Abstract
Despite the health-related benefits of exercise, many people do not engage in enough activity to realize the rewards, and little is known regarding the genetic or environmental components that account for this individual variation. We created and phenotyped a large G4 advanced intercross line originating from reciprocal crosses between mice with genetic propensity for increased voluntary exercise (HR line) and the inbred strain C57BL/6J. G4 females (compared to males) ran significantly more when provided access to a running wheel and were smaller with a greater percentage of body fat pre- and postwheel access. Change in body composition resulting from a 6-day exposure to wheels varied between the sexes with females generally regulating energy balance more precisely in the presence of exercise. We observed parent-of-origin effects on most voluntary wheel running and body composition traits, which accounted for 3–13% of the total phenotypic variance pooled across sexes. G4 individuals descended from progenitor (F0) crosses of HR♀ and C57BL/6J♂ ran greater distances, spent more time running, ran at higher maximum speeds/day, and had lower percent body fat and higher percent lean mass than mice descended from reciprocal progenitor crosses (C57BL/6J♀ × HR♂). For some traits, significant interactions between parent of origin and sex were observed. We discuss these results in the context of sex dependent activity and weight loss patterns, the contribution of parent-of-origin effects to predisposition for voluntary exercise, and the genetic (i.e., X-linked or mtDNA variations), epigenetic (i.e., genomic imprinting), and environmental (i.e., in utero environment or maternal care) phenomena potentially modulating these effects.
Keywords: advanced intercross line, body composition, mouse, voluntary wheel running
Complex traits and parent-of-origin effects.
Complex traits, such as human disease and behavior, typically involve a number of genetic determinants and environmental factors that contribute jointly to the total phenotypic variation among individuals (31, 66). Potential sources of variation underlying the distribution of complex traits include the inherited genetic contributions and/or environmental stimuli (acting at any time after formation of the zygote, or in some cases even before). In particular, parent-of-origin effects are defined by cases in which a phenotype follows either a maternal or paternal line, as opposed to standard Mendelian inheritance patterns (73, 87). Parent-of-origin-dependent effects have been demonstrated to be important in modulating a variety of complex traits, including, but not limited to, growth and development (26, 38, 40, 41), cognitive abilities (48, 50), adult body composition (15, 36), cardiac response (2), such human diseases as Prader-Willi and Angelman syndromes, and some cancers (see Fig. 1 in Ref. 19, also Refs. 24, 62, 82). Furthermore, parent-of-origin effects have been demonstrated to play a role in the regulation of behavioral predispositions such as attention-deficit hyperactivity disorder (35), maternal behavior, learning and memory deficits, and altered rest/activity cycles (reviewed in Ref. 48). Effects are typically manifested through a broad range of possible genetic (e.g., sex-linkage) and epigenetic (e.g., x-chromosome inactivation, genomic imprinting) phenomena (37, 82, 86).
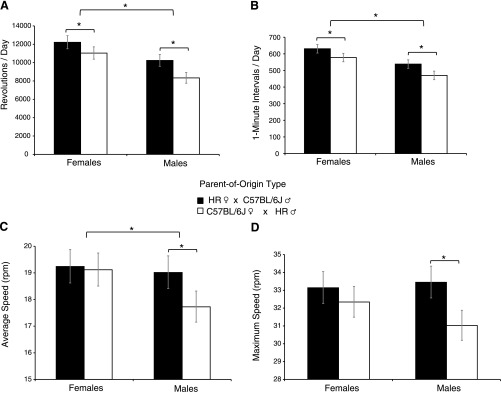
Fig. 1.
