Abstract
To evaluate the transplacental transfer of Theileria sergenti infection in cattle, we used DNA probes to detect T. sergenti in 6 pregnant cows and their calves. All the animals were monitored by parasitologic, serologic, and polymerase chain reaction (PCR) assays for a predicted 875-base-pair (bp) DNA product and a 684-bp amplicon detected by nested PCR in the blood and spleens of aborted fetuses. An open reading frame (ORF) starting at nucleotide 170 and terminating at position 1021 was shown to code for a polypeptide of 283 amino acid residues. All 6 dams and 5 calves were positive for T. sergenti in all tests. One calf was positive only with nested PCR. We conclude that transplacental transmission of T. sergenti is a significant problem. The relevance of the data in the programmed introduction of new (especially pregnant) animals into established clean herds needs serious consideration with regard to control of theileriosis and other tickborne diseases.
Introduction
Theileriosis caused by Theileria sergenti is one of the most economically devastating diseases of livestock in Korea. The etiologic agent is a tickborne protozoan parasite of cattle that multiplies in erythrocytes, causing mild hyperthermia and anemia. When infected calves are under stress, such as when they are infected with other parasites or viruses, they show severe clinical signs associated with high morbidity but low mortality.
A number of hematotropic parasites (Plasmodium falciparum in human and Anaplasma and Theileria spp. in cattle) are known to cause transplacental infection (1,2,3). The diagnosis of T. sergenti infection has traditionally relied on laborious microscopic examination of Giemsa-stained blood smears. Various investigators have demonstrated that molecular and immunologic methods complement each other in the definitive and specific identification of these pathogens (4,5).
We describe the use of the polymerase chain reaction (PCR) to amplify T. sergenti DNA from blood and tissues of dams and their calves. We used oligonucleotide primers of a region encoding the 32-kDa surface protein of T. sergenti. The specificity of this PCR for T. sergenti was validated by Southern blot hybridization with a complementary DNA fragment labelled with radioactive phosphorus (32P).
Materials and methods
Animals and tick challenge
The ticks (Hemaphysalis longcornis) used throughout the study were reared on the ears of specific-pathogen-free rabbits at the Chonbuk animal facility. The ticks were infected by feeding on highly parasitemic cattle and screened for infection status by Giemsa staining of a squash preparation of each tick.
Six 2-year-old heifers were artificially inseminated, confirmed to be pregnant by a staff veterinarian, and determined to be free of T. sergenti infection parasitologically, serologically, and by PCR. One week after insemination, the animals were exposed to T. sergenti by tick challenge under controlled field conditions in which the final tick infestation per cow was estimated at 200. The ticks were allowed to feed ad libitum until they either dropped off or died on the cattle host. Two of the cows aborted at 6 and 7 mo of gestation. One calf was delivered by cesarean section at 8 mo of gestation. The other dams calved naturally. The dams were euthanized 6 mo after calving by intravenous administration of sodium pentobarbital, 15 mg/kg. All animal-handling procedures were strictly compliant with the guidelines from the Korean government, which were consistent with those of the Canadian Council on Animal Care.
Parasitologic methods
All the cattle were monitored for intraerythrocytic inclusions by Giemsa and acridine orange staining of peripheral blood smears at weekly intervals. The structures of the inclusions were compared with prototype T. sergenti parasites. In addition, serum was tested by the indirect fluorescent antibody test (IFA) at weekly intervals.
Preparation of DNA from blood and organs
Samples of blood, liver, spleen, and lymph nodes were collected from the 2 aborted fetuses, the calves, and the 6 dams. To prevent autolysis, fetal and neonatal specimens were preserved in 10% formalin until analyzed. Before further processing, the formalin-fixed tissues were deformalinized for 7 d, as described by Greer and colleagues (6). Tissue samples from the dams were processed without formalin. In both cases, the tissues were sliced into 50-μm-thick pieces and homogenized in a blender. Blood samples and tissue homogenates were washed twice in phosphate-buffered saline (PBS; pH 7.3; 0.137 M NaCl, 10 mM Na2HPO4, and 3.2 mM KH2PO4) by centrifugation at 3000 × g for 10 min. The erythrocytes were resuspended to the original volume in PBS. Finally, DNA was extracted from the erythrocytes and tissue homogenates by means of published protocols (7,8,9,10) and analyzed by PCR.
