Abstract
In rice panicle development, new meristems are generated sequentially in an organized manner and acquire their identity in a time- and position-dependent manner. In the panicle of the panicle phytomer2-1 (pap2-1) mutant, the pattern of meristem initiation is disorganized and newly formed meristems show reduced competency to become spikelet meristems, resulting in the transformation of early arising spikelets into rachis branches. In addition, rudimentary glumes and sterile lemmas, the outermost organs of the spikelet, elongate into a leafy morphology. We propose that PAP2 is a positive regulator of spikelet meristem identity. Map-based cloning revealed that PAP2 encodes OsMADS34, a member of the SEPALLATA (SEP) subfamily of MADS-box proteins. PAP2/OsMADS34 belongs to the LOFSEP subgroup of MADS-box genes that show grass-specific diversification caused by gene duplication events. All five SEP subfamily genes in rice are expressed exclusively during panicle development, while their spatial and temporal expression patterns vary. PAP2 expression starts the earliest among the five SEP genes, and a low but significant level of PAP2 mRNA was detected in the inflorescence meristem, in branch meristems immediately after the transition, and in glume primordia, consistent with its role in the early development of spikelet formation. Our study provides new evidence supporting the hypothesis that the genes of the LOFSEP subgroup control developmental processes that are unique to grass species.
Keywords: LOFSEP, OsMADS34, PANCILE PHYTOMER2 (PAP2), Rice inflorescence, SEPALLATA (SEP), Spikelet
Introduction
The basic architecture of the inflorescence is determined by the identity and activity of meristems. Studies in the model eudicot species, mostly Arabidopsis, have allowed the basic model of the genetic control of flower initiation to be built (for a review, see Blázquez et al. 2006). LEAFY (LFY) and APETALA1 (AP1) are key regulators of floral meristem identity in Arabidopsis (Mandel et al. 1992, Weigel et al. 1992, Wagner et al. 1999). Other genes, such as CAULIFLOWER (CAL) and SEPALLATA (SEP), together with LFY and AP1, control floral meristem identity (Ferrándiz et al. 2000, Ditta et al. 2004). Once the floral meristem fate is established, the meristem starts to produce floral organs according to the mechanism known as the ABCE model (Goto et al. 2001).
Grass species show unique features of inflorescence development (Clark and Pohl 1969). A conspicuous characteristic of grass inflorescence is the spikelet, a small branch containing a variable number of flowers called florets. Because the spikelet is a terminal structure, the arrangement of spikelets rather than flowers is considered to define inflorescence architecture in grass species. A schematic of the rice inflorescence, called the panicle, is shown in Fig. 1A. During rice panicle formation, new meristems are sequentially generated. Newly formed meristems acquire the identity of a rachis branch meristem or terminate as a spikelet meristem, depending upon their position and the timing of their occurrence. The meristem that becomes a rachis branch meristem continues to generate next order lateral meristems that will eventually acquire the spikelet identity and terminate (Itoh et al. 2005). The key factor that determines the grass inflorescence form is the spatial and temporal regulation of spikelet meristem fate.
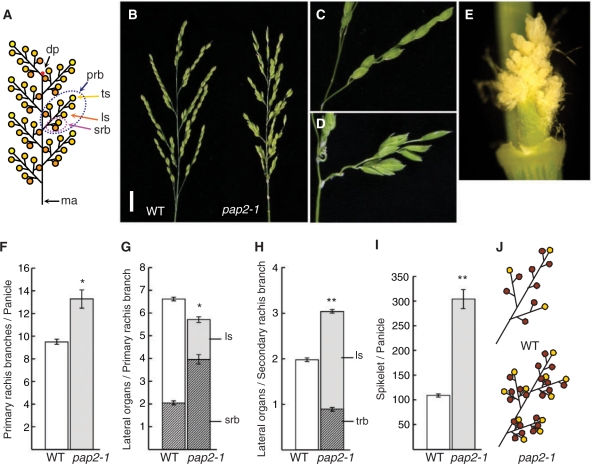
Fig. 1.
Phenotype of the pap2-1 mutant in panicle development. (A) Schematic of a rice panicle. ma, main axis; prb, primary rachis branch; srb, secondary rachis branch; ls, lateral spikelet; ts, terminal spikelet; dp, degeneration point of the IM. (B) Morphology of panicles in the wild-type (left) and pap2-1 (right). Scale bar = 2 cm. (C and D) Spikelets on the primary rachis branch in the wild-type (C) and pap2-1 (D). (E) Magnified view of aggregated branches on the pap2-1 primary rachis branch shown in D. (F) Number of primary branches per panicle. Sample size, n = 10. (G) Number of lateral organs generated on a primary branch. srb, secondary rachis branch; ls, lateral spikelet. Sample size, n = 10. (H) Number of lateral organs generated on a secondary branch. trb, tertiary rachis branch; ls, lateral spikelet. Sample size, n = 10. (I) Number of spikelets per panicle. All spikelets generated were counted under the stereomicroscope irrespective of whether they were normal or abortive. Sample size, n = 10. (J) Schematics of a spikelet of the wild-type (top) and pap2-1 mutant (bottom). The asterisks show that the difference is significant at the *1% and **0.1% level, respectively.