Sex (female vs. male) and parent-of-origin group differences. A: running distance (revolutions/day); B: time spent running (i.e., cumulative 1 min intervals in which at least 1 revolution was recorded); C: average speed (total revolutions/time spent running); and D: maximum speed (highest number of revolutions in any 1 min interval within a 24 h period). Bars [HR♀ × C57BL/6J♂ (solid bars) vs. C57BL/6J♀ × HR♂ (open bars)] represent adjusted least-squares means (from 2-way nested ANCOVAs, as shown in Table 1), back transformed along with back-transformed 95% confidence intervals. *Significant differences between groups. Note that for C and D there were significant interactions between sex and parent of origin (Table 1). So, for C and D, *significance based on analyses shown in Tables 1 and 2.
Physical activity.
A robust association exists between at least one environmental factor, physical inactivity, and the emergence and prevalence of modern chronic diseases. And, as Booth and colleagues (7) and others have hypothesized, physical inactivity may be at the very “environmental roots” of an ever-increasing list of chronic human health conditions. Variation in voluntary physical activity has been shown to be an important component of health, and regular physical activity has been positively correlated with the prevention and treatment of chronic human-health conditions including obesity and some cancers (6, 14, 28, 47, 59; but also see 84, 85). The benefits of voluntary exercise have been in part attributed to the regulation of energy balance, but there are few mechanistic studies that explain how exercise protects against disease pathogenesis. Regardless, there is extensive support at a descriptive level for the benefits of exercise on many aspects of human well being. Despite the health-related benefits of voluntary activity, more than 50% of U.S. adults do not engage in enough exercise to realize the rewards (80), and little is known regarding the genetic or environmental components that account for individual variation in predisposed voluntary activity levels. Broadly, in humans and rodents, it has been suggested that genetic architecture may play an important role in the regulation of voluntary activity levels, and empirical support for these suggestions is rooted in the demonstration of heritability (52, 55, 65, 70, 75).
Although the predisposition to engage in voluntary physical activity is heritable, used as a common therapeutic intervention for health-related disease, and may be a primary factor in the prevention of chronic health conditions, the location or nature of underlying genetic variation is still an emerging field in humans (9, 13, 22, 72) and rodents (49, 51, 56, 71, 79, 88). Thus, our long-term goal was to create a mapping population to detect quantitative trait loci (QTL) underlying the predisposition to engage in voluntary exercise. Toward this goal, we created a large advanced intercross line (AIL) of mice. Given the demonstration of the role of parent-of-origin effects on QTL for traits related to energy balance (e.g., 15) we utilized a reciprocal cross design to allow for detection of similar effects on voluntary exercise levels. This G4 population originated from a cross between mice with genetic propensity for increased voluntary exercise (HR line) and the inbred strain C57BL/6J.
Beginning in 1993, Garland and colleagues initiated an artificial selection experiment for voluntary wheel running in mice (reviewed in Refs. 30, 75). By generation 16, and continuing through generation 50 and beyond, the HR lines had diverged significantly from the control lines (C lines) with an approximate 2.5- to 3.0-fold increase in total revolutions/day. Selection history has caused the HR lines to diverge in a number of morphological, physiological, and behavioral traits. Examples include but are not limited to reduced body fat (25, 76), increased circulating adiponectin (81), decreased circulating leptin (33), and increased in home-cage activity (57, 58) and altered aspects of open-field behavior (10).
Here, we report on phenotypic measures of the G4 and present results in the context of strong differential sex and parent-of-origin effects on voluntary wheel running and body composition traits. We also explore the potential of the parent-of-origin effects to be differentially expressed in the two sexes. Finally, we discuss the mechanisms whereby these parent-of-origin effects could be transgenerationally inherited through direct genetic and/or epigenetic mechanisms.
MATERIALS AND METHODS
Animals.
Two strains of mice underwent a reciprocal intercross breeding protocol leading to a G4 AIL. The first of these two strains has been selectively bred for high voluntary wheel running for >40 generations (HR mice). Full details of this selection experiment have been provided elsewhere (75), so we will only offer a brief overview here. The original progenitors of the selection experiment were outbred, genetically variable house mice (Mus domesticus) of the Hsