Abstract
Angiopoietin-1 (Ang1) and Ang2 are ligands for the receptor tyrosine kinase Tie2. Structural data suggest that the two ligands bind Tie2 similarly. However, in endothelial cells Ang1 activates Tie2 whereas Ang2 can act as an apparent antagonist. In addition, each ligand exhibits distinct kinetics of release following binding. These observations suggest that additional factors influence function and binding of angiopoietins with receptors in the cellular context. Previous work has shown that Ang1 binding and activation of Tie2 are inhibited by Tie1, a related receptor that complexes with Tie2 in cells. In this study we have investigated binding of Ang1 and Ang2 to Tie2 in endothelial cells. In contrast to Ang1, binding of Ang2 to Tie2 was found to be not affected by Tie1. Neither PMA-induced Tie1 ectodomain cleavage nor suppression of Tie1 expression by siRNA affected the ability of Ang2 to bind Tie2. Analysis of the level of Tie1 co-immunoprecipitating with angiopoietin-bound Tie2 demonstrated that Ang2 can bind Tie2 in Tie2:Tie1 complexes whereas Ang1 preferentially binds non-complexed Tie2. Stimulation of Tie1 ectodomain cleavage did not increase the agonist activity of Ang2 for Tie2. Similarly, the Tie2-agonist activity of Ang2 was not affected by siRNA suppression of Tie1 expression. Consistent with previous reports, loss of Tie1 ectodomain enhanced the agonist activity of Ang1 for Tie2. Importantly, Ang2 was still able to antagonize the elevated Ang1-activation of Tie2 that occurs on Tie1 ectodomain loss. Together these data demonstrate that Ang1 and Ang2 bind differently to Tie2 at the cell surface and this is controlled by Tie1. This differential regulation of angiopoietin binding allows control of Tie2 activation response to Ang1 without affecting Ang2 agonist activity and maintains the ability of Ang2 to antagonize even the enhanced Ang1 activation of Tie2 that occurs on loss of Tie1 ectodomain. This provides a mechanism by which signalling through Tie2 can be modified by stimuli in the cellular microenvironment.
Keywords: Receptor tyrosine kinase, Angiopoietin, Endothelial, Angiogenesis
1. Introduction
The receptor tyrosine kinase Tie2 is essential for vascular development and maintenance and interacts with a family of ligands known as the angiopoietins [1]. Signalling through Tie2 is tightly controlled. The angiopoietin family contains members with full agonist activity, the best characterized of which is Ang1, as well as, unusually, members with low or partial activity, notably Ang2 [2,3]. This partial agonist activity of Ang2 allows the ligand to act as an apparent antagonist to Ang1, by competing with Ang1 for Tie2 binding and thereby replacing the full agonist activity of Ang1 with the much lower activity of Ang2 [2,3]. The activation state of Tie2 is therefore determined by the relative balance between Ang1 and Ang2 [4].
Ang1 and Ang2 have a similar overall structure, both are glycoproteins of approximately 70 kDa in size with an amino-terminal half comprising of coiled:coil domains important for ligand oligomerization [4,5]. Downstream of this is a short linker sequence of approximately 20 residues followed by a fibrinogen related domain (FReD). Binding of angiopoietins to Tie2 is mediated by the FReD [6,7]. This domain is found in a number of proteins and comprises of three subdomains, designated A, B and P [8]. The P-domain mediates interaction of FReD containing proteins such as fibrinogen and tachylectin 5A with their ligands and it is this region of the Ang FReD that interacts with Tie2 [9]. The structure of Ang2 bound to Tie2 ectodomain has been solved and shows the Ang2 P-domain interacting with the second Ig domain of the receptor which is within a globular fold containing three Ig and three EGF-like domains at the amino-terminal head of the receptor [9–11]. Comparison of un-liganded and Ang2-bound Tie2 ectodomain suggests no significant conformational change in Tie2 on ligand binding [10]. Ang1 and Ang2 have approximately 73% amino acid identity in their P-domains. The strong sequence similarity between Ang2 and Ang1 in the amino acid residues critical for Tie2 ectodomain interaction suggests that the two ligands bind similarly with Tie2 under in vitro conditions with purified proteins [9,10]. However, recent data indicate that interaction of the ligands with their receptor on the cell surface may be more complex. Following binding in endothelial cells Ang1 and Ang2 are released prior to receptor internalization and the kinetics of release are different for each ligand, with Ang2 release being more than three-fold faster than Ang1 [2]. The mechanism responsible for this differential behaviour of the ligands is not known.
In addition to its ligands, Tie2 physically interacts with the related receptor tyrosine kinase Tie1 and the two receptors exist as pre-formed hetero-oligomeric complexes at the endothelial surface [12,13]. Tie1 ectodomain is not able to bind angiopoietins [5] and in the Tie2:Tie1 complex it appears to partially occlude Ang1 binding to Tie2 [14]. However, Tie1 ectodomain can undergo regulated cleavage in which the extracellular domain of the receptor is removed by metalloprotease activity in response to vascular endothelial growth factor (VEGF), inflammatory stimuli, changes in shear stress and stimulation of cells with phorbol esters [13,15–17]. This cleavage removes Tie1 ectodomain from the Tie2:Tie1 complexes and releases it as an intact fragment from the cell [15]. Regulated removal of Tie1 ectodomain increases access of Ang1 to Tie2 on the surface of endothelial cells and enhances Ang1-activation of Tie2 and Tie2-mediated signalling [14]. This provides a mechanism whereby responsiveness of Tie2 to its activating ligand can be controlled by VEGF and other stimuli.
Given the strong similarity between Ang1 and Ang2 in their Tie2-binding domain it is possible that Ang2 binding to Tie2 is regulated by Tie1 in the same way as occurs for Ang1. This would have important consequences for the partial agonist activity of Ang2, allowing Tie1 ectodomain release to increase the number of Tie2 molecules able to bind Ang2 and thereby increasing the net agonist activity of Ang2 on the cell. The effects of Tie1 on Ang2 interaction with Tie2 have not yet been defined. In the present study, therefore, we investigate whether Ang2 binding to Tie2 is influenced by Tie1 and compare interaction of the two ligands with Tie2 in the Tie2:Tie1 complexes in endothelial cells.
2. Materials and methods
2.1. Materials
Human umbilical vein endothelial cells (HUVEC) were obtained from Promocell and were maintained in Medium 199 containing 20% fetal calf serum, 5 units/ml heparin, and 50 µg/ml endothelial cell growth supplement. Ang1 and Ang2 were obtained from R & D Systems (Abingdon, UK). Antibodies recognizing Tie2 extracellular domain were obtained from R & D Systems and those recognizing Tie1 and Tie2 intracellular domains and phosphotyrosine were from Santa Cruz Biotechnology (from Autogen Bioclear, Calne, UK). Anti-His6 antibodies were from Sigma (Dorset, UK). Annealed, purified and de-salted double-stranded siRNA oligonucleotides have been previously detailed [18] and were obtained from MWG Biotech (London, UK). All other reagents were as previously described [14].
2.2. Cell treatments
Before the experiments cells were washed in PBS and incubated in serum-free medium for 30 min. As appropriate, and indicated in Results, cells were treated with the following concentrations of reagents for the indicated times; PMA, 10 ng/ml, 30 min; TAPI-2, 100 μM, 2 h; Ang2, 200 ng/ml, 30 min and Ang1 200 ng/ml, 30 min, unless otherwise indicated. In some experiments, indicated in Results, HUVEC were transfected with control scrambled siRNA or siRNA directed against Tie1 or Tie2. For transfection HUVEC were grown to approximately 80% confluence and then transfected with 100 nM siRNA using Lipofectamine as directed in the manufacturer's protocol 24 h before treatments and cell lysis.
2.3. Immunoprecipitation and immunoblotting
Cells were washed with PBS and lysed with ice-cold lysis buffer (50 mM Tris, pH 7.4, 50 mM NaCl, 1 mM sodium fluoride, 1 mM EGTA, 1 mM sodium orthovanadate, 1% TritonX-100, complete protease inhibitor mixture), cleared by centrifugation at 13,000 × g for 5 min, and assayed for protein content. In binding experiments, the cell-impermeable cross-linker 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP) was added to a final concentration of 0.5 mM in PBS for 30 min, cross-linking was terminated by addition of 20 mM Tris in PBS followed by washing, and cell lysis.
For analysis of whole cell lysates, Laemmli sample buffer containing 100 mM dithiothreitol was mixed with cleared cellular lysates and boiled for 5 min before loading equal amounts of protein onto SDS-PAGE and resolving. For immunoprecipitates, supernatants cleared of particulate material were immunoprecipitated by the addition of 2 µg of the indicated antibody for 2–3 h in the presence of protein-A- or protein-G-agarose. Immunoprecipitated proteins were recovered by centrifugation at 13,000 × g for 5 min and washed 3 times with wash buffer (as lysis buffer but with 0.1% Triton X-100). Proteins were eluted from beads by the addition of Laemmli sample buffer containing 100 mM dithiothreitol and boiling for 5 min before SDS-PAGE. For immunoblotting proteins were transferred to nitrocellulose membranes electrophoretically and membranes probed with the relevant antibodies. Immunoreactive proteins were visualized with peroxidase-conjugated secondary antibodies and chemiluminescent detection [19].
2.4. Data analysis
Bands on Western blots were quantified by densitometric scanning of films. Graphs were derived from 3 or more independent experiments and data is plotted as means and standard error. Statistical analysis was performed using Student's t test and differences between means were judged statistically significant for p < 0.05.
3. Results
3.1. Tie1 differentially regulates binding of Ang1 and Ang2 to Tie2
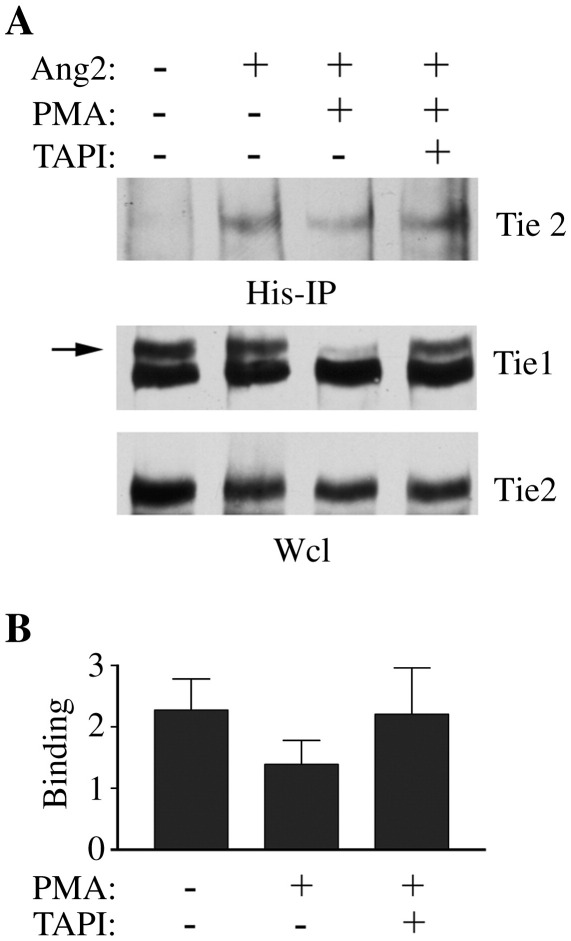
Tie1 regulates Ang1 signalling by limiting access of the ligand to its receptor, Tie2, in the Tie1:Tie2 complexes found on the surface of endothelial cells [14]. It is not known whether Tie1 also affects the ability of the related ligand Ang2 to bind and activate Tie2. To begin investigating this question we examined the effects of Tie1 ectodomain cleavage on interaction of Ang2 with Tie2 at the surface of endothelial cells. As previously reported, addition of phorbol ester to endothelial cells activates Tie1 ectodomain cleavage resulting in loss of the extracellular domain. This effect has been well characterized and is shown in Fig. 1A by the effects of PMA on loss of the upper band of the 145 kDa doublet which corresponds to the surface expressed full-length Tie1 [15]. Inclusion of the metalloproteinase inhibitor TAPI-2 blocked Tie1 ectodomain cleavage (Fig. 1A). To examine the effect of Tie1 cleavage on Ang2 binding endothelial cells were treated with control vehicle, PMA, or PMA plus TAPI-2 before addition of His6-tagged Ang2, followed by cross-linking and recovery of Ang2 together with its bound receptor by immunoprecipitation with anti-His6 antibodies (Fig. 1A). As expected, immunoprecipitation in the absence of His6-tagged Ang2 did not recover any Tie2. However, in the presence of Ang2 immunoprecipitation resulted in recovery of bound Tie2, confirming interaction between Ang2 and its receptor on endothelial cells (Fig. 1A, B). Induction of Tie1 cleavage by PMA did not affect the amount of Tie2 bound to Ang2 either in the absence or presence of TAPI-2 (Fig. 1A). Quantitation of the amounts of Tie2 bound to Ang2 was performed in three independent experiments by densitometric scanning of blots, this confirmed no statistically significant change in Ang2 binding to Tie2 in response to PMA activated Tie1 cleavage (Fig. 1B).
Fig. 1.
Binding between Ang2 and Tie2 is not regulated by Tie1 ectodomain cleavage. (A) Endothelial cells were treated with 10 ng/ml PMA or PMA plus 100 μM TAPI-2 before addition of 200 ng/ml Ang2 for 30 min, as indicated, followed by cross-linking with the cell-impermeable cross-linker DTSSP. 20 mM Tris was added to quench cross-linking before washing and cell lysis. Ang2 was immunoprecipitated via its His-tag and immunoprecipitates or whole cell lysates (Wcl) were resolved by SDS/PAGE. Tie2 bound to Ang2 and Tie1 and Tie2 in whole cell lysates were detected by immunoblotting as indicated. Tie1 ectodomain cleavage is indicated by loss of the higher molecular mass Tie1 immunoreactive band corresponding to surface expressed Tie1 (arrow). (B) Tie2 bound to Ang2 was determined by immunoblotting in three independent experiments. Data is presented as means and SEM. No significant effect of PMA on Tie2 binding was observed (Student's t test).