Because the spikelet is unique to grasses, unveiling the molecular and genetic control of spikelet initiation and development has been of great interest from both evolutionary and developmental points of view. So far, several genes that control panicle and spikelet development have been identified in grass species such as maize and rice (Bommert et al. 2005, Bortiri and Hake 2007). They can be classified based on the developmental steps they control. Genes of the first class, such as ABERRANT PANICLE ORGANIZATION1 (APO1), RICE CENTROLADIALLIS (RCN), GRAIN NUMBER1 (GN1) and DENSE AND ERECT PANICLE1 (DEP1), control the initiation of meristem identity (Ashikari et al. 2005, Ikeda et al. 2005, Ikeda et al. 2007, Huang et al. 2009, Ikeda et al. 2009). APO1, which encodes an F-box protein related to Arabidopsis UNUSUAL FLORAL ORGANS (UFO), negatively regulates the spikelet meristem identity (Ikeda et al. 2005, Ikeda et al. 2007, Ikeda et al. 2009). In apo1 mutants, specification of the spikelet identity is accelerated, resulting in the formation of a small panicle. In contrast, an increased expression of APO1 caused the delay of spikelet identity determination, leading to an increased number of branches in the panicle. Constitutive expression of RCN, a TERMINAL FLOWER1 (TFL1) ortholog, delays spikelet meristem identity and causes an extreme increase in the number and in the level of ramification of rachis branches, indicating that RCN can suppress the spikelet meristem identity (Nakagawa et al. 2002). GN1 and DEP1 were identified as quantitative trait loci that modulate grain yield through the control of the meristem phase transition to a spikelet meristem (Ashikari et al. 2005, Huang et al. 2009). GN1 encodes a cytokinin oxidase, an enzyme that catalyzes the degradation of cytokinins, implying an involvement of cytokinins in the control of spikelet meristem identity. Arrangement of spikelets in the panicle is disturbed in panicle phytomer1 (pap1) mutants, suggesting that the PAP1 gene may play a role in the initiation of the spikelet meristems although its molecular nature remains to be determined (Takahashi et al. 1998).
Genes of the second class are required to maintain the spikelet meristem identity and/or to control the transition from spikelet to floret meristem identity. Mutations of these genes do not affect the arrangement of the spikelets but spikelet development is impaired. SUERNUMEROUS BRACT (SNB), FRIZZY PANICLE (FZP), LEAFY HULL STERILE1 (LHS1) and EXTRA GLUME1 (EG1) belong to this class (Jeon et al. 2000, Komatsu et al. 2003, Agrawal et al. 2005, Chen et al. 2006, Lee et al. 2006, Li et al. 2009). Finally, a third group of genes including SUPERWOMAN1, OsMADS3, OsMADS58 and DROOPING LEAF is required for floral organ development (Nagasawa et al., 2003, Yamaguchi et al. 2004, Yamaguchi et al. 2006).
Despite successful isolation and characterization of all the genes mentioned above, our understanding of the molecular basis of rice panicle development remains fragmented, and more molecular studies are needed for a full understanding of the process. In this study, we isolated a new mutant named panicle phytomer2 (pap2) that shows abnormal panicle development including a disordered arrangement of spikelets, an increase of rachis branches and the elongation of rudimentary glumes and sterile lemmas. We show that PAP2 encodes OsMADS34, which belongs to the SEP subfamily of the MADS-box proteins. We suggest that PAP2 is one of the key regulators of spikelet meristem identity in rice.
Results
The panicle of the pap2-1 mutant produces more rachis branches
A wild-type panicle contains several primary rachis branches that are produced by the inflorescence meristem (IM). After producing several rachis branch meristems the IM loses its activity and is left as a vestige called a degenerate point at the base of the uppermost primary branch (Fig. 1A). The primary rachis branch contains a few secondary rachis branches at the bottom, several lateral spikelets and a terminal spikelet. Secondary rachis branches produce a few spikelets.
A recessive mutant with abnormal panicle morphology was identified through a screening of a rice population derived from tissue culture regenerated plants (Hirochika 2001). The morphological resemblance between the panicle of the mutant identified and that of a previously reported pap1 mutant (Takahashi et al. 1998) prompted us to test the genetic relationship between the two mutations. Because the genetic complementation test indicated that the mutation was not located in the PAP1 locus (data not shown), the mutant was named pap2-1. In the pap2-1 mutant, the arrangement of lateral organs, such as rachis branches and spikelets, was disordered and the panicle became irregularly denser, in sharp contrast to the well-defined phyllotaxy of the wild-type panicle (Fig. 1B–D). Furthermore, agglomerates of a large number of branches and spikelets with arrested development were often observed (Fig. 1B, D, E).
The pap2-1 panicle appears smaller than that of the wild-type due to the suppressed elongation of nodes in the panicle (Fig. 1B), but it contains more branches. We examined the branching pattern of pap2-1 in more detail. In this analysis, all spikelets were counted regardless of their development. The number of primary rachis branches in a rice panicle depends on the timing of IM deactivation. A significant increase in the number of primary rachis branches per panicle was observed in pap2-1, indicating that the IM activity was maintained longer in pap2-1 than in the wild-type (Fig. 1F). The total number of lateral organs generated on primary rachis branches is slightly reduced in pap2-1 while it is significantly increased on the secondary branches (Fig. 1G, H). In rice panicles, a few basal lateral organs on the rachis branches grow as next order branches and distal ones grow as spikelets. The shift from rachis branch formation to spikelet formation is delayed in pap2-1, resulting in the transformation of spikelets that arise early into rachis branches (Fig. 1G, H). In the wild-type panicle, 31.0% of lateral organs produced in the primary rachis branches grow as secondary rachis branches, whereas 70.8% become secondary branches in pap2-1 (Fig. 1G). This characteristic, namely production of more rachis branches, was again observed on the secondary rachis branches in pap2-1, leading to the formation of tertiary branches, which are not usually seen in the wild-type panicle (Fig. 1H). The increase in the number of branches resulted in an increase in the total number of spikelets initiated per panicle (Fig. 1I). Defects observed in pap2-1 panicles are schematically summarized in Fig. 1J. The panicle phenotypes observed in pap2-1 suggest that PAP2 positively controls spikelet meristem identity.
