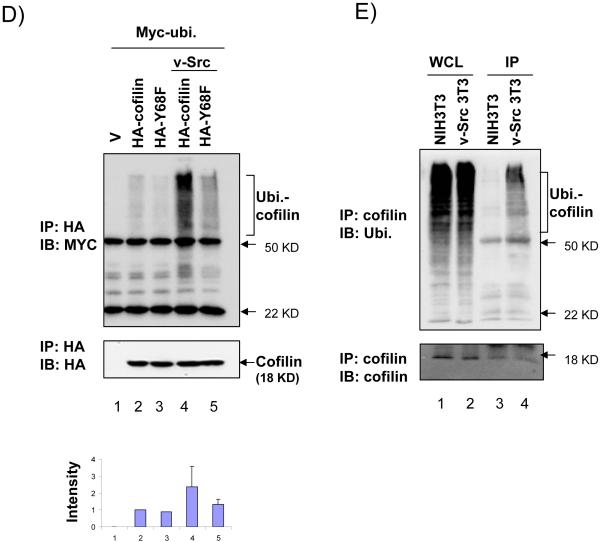
(A, B) 293T cells were co-transfected with Myc-tagged cofilin, Y68F, or S3A mutant, and HA-tagged v-Src or vector control as indicated. Lysates were then prepared and analyzed by western blotting with anti-Myc, anti-HA, or anti-vinculin, as indicated. In panel B, cells were treated with cycloheximide and MG132 or DMSO as control for 4 hrs before lysis as described in Experimental Procedures. (C) NIH3T3 and v-Src-transformed-NIH3T3 (v-Src-3T3) cells were treated with nothing (lanes 1 and 2), cycloheximide with DMSO (lanes 3 and 4) or with MG132 (lanes 5 and 6) for 4 hrs. Lysates were then prepared and analyzed by western blotting with anti-cofilin (upper) and anti-vinculin (lower). Molecular weight markers are indicated on the right. (D) 293T cells were co-transfected with HA-tagged cofilin or Y68F mutant, and Myc-tagged ubiquitin and Flag-tagged v-Src or vector control as indicated. Cells were treated with MG132 for 4 hrs and then lysates were prepared and immunoprecipitated with anti-HA antibody, followed by western blotting with anti-Myc (top) or anti-HA (middle). The Ubiquitinated cofilin bands (marked on the right) were quantified by densitometer from 3 independent experiments and relative intensity was shown with the mean + S.E (bottom). (E) NIH3T3 and v-Src-3T3 cells were treated with MG132 for 4 hr and lysates were prepared and immunoprecipitated with anti-cofilin antibody, followed by western blotting with anti-ubiquitin (upper) or anti-cofilin (lower). The ubiquitinated cofilin bands are marked on the right. Molecular weight markers are indicated on the right.
The proteosome-mediated degradation of ubiquitinated proteins is a major pathway that regulates expression of various proteins in cells. To examine whether the effect of v-Src phosphorylation of cofilin on its degradation could be through such a mechanism, we first tested the influence of the proteosome inhibitor, MG132, on the expression level of cofilin and Y68F mutant in the presence of v-Src. As shown in Fig. 2B, treatment of cells with MG132 reversed the decrease of cofilin expression by v-Src (lanes 4–6). Similarly, treatment of cells with MG132 reversed the degradation of endogenous cofilin induced by v-Src in NIH 3T3 (Fig. 2C, lanes 5 and 6) and 293T cells (data not shown). We then examined potential ubiquitination of cofilin and the Y68F mutant with or without v-Src co-expression in 293T cells. Fig. 2D shows that cofilin could be ubiquitinated in the cells, which was significantly increased by co-expression of v-Src (lanes 2 and 4). Co-expression of v-Src with the Y68F mutant also increased its ubiquitination slightly but at a much reduced extent compared to the induction for the wild type cofilin (lanes 3 and 5), suggesting that v-Src-mediated phosphorylation at Y68 may stimulate ubiquitination of cofilin. Furthermore, we also found an elevated ubiquitination of endogenous cofilin in the NIH 3T3 cell line stably expressing v-Src compared to the control NIH 3T3 cells (Fig. 2E). Taken together, these results suggest that v-Src-mediated tyrosine phosphorylation of cofilin at 68Y can induce its ubiquitination resulting in its degradation through the proteosomes.