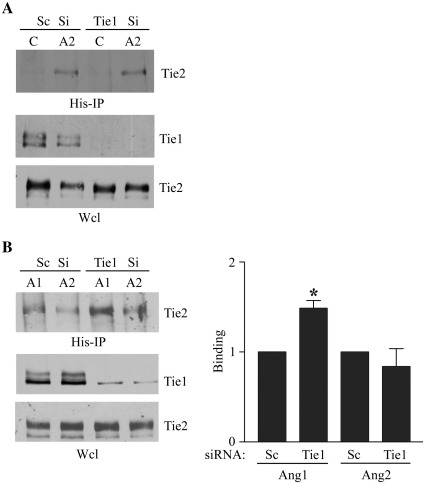
The effects of Tie1 on binding of Ang2 to Tie2 were explored further using an siRNA approach to generate endothelial cells lacking Tie1. Cells were transfected with control siRNA or siRNA directed against Tie1 and expression of Tie1 determined by immunoblotting. Tie1 siRNA effectively suppressed Tie1 expression (Fig. 2A). Interaction of Ang2 with Tie2 in endothelial cells expressing Tie1, and in which Tie1 expression was inhibited by siRNA, was determined by addition of the ligand and immunoprecipitation as before. Ang2 was able to bind Tie2 equally well in the absence and presence of Tie1 (Fig. 2A). The different effects of Tie1 on interaction of Ang1 and Ang2 with Tie2 were directly compared by examining the ability of Ang1 and Ang2 to bind and recover Tie2 from control cells and cells lacking Tie1. Loss of Tie1 did not affect the binding of Ang2 to Tie2, however, binding of Ang1 to Tie2 was increased in the absence of Tie1 (Fig. 2B).
Fig. 2.
Suppression of Tie1 expression differentially affects binding of Ang1 and Ang2 to Tie2 in endothelial cells. (A) Endothelial cells were transfected with siRNA directed against Tie1 or control randomised siRNA (Sc) and cultured for 24 h before addition of control vehicle (C) or 200 ng/ml Ang2 (A2) for 30 min as indicated followed by cross-linking with the cell-impermeable cross-linker DTSSP. 20 mM Tris was added to quench cross-linking before washing and cell lysis. Ang2 was immunoprecipitated and immunoprecipitates or whole cell lysates (Wcl) were resolved by SDS/PAGE. Tie2 bound to Ang2 and Tie1 and Tie2 in whole cell lysates were detected by immunoblotting as indicated. (B) Endothelial cells were transfected with siRNA directed against Tie1 or control randomised siRNA (Sc) and cultured for 24 h before addition of 200 ng/ml Ang1 (A1) or Ang2 (A2) for 30 min as indicated. Cross-linking, quenching and immunoprecipitation were performed as above and Tie2 bound to Ang1 and Ang2 and Tie1 and Tie2 in whole cell lysates were detected by immunoblotting as indicated. The effect of suppression of Tie1 expression by siRNA on Ang1 and Ang2 binding to Tie2 on cells was determined in three independent experiments by immunoblotting and densitometric quantification of blots. Data are presented as means and SEM. ⁎ indicates Ang1 binding to Tie2 was significantly increased by loss of Tie1 (p < 0.05, Student's t test).
3.2. Ang1 preferentially binds free Tie2
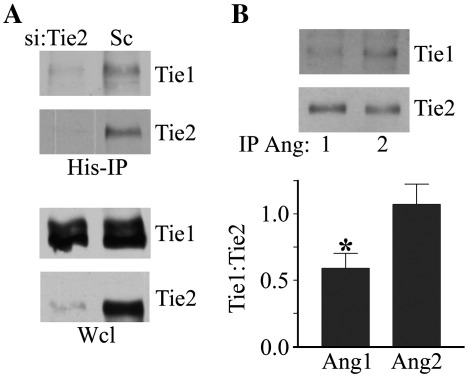
As Ang1 binds better to Tie2 in the absence of Tie1, and Ang2 binds equally well to Tie2 irrespective of Tie1, we hypothesised that Ang2 can bind Tie2 in the Tie1:Tie2 complex whereas Ang1 preferentially binds to non-complexed Tie2. To test this we examined the ability of Ang1 and Ang2 to bind and recover Tie2 and Tie1:Tie2 complexes. It has previously been reported [4] and we have confirmed (data not shown) that Ang2 does not bind to Tie1 ectodomain directly. As shown in Fig. 3, Ang2 immunoprecipitates contain both Tie2 and Tie1. However, in cells lacking Tie2, as a result of suppression of Tie2 expression by siRNA, Ang2 is unable to recover Tie1 (Fig. 3A) consistent with the inability of Ang2 to bind directly to Tie1 but to bind Tie2 in the Tie1:Tie2 complex. The binding of Ang1 and Ang2 to the Tie1:Tie2 complex was tested by examining recovery of Tie1 and Tie2 by each of the ligands. Tie2 bound to Ang2 had approximately two-fold more Tie1 associated with it than the Tie2 bound to Ang1 (Fig. 3B). These findings are consistent with the preferential binding of Ang1 to Tie2 in the absence of Tie1 and suggest that Ang1 preferentially binds a free or Tie1-poor pool of Tie2.
Fig. 3.
Ang1 and Ang2 bind differentially to Tie2 in Tie2:Tie1 complexes. (A) Endothelial cells were transfected with siRNA directed against Tie2 or control randomised siRNA (Sc) and cultured for 24 h before addition of 200 ng/ml Ang2 for 30 min followed by cross-linking with the cell-impermeable cross-linker DTSSP. 20 mM Tris was added to quench cross-linking before washing and cell lysis. Ang2 was immunoprecipitated and immunoprecipitates or whole cell lysates (Wcl) were resolved by SDS/PAGE. Tie1 and Tie2 recovered from immunoprecipitates and in whole cell lysates were detected by immunoblotting as indicated. (B) Endothelial cells were treated with 200 ng/ml Ang1 or 200 ng/ml Ang2 for 30 min before cross-linking, quenching and immunoprecipitation, as above, and Tie1 and Tie2 recovered from immunoprecipitates were detected by immunoblotting as indicated. Tie1 and Tie2 bound to angiopoietins were determined by immunoblotting in three independent experiments. Data is presented as means and SEM of the ratio of Tie1:Tie2 recovered. ⁎ indicates statistically significant difference between Tie1:Tie2 ratio recovered by Ang1 and Ang2 (p < 0.05, Student's t test).