In contrast to the defects in the panicle, no alteration in the overall growth phenotype, including plant height, growth of tillers, the plastochron and the total number of leaves produced, was observed in pap2-1 during vegetative development (Supplementary Fig. S1). Although flag leaves of the wild-type and pap2-1 emerged at the same time, the pap2-1 panicle appeared approximately 5 d later than that of the wild-type (82.6 ± 0.7 days in the wild-type, vs. 87.5 ± 0.7 d in pap2-1), most probably due to a delay in the elongation of the panicle node.
Abnormal morphology of the pap2-1 spikelet
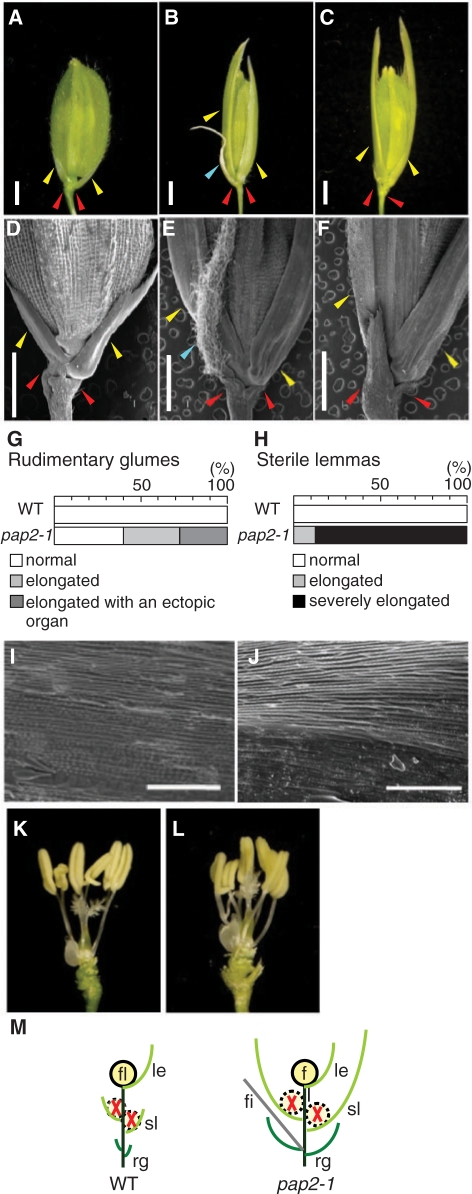
The spikelet meristem produces lateral organs in a distichous phyllotaxy. The first set is two bracts called glumes. Then, one or more bracts known as lemmas are generated and a floral meristem initiates in the axil of the lemma. Because glumes in the rice spikelet are reduced in size, they are called rudimentary glumes. Inside of the rudimentary glumes are empty glumes and a floret containing a lemma, a palea, two lodicules, six stamens and a pistil (Yamaguchi and Hirano 2006). Only one floret is formed in a rice spikelet, but it is interpreted that a rice spikelet originally generated three florets and empty glumes are lemmas of two florets that have been lost during the course of evolution (Komatsu et al. 2003, Bommert et al. 2005). According to this interpretation, empty glumes would correspond to sterile lemmas in other grass species. Herein we use the term sterile lemmas instead of empty glumes to better compare the spikelet structure with that of other grass species.
The most conspicuous abnormality in pap2-1 spikelets is the elongation of the sterile lemmas (Fig. 2A–F). Rudimentary glumes are also elongated, and an ectopic filamentous organ often develops in the axils of the rudimentary glumes, which are usually barren (Fig. 2B, E, blue arrowhead). Approximately 60% of the spikelets in a pap2-1 panicle exhibited elongated rudimentary glumes, and 40% of them developed an ectopic filamentous organ (Fig. 2G). Elongation of sterile lemmas was observed in all spikelets examined (Fig. 2H). Approximately 13% of the sterile lemmas elongated to about half the size of the lemma, 65% grew to a similar size and 23% grew longer. In spite of the elongation, the appearance of the surface of the elongated sterile lemmas remained unchanged, indicating that their identity as sterile lemmas was maintained (Fig. 2I, J). No detectable abnormality was observed in the floret (Fig. 2K, L), including the morphology of the lemma and palea (data not shown). The abnormalities in pap2-1 spikelets are illustrated in Fig. 2M.
Fig. 2.
Phenotype of pap2-1 in spikelet development. (A–C) Spikelets in the wild-type (A) and pap2-1 (B, C). Rudimentary glumes (red arrowheads) and sterile lemmas (yellow arrowheads) elongate in pap2-1. An ectopic organ (blue arrowhead) often developed in the axil of the rudimentary glume in pap2-1 (B, C). Scale bars = 1 mm. (D–F) SEM analysis of the spikelet of wild-type (D) and pap2-1 (E, F). Scale bars = 500 μm. (G and H) Number of rudimentary glumes (G) and sterile lemmas (H) in the wild-type and pap2-1. Sample size, n = 40. (I and J) Surface of sterile lemmas in the wild-type (I) and pap2-1 (J). Scale bars = 100 μm. (K and L) Floral organs in the wild-type (K) and pap2-1 (L) spikelets. (M) Schematics of spikelets from the wild-type (left) and pap2-1 (right). rg, rudimentary glume; sl, sterile lemma; fi, filamentous organ; le, lemma; fl, floret.