v-Src-mediated phosphorylation inhibits cofilin activity in vivo
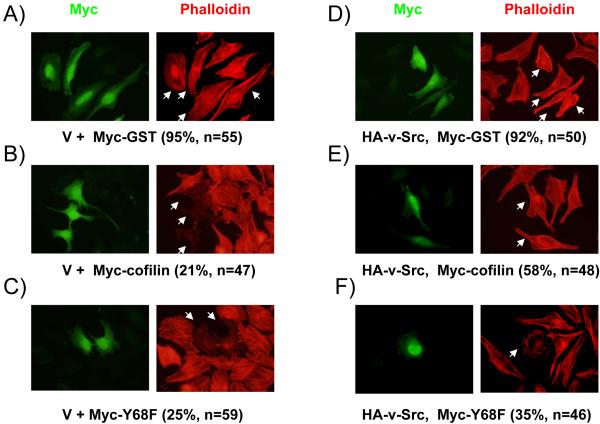
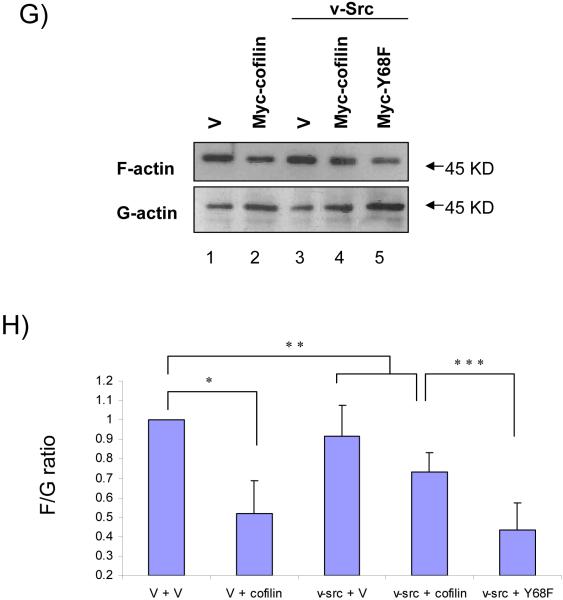
To study the physiological role of v-Src-mediated degradation of cofilin in vivo, we first analyzed the effect of co-expression of v-Src on the function of cofilin to reduce the cellular F-actin contents. HeLa cells were co-transfected with Myc-tagged cofilin or Y68F mutant and HA-tagged v-Src, and examined by immunofluorescence staining with Texas-Red conjugated phalloidin to detect cellular F-actin in stress fibers. Consistent with a previous report (Yap et al., 2005), over-expression of cofilin reduced the cellular F-actin contents compared with the control cells while transfection of the control vector did not affect it (Figs. 3A and 3B). Over-expression of the Y68F mutant also reduced F-actin contents to a similar extent (Fig. 3C), suggesting that mutation at this residue did not affect the cofilin activity per se in reducing F-actin contents. Interestingly, co-expression of v-Src with cofilin reversed its function to reduce the F-actin contents (Fig. 3E) whereas expression of v-Src alone did not affect stress fiber formation under the experimental conditions (Fig. 3D). In contrast, co-expression of v-Src with the Y68F mutant did not prevent the reduction of stress fibers by the Y68F mutant (Fig. 3F). We next measured the ratio of F-actin and G-actin (F/G actin) directly to quantify the effect of v-Src-mediated phosphorylation of cofilin on its activity to reduce F-actin contents. Consistent with data from the immunofluorescence, expression of cofilin reduced F/G actin significantly whereas expression of v-Src alone did not affect it under the experimental conditions (Figs. 3G and 3H). Furthermore, co-expression of v-Src with cofilin, but not the Y68F mutant, inhibited cofilin function to reduce F/G actin in cells. These results suggest that v-Src inhibits cofilin function to reduce F-actin contents through phosphorylation-mediated protein degradation in vivo.
Figure 3. Phosphorylation of cofilin by v-Src reduces its activity in inducing stress fiber disassembly and actin depolymerization.
(A-F) HeLa cells were co-transfected with HA-tagged v-Src and Myc-tagged cofilin or Y68F mutant or control protein (GST) as indicated. Cells were fixed and subjected to immunofluorescence staining with anti-Myc or phalloidin to view filamentous actin. The transfected cells are marked by arrows. The percentage of transfected cells with positive phalloidin staining are indicated in the parentheses. (G) 293T cells were co-transfected with Myc-tagged cofilin, Y68F mutant or vector control and HA-tagged v-Src or vector control, as indicated. The cellular F- and G-actin contents were determined by western blotting after fractionation as described in the Experimental Procedures. Molecular weight markers are indicated on the right. (H) Relative ratio of F/G actin were obtained from three independent experiments and shown with the mean + S.E. after normalization to that of control cells (V+V, lane 1). *P<0.05; **P>0.05 in comparison with values from control cells. ***P<0.05 in comparison with values from cells transfected with v-Src and cofilin (v-Src + cofilin).
