Abstract
The deg-3 gene from the nematode Caenorhabditis elegans encodes an α subunit of a nicotinic acetylcholine receptor that was first identified by a dominant allele, u662, which produced neuronal degeneration. Because deg-3 cDNAs contain the SL2 trans-spliced leader, we suggested that deg-3 was transcribed as part of a C. elegans operon. Here we show that des-2, a gene in which mutations suppress deg-3(u662), is the upstream gene in that operon. The des-2 gene also encodes an α subunit of a nicotinic acetylcholine receptor. As expected for genes whose mRNAs are formed from a single transcript, both genes have similar expression patterns. This coexpression is functionally important because (i) des-2 is needed for the deg-3(u662) degenerations in vivo; (ii) an acetylcholine-gated channel is formed in Xenopus oocytes when both subunits are expressed but not when either is expressed alone; and (iii) channel activity, albeit apparently altered from that of the wild-type channel, results from the expression of a u662-type mutant subunit but, again, only when the wild-type DES-2 subunit is present. Thus, the operon structure appears to regulate the coordinate expression of two channel subunits.
Twenty-five percent of the genes in the nematode Caenorhabditis elegans are organized into operons (1, 2), where adjacent genes are transcribed as a single primary transcript that is processed by 3′ end formation and trans-splicing to yield separate mRNAs (3). Often the existence of an operon can be detected because mRNA that is encoded by an internal gene in a polycistronic operon possesses an SL2 trans-spliced leader (3). The functional importance of the coexpression of mRNAs from operons, however, is not clear, usually because the function of every gene in an operon is not known (1, 2). Examples that suggest that operons encode proteins required for the same process are the cyp-9 pdi-1 operon (4), which encodes two enzymes needed for protein folding, and the lin-15 operon, which encodes two genes needed for the negative regulation of vulval development (5, 6). Although these latter genes are needed for the same activity, their function in this activity is unknown, because both encode novel proteins.
During the course of studying the deg-3 gene, a gene that encodes a subunit of a nicotinic acetylcholine receptor (nAChR) (7), we found evidence for the functional connection of genes within an operon. The deg-3 gene was originally identified by a dominant allele, u662, which resulted in neuronal degeneration (the Deg phenotype) (7). Because the u662 mutation was in the presumptive pore-forming region, degeneration presumably occurred by deregulation of channel activity. Although the presumptive second membrane spanning domain of DEG-3 looked similar to that of the α7 type subunits, which are known to form homopolymeric channels in Xenopus oocytes (8, 9), expression of cRNA in frog oocytes failed to produce a detectable current (7). Furthermore, because mutations in another gene, des-2, prevented the deg-3(u662) degenerations, at least one other component might be needed for DEG-3 to form a functional channel.
Because deg-3 mRNAs had SL2 trans-spliced 5′ ends, we suggested that deg-3 was cotranscribed with mRNA from at least one other gene in a C. elegans operon (7). In this paper, we show that des-2, which also encodes a nAChR subunit, is the upstream gene in an operon with deg-3. Coexpression of cRNAs for both genes, but not the single cRNAs, in Xenopus oocytes results in a functional channel. These data suggest that the operon structure is used in C. elegans for the coordinate and stoichiometric expression of interacting proteins.
MATERIALS AND METHODS
Strain Maintenance and Genetics.
Strains were grown as described by Wood et al. (10). The des-2 mutations were obtained by the reversion of the Unc phenotype of deg-3(u662) (7). In addition to the original screen (7) that identified one des-2 mutation, u695, we examined F2 progeny representing 20,000 haploid genomes from ethyl methanesulfonate-mutagenized deg-3(u662) animals and obtained two additional des-2 alleles. All des-2 mutations partially suppressed the uncoordination produced by the deg-3(u662) mutation. The des-2(u695) mutation was used for mapping and could not be separated from deg-3(u662). The presence of the u695 mutation was tested by mating animals to des-2(u695) deg-3(u662) males and examining the male progeny. None of the nine non-Unc Sqt recombinants from unc-42(e270) + + sqt-3(sc63)/+ des-2(u695) deg-3(u662) + hermaphrodites contained the des-2 mutation. All 14 Dpy Unc progeny derived from dpy-11(e224) + +/+ des-2(u695) deg-3(u662) animals and all 6 Unc Sqt progeny derived from + + sqt-3(sc63)/des-2(u695) deg-3(u662) + animals contained the des-2 mutation (and were partially uncoordinated).
Molecular Analysis.