PAP2 encodes a MADS-box transcription factor similar to SEPALLATA (SEP)
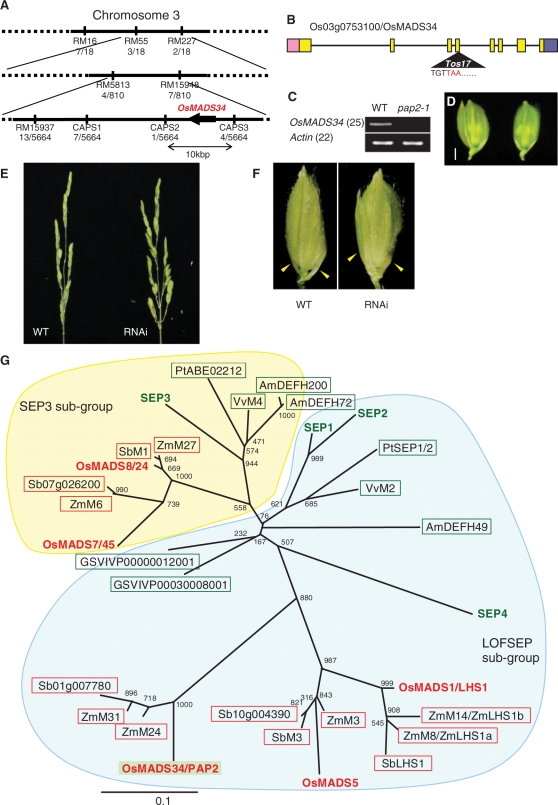
We isolated the PAP2 gene by map-based cloning. Fine mapping using 2,832 F2 plants allowed the PAP2 locus to be mapped to a 10,000 bp interval on chromosome 3, where only one gene, OsMADS34 (Os03g0753100), was predicted (Fig. 3A). Sequence analysis revealed an insertion of Tos17, an endogenous retrotransposon, in the fourth exon of OsMADS34 in pap2-1 (Fig. 3B). This insertion introduces a termination codon in the K-box of the protein (Supplementary Fig. S2). The OsMADS34 mRNA accumulation was decreased to an undetectable level in pap2-1 (Fig. 3C). These data suggest that PAP2 is OsMADS34 and that the defects in pap2-1 are most probably caused by the insertion of Tos17. We confirmed this hypothesis by introducing a wild-type genomic DNA fragment containing the OsMADS34 region into pap2-1 and achieving full complementation of the pap2-1 phenotype (Fig. 3D). Finally, a reduction of OsMADS34 expression by RNA interference (RNAi) mimicked the pap2-1 phenotype (Fig. 3E, F). Based on these results, we concluded that PAP2 is Os03g0753100, encoding a MADS-box protein, OsMADS34.
Fig. 3.
Isolation of PAP2 by positional cloning. (A) Location of the PAP2 locus on rice chromosome 3. The numbers of recombinants are indicated under each marker. (B) Structure of the PAP2/OsMADS34 gene. The boxes indicate exons, the line indicates introns and the triangle shows the insertion of Tos17. (C) Steady-state level of PAP2/OsMADS34 mRNA in the shoot apex of the wild-type and pap2-1. (D) Complementation test of pap2-1 by PAP2/OsMADS34. The elongated sterile lemma phenotype of pap2-1 (left) was rescued in the complemented plant (right). (E) Reduction of OsMADS34 mRNA accumulation phenocopied the pap2-1 phenotype. Panicles in an untransformed wild-type plant (left) and the OsMADS34 knock-down plant (right). (F) Spikelets in an untransformed wild-type plant (left) and the OsMADS34 knock-down plant (right). Yellow arrows show sterile lemmas. Sterile lemmas are elongated in spikelets of OsMADS34 knock-down plants, resembling those of the pap2-1 mutant. (G) Phylogenetic tree of SEP genes in plants. Amino acid sequences in the M-box and K-box were used for the analysis. The phylogenetic analysis was conducted using the Clustal W program, and the phylogenetic tree was constructed by the Neighbor–Joining method. Bootstrap values from 1,000 replicates are indicated at each node. Letters in red indicate rice genes and letters in green indicate Arabidopsis genes. Genes shown in the red boxes are from grass species, whereas genes in the green boxes are from eudicots. Os, rice; Zm, maize; Vv or GSVIVP, grape; Pt, poplar; Sb, sorghum (Sorghum bicolor); At, Arabidopsis; Am, snapdragon.
OsMADS34 is a member of the MADS-box gene family that regulates many aspects of plant growth and development (Malcomber and Kellogg 2005, Kater et al. 2006). PAP2/OsMADS34 is a type II MADS-box gene (there are >44 known type II MADS-box genes), a member of the SEPALLATA (SEP) subfamily (Fig. 3G). SEP genes are further divided into two major subgroups, namely the SEP3 and LOFSEP subgroups (Malcomber and Kellogg 2005, Zahn et al. 2005, Arora et al. 2007). Among the five genes of the SEP subfamily in rice, LHS1/OsMADS1, OsMADS5 and PAP2/OsMADS34 belong to the LOFSEP subgroup, and OsMADS7 and OsMADS8 are members of the SEP3 subgroup.
Expression pattern of the five rice SEP genes during early stages of panicle development
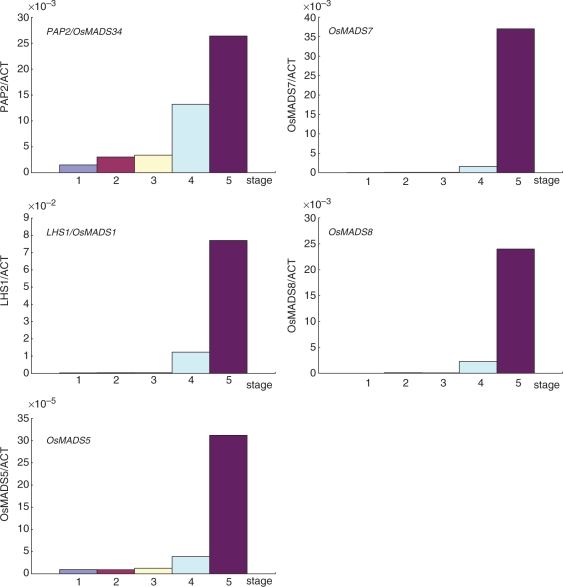
For a better understanding of the functions of the rice SEP genes in panicle development, the expression patterns of the five SEP genes during early stages of panicle development were examined (Fig. 4). Although a comprehensive study of MADS-box gene expression of rice was reported previously, the earliest stage examined in the study was at the 0–3 cm panicle stage (Arora et al. 2007). Because all critical steps of spikelet and floral organ differentiation end before the panicle reaches 2 mm in size, and maturation and elongation of organs occur in the later stages, we decided to examine expression of the five SEP genes during much earlier stages of panicle development. We collected, under the microscope, panicles that were approximately 0.1–1 mm in length and further categorized them into five developmental stages as reported by Furutani et al. (2006). The expression of the five SEP genes started at various time during panicle development. A low level of PAP2/OsMADS34 mRNA accumulation could be observed from stage 1 when the primary rachis branches differentiate. The expression of PAP2/OsMADS34 continued through stages 2 and 3 and increased at stage 4 when spikelet organs differentiate. In contrast to the early onset of PAP2/OsMADS34, LHS1/OsMADS1 expression started from the spikelet meristem initiation stage (stage 4). High levels of LHS1/OsMADS1 expression were observed at the floral organ differentiation stage (stage 5). OsMADS5 showed a pattern of mRNA accumulation similar to that of PAP2/OsMADS34 but at relatively lower levels. Both OsMADS7 and OsMADS8 mRNAs are expressed predominantly at stage 5 as previously reported (Pelucchi et al. 2002).
Fig. 4.
Expression pattern of five rice SEP genes during panicle development. Levels of mRNA accumulation during panicle development were examined by quantitative RT–PCR. Panicle development was divided into five stages according to Furutani et al. (2006). Stage 1, IM after the transition to the reproductive phase to the panicle at primary branch differentiation; stage 2, late primary branch differentiation to early secondary branch initiation; stage 3, young panicles at secondary branch differentiation; stage 4, spikelet differentiation; stage 5, floral organ differentiation. Values are averages of three biological repeats.
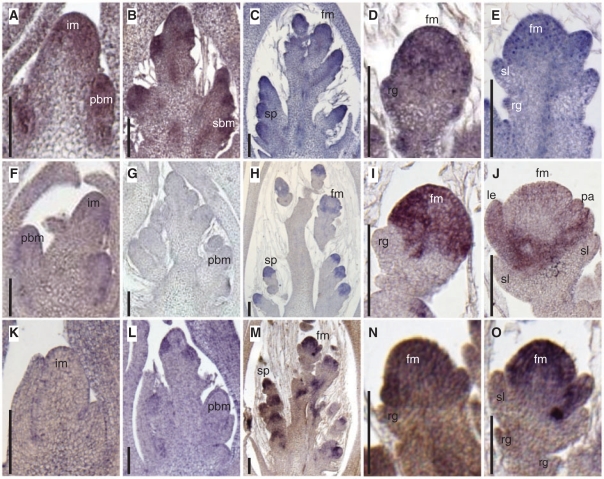
Finally, we examined the spatial distribution of the expression of the three LOFSEP subgroup genes by in situ hybridization (Fig. 5). PAP2/OsMADS34 mRNA accumulation was first observed in the IM and initiating primary rachis branches (Fig. 5A, B), and continued in the lateral spikelet meristem and the floret meristem (Fig. 5C, D). PAP2/OsMADS34 mRNA was also detected at the tips of the rudimentary glumes and sterile lemmas, as previously described by Pelucchi et al. (2002) (Fig. 5E). LHS1/OsMADS1 expression was first observed in the entire region of the floret meristem (Fig. 5H), and it started to be excluded from the meristem region as the lemma and palea primordial are formed, as previously reported (Fig. 5I, J) (Prasad et al. 2001). OsMADS5 expression started slightly earlier than that of LHS1/OsMADS1 and was localized in the floret meristem (Fig. 5L, M) and the palea primordia (Fig. 5N, O).
Fig. 5.
In situ mRNA accumulation of LOFSEP genes. (A–E) Expression patterns of PAP2/OsMADS34. A, stage 1; B, stage 3; C, stage 4; close-up views of a spikelet meristem and a floret meristem are shown in D and E, respectively. (F–J) Expression patterns of LHS1/OsMADS1. F, stage 2; G, stage 3; H, stage 5; close-up views of a spikelet meristem and a floret meristem are shown in I and J, respectively. (K–O) Expression patterns of OsMADS5. K, stage 1; L, stage 3; M, stage 4; close-up views of floret meristems are shown in N and O. Scale bar = 100 μm. im, inflorescence meristem; pbm, primary branch meristem; sbm, secondary branch meristem; sp, spikelet meristem; fm, floret meristem; rg, rudimentary glume; sl, sterile lemma; le, lemma; pa, palea.
Discussion
PAP2 is a positive regulator of spikelet meristem identity in rice
Here we describe pap2, a new rice mutant that exhibits abnormal development of the panicle. In pap2-1, the early arising spikelet meristems are converted to rachis branch meristems, resulting in an increase in the number and the ramification level of rachis branches. Based on this observation, we propose that PAP2 is a spikelet meristem identity gene of rice. So far, only a few rice genes that control spikelet meristem identity have been reported. APO1 and GN1 work in the control of spikelet meristem identity, however, they suppress the spikelet’s identity, an action opposite to that of PAP2 (Ashikari et al. 2005, Ikeda et al. 2005). On the other hand, DEP1 controls spikelet identity in a positive manner, as does PAP2 (Huang et al. 2009). RFL, a rice LFY ortholog, is another gene that negatively regulates spikelet identity (Rao et al. 2008). Elucidation of the genetic interactions among these genes is required in order to understand the mechanisms controlling rice panicle development. In addition to the conversion of spikelets to rachis branches, rudimentary glumes and sterile lemmas showed leafy morphology in pap2-1. This may be an indication of reduced floral fate, but we cannot rule out the possibility that PAP2 controls the identity of rudimentary glumes and sterile lemmas.