v-Src inhibits cell spreading through phosphorylation of cofilin at Y68
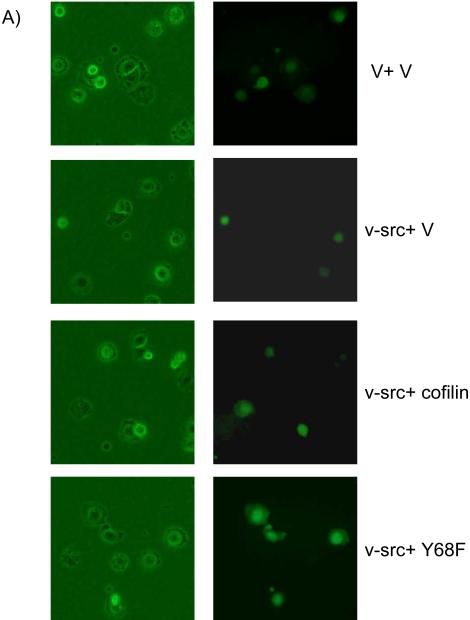
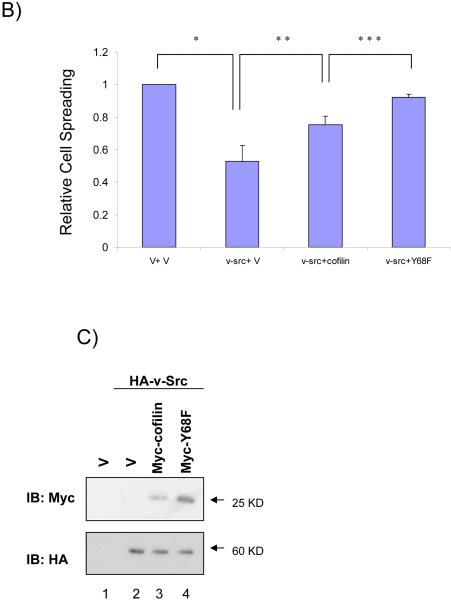
Cofilin has been shown to promote cell spreading by stimulating actin cytoskeleton dynamics and membrane protrusion (Bamburg, 1999; Bamburg et al., 1999; Carlier et al., 1999; Huang et al., 2006; Ichetovkin et al., 2002). V-Src is also known to reduce cell spreading in many cell types through phosphorylation of target cellular proteins, even though the precise mechanisms are still not well understood (Brown & Cooper, 1996; Frame, 2002; Lin et al., 2006). Therefore, we investigated whether v-Src could regulate cell spreading through its phosphorylation of cofilin at Y68 by examining the effects of co-expression of v-Src with cofilin or its Y68F mutant on cell spreading. CHO cells were co-transfected with Myc-tagged cofilin or the Y68F mutant, and HA-tagged v-Src or control vector, along with a plasmid encoding GFP as a transfection marker. Cell spreading was assessed for the transfected cells after plating on fibronectin (FN), as described previously (Yoo et al., 2006). As shown in Figs. 4A and 4B, expression of v-Src in CHO cells reduced its spreading as expected. Consistent with the possibility that v-Src inhibits cell spreading through its induction of degradation of cofilin which plays a positive role in cell spreading, co-expression of cofilin with v-Src partially reversed the decrease in cell spreading induced by v-Src. Interestingly, co-expression of the Y68F mutant, which showed reduced phosphorylation by v-Src and consequent degradation (see Figs. 1 and 2), almost completely abolished the inhibitory effects by v-Src. Western blotting analysis of aliquots of the transfected cells confirmed the higher expression of the Y68F mutant (due to less degradation) compared to the wild type, when co-expressed with v-Src (Fig. 4C). Together, these results suggest that inhibition of cell spreading by v-Src, at least in part, is through its phosphorylation of cofilin at Y68 and subsequence induction of cofilin degradation via ubiquitination-proteosome pathway.
Figure 4. Reversion of v-Src inhibition of cell spreading by cofilin and Y68F mutant.
(A and B) CHO cells were transfected with Myc-tagged cofilin or Y68F mutant, and HA-tagged v-Src or vector control along with a plasmid encoding GFP as a transfection marker, as indicated. Cell spreading assays were performed as described in the Experimental Procedures. Representative micrographs are shown in A. Panel B shows the mean + S.E. of percentage of spread cells (among transfected cells as identified by GFP expression) from three independent experiments are shown as relative spreading after normalization to that in mock transfected cells. *P<0.05 in comparison with value from mock cells. **P<0.05 in comparison with value from cells transfected with v-Src and vector (v-Src + V). ***P<0.05 in comparison with value from cells transfected with v-Src and cofilin (v-Src + cofilin). (C) Aliquots of WCL were analyzed by western blotting with anti-Myc (upper) or anti-HA (lower). Molecular weight markers are indicated on the right.
Discussion
As a major actin regulatory protein with depolymerizing and severing activity, cofilin plays a key role in many cellular processes in which reorganization or rapid turnover of actin filaments are required. Consistent with its important role in actin cytoskeleton dynamics, cofilin is tightly regulated by multiple mechanisms in the cell (Bamburg, 1999; Carlier et al., 1999; Ono, 2003). Phosphorylation of cofilin at S3 is well established as a major regulatory mechanism that inhibits its cellular activities (Agnew et al., 1995; Moriyama et al., 1996). In the present study, we report a novel regulatory mechanism of cofilin through its tyrosine phosphorylation at Y68 that triggers degradation of cofilin via ubiquitination-proteosome pathway and consequently inhibits cellular function of cofilin in reducing cellular F-actin contents and cell spreading.
A previous proteomic analysis showed phosphorylation of Y140 in v-Src transformed cells (Rush et al., 2005). However, the potential biological significance of this potential event was not assessed. Indeed, our mutational analysis suggested that changing Y140 to F did not affect the overall tyrosine phosphorylation of cofilin to any significant extent, suggesting that Y140 is probably a minor phosphorylation site of cofilin. While the potential biological significance of Y140 phosphorylation remains to be established, our results suggest that Y68 is a major phosphorylation site by v-Src whose phosphorylation is critical in the regulation of cellular function of cofilin. First, mutation of Y68 to F almost completely abolished tyrosine phosphorylation of cofilin by v-Src. Secondly, co-expression of v-Src with the wild type cofilin, but not the Y68F mutant, led to increased cofilin ubiquitination, degradation, and reduced its function to decrease cellular F-actin contents. Lastly, co-expression of wild type cofilin, but not the Y68F mutant, partially reversed inhibition of cell spreading by v-Src. Therefore, to our knowledge, this report is the first to demonstrate a role for tyrosine phosphorylation in the regulation of cofilin and its cellular functions.
Both protein phosphorylation and ubiquitination are important posttranslational regulatory mechanisms for a variety of proteins. Increasing evidence suggest that these two proteins modifications may crosstalk to each other in either a positive or negative manner (Hunter, 2007). Protein phosphorylations have been shown to serve as markers to target the modified proteins to the ubiquitination-proteosome pathway (Bao et al., 2003; Levkowitz et al., 1999; Martinez-Moczygemba et al., 2007; Scaglioni et al., 2006; Yan et al., 2006). For example, Src-induced tyrosine phosphorylation of cbl could lead to its ubiquitination and subsequent degradation (Bao et al., 2003; Levkowitz et al., 1999). It has also been reported that Src directly phosphorylates FHIT, a tumor suppressor, which induces its ubiquitination and protein degradation (Bianchi et al., 2006). Jak-mediated tyrosine phosphorylation of IL-5RβC-sharing receptor also induces its degradation through ubiquitination (Martinez-Moczygemba et al., 2007). Our findings of increased ubiquitination and degradation of cofilin upon Src phosphorylation at Y68 suggested that such a positive crosstalk between protein phosphorylation and ubiquitination is also used in the cellular regulation of actin dynamics. Interestingly, LIMK, the upstream kinase for cofilin phosphorylation at S3, has also been shown to be regulated by ubiquitination-proteosome pathway, although it is not clear whether polyubiquitination of LIMK by its ubiquitin ligase RNF6 is affected by potential phosphorylation of LIMK itself (Tursun et al., 2005).
Phosphorylation of cofilin at S3 reduces its actin binding and depolymerization activity (Agnew et al., 1995; Moriyama et al., 1996). In contrast, tyrosine phosphorylation of cofilin at Y68 by v-Src appears not to affect its activity per se. We found that the Y68F mutant exhibited similar activity as the wild type cofilin in reducing cellular F-actin contents (see Fig. 3). Furthermore, neither the mutation of Y68 to F nor co-expression of v-Src with cofilin affected the status of S3 phosphorylation of cofilin (data not shown), suggesting that tyrosine phosphorylation at Y68 did not influence serine phosphorylation at S3 or cofilin activity indirectly. Instead of affecting cofilin activity itself, our data suggested that tyrosine phosphorylation at Y68 induces cofilin degradation through the ubiquitination-proteosome pathway thus affecting its cellular functions in actin dynamics. The precise control of the protein level of cofilin is important for its regulation of cell spreading and migration. RNAi-mediated knockdown of cofilin and slingshot, a phosphatase that activates cofilin, resulted in inhibition of cell spreading of Drosophila S2 cells (Rogers et al., 2003), suggesting that cofilin activity is required for cell spreading. On the other hand, inhibition of a negative regulator of cofilin, TESK1, by either actopaxin (LaLonde et al., 2005; Tsumura et al., 2005) or Spry4 (Tsumura et al., 2005), resulted in the inhibition of cell spreading, suggesting that over-activation of cofilin may also lead to reduced cell spreading. A biphasic relationship between cofilin protein level and cell migration has also been reported (Yap et al., 2005). Cells producing a moderate amount of cofilin increase the locomotion rate, whereas cells displaying higher amount of cofilin decrease the locomotion speed. The critical importance of cellular levels of cofilin suggests that both its regulation by phosphorylation-dependent ubiquitination-proteosome pathway as well as that by the phosphorylation at S3 are likely to serve as a major, although independent, regulatory mechanisms.