General molecular biology methods followed the protocols of Wood et al. (10) and Sambrook et al. (11). We had found that a 6-kb XbaI–StuI fragment (the StuI site was introduced between the SL2 splice site and the first AUG of deg-3) contained all of the upstream sequences needed for deg-3 expression (7). A 2-kb StuI–BamHI fragment, which covers the region immediately upstream of deg-3, was labeled by random priming and used as a probe on a mixed stage cDNA library (12). A screen of 106 plaques yielded two cDNA clones (2.2 and 1.6 kb) for the same transcript. The cDNA inserts in these clones were cloned into pBluescript SKII(−) (Stratagene). The cDNAs and the corresponding genomic fragment (5.8 kb) were sequenced by using nested deletions and internal primers. Sequencing was done on an Applied Biosystems 370A DNA sequencer (Columbia University DNA facility). Sequencing of des-2 alleles was done directly on PCR amplified genomic DNA [purified by using Qiagen gel extract (Chatsworth, CA)], in the Life Science Sequencing Facilities (The Hebrew University). For oocyte expression, deg-3 and des-2 were cloned in the pBGT vector (courtesy of E. Honore, Institute de Pharmacologie Moleculaire et Cellulaire, CNRS, Valbonne, France), a pBluescript derivative that contains the Xenopus globin untranslated regions (13). The des-2 expression pattern was visualized by using either gfp (14) or lacZ (15) expression. The gfp construct has a 5-kb BamHI–XbaI fragment [from a plasmid containing the deg-3 operon (7)] inserted into plasmid pPD95.75 (courtesy of A. Fire, Carnegie Institution of Washington, Department of Embryology, Baltimore). The gfp gene is fused to the intracellular loop of des-2 in this construct. The lacZ construct was obtained after insertion of an EcoRV site upstream of the des-2 AUG by using in vitro mutagenesis with the primer CACAACACAGATATCCTTTTACTAC. A 2-kb EcoRV–XbaI fragment from this plasmid was inserted into plasmid pPD22.07, which adds sequences for a nuclear localization signal (15). The lacZ transgene was integrated by the procedure of Mitani (16) to produce integrated construct hmIs1. Cells were identified by their positions and by their elimination in mec-3(e1338) (17), unc-86(e1416) (18), or deg-1(u506) (19, 20) backgrounds.
Oocyte Injection and Physiology.
Xenopus laevis (African Xenopus Facility, Noordhoek, South Africa) were maintained at 18–20°C in a 15 hr day/9 hr night cycle and fed with either chopped liver or frog pellets twice a week. A section of ovary was removed surgically from female frogs under tricaine anesthesia, and the oocytes defolliculated for 2 hr with 2 mg/ml collagenase in calcium-free ND96 solution (96 mM NaCl/2 mM KCl/5 mM MgCl2/5 mM Hepes, pH 7.5) (21). Oocytes were examined under a dissecting microscope, and healthy looking stage V and stage VI cells (22) were selected and maintained at 19°C in enriched ND96 (supplemented with 1.8 mM CaCl2/2.5 mM sodium pyruvate/100 units/ml penicillin/100 μg/ml streptomycin/0.1% BSA fraction V) for 1–3 days before recording.
A microdispenser (Drummond Scientific, Broomall, PA) was used for intracellular injections. A pulled glass capillary was broken to a 15–20 μm diameter tip. A drop of mineral oil was added to the tube before assembly of the microdispenser plunger. Solution (1–2 μl) was introduced via the tip by pulling. The cells were penetrated and 50 nl of cRNA solution (0.2 μg/μl of each cRNA) was introduced.
Cells were placed in a 0.5 ml bath that was constantly perfused with medium and penetrated with two 0.5–1 MΩ 3 M KCl filled glass microelectrodes attached to a GeneClamp 500 amplifier (Axon Instruments, Foster City, CA), by using an HS-2A headstage (Axon Instruments). A reference electrode was connected to ground by means of a 3 M KCl agar bridge. An IBM PC/486 system employing the TL-1 interface and pclamp software, version 6 (Axon Instruments) was used for maintaining voltage clamp. Cells were held at −70 mV. The current and the voltage in the voltage-clamp circuit were recorded simultaneously on a chart recorder and were saved directly onto the computer. Results are presented as the mean ± SEM with n equal to the number of oocytes tested and N equal to number of different frogs in each experiment. In all experiments, the oocytes were incubated and perfused with “calcium-free medium” (96 mM NaCl/2 mM KCl/5 mM MgCl2/5 mM Hepes, pH 7.5).