3.3. Effects of Tie1 on ability of Ang1 and Ang2 to activate Tie2
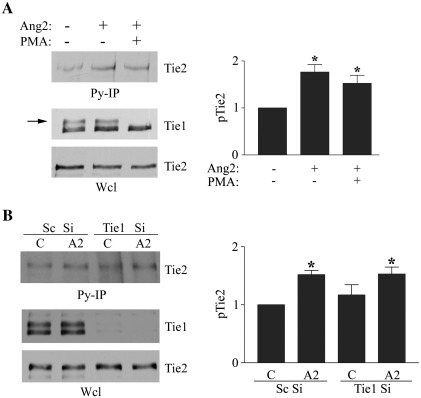
The reason why Ang2 is a partial rather than full agonist is not known. It is possible that Tie1 could suppress full agonist activity of Ang2. Therefore to examine the influence of Tie1 ectodomain and full-length Tie1 on the agonist activity of Ang2 we examined the effects of Tie1 ectodomain cleavage on the ability of Ang2 to activate Tie2 phosphorylation. Endothelial cells were treated with Ang2 in the presence or absence of PMA and phosphorylated Tie2 immunoprecipitated. Ang2 induced only a marginal increase in the amount of Tie2 immunoprecipitated by anti-phosphotyrosine antibodies demonstrating a mild agonist activity. However, induction of Tie1 ectodomain cleavage did not increase the ability of Ang2 to activate Tie2 (Fig. 4A). The impact of Tie1 on Ang2 activation of Tie2 was further examined by suppression of Tie1 expression using siRNA. Endothelial cells transfected with siRNA directed against Tie1 expressed undetectable levels of this receptor (Fig. 4B). Again, Ang2 exhibited very low agonist activity and this was not increased by removal of Tie1 (Fig. 4B). These data demonstrate that Tie1 does not affect the agonist activity of Ang2.
Fig. 4.
Tie1 does not affect the agonist activity of Ang2. (A) Endothelial cells were treated with control vehicle or 10 ng/ml PMA before addition of 200 ng/ml Ang2 for 30 min as indicated. Cells were lysed and immunoprecipitated with anti-phosphotyrosine antibodies (Py-IP). Tie2 in immunoprecipitates and Tie2 and Tie1 in whole cell lysates (Wcl) were detected by immunoblotting following SDS/PAGE. Tie1 ectodomain cleavage is indicated by loss of the higher molecular mass Tie1 immunoreactive band corresponding to surface expressed Tie1 (arrow). The effect of PMA on Ang2-induced Tie2 phosphorylation was determined in three independent experiments by immunoblotting and densitometric quantification of blots. Data are presented as means and SEM. ⁎ indicates Ang2 significantly increased Tie2 phosphorylation (p < 0.05, Student's t test). (B) Endothelial cells were transfected with siRNA directed against Tie1 or control randomised siRNA (Sc) and cultured for 24 h before addition of 200 ng/ml Ang2 for 30 min. Cells were lysed and immunoprecipitated with anti-phosphotyrosine antibodies (Py-IP). Tie2 detected in immunoprecipitates and Tie2 and Tie1 in whole cell lysates (Wcl) were detected by immunoblotting following SDS/PAGE. The effect of control and Tie1 siRNA on the ability of Ang2 to stimulate Tie2 phosphorylation was determined in three independent experiments by immunoblotting and densitometric quantification of blots. Data are presented as means and SEM. ⁎ indicates Ang2 significantly increased Tie2 phosphorylation compared with control Sc Si transfected cells (p < 0.05, Student's t test), however loss of Tie1 did not enhance Ang2-activation of Tie2.