Among the spikelet meristem identity genes of rice, PAP2 is unique. pap2-1 is the only mutant in which disorganized positioning of branches and spikelets was observed in addition to the increase in the number of branches. In the other mutants, although the patterns of meristem identity are altered, the arrangement of lateral organs is well organized as in the wild-type panicle. Considering that all meristems in the rice panicle initiate as axillary meristems in the axils of bracts, the irregular pattern of branch formation in pap2-1 suggests that PAP2 may play a role in the control of phyllotaxy. This hypothesis should be examined in future studies.
Function of SEP subfamily MADS-box genes in rice panicle development
A large number of studies have shown that molecular mechanisms controlling the development of inner floral organs are relatively well conserved between grasses and eudicots (Kater et al. 2006, Yamaguchi and Hirano 2006). In contrast, little is known about the regulatory mechanisms that control the formation of outside organs that are unique to grass species and yet show a great variation among grass species. In this study, we show that PAP2/OsMADS34, belonging to the LOFSEP subgroup of the SEP family MADS-box genes, regulates the identity of the spikelet meristem and development of rudimentary glumes and sterile lemmas, the outermost organs in the spikelet. PAP2/OsMADS34 branches out with a group of monocot genes on a phylogenetic tree (Fig. 3E). Phylogenetic analyses have shown that duplications of LOFSEP genes occurred near the origin of grasses to produce PAP2/OsMADS34, LHS1/OsMADS1 and OsMADS5 clades (Malcomber and Kellogg 2005, Zahn et al. 2005). The early diversification of monocot LOFSEP genes and their distinct expression patterns suggests that they might have contributed to generate diversity in the reproductive development among monocots. Notably, PAP2/OsMADS34 is required for spikelet meristem identity while LHS1/OsMADS1 is essential for normal development of the lemma and palea. Lemma- and palea-like organs are reiteratively generated in lhs1 loss-of-function mutants, and glumes are transformed into lemma- and palea-like organs by constitutive expression of LHS1/OsMADS1 (Prasad et al. 2001, Agrawal et al. 2005, Chen et al. 2006). There is a clear distinction between the expression patterns of PAP2/OsMADS34 and LHS1/OsMADS1 (Prasad et al. 2001, Arora et al. 2007, this study). Based on these observations, we postulate that grass-specific LOFSEP genes sequentially control spikelet development, in particular leafy organs in the spikelet. First, PAP2/OsMADS34 works to initiate spikelet meristem identity and to control the development of rudimentary glumes and sterile lemmas. Then, LHS1/OsMADS1 acts to maintain the spikelet meristem identity and controls palea/lemma organ identity, which form inside of the rudimentary glumes and sterile lemmas. No obvious defect was observed in the loss-of-function mutants of OsMADS5, the other member of the LOFSEP subgroup (Agrawal et al. 2005). High sequence similarity between OsMADS1 and OsMADS5 suggests that they may have redundant functions. Although loss-of-function phenotypes of OsMADS7 and OsMADS8, members of the SEP3 subgroup in the SEP subfamily, are yet to be described, the spatial distribution of their mRNA suggests that they probably control the identity of the inner three floral organs in a similar fashion to the Arabidopsis SEP3 subgroup genes (Pelucchi et al. 2002, this study). The five rice MADS-box genes are expressed sequentially from the initiation of spikelet meristem to floral organ development, so that they may control the entire process of spikelet development. This is in contrast to the four SEP genes of Arabidopsis, which redundantly act to regulate floral meristem identity and organ identities of all four whorls (Pelaz et al. 2000, Ditta et al. 2004).
Previous analysis of LHS1/OsMADS1 mRNA expression patterns and spikelet development in grass species has suggested that changes in the expression pattern of LHS1/OsMADS1 contributed to the diversification of spikelets (Malcomber and Kellogg 2004, Reinheimer et al. 2006). Our study also supports the idea that the LOFSEP subgroup of SEP genes, a class of genes specific to grasses, plays a crucial role in the control of spikelets that are unique to grasses. Analysis of the function of PAP2 orthologs in other grasses will facilitate our understanding of the molecular basis of the evolution of spikelet development.
Molecular network controlling rice spikelet development
SEP proteins function as transcription factors in the form of a tetrameric complex (Theißen 2001, Kaufmann et al. 2005). Specific functions of SEP genes depend on the combination of MADS-box proteins in the complex. For instance, the combination of class-E SEP and class-A AP1 determines sepal formation in whorl 1 (Pelaz et al. 2001). Similarly, rice SEP proteins could form a tetrameric complex to regulate spikelet and floret development. Indeed, interactions between LHS1/OsMADS1 and RAP1A/OsMADS15 or RAP1B/OsMADS14, both orthologs of Arabidopsis AP1, were reported (Lim et al. 2000). The significance of an AP1 and LHS1 combination for the determination of glumes and florets in grasses has been demonstrated (Preston and Kellogg 2007). Identifying partners of PAP2 in the transcription factor complex will be a prerequisite for further elucidation of the role of PAP2. Moreover, isolation of the PAP1 gene and determination of genetic and molecular interactions between PAP1 and PAP2 will lead to a more comprehensive understanding of rice spikelet development.