RESULTS AND DISCUSSION
By using a fragment corresponding to sequences upstream of deg-3, we identified two cDNAs from this upstream gene. These cDNAs and the corresponding 5 kb genomic DNA were sequenced. The longer cDNA (2.2 kb) encoded 260 bp of upstream sequence, an ORF for a 548 amino acid polypeptide, and 329 bp of downstream sequence before the site of polyadenylation (the shorter cDNA is a truncated version). Although the cDNAs do not contain spliced leader sequences, reverse transcription–PCR reveals that this mRNA contains the SL1 trans-spliced leader, an indication that no other mRNA precedes it in an operon (3). The small distance (112 bp) between the sites for polyadenylation and SL2 trans-splicing for deg-3 is common for genes in C. elegans operons (1, 2) (Fig. 1A). No other gene predicted by the C. elegans Genome Project (23) is close enough to be included in this operon.
Figure 1.
The des-2 deg-3 operon. (A) Genomic organization. Arrows mark the site of spliced leader addition. Although the longer des-2 cDNA lacks a spliced leader, it starts at the same place as SL1-spliced variants that were identified from a random primed cDNA library. deg-3 is trans-spliced to the SL2 leader (7) (B) Alignment of DES-2 and DEG-3. The positions of the des-2 mutations (all of which result in premature termination) are indicated by asterisks. Codon changes are: hm5 (TGG to TGA), hm6 (TGG to TAG), and u695 (CAG to TAG). Thin underlining indicates the neurotransmitter gated-channel signature (24) and thick underlining indicates the putative transmembrane domains (see ref. 7). The two adjacent cysteines indicative of nAChR α subunits (25) are at amino acids 220 and 221 in DES-2.
Because this new gene encoded a nAChR A subunit (see below), we originally named it acr-4 (acetylcholine receptor gene 4); however, subsequent analysis showed it to be the des-2 gene. The des-2 (degeneration suppression) gene was identified by mutations that prevent the uncoordination caused by the dominant deg-3(u662) mutation (7). The des-2 mutations are genetically inseparable from deg-3 (less than 0.2 map unit), but unlike intragenic suppressor mutations of deg-3, which are cis dominant (7), des-2 mutations are recessive suppressors of the uncoordination and touch insensitivity phenotypes of deg-3(u662). As expected for genes from the same operon, the expression patterns of a des-2 lacZ fusion (Fig. 2) and a des-2 gfp fusion (data not shown) were essentially the same as that of a deg-3 gfp fusion (7). The only difference we have seen is the lack of expression in the touch receptor neurons. However, this expression was very difficult to see in the deg-3 constructs (7). Nonetheless, we believe that des-2 is expressed in these cells, since des-2 mutations suppress the touch insensitivity produced by the deg-3(u662) degeneration-causing mutation. The des-2 constructs in lacking introns 8 and 9 and the 3′ untranslated regions of des-2 may lack the necessary elements for touch cell expression.
Figure 2.
Expression of a des-2 lacZ fusion. This pattern is similar to that for deg-3 expression (7). Identified cells include the M1 head muscles (the staining in front of the IL2 neurons; these cells were identified by their structure in a des-2 gfp expressing strain), IL2 neurons (these cells disappear in unc-86 animals), FLP neurons (these cells disappear in mec-3 animals), PVD neurons (identified by the pattern of their processes using a des-2 gfp fusion and because they disappear in mec-3 and unc-86 animals), and PVC neurons (these cells disappear in deg-1 animals).
DES-2 has all the hallmarks of nAChR α subunits (Fig. 1B): a signal sequence, four hydrophobic regions, the signature sequence for neurotransmitter-gated channels (24), and the two adjacent cysteine residues needed for acetylcholine binding (25). We cannot assign the DES-2 subunit to any particular class, because in blast (26) searches of GenBank several α6 and α7 subunits and one α2 subunit show virtually equivalent similarity, which is limited to the N terminus and the first two putative transmembrane regions. The positions of introns are completely different in des-2 and deg-1, suggesting that these genes are evolutionarily distant and not the result of a recent gene duplication.
Acetylcholine (ACh; up to 1 mM) did not produce detectable currents when deg-3 cRNA [7 (n) oocytes from 2 (N) frogs] or des-2 cRNA (n = 11, N = 4) was expressed in Xenopus oocytes. ACh did produce currents when oocytes expressed both deg-3 and des-2 cRNAs (Fig. 3). For example, application of 300 μM ACh to oocytes injected with 10 ng of each cRNA produced currents of 349 ± 70 nA (n = 29, N = 6) at 3–5 days after injection.