Cloning and PCR amplification
The cDNA clone C-2, with 1100 base pairs (bp), from a CDM8 library, was subcloned into the Xba1 site of the pBluescript SK+vector, and the nucleotide sequence was determined (5). The gene for the 32-kD surface protein of T. sergenti was sequenced. An open reading frame (ORF) starting at nucleotide 170 and terminating at position 1021 was shown to code for a polypeptide of 283 amino acid residues (5). A pair of oligonucleotide primers — a forward primer (23-mer), 5'-CACGCTATGTTGTCCAAGAG-3' (CAC + nucleotide positions 167 to 183), and a reverse primer, 5'-TGTGAGACT CAATGCGCCCTA-3' (nucleotide positions 1019 to 1038) (8) — was synthesized (Applied Biosystems 391 DNA synthesizer; Applied Biosystems, Philadelphia, Pennsylvania, USA).
Since no amplification with the use of these primers was observed in DNA from formalin-fixed samples of liver, spleen, and lymph nodes from the fetuses of infected dams, a nested PCR approach was adopted. A primer set was designed such that G + C and A + T content averaging 50% was maintained for both the forward primer (5'-TGCAAAGGCTGATGA-3'; nucleotide positions 258 to 272) and the reverse primer (5'-AGTCCAAAGGCCAAGCA-3'; nucleotide positions 925 to 941).
We chose to target the ORF to maximize differentiation of T. sergenti from other bovine pathogens with DNA and protein homologies published in public databases. Specific primers were designed to target the conserved sequences in a species-specific manner. A PCR assay was carried out on all blood samples, fetal livers and spleens from dams, and calves. All samples were subjected to nested PCR (10), whose protocols were checked with maximum stringency against common hematotropic pathogens (Babesia ovata, Theileria buffeli, and Anaplasma centrale) (11,12).
In the initial reaction of the PCR, the final volume of each reactant in 100 μL of PCR total reaction mixture was as follows: 5 μg of extracted DNA as a template, 1 μM of each primer in the 1st set, 200 μM of each deoxynucleotide triphosphate (dNTP), 5 U of Taq polymerase (Applied Biosystems), and 1 × PCR buffer [10 mM Tris-HCl, pH 8.3; 50 mM KCl; 0.001% (w/v) gelatin; and 1.5 mM MgCl2].
The nested PCR for the assessment of vertical transmission of T. sergenti consisted of 5 μL of the 1st PCR product as a template, 1 μM of each primer of the 2nd set, 200 μM of each dNTP, and 2.5 U of Taq polymerase in 1 × PCR buffer.
The amplification reactions were performed in an automatic DNA thermal cycler (PerkinElmer/Cetus, Wellesley, Massachusetts, USA) for 35 cycles. The initial PCR consisted of 2 min of denaturation at 95°C (3 min for the 1st cycle), 2 min of annealing at 63°C, and 3 min of extension at 73°C, with a final extension step of 3 min at 73°C. The reamplification step consisted of 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min of extension at 70°C, with a final extension step of 3 min at 73°C. Positive controls were T. sergenti piroplasm DNA; negative controls were derived from uninfected erythrocytes, normal bovine kidney cells, and heterologous sources of common bovine parasites, such as B. ovata and A. centrale.
Southern blotting
Definitive verification by Southern blot was carried out as previously described (5). The PCR products were electrophoresed onto 1% agarose, transferred to a Hybond-N nylon membrane (Amersham Biosciences, Little Chalfont, Buckinghamshire, England), and cross-linked by exposure to long-wave ultraviolet light. The membranes were hybridized overnight at 42°C with 1 to 2 × 106 cpm of 32P-labelled T. sergenti probes per millilitre in a solution containing 50% formamide. The membranes were washed 3 times at room temperature with 2 × saline sodium citrate (SSC) containing 0.1% sodium dodecyl sulfate (SDS) and once at 60%C with 0.1 × SSC containing 0.1% SDS, for 30 min each time (5).
ResultsResults
Microscopic observation
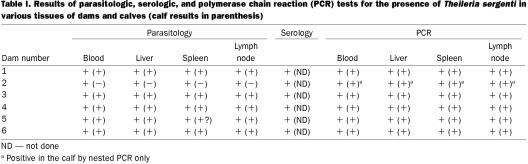
Peripheral blood smears from the 6 dams were all positive for T. sergenti by 7 to 10 d after exposure. After parasitemia was observed, all the animals seroconverted (titre 320 to 1280). Parasites were detectable in the peripheral blood throughout pregnancy and persisted for up to 6 mo after calving or abortion, at which point the dams were euthanized. Peripheral blood smears from 3 calves were also consistently parasitologically positive according to Giemsa and acridine orange staining. The parasite structure was compatible with that of T. sergenti. One calf had negative results of blood examination in spite of the positive status of the dam. Table I summarizes the results.
Table I.
DNA amplification and Southern blot results
All of the animals with positive peripheral blood smears also had positive PCR results (Table I). The predicted 875-bp amplicon, which included an ORF region (849 bp), was detected by agarose gel electrophoresis (Figure 1). The DNA from the formalin-fixed tissues of the aborted fetuses, which failed to react with the 1st PCR primer set when amplified with the 2nd primer set, yielded a 684-bp amplification product, as predicted for T. sergenti (Figure 2). A distinct amplicon was generated with nested PCR. Both bands were validated to be specific for T. sergenti by Southern blot (Figure 3). There were no detectable bands in the negative-control DNA prepared from uninfected erythrocytes (Figure 3), normal bovine kidney cells (Figure 3), and heterologous sources of common bovine parasites (data not shown). Positive PCR was established in fetuses aborted at 6 mo of gestation.
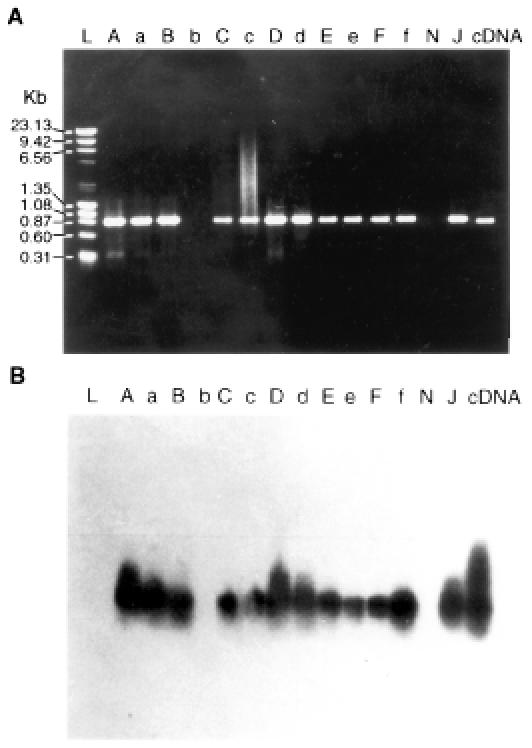
Figure 1. A: Polymerase chain reaction (PCR) profile of DNA extracted from 6 dams (A, B, C, D, E, and F) and the blood of their respective calves (a, b, c, d, e, and f). L — ladder DNA; N — blank (distilled water); J — positive-control DNA (from Japanese Theileria sergenti strain). B: Southern blot hybridization to verify that the PCR products in A are specific for T. sergenti; a T. sergenti probe labelled with radioactive phosphorus was used.
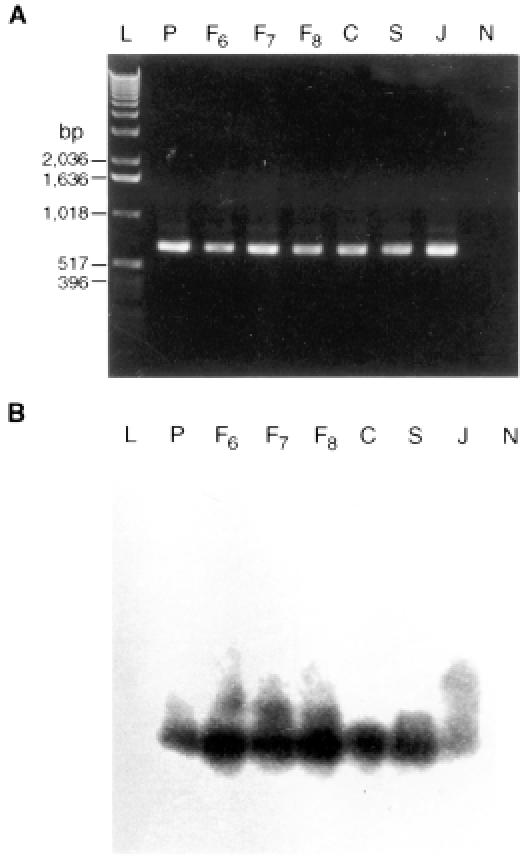
Figure 2. A: Amplification products from DNA extracted from formalin-fixed tissues. The 684-base pair products were obtained after 2 amplification steps involving 2 sets of primers. L — ladder DNA for size range; P — positive control (T. sergenti DNA); F6, F7, and F8 — DNA from spleens of 2 fetuses aborted at 6 and 7 mo of gestation and 1 calf born at 8 mo of gestation; C — DNA from positive dam; S — DNA from spleen of adult cow; J — positive-control DNA (from Japanese T. sergenti strain); N — DNA from normal tissue. B: Southern blot validation of PCR products in A.
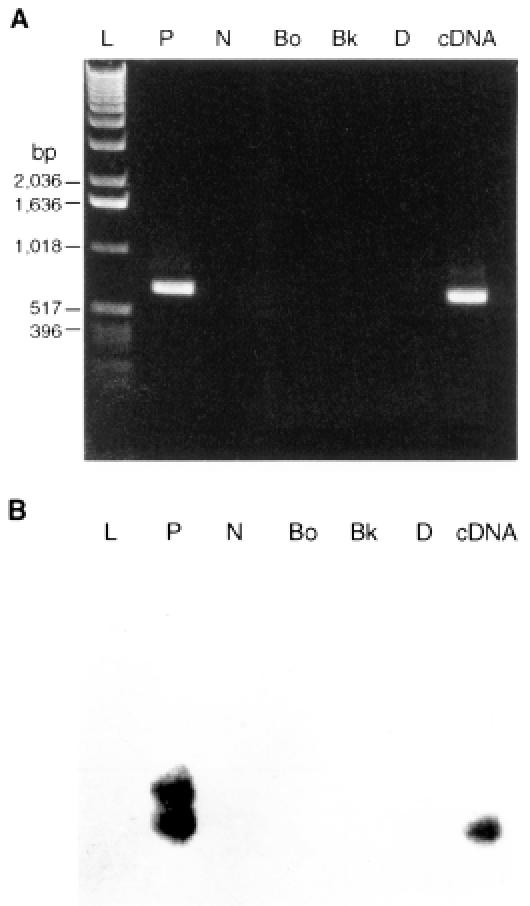
Figure 3. A: Assessment of the specificity of the PCR protocol. L — ladder DNA for size range; P — positive-control DNA from blood sample of calf infected with T. sergenti; N — negative-control DNA from normal bovine erythrocyte; Bo — DNA from Babesia ovata; Bk — DNA from normal bovine kidney cell; D — distilled water with all components but no template; cDNA — reference cDNA from T. sergenti 34 kDa expressed E. coli. B: Southern blot validation of PCR products generated during the specificity testing and 2-step amplification with 2 primer sets, as described in A.