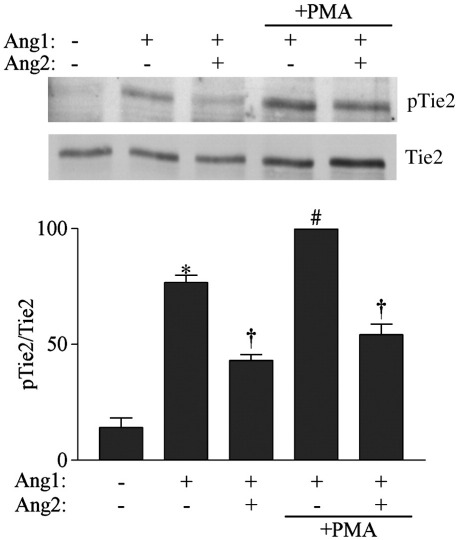
Our data demonstrates that Tie1 does not affect Ang2 access to Tie2 or the partial agonist activity of Ang2, in contrast to the situation for Ang1 where Tie1 loss increases the number of Tie2 receptors accessed by Ang1 and therefore activated by Ang1 [14]. As Ang2 is able to bind Tie2 irrespective of the presence of Tie1 ectodomain it would be expected that Ang2 would still be able to act as an apparent antagonist of Ang1 on Tie2 receptors made newly available to it by loss of Tie1. We directly tested the apparent antagonist effects of Ang2 on Ang1 activity in the presence and absence of Tie1 ectodomain. To do this, endothelial cells were treated with control or PMA before being activated with Ang1 in the presence or absence of Ang2 and the net effect on Tie2 activation determined. As previously described, Ang1 activated Tie2 and this was antagonized by Ang2 (Fig. 5). Induction of Tie1 cleavage by addition of PMA caused an increase in Tie2 activation by Ang1 however even under these conditions Ang2 was still able to antagonize Ang1-activation of Tie2 (Fig. 5).
Fig. 5.
Ang2 inhibits enhanced Ang1-activation of Tie2. Endothelial cells were treated with control vehicle or 10 ng/ml PMA before addition of 50 ng/ml Ang1 in the absence and presence of 1000 ng/ml Ang2 for 30 min as indicated. Cells were lysed and immunoprecipitated with anti-Tie2 antibodies. Phosphorylated Tie2 (pTie2) was detected by immunoblotting with anti-phosphotyrosine antibodies following SDS/PAGE. Blots were stripped and re-probed with anti-Tie2 antibodies. Tie2 phosphorylation was determined in three independent experiments. Data is presented as means and SEM of pTie2/Tie2 ratio normalized to that of PMA plus Ang1. ⁎ indicates statistically significant difference between control and Ang1 stimulated cells, † indicates significant inhibition of Ang1 induced phosphorylation by Ang2 and # indicates significant enhancement of Ang1-induced phosphorylation by PMA (p < 0.05, Student's t test).
4. Discussion
In this study we demonstrate that Ang1 and Ang2 interact differently with Tie2 on endothelial cells. Ang2 binding to Tie2 is not affected by the presence of Tie1 and neither Tie1 ectodomain cleavage nor suppression of Tie1 expression influence Ang2 binding. This contrasts with the situation for Ang1 where binding to Tie2 is regulated by the presence of Tie1 ectodomain, and cleavage of Tie1 or suppression of Tie1 expression increases Ang1 binding to Tie2 and the ability of the ligand to activate the receptor [14,20]. Our data also demonstrate that Ang1 binds preferentially to Tie2 that is either not complexed to Tie1 or in Tie1-poor complexes, consistent with lower binding in the presence of Tie1 ectodomain. Loss of Tie1 ectodomain occurs in vivo and is activated by VEGF, inflammatory stimuli and changes in shear stress over a period of minutes [13,16]. The effect of Tie1 ectodomain on Ang binding to Tie2, therefore, provides a mechanism for acute regulation and integration of Ang signalling with that of other signalling inputs to the cell at the level of the Tie2 receptor, with enhanced Ang1 signalling being favoured in endothelial cells experiencing increased VEGF or receiving inflammatory or shear stimuli. In addition, it would be expected that over the longer term factors altering the expression level of Tie1 and Tie2:Tie1 ratio in the cell would also be expected to modify the endothelial response to Ang1.
The crystal structure of Ang2 bound to Tie2 ectodomain has been solved and reveals the Tie2 ectodomain to exist as a globular head comprising of three Ig and three EGF-like domains followed by a rod-like structure incorporating the three FnIII motifs [9–11]. Ang2 binding occurs at the second of the amino-terminal Ig domains and does not appear to induce any significant conformational change in either receptor ectodomain or ligand [10]. It is the most carboxy-terminal domain in Ang2, the P-domain, that binds Tie2 [9–11]. Based on the structural determinations and sequence similarities it is highly likely that all the angiopoietins bind in a structurally similar manner to Tie2 [10]. Our data indicates that in the cellular context Tie1 ectodomain can modify this interaction differentially between Ang1 and Ang2. It will be important to gain structural details of how Tie1 complexed with Tie2 differentially modifies this binding.
The present study has focussed on the impact of Tie1 ectodomain on the ability of Ang1 and Ang2 to interact with Tie2. In addition to affecting Ang1 binding, it is possible that Tie1 could also have other effects on Tie2 signalling, for example Tie1 intracellular domain may influence the profile of sites phosphorylated on Tie2 intracellular domain and thus modify downstream signalling pathways. Further work will be required to examine such potential effects of Tie1 on Tie2 intracellular domain phosphorylation profile and recruitment of signalling intermediates.