Materials and methods
Plant materials
Plants grown in a glasshouse under natural conditions were used for the analysis of pap2-1 phenotypes. For mRNA extraction and in situ hybridization analysis, plants (cv. Nipponbare) were grown in a growth chamber under short day conditions (12 h light 28°C, 12 h dark 24°C).
Scanning electron microscope analysis
Spikelets in wild-type (cv. Nipponbare) and pap2-1 were fixed in 2.5% glutaraldehyde overnight at 4°C, dehydrated in a series of ethanol solutions and substituted with 3-methylbuthyl acetate. Subsequently, samples were dried at critical point, sputter-coated with platinum and observed under a scanning electron microscope (S-4000; Hitachi, Japan) at an accelerating voltage of 10 kV.
Molecular cloning of the PAP2 gene
Rough mapping of the PAP2 locus was performed with simple sequence repeat (SSR) markers using the mutant F2 plants obtained from a cross between pap2-1 and wild-type (cv. Kasalath) plants. For fine mapping, cleaved amplified polymorphic sequence (CAPS) markers were generated based on single nucleotide polymorphisms identified in nucleotide sequences between Nipponbare and Kasalath.
For the complementation test, a genomic fragment of 6,420 bp containing the OsMADS34 gene was generated by BglII and EcoRV digestions from a bacterial artificial chromosome (BAC) clone OSJNBa0047E24, and subcloned into pBluescript to generate pBSMADS34. A 1,330 bp fragment containing the promoter region of OsMADS34 was amplified by PCR using TArget Clone-Plus- (TOYOBO, Japan) polymerase with the following set of primers: 5′-CACCATTGCAGCTACAGTA CACCC-3′ and 5′-CTTGTTCTCGATCCGCTGAA-3′. Another genomic fragment of 2,504 bp containing the OsMADS34 3′-untranslated region and a terminator was generated by HindIII digestion of BAC clone OSJNBa0047E24, and cloned into pBluescript to generate pBSMADS34Term. The amplified fragment of the OsMADS34 promoter was cloned into pBSMADS34Term to generate pBSPMADS34. The fragment containing the whole region of the OsMADS34 gene including the promoter, the coding region and the terminator was cloned into the binary vector pPZP (Fuse et al. 2001).
Suppression of OsMADS34 expression by RNAi
To suppress the expression of OsMADS34, a part of the coding region amplified with the set of primers 5′-CACCTTATGCTTCG CAAGATGCTG-3′ and 5′-AGGTCGCAGAGTTCATCAAG-3′ was cloned into pANDA (Miki and Shimamoto 2004, Miki et al. 2005) and transferred into wild-type rice (cv. Nipponabare).
Real-time reverse transcription–PCR (RT–PCR) analysis
Total RNA was extracted from very young panicles by using a Plant RNA Isolation Mini Kit (Agilent) according to the manufacture’s protocol. After the DNase I treatment, the first-strand cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen, USA). RT–PCR was performed using the following primer sets: PAP2/OsMADS34, 5′-TTGATGAA CTCTGCGACCTAAA-3′ and 5′-TGCTGCAGTTTCCGTTCC-3′; LHS1/OsMADS1, 5′-GTGACCATTCCCTGCAGATT-3′ and 5′-GTCTGCTGCTTCATTGCTCA-3′; OsMADS5, 5′-TTGCAACT ACAACCTTAACTCATGT-3′ and 5′-GCAGCATTCATCAATC AAACC-3′; OsMADS7, 5′-GTGGCAACGGATTCTTCC-3′ and 5′-CGCATGAGTTGTTCATCTGC-3′; OsMADS8, 5′-ACGGAGC TTCAGAGAAAGGA-3′ and 5′-CCTGCTCCCACACTTGCT-3′; and ACTIN, 5′-GCCGTCCTCTCTCTGTATGC-3′ and 5′-GG GGACAGTGTGGCTGAC-3′. The relative abundances of mRNAs were measured with a LightCycler 480 system (Roche Applied Science, Germany) using ACTIN gene expression as a reference for normalization.
In situ hybridizations
In situ hybridizations were carried out as previously described (Kyozuka et al. 1998). To synthesize a digoxigenin-labeled antisense probe of PAP2/OsMADS34, a fragment containing the C-terminal region was amplified by PCR, cloned into pGEMT-easy vector (Promega, USA) and used as the template. The primer set used to amplify PAP2/OsMADS34 was 5′-GTAGA GGCAGCTCCCCCAC-3′ and 5′-GCTAGGCCATCCACTCAGG AGG-3′. Digoxigenin-labeled antisense probes of LHS1/OsMADS1 and OsMADS5 were synthesized by using full-length cDNAs, AK070981 and AK064184, respectively, as templates.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Ministry of Agriculture, Forestry and Fisheries [Genomics for Agricultural Innovation, IPG-0001 to J.K.]; Ministry of Education, Culture, Sports, Science and Technologies [Grant-in-Aid for Scientific Research on Priority Areas to J.K.]; Ministry of Education, Culture, Sports, Science and Technology, Japan [Grant in-Aid for Scientific Research for Plant Graduate Student from Nara Institute Science and Technology to K.K].
Supplementary Material
Glossary
Abbreviations
- BAC
bacterial artificial chromosome
- IM
inflorescence meristem
- RNAi
RNA interference
- RT–PCR
reverse transcription–PCR.