Discussion
Theileria sergenti is an economically important tickborne hematotropic parasite of cattle in Korea and Japan. Theileriosis is routinely diagnosed by direct microscopic observation of parasites in Giemsa-stained blood smears from animals showing clinical signs consistent with theileriosis. These criteria are not optimal or stringent for the detection of T. sergenti infection, especially among carriers or animals that have an early infection and low blood levels of parasites. There are no pathognomonic lesions or signs caused by T. sergenti. In a previous study (3), 29 of 67 neonatal calves were shown to be infected with T. sergenti by routine staining methods. We have now complemented the parasitologic methods for T. sergenti diagnosis with molecular biologic tools, in line with developments for various hematoparasites (7,12).
Advances in molecular biology have led to the development of diagnostic assays that use nucleic acid probes to detect pathogens directly, with greater specificity and sensitivity. We found the PCR method to be specific: no amplification was detected with DNA from other common bovine parasites (A. centrale and B. ovata) or from uninfected control leukocytes and erythrocytes (8). The detection limit of this method was approximately 4.5 parasites per microlitre of blood. Thus, the PCR method provided a useful technique for verifying vertical transmission of T. sergenti. The relative sensitivity of the method, however, needs further investigation. Previous clinical observations suggested congenital and/or neonatal infection with T. sergenti (3). This phenomenon has now been verified by amplification of specific DNA from precolostral blood samples from calves and from spleen and lymph node samples from aborted fetuses, consistent with the view that most Korean cattle may be carriers of T. sergenti (13). We hypothesized that the aborted fetuses were infected with T. sergenti in utero by the experimentally infected dams. Using PCR, we have been able to show unequivocally that vertical transmission occurs under strict experimental conditions.
Theileria sergenti is similar to many other protozoan parasites in having a complex, multistage life cycle that involves 2 hosts (cattle and ticks) and has genomic diversity comparable to that of P. falciparum (13,14), Theileria parva (15,16,17), and Trypanosoma brucei rhodesiense (18). We have demonstrated that the gene encoding for the 32-kDa surface protein of T. sergenti, targetted by synthetic oligonucleotide primers and the cDNA probe, facilitates the detection of congenital infection with T. sergenti in a highly specific way. We estimate that this system is sufficiently sensitive to detect the equivalent of parasitemia of 0.00009%, corresponding to 4.5 parasites per microlitre of peripheral blood (8). Therefore, it would most predictably detect parasites in the invertebrate host and in carrier animals with invariably low levels of parasites. Its potential for detecting live parasites in invertebrate vectors is beyond the scope of our current investigation.
Of special interest is the observation that PCR is applicable to formalin-fixed tissues after prolonged storage or transportation, thus confirming previous reports of the efficacy of using PCR on fixed tissues (6,19). However, in many cases single-step amplification failed to occur as predicted until nested PCR was performed. Similarly, our method required nested PCR to reamplify the putatively positive formalin-fixed preparations. The nested PCR results correlated well with the parasitologic data. The modified PCR protocol was optimized by the highly specific primer pair that in previous testing was found to be specific to the ORF DNA domain of T. sergenti (5). These data are consistent with those of previous workers (10,11). We designed and used a nested PCR protocol with heterogeneous primer sequences. Primer sets targetting the ORF DNA domain have the advantage of enhanced sensitivity without compromised specificity. This observation may be particularly important in cases of low-level parasitemia associated with the early phase of the disease, recovery, or the carrier state so typical of bovine theileriosis due to T. sergenti. In particular, we feel that routine use of this method would minimize inadvertent introduction of infected animals into an immunologically naïve population of cattle. Exotic cattle breeds from nonendemic countries would also need to be protected against introduction of or exposure to these agents.
Footnotes
Acknowledgments
The technical discussions with Dr. Michael Vodkin of the University of Illinois were greatly appreciated, as was the generous financial support of Chonbuk National University, Biosafety Research Institute.
Address all correspondence and reprint requests to Dr. Ibulaimu Kakoma; telephone: (217) 333-1859; fax: (217) 333-0346; e-mail: kakomai@uiuc.edu
Received January 27, 2003. Accepted June 4, 2003.