Ang1 is an agonist in endothelial cells whereas Ang2 can act as an antagonist or partial agonist. The finding that Ang1 and Ang2 interact differently with Tie2 on the endothelium and that this is differentially regulated by Tie1 suggested that Tie1 may have a role in determining the agonist status of each ligand. We hypothesised that the lack of full agonist activity of Ang2 was due to Tie1. To test this we examined the effects of Ang2 on Tie2 activation in cells in which Tie1 ectodomain cleavage had occurred and also in cells in which expression of Tie1 was suppressed by siRNA. Contrary to our hypothesis we found that neither Tie ectodomain cleavage nor lack of Tie1 influenced the ability of Ang2 to activate Tie2 phosphorylation. These data demonstrate that the presence of Tie1 is not the sole determinant of Ang2 agonist activity. The ability of Ang2 to bind Tie2 in Tie2:Tie1 complexes raises the possibility that Ang2 antagonizes Ang1 by favouring Tie2:Tie1 complex formation and thereby inhibiting access of Ang1 to Tie2. However, the finding that Ang2 can still suppress Ang1-activation of Tie2 even when Tie1 ectodomain is cleaved, and therefore unable to restrict Ang1 access (Fig. 5), indicates Ang2 can act as a direct Ang1 antagonist without involvement of the Tie1 ectodomain.
The differential binding of Ang1 and Ang2 to Tie2 has important consequences for regulation of Tie2 responsiveness and signalling. If Tie1 did restrict Ang2 binding, as it does for Ang1, then in the presence of Ang2, or a high Ang2/Ang1 ratio, Tie1 cleavage would lead to an increase in the number of Tie2 receptors binding Ang2. As Ang2 is a partial agonist for Tie2 [2,3] this would increase the number of partially activated Tie2 receptors in each cell causing a net increase in Ang2 agonist activity. In this case Tie1 cleavage would increase both Ang1-mediated Tie2 activation and Ang2-mediated Tie2 activation. However, Ang1 and Ang2 were found to bind differently and Tie1 does not affect Ang2 binding or total number of Tie2 receptors (partially) activated by Ang2. This therefore allows an increase in total Tie2 signalling at high Ang1/Ang2 ratios but retains the apparent inhibitory effect of Ang2 as the ratio shifts towards lower Ang1/Ang2.
Recently it has been suggested that Ang1 and Ang2 interaction with the cell surface is more complicated than that of many ligand:receptor interactions and that the two ligands interact differently with receptors on the endothelial plasma membrane, as evidenced by their release following initial binding and the more rapid release of Ang2 than Ang1 [2]. Our data supports this assertion of differential and complex binding at the cell surface and implicates Tie1 as a differential regulator, though not direct binder, of the ligands. It is clear that the angiopoietin signalling mechanisms are multifaceted and likely to involve other regulatory influences in addition to the mechanism of differential regulation of Ang responsiveness via Tie1. The angiopoietins have key roles in development and maintenance of the vascular system. It is important, therefore, to elucidate how signalling by these ligands is controlled both to provide new insight into general principles of signal integration and regulation, as well as to improve understanding of how the angiopoietins contribute to vascular maintenance and disease.
The ability of cells to integrate multiple signalling inputs and transduce these into appropriate functional responses is essential for normal development and homeostasis. Integration and cross-talk between signalling pathways occur at intracellular nodes where different signalling cascades intersect [21], but can also arise at the level of receptor activation. For example in the endothelium, the Notch signalling pathway regulates responsiveness to VEGF, at least partly by modifying expression of soluble- and full-length-VEGFR1 which binds VEGF preventing its interaction and activation of VEGFR2 and inhibiting downstream signalling [22,23]. This cross-talk between Notch and VEGF signalling is important in maintaining the distinction between leading tip cells and trailing stalk cells during angiogenesis and without this normal vessel formation is disrupted [24]. In the case of the receptor tyrosine kinase Tie2, signalling is tightly controlled by the balance of Ang1:Ang2. We suggest it is also regulated by Tie1 ectodomain, which itself is acutely controlled by stimuli distinct from angiopoietins including VEGF and changes in shear stress. The result of the differential effects of Tie1 on binding of Ang1 and Ang2 to Tie2 shown in this study is that stimuli modulating Tie1 cleavage control the amplitude of the Tie2 response to Ang1 without increasing the net Ang2 agonist activity and maintaining the ability of Ang2 to antagonize Ang1. The combination Ang1:Ang2 ratio, Tie1:Tie2 balance and Tie1 cleavage status provide a complex multilayered control over signalling through Tie2 that can determine the Tie2 signal in response to its ligands in the context of the ambient angiogenic, inflammatory and shear status of the cellular environment.
Acknowledgement
We are grateful to the British Heart Foundation (PG/07/029/22656) for financial support of this work.