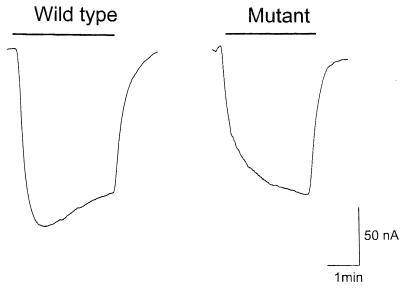
Figure 3.
ACh response of the normal and mutant DES-2/DEG-3 channel. Representative current responses were elicited by 1 mM ACh in voltage-clamped oocytes during the time indicated by the solid bar.
As with wild-type deg-3 cRNA, no channel activity was found when deg-3 cRNA with the u662 change was expressed in frog oocytes (n = 3, N = 2). However, channel activity was found when this mutant deg-3 cRNA was coexpressed with wild-type des-2 cRNA (Fig. 3). Three days after injection, the mutant channel response to 1 mM ACh was 202 ± 39 nA (n = 23, N = 5) compared with wild-type 336 ± 111 nA (n = 15, N = 5). An apparent difference from the response of channels formed from wild-type subunits is that the response of the mutant channel more slowly rises to peak depolarization. This difference is very reproducible when we compare similar, low amplitudes responses in 50 oocytes (from 10 frogs) for both the mutant and wild-type channel; the response with the mutant channel always shows a slow rise time compared with wild type and no decay in amplitude, whereas the response of the wild-type channel always shows a decay in the amplitude. For example, for a 1 min pulse of 1 mM ACh, current carried by wild-type channels reached a peak in 9 ± 3 sec (n = 10, N = 3) and then decayed, whereas that carried by mutant channels failed to reach a peak in the same time interval (n = 10, N = 5). In addition to these differences, many of the oocytes injected with cRNAs to produce the mutant channel died or had very low membrane potentials. We suspect that the poor health of the cells was a consequence of the coexpression.
These observations in frog oocytes demonstrate that DEG-3 and DES-2 form a functional channel, but they do not indicate whether these proteins interact in vivo in C. elegans. Support for in vivo interaction comes from our finding that the upstream gene in this operon is des-2. All three ethyl methanesulfonate-derived alleles of this gene were identified as suppressors of the deg-3(u662) uncoordination (ref. 7; this work). These alleles, all of which contain nonsense mutations (Fig. 1), are recessive with regard to suppression, suggesting that the suppression is not the result of a cis-acting polar effect on deg-3 of the des-2 mutations. These results support the hypothesis that the products of these genes function together, but they do not say whether the interaction is direct, i.e., that DES-2 and DEG-3 form into the same channel in vivo. Although we believe that a single DES-2 DEG-3 channel is likely, conceivably these subunits could contribute to different channels in vivo, such that deg-3-induced neurodegeneration is a consequence of additive ion flux through both channels.
Moreover, although all three mutations may be null alleles (two terminate the protein before the first putative transmembrane domain), suppression by these mutations is incomplete. Some cell swelling (the hallmark of the degeneration process) is still seen, although greatly reduced, and the number of adult deg-3lacZ-expressing cells, i.e., surviving cells, is less than seen in wild-type animals [set at 100%; number of animals (N) = 28; number of cells (n) = 296]; 32% of these cells survive in deg-3(u662) adults (N = 50; n = 168) compared with 68% and 53% for des-2(u695) deg-3(u662) (N = 15; n = 108) and des-2(hm5) deg-3(u662) (N = 10; n = 56) adults, respectively. These results suggest that whereas des-2 is important for deg-3-induced deaths, it is not essential. Presumably, DEG-3(u662)-containing channels form in vivo in the absence of des-2 activity. In addition, the channel formed in vivo in the nematodes need not contain only DES-2 and DEG-3 subunits. Genes coding for additional subunits of the channel or proteins needed for its formation and maintenance may have been identified through mutations that suppress the deg-3(u662) degenerations (ref. 7; M.T., unpublished data).
Our results indicate that functional activity in frog oocytes and in C. elegans requires the coexpression of des-2 and deg-3. Their linkage in a single operon allows their coordinated and stoichiometric production in the same cells at the same time. This genomic organization is probably not a result of a recent gene duplication as both protein sequences and intron locations are highly diverged. DES-2 and DEG-3 are, so far, unique among nAChR subunits in being able to form a heteromeric nAChR composed of two different α subunits without non-α subunits in frog oocytes. This finding suggests that the combinatorial possibilities in the formation of these receptors are larger than previously expected.
Our results with the des-2 deg-3 operon and the functional interaction seen in the cyp-9 pdi-1 (4) and the lin-15 operon (5, 6) suggest that other operons in C. elegans will encode genes whose products interact or participate in the same biological process. We suggest that operon organization in C. elegans can be used as a means of identifying interacting proteins.
Acknowledgments
We thank S. Cohen for helpful comments, M. Eshel for technical assistance, and B. Minke for his generosity in providing us with frogs and electrophysiological facilities. We also wish to thank the C. elegans Genetics Center for providing strains used in this work. This work was supported by National Institutes of Health (NIH) Grant GM34775 and part of NIH Alzheimer’s Disease Research Center Grant AG08702 to M.C., by a European Molecular Biology Organization long-term fellowship, Ministry of Science (Israel) Grant 6725-1-95, and Binational Science Foundation Grant 95-0133/2 to M.T., and by funding from the Kuhne Minerva Center for Studies of Visual Transduction to B.G.
ABBREVIATIONS
- ACh
acetylcholine
- nAChR
nicotinic acetylcholine receptor
Footnotes
Data deposition: The sequence reported in this paper, des-2 cDNA, has been deposited in the GenBank database (accession no. AF077307).
References
- 1.Blumenthal T, Spieth J. Curr Opin Genet Dev. 1996;6:692–698. doi: 10.1016/s0959-437x(96)80022-0. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal T, Steward K. In: Caenorhabditis elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 117–145. [Google Scholar]
- 3.Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 4.Page A P. DNA Cell Biol. 1997;16:1335–1343. doi: 10.1089/dna.1997.16.1335. [DOI] [PubMed] [Google Scholar]
- 5.Clark S G, Lu X, Horvitz H R. Genetics. 1994;137:987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L S, Tzou P, Sternberg P W. Mol Biol Cell. 1994;5:395–411. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treinin M, Chalfie M. Neuron. 1995;14:871–877. doi: 10.1016/0896-6273(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 8.Couturier S, Bertrand D, Matter J-M, Hernandez M-C, Bertrand S, Millar N, Valera S, Barkas T, Ballivet M. Neuron. 1990;5:847–856. doi: 10.1016/0896-6273(90)90344-f. [DOI] [PubMed] [Google Scholar]
- 9.Séguéla P, Wadiche J, Dineley-Miller K, Dani J A, Patrick J W. J Neurosci. 1993;13:596–604. doi: 10.1523/JNEUROSCI.13-02-00596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood W B the Community of Caenorhabditis elegans Researchers. The Nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Okkema P G, Fire A. Development (Cambridge, UK) 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- 13.Krieg P A, Melton D A. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 15.Fire A, Harrison S W, Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 16.Mitani S. Dev Growth Differ. 1995;37:551–557. doi: 10.1046/j.1440-169X.1995.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- 17.Way J C, Chalfie M. Genes Dev. 1989;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- 18.Finney M, Ruvkun G. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 19.Chalfie M, Wolinsky E. Nature (London) 1990;345:410–416. doi: 10.1038/345410a0. [DOI] [PubMed] [Google Scholar]
- 20.García-Añoveros J, Ma C, Chalfie M. Curr Biol. 1995;4:441–448. doi: 10.1016/s0960-9822(95)00085-6. [DOI] [PubMed] [Google Scholar]
- 21.Gillo B, Lass Y, Nadler E, Oron Y. J Physiol (London) 1987;392:349–361. doi: 10.1113/jphysiol.1987.sp016784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dumont J N. J Morphol. 1972;136:153–179. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- 23.Sulston J, Du Z, Thomas K, Wilson R, Hillier L, Staden R, Halloran N, Green P, Thierry-Mieg J, Qiu L, et al. Nature (London) 1992;356:37–41. doi: 10.1038/356037a0. [DOI] [PubMed] [Google Scholar]
- 24.Galzi J-L, Revah F, Bessis A, Changeux J-P. Annu Rev Pharmacol. 1991;31:37–72. doi: 10.1146/annurev.pa.31.040191.000345. [DOI] [PubMed] [Google Scholar]
- 25.Kao P N, Karlin A. J Biol Chem. 1986;261:8085–8088. [PubMed] [Google Scholar]
- 26.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]