References
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H. Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol. Biol. 2005;59:125–135. doi: 10.1007/s11103-005-2161-y. [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Ferrándiz C, Madueňo F, Parcy F. How floral meristems are built. Plant Mol. Biol. 2006;60:855–870. doi: 10.1007/s11103-006-0013-z. [DOI] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano H-Y. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 2005;46:69–78. doi: 10.1093/pcp/pci504. [DOI] [PubMed] [Google Scholar]
- Bortiri E, Hake S. Flowering and determinacy in maize. J. Exp. Bot. 2007;58:909–916. doi: 10.1093/jxb/erm015. [DOI] [PubMed] [Google Scholar]
- Chen Z-X, Wu J-G, Ding W-N, Chen H-M, Wu P, Shi C-H. Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta. 2006;223:882–890. doi: 10.1007/s00425-005-0141-8. [DOI] [PubMed] [Google Scholar]
- Clark LG, Pohl RW. Agnes Chase’s First Book of Grasses. The Structure of Grasses Explained for Beginners. Washington, DC: Smithsonian Institute Press; 1969. 4th edn. [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Furutani I, Sukegawa S, Kyozuka J. Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J. 2006;46:503–511. doi: 10.1111/j.1365-313X.2006.02703.x. [DOI] [PubMed] [Google Scholar]
- Fuse T, Sasaki T, Yano M. Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 2001;18:219–222. [Google Scholar]
- Goto K, Kyozuka J, Bowman JL. Turning floral organs into leaves, leaves into floral organs. Curr. Opin. Genet. Dev. 2001;11:449–456. doi: 10.1016/s0959-437x(00)00216-1. [DOI] [PubMed] [Google Scholar]
- Hirochika H. Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant Biol. 2001;4:118–122. doi: 10.1016/s1369-5266(00)00146-1. [DOI] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009;41:494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ito M, Nagasawa N, Kyozuka J, Nagato Y. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 2007;51:1030–1040. doi: 10.1111/j.1365-313X.2007.03200.x. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa N, Nagato Y. ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev. Biol. 2005;282:349–360. doi: 10.1016/j.ydbio.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K, Yasuno N, Oikawa T, Iida S, Nagato Y, Maekawa M, et al. Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescense form through control of cell proliferation in the meristem. Plant Physiol. 2009;150:736–747. doi: 10.1104/pp.109.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, et al. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- Jeon J-S, Jang S, Lee S, Nam J, Kim C, Lee S-H, et al. leafy hull sterileq is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell. 2000;12:871–884. doi: 10.1105/tpc.12.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater MM, Dreni L, Colombo L. Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 2006;57:3433–3444. doi: 10.1093/jxb/erl097. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Melzer E, Theißen G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene. 2005;347:183–198. doi: 10.1016/j.gene.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development. 2003;130:3841–3850. doi: 10.1242/dev.00564. [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K. Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc. Natl Acad. Sci. USA. 1998;95:1979–1982. doi: 10.1073/pnas.95.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-Y, Lee J, Moon S, Park SY, An G. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 2006;49:64–78. doi: 10.1111/j.1365-313X.2006.02941.x. [DOI] [PubMed] [Google Scholar]
- Li H, Xue D, Gao Z, Yan M, Xu W, Xing Z, et al. A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J. 2009;57:593–605. doi: 10.1111/j.1365-313X.2008.03710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Moon Y-H, An G, Jang SK. Two rice MADS domain proteins interact with OsMADS1. Plant Mol. Biol. 2000;44:513–527. doi: 10.1023/a:1026517111843. [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell. 2004;16:1692–1706. doi: 10.1105/tpc.021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 2005;9:427–435. doi: 10.1016/j.tplants.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K. RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 2005;138:1903–1913. doi: 10.1104/pp.105.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, et al. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development. 2003;130:705–718. doi: 10.1242/dev.00294. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313x.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF. APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 2001;26:385–394. doi: 10.1046/j.1365-313x.2001.2641042.x. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-Lόpez R, Alvarez-Buylla ER, Yanofsky MF. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 2000;11:182–184. doi: 10.1016/s0960-9822(01)00024-0. [DOI] [PubMed] [Google Scholar]
- Pelucchi N, Fornara F, Favalli C, Masíero S, Lago C, Pè E, et al. Comparative analysis of rice MADS-box genes expressed during flower development. Sex. Plant Reprod. 2002;15:113–122. [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U. Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev. Genes Evol. 2001;211:281–290. doi: 10.1007/s004270100153. [DOI] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. Conservation and divergence of APETALA1/FRUITFULL-like gene function in grasses: evidence from gene expression analysis. Plant J. 2007;52:69–81. doi: 10.1111/j.1365-313X.2007.03209.x. [DOI] [PubMed] [Google Scholar]
- Rao NN, Prasad K, Kumar PR, Vijayraghavan U. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc. Natl Acad. Sci. USA. 2008;105:3646–3651. doi: 10.1073/pnas.0709059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheimer R, Malcomber ST, Kellogg EA. Evidence for distinct roles of the SEPALLATA gene LEAFY HULL STERILE1 in Eleusine indica and Megathyrsus maximus (Poaceae) Evol. Dev. 2006;8:293–303. doi: 10.1111/j.1525-142X.2006.00100.x. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Nagasawa N, Kitano H, Nagato Y. panicle phytomer 1 mutations affect the panicle architecture of rice. Theor. Appl. Genet. 1998;96:1050–1056. [Google Scholar]
- Theißen G. Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 2001;4:75–85. doi: 10.1016/s1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- Wagner D, Sablowski RWM, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285:582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvalez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Hirano H-Y. Function and diversification of MADS-box genes in rice. TSW Dev. Embryol. 2006;1:99–108. doi: 10.1100/tsw.2006.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell. 2004;16:500–509. doi: 10.1105/tpc.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Miyao A, Hirochika H, An G, Hirano HY. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell. 2006;18:15–28. doi: 10.1105/tpc.105.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebans-Mack JH, Kim S, Soltis PS, Landherr LL, et al. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005;169:2209–2223. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.