References
- 1.Jones N., Iljin K., Dumont D.J., Alitalo K. Nat. Rev., Mol. Cell Biol. 2001;2:257. doi: 10.1038/35067005. [DOI] [PubMed] [Google Scholar]
- 2.Bogdanovic E., Nguyen V.P.K.H., Dumont D.J. J. Cell Sci. 2006;119:3551. doi: 10.1242/jcs.03077. [DOI] [PubMed] [Google Scholar]
- 3.Yuan H.T., Khankin E.V., Karumanchi S.A., Parikh S.M. Mol. Cell Biol. 2009;29:2011. doi: 10.1128/MCB.01472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maisonpierre P.C., Suri C., Jones P.F., Bartunkova S., Wiegand S.J., Radziejewski C., Compton D., McClain J., Aldrich T.H., Papadopoulos N., Daly T.J., Davis S., Sato T.N., Yancopoulos G.D. Science. 1997;277:55. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 5.Davis S., Aldrich T.H., Jones P.F., Acheson A., Compton D.L., Jain V., Ryan T.E., Bruno J., Radziejewski C., Maisonpierre P.C., Yancopoulos G.D. Cell. 1996;87:1161. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 6.Procopio W.N., Pelavin P.I., Lee W.M., Yeilding N.M. J. Biol. Chem. 1999;274:30196. doi: 10.1074/jbc.274.42.30196. [DOI] [PubMed] [Google Scholar]
- 7.Davis S., Papadopoulos N., Aldrich T.H., Maisonpierre P.C., Huang T., Kovac L., Xu A., Leidich R., Radziejewska E., Rafique A., Goldberg J., Jain V., Bailey K., Karow M., Fandl J., Samuelsson S.J., Ioffe E., Rudge J.S., Daly T.J., Radziejewski C., Yancopoulos G.D. Nat. Struct. Biol. 2003;10:38. doi: 10.1038/nsb880. [DOI] [PubMed] [Google Scholar]
- 8.Yee V.C., Pratt K.P., Cote H.C., Trong I.L., Chung D.W., Davie E.W., Stenkamp R.E., Teller D.C. Structure. 1997;5:125. doi: 10.1016/s0969-2126(97)00171-8. [DOI] [PubMed] [Google Scholar]
- 9.Barton W.A., Tzvetkova D., Nikolov D.B. Structure. 2005;13:825. doi: 10.1016/j.str.2005.03.009. Camb. [DOI] [PubMed] [Google Scholar]
- 10.Barton W.A., Tzvetkova-Robev D., Miranda E.P., Kolev M.V., Rajashankar K.R., Himanen J.P., Nikolov D.B. Nat. Struct. Mol. Biol. 2006;13:524. doi: 10.1038/nsmb1101. [DOI] [PubMed] [Google Scholar]
- 11.Macdonald P.R., Progias P., Ciani B., Patel S., Mayer U., Steinmetz M.O., Kammerer R.A. J. Biol. Chem. 2006;281:28408. doi: 10.1074/jbc.M605219200. [DOI] [PubMed] [Google Scholar]
- 12.Marron M.B., Hughes D.P., Edge M.D., Forder C.L., Brindle N.P.J. J. Biol. Chem. 2000;275:39741. doi: 10.1074/jbc.M007189200. [DOI] [PubMed] [Google Scholar]
- 13.Chen-Konak L., Guetta-Shubin Y., Yahav H., Shay-Salit A., Zilberman M., Binah O., Resnick N. FASEB J. 2003;17:2121. doi: 10.1096/fj.02-1151fje. [DOI] [PubMed] [Google Scholar]
- 14.Marron M.B., Singh H., Tahir T.A., Kavumkal J., Kim H.-Z., Koh G.Y., Brindle N.P.J. J. Biol. Chem. 2007;282:30509. doi: 10.1074/jbc.M702535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yabkowitz R., Myer S., Yanagihara D., Brankow D., Staley T., Elliot G., Hu S., Ratzkin B. Blood. 1997;90:706. [PubMed] [Google Scholar]
- 16.Yabkowitz R., Meyer S., Black T., Elliott G., Merewether L.A., Yamane H.K. Blood. 1999;93:1969. [PubMed] [Google Scholar]
- 17.McCarthy M.J., Burrows R., Bell S.C., Christie G., Bell P.R.F., Brindle N.P.J. Lab. Invest. 1999;79:889. [PubMed] [Google Scholar]
- 18.Singh H., Milner C.S., Aguilar Hernandez M.M., Patel N., Brindle N.P. Cell Signal. 2009;21:1346. doi: 10.1016/j.cellsig.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Matthews J.A., Batki A., Hynds C., Kricka L.J. Anal. Biochem. 1985;151:205. doi: 10.1016/0003-2697(85)90073-9. [DOI] [PubMed] [Google Scholar]
- 20.Yuan H.T., Venkatesha S., Chan B., Deutsch U., Mammoto T., Sukhatme V.P., Woolf A.S., Karumanchi S.A. FASEB J. 2007;21:3171. doi: 10.1096/fj.07-8487com. [DOI] [PubMed] [Google Scholar]
- 21.Kestler H.A., Wawra C., Kracher B., Kuhl M. Bioessays. 2008;30:1110. doi: 10.1002/bies.20834. [DOI] [PubMed] [Google Scholar]
- 22.Suchting S., Freitas C., le Noble F., Benedito R., Breant C., Duarte A., Eichmann A. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3225. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington L.S., Sainson R.C., Williams C.K., Taylor J.M., Shi W., Li J.L., Harris A.L. Microvasc. Res. 2008;75:144. doi: 10.1016/j.mvr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Siekmann A.F., Covassin L., Lawson N.D. Bioessays. 2008;30:303. doi: 10.1002/bies.20736. [DOI] [PubMed] [Google Scholar]