Activated Src family members such as v-Src are potent oncogenes that induce cellular transformation by tyrosine phosphorylation of multiple cellular proteins (Bjorge et al., 2000; Brown & Cooper, 1996; Thomas & Brugge, 1997). Altered cell adhesion and migration are likely important determinants of cancer invasion and metastasis and tyrosine phosphorylation of several cellular proteins by v-Src has been implicated to mediate these activities of v-Src (Brown & Turner, 2004; Defilippi et al., 2006; Mitra & Schlaepfer, 2006; Parsons, 2003). Our results here suggest an additional mechanism by which v-Src may regulate actin dynamics and cell spreading and migration. Consistent with previous studies that showed a negative effect of v-Src on cell spreading (Boschek et al., 1981; Kellie et al., 1986; Shriver & Rohrschneider, 1981), we found that v-Src over-expression inhibited spreading of CHO cell on FN. This inhibition was rescued by co-expression with cofilin, and to a greater extent by the Y68F mutant which is more stable due to its resistance of phosphorylation by v-Src, suggesting that v-Src down-regulates cell spreading, at least in part, through its phosphorylation of cofilin.
Our findings of tyrosine phosphorylation of cofilin at Y68 by v-Src raise the interesting question on whether this site could be a target for other tyrosine kinases. We did not detect tyrosine phosphorylation of endogenous cofilin in 293T cells (data not shown) and cofilin was not phosphorylated by FAK in vitro (see Fig. 1A). Nevertheless, it will be interesting to determine whether cofilin could be phosphorylated and regulated by other cellular tyrosine kinases under appropriate conditions and how such regulation is involved in normal physiological processes besides transformation by an active oncogene like v-Src. Likewise, it would also be interesting to investigate the role and mechanisms of cofilin phosphorylation by activated Src in tumor development and progression in vivo in future studies.
Materials and Methods
Plasmid construction
Mammalian expression vectors pKH3, pHan, pKH3-FAK and pKH3-v-Src have been described previously (Abbi et al., 2002; Chen et al., 1996; Cooper et al., 2003; Gan et al., 2005). To clone the cDNA of cofilin, 5′-CGGGATCCAAGCTTATGGCCTCCGGTGTGGCTGTCTCTGATGGT-3′ and 5′-GGAATTCCTCACAAAGGCTTGCCCTCCAGGGAGATGAC-3′ were used for the PCR from cDNA library prepared from 293T cells using the Qiagen RNeasy kit and the Superscript III reverse transcription -PCR kit.(Qiagen, valencia,CA, Invitrogene) according to the manufacturer’s manuals. The PCR product was digested with BamH1 and EcoR1, and then ligated to a linearlized pKH3 vector ad sequenced to generate pKH3-cofilin. BamH1-EcoR1 fragment digested from pKH3-cofilin was ligated to a linearlized pHan and pGex2T to generate pHan-cofilin and pGex2T-cofilin, respectively. To generate point mutant of cofilin at each tyrosine residue, site-directed mutagenesis was performed using pKH3-cofilin as template and primers, 5′-ACTGTCGACGACCCCTTCGCCACCTTTGTCAAGATG-3′ and 5′-CATCTTGACAAAGGTGGCGAAGGGGTCGTCGACAGT-3′ for pHA-cof-Y68F, 5′-GATAAGGACTGCCGCTTTGCCCTCTATGATGCAACC-3′ and 5′-GGTTGCATCATAGAGGGCAAAGCGGCAGTCCTTATC-3′ for pHA-cof-Y82F, 5′-TGCCGCTATGCCCTCTTTGATGCAACCTATGAGACC-3′ and 5′-GGTCTCATAGGTTGCATCAAAGAGGGCATAGCGGCA-3′ for pHA-cof-Y85F, 5′-CTCTATGATGCAACCTTTGAGACCAAGGAGAGCAAG-3′ and 5′-CTTGCTCTCCTTGGTCTCAAAGGTTGCATCATAGAG-3′ for pHA-cof-Y89F, 5′-AAGAGCAAAATGATTTTTGCCAGCTCCAAGGACGCC-3′ and 5′-GGCGTCCTTGGAGCTGGCAAAAATCATTTTGCTCTT-3′ pHA-cof-Y117F, 5′-TTGCAAGCAAACTGCTTCGAGGAGGTCAAGGACCGC-3′ and 5′-GCGGTCCTTGACCTCCTCGAAGCAGTTTGCTTGCAA-3′ for pHA-cof-Y140F, 5′-CGGGATCCATGGCCGCCGGTGTGGCTGTCT-3′ and 5′-CAGCCACACCGGCGGCCATGGATCCCG-3′ for pHA-cof-S3A. All primers were used sequentially for site-directed mutagenesis to make pHA-cof-YallF. Each PCR product was cloned to pKH3 vector with the same method as pKH3-cofilin. BamH1-Cla1 fragment from pKH3-v-Src was ligated to pCDNA3.1-N-Flag to generate pFlag-v-Src.
Antibodies
The mouse monoclonal α-phosphotyrosine antibody (4G10) was obtained from upstate Biotechnology (Lake plasmid, NY). The rabbit polyclonal α-HA (Y11), the mouse monoclonal α-c-Myc (9E10), the goat polyclonal α-cofilin, the goat polyclonal α-actin and the mouse monoclonal α-ubiquitin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz,CA). The rabbit polyclonal α-GST antibody was described previously (Wu et al., 2004; Yoo et al., 2006). The mouse monoclonal α-Flag and the mouse monoclonal α-vinculin antibodies were obtained from Sigma. Texas-red conjugated phalloidin was obtained from Molecular Probes.
Cell culture and transfection
293T and HeLa cells were cultured in DMEM supplemented with 10% FBS. NIH3T3 and v-Src-NIH3T3 cells were maintained in DMEM with 10% CS. CHO cells were cultured in Ham’s F-12 with 10% FBS. Transient transfections were performed using lipofectamin (invitrogen) according to the manufacturer’s manual. In case of co-transfection, each plasmid was used in a 1:1 ratio. In cell spreading assay, GFP-encoding plasmid was used for transfection marker. The ratio between GFP, HA-tagged-, and Myc-tagged protein-coding plasmid was 1:3:3. For some experiments, cells were incubated with MG132 (5μM) and cycloheximide (10μg/ml) for 4–5 hours before lysis.
Immunoprecipitation and western blotting
Subconfluent cells were washed two times with cold PBS and lysed with Nonidet P-40 lysis buffer (20mM Tris, pH8.0, 137mM Nacl, 1% Nonidet P-40, 10% Glycerol, 1mM phenylmethylsulfonyl fluoride, 10mg/ml aprotinine and 20mg/ml leupeptin). Lysates were centrifuged for 20 min. at 4 °C, and protein concentration was determined by Bio-Rad protein assay. Immunoprecipitations were performed by incubating the lysate with appropriate antibody for >2hrs at 4°C. For detection endogenous protein, immunoprecipitation was followed by incubation with protein A-sepharose for another 2hrs. After washing five times with lysis buffer, the immune complex was resolved by SDS-PAGE. Western blotting was carried out using horseradish peroxidase-conjugated IgG and the ECL system for detection.
Preparation of GST fusion protein and in vitro kinase assay
GST-fusion proteins were produced and purified as described previously (Reiske et al., 1999). HA-tagged v-Src and FAK were prepared by immunoprecipitation from 293T cell lysates that was transfected with pKH3-v-Src and pKH3-FAK, respectively. Purified GST-fusion protein and HA-tagged v-Src or FAK protein were incubated in the kinase reaction buffer (10mM Hepes, pH7.4, 3mM MnCl2, 3 mM MgCl2, 1mM Na3VO4) with or without 100 mM ATP for 20 min. at room temperature. The reactions were then stopped by addition of SDS sample buffer. The samples were resolved on SDS-PAGE and analyzed by western blotting using anti-phosphotyrosine antibody 4G10 to detect tyrosine-phosphorylated proteins.
Fluorescent Microscopy
Cells were processed for immunofluorescent staining as described previously (Wu et al., 2004). Briefly, cells were fixed with 3.7% formaldehyde and treated with 0.5% triton X-100 for 10 min. mouse monoclonal anti-Myc and FITC-conjugated rabbit anti-mouse antibody were used as primary and secondary antibody, respectively. Texas-red conjugated phalloidin was used to detect F-actin.
F- and G-actin fractionation
F/G actin fractionation was performed as described previously (Tu et al., 2003). Briefly, cells were washed with PBS twice and lysed in F-actin stabilizing (LAS) buffer (50mM Pipes, pH6.9, 50 mM NaCl, 5mM MgCl2, 5mM EGTA, 5% glycerol, 0.1% NP-40, 0.1% triton X-100, 0.1% Tween20, 0.1% 2-mercaptoethanol, 0.001% antifoam, protease inhibitor cocktail, 1mM ATP) by homogenizing with 26.5G syringe. The lysates were centrifuged at 100,000g for 1 hr at 37°C. The supernatants (G-actin) were collected and placed on ice. The pellets (F-actin) were resuspended with the same volume as the supernatant of dH2O containing 2μM cytochalysin D and incubated on ice for 1 hr. Equal amount of G and F actin were subjected to immunoblotting assay with anti-actin antibody. The intensity of bands were quantified with image J software. Three independent experiments were performed, and the student’s t test was used to determine the statistical significance.