References
- 1.Woods WG, Mills E, Ferrieri P. Neonatal malaria due to Plasmodium vivax. J Pediatr 1982;85:669. [DOI] [PubMed]
- 2.Bird JE. Neonatal anaplasmosis in a calf. J S Afr Vet Med Assoc 1973;44:69–70. [PubMed]
- 3.Baek BK, Rim BM, Lee WJ, et al. Study on infection of Theileria sergenti in neonatal calves. Korean Vet Res 1993;3:665–671.
- 4.Kajiwara N, Kirisawa R, Onuma M, Kawakami Y. Specific DNA probe for the detection of Theileria sergenti infection in cattle. Jpn J Vet Sci 1990;52:1199–1204. [DOI] [PubMed]
- 5.Matsuba T, Kubota S, Tanaka M, et al. Analysis of mixed parasite populations of Theileria sergenti using cDNA probes encoding a major piroplasm surface protein. Parasitology 1993;107: 369–377. [DOI] [PubMed]
- 6.Greer CE, Peterson SL, Kiviat NB, Manos MM. PCR amplification from paraffin-embedded human tissue. Effects of fixatives and fixation time. Am J Clin Pathol 1991;95:117–124. [DOI] [PubMed]
- 7.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 1989.
- 8.Tanaka M, Matsuba T, Onoe S, et al. Biotin-labeled genomic DNA probe for the detection of Theileria sergenti and its nucleotide sequence. J Protozool Res 1992;2:34–39.
- 9.Sugimoto C, Sato M, Kawazu S, Kamio T, Fujisaki K. Purification of merozoites of Theileria sergenti from infected bovine erythrocytes. Parasitol Res 1991;77:129–131. [DOI] [PubMed]
- 10.Tanaka M, Onoe S, Matsuba T, et al. Detection of Theileria sergenti infection in cattle by polymerase chain reaction amplification of parasite-specific DNA. J Clin Microbiol 1993;31:2565–2569. [DOI] [PMC free article] [PubMed]
- 11.Innis MA, Gelfand DH, Sninsky JJ, White TJ. PCR protocols. New York, NY: Academic Press, 1982.
- 12.McLaughlin G, Ssebtibgam SS, Nantez E, et al. PCR-based detection and typing of parasites. In: Ozcel MA, Alkan MZ, eds. Parasitology for the 21st century: proceedings of ICOPA VIII. Wallingford, England: CAB International, 1996:261–287.
- 13.Cappel RL, Saint RB, Stahl HD, et al. Plasmodium falciparum: differentiation of isolates with DNA hybridization using antigen gene probes. Exp Parasitol 1985;60:82–89. [DOI] [PubMed]
- 14.Vernick KD, Walliker D, McCuchan TE. Genetic hypervariability of telomere-related sequence is associated with meiosis in Plasmodium falciparum. Nucleic Acids Res 1988;16:6973–6985. [DOI] [PMC free article] [PubMed]
- 15.Bishop R, Sohanpal B, Kariuki DP, et al. Detection of a carrier state in Theileria parva-infected cattle by the polymerase chain reaction. Parasitology 1992;104:215–232. [DOI] [PubMed]
- 16.Chang DH. Epidemiological study of theileriosis (East Coast Fever). Korean J Parasitol 1974;12:14–20.
- 17.Allsopp BA, Allsopp MTE. Theileria parva: genomic DNA studies reveal intraspecific sequence diversity. Mol Biochem Parasitol 1988;28:77–84. [DOI] [PubMed]
- 18.Hide G, Buchanan N, Welburn S, Maudlin I, Barry JD, Tait A. Trypanosoma brucei rhodesiense: characterization of stocks from Zambia, Kenya, and Uganda using repetitive DNA probes. Exp Parasitol 1991;72:430–439. [DOI] [PubMed]
- 19.Tokuda Y, Nakamura T, Satonaka K, et al. Fundamental study on the mechanism of DNA degradation in tissue fixed in formaldehyde. J Clin Pathol 1990;43:748–751. [DOI] [PMC free article] [PubMed]