Cell spreading assay
Cell spreading assays were performed as described previously (Yoo et al., 2006). Cells were lifted by trypsin treatment, pelleted, resuspended in serum-free media and incubated at 37 °C for 1hr. The cells were replated and allowed to spread on the fibronectin (10μg/μl)-coated plates. After 1 hr, cells were fixed with 3.7% formaldehyde and photographed in random field. Spread cells were defined as cells with irregular morphology and lacking phase brightness, whereas non-spread cells were rounded shape and phase-bright under the microscope. Multiple fields were monitored and more than 200 transfected cells were counted blindly for each experiment. Three independent experiments were performed, and the student t test was used for statistical significance.
Supplementary Material
Acknowledgements
We are grateful to Dr. Xiaoyang Wu for discussions during the initial stages of the project. We thank our colleagues Huijun Wei, Ming Luo, Huaping Fan, Fei Liu, Chenran Wang, Richard Liang, Ann Park and Xiaofeng Zhao for their critical reading of the manuscript and helpful comments. This research was supported by NIH grant GM48050 to J.-L. Guan.
References
- Abbi S, Ueda H, Zheng C, Cooper LA, Zhao J, Christopher R, Guan JL. Mol Biol Cell. 2002;13:3178–91. doi: 10.1091/mbc.E02-05-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnew BJ, Minamide LS, Bamburg JR. J Biol Chem. 1995;270:17582–7. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- Bailly M, Jones GE. Curr Biol. 2003;13:R128–30. doi: 10.1016/s0960-9822(03)00072-1. [DOI] [PubMed] [Google Scholar]
- Bamburg JR. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, McGough A, Ono S. Trends Cell Biol. 1999;9:364–70. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Wiggan OP. Trends Cell Biol. 2002;12:598–605. doi: 10.1016/s0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- Bao J, Gur G, Yarden Y. Proc Natl Acad Sci U S A. 2003;100:2438–43. doi: 10.1073/pnas.0437945100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi F, Magnifico A, Olgiati C, Zanesi N, Pekarsky Y, Tagliabue E, Croce CM, Menard S, Campiglio M. Proc Natl Acad Sci U S A. 2006;103:18981–6. doi: 10.1073/pnas.0605821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorge JD, Jakymiw A, Fujita DJ. Oncogene. 2000;19:5620–35. doi: 10.1038/sj.onc.1203923. [DOI] [PubMed] [Google Scholar]
- Boschek CB, Jockusch BM, Friis RR, Back R, Grundmann E, Bauer H. Cell. 1981;24:175–84. doi: 10.1016/0092-8674(81)90513-4. [DOI] [PubMed] [Google Scholar]
- Brown MC, Turner CE. Physiol Rev. 2004;84:1315–39. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- Brown MT, Cooper JA. Biochim Biophys Acta. 1996;1287:121–49. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. J Cell Biol. 1997;136:1307–22. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Ressad F, Pantaloni D. J Biol Chem. 1999;274:33827–30. doi: 10.1074/jbc.274.48.33827. [DOI] [PubMed] [Google Scholar]
- Caswell PT, Norman JC. Traffic. 2006;7:14–21. doi: 10.1111/j.1600-0854.2005.00362.x. [DOI] [PubMed] [Google Scholar]
- Chaar Z, O’Reilly P, Gelman I, Sabourin LA. J Biol Chem. 2006;281:28193–9. doi: 10.1074/jbc.M605665200. [DOI] [PubMed] [Google Scholar]
- Chen HC, Appeddu PA, Isoda H, Guan JL. J Biol Chem. 1996;271:26329–34. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- Christopher RA, Guan JL. Int J Mol Med. 2000;5:575–81. doi: 10.3892/ijmm.5.6.575. [DOI] [PubMed] [Google Scholar]
- Condeelis J. Trends Cell Biol. 2001;11:288–93. doi: 10.1016/s0962-8924(01)02008-6. [DOI] [PubMed] [Google Scholar]
- Cooper LA, Shen TL, Guan JL. Mol Cell Biol. 2003;23:8030–41. doi: 10.1128/MCB.23.22.8030-8041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe HR, Minamide LS, Bamburg JR, Cramer LP. Curr Biol. 2003;13:252–7. doi: 10.1016/s0960-9822(03)00040-x. [DOI] [PubMed] [Google Scholar]
- Defilippi P, Di Stefano P, Cabodi S. Trends Cell Biol. 2006;16:257–63. doi: 10.1016/j.tcb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Frame MC. Biochim Biophys Acta. 2002;1602:114–30. doi: 10.1016/s0304-419x(02)00040-9. [DOI] [PubMed] [Google Scholar]
- Gan B, Melkoumian ZK, Wu X, Guan KL, Guan JL. J Cell Biol. 2005;170:379–89. doi: 10.1083/jcb.200411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gungabissoon RA, Bamburg JR. J Histochem Cytochem. 2003;51:411–20. doi: 10.1177/002215540305100402. [DOI] [PubMed] [Google Scholar]
- Huang C, Ni Y, Wang T, Gao Y, Haudenschild CC, Zhan X. J Biol Chem. 1997;272:13911–5. doi: 10.1074/jbc.272.21.13911. [DOI] [PubMed] [Google Scholar]
- Huang TY, DerMardirossian C, Bokoch GM. Curr Opin Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Hunter T. Mol Cell. 2007;28:730–8. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Ichetovkin I, Grant W, Condeelis J. Curr Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- Kellie S, Patel B, Wigglesworth NM, Critchley DR, Wyke JA. Exp Cell Res. 1986;165:216–28. doi: 10.1016/0014-4827(86)90546-x. [DOI] [PubMed] [Google Scholar]
- LaLonde DP, Brown MC, Bouverat BP, Turner CE. J Biol Chem. 2005;280:21680–8. doi: 10.1074/jbc.M500752200. [DOI] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell. 1996;84:359–69. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Mol Cell. 1999;4:1029–40. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Lin R, Martyn KD, Guyette CV, Lau AF, Warn-Cramer BJ. Cell Commun Adhes. 2006;13:199–216. doi: 10.1080/15419060600848516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Moczygemba M, Huston DP, Lei JT. J Leukoc Biol. 2007;81:1137–48. doi: 10.1189/jlb.0706465. [DOI] [PubMed] [Google Scholar]
- Meberg PJ. Mol Neurobiol. 2000;21:97–107. doi: 10.1385/MN:21:1-2:097. [DOI] [PubMed] [Google Scholar]
- Mitra SK, Schlaepfer DD. Curr Opin Cell Biol. 2006;18:516–23. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Moriyama K, Iida K, Yahara I. Genes Cells. 1996;1:73–86. doi: 10.1046/j.1365-2443.1996.05005.x. [DOI] [PubMed] [Google Scholar]
- Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Cell. 2002;108:233–46. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- Ono S. Biochemistry. 2003;42:13363–70. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- Parsons JT. J Cell Sci. 2003;116:1409–16. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cell. 2003;112:453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Reiske HR, Kao SC, Cary LA, Guan JL, Lai JF, Chen HC. J Biol Chem. 1999;274:12361–6. doi: 10.1074/jbc.274.18.12361. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD. J Cell Biol. 2003;162:1079–88. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Sabourin LA, Tamai K, Seale P, Wagner J, Rudnicki MA. Mol Cell Biol. 2000;20:684–96. doi: 10.1128/mcb.20.2.684-696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstag Y, Dreizler EM, Ambach A, Sczakiel G, Meuer SC. J Immunol. 1996;156:4167–73. [PubMed] [Google Scholar]
- Scaglioni PP, Yung TM, Cai LF, Erdjument-Bromage H, Kaufman AJ, Singh B, Teruya-Feldstein J, Tempst P, Pandolfi PP. Cell. 2006;126:269–83. doi: 10.1016/j.cell.2006.05.041. [DOI] [PubMed] [Google Scholar]
- Shriver K, Rohrschneider L. J Cell Biol. 1981;89:525–35. doi: 10.1083/jcb.89.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Nat Rev Mol Cell Biol. 2001;2:138–45. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Thomas SM, Brugge JS. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- Tsumura Y, Toshima J, Leeksma OC, Ohashi K, Mizuno K. Biochem J. 2005;387:627–37. doi: 10.1042/BJ20041181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Wu S, Shi X, Chen K, Wu C. Cell. 2003;113:37–47. doi: 10.1016/s0092-8674(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Tursun B, Schluter A, Peters MA, Viehweger B, Ostendorff HP, Soosairajah J, Drung A, Bossenz M, Johnsen SA, Schweizer M, Bernard O, Bach I. Genes Dev. 2005;19:2307–19. doi: 10.1101/gad.1340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatsuki T, Wysolmerski RB, Elson EL. J Cell Sci. 2003;116:1617–25. doi: 10.1242/jcs.00340. [DOI] [PubMed] [Google Scholar]
- Wu H, Parsons JT. J Cell Biol. 1993;120:1417–26. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Suetsugu S, Cooper LA, Takenawa T, Guan JL. J Biol Chem. 2004;279:9565–76. doi: 10.1074/jbc.M310739200. [DOI] [PubMed] [Google Scholar]
- Yan D, Guo L, Wang Y. J Cell Biol. 2006;174:415–24. doi: 10.1083/jcb.200511028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap CT, Simpson TI, Pratt T, Price DJ, Maciver SK. Cell Motil Cytoskeleton. 2005;60:153–65. doi: 10.1002/cm.20053. [DOI] [PubMed] [Google Scholar]
- Yeatman TJ. Nat Rev Cancer. 2004;4:470–80. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- Yoo Y, Wu X, Egile C, Li R, Guan JL. J Biol Chem. 2006;281:15352–60. doi: 10.1074/jbc.M511825200. [DOI] [PubMed] [Google Scholar]
- Zigmond SH. Curr Opin Cell Biol. 2004;16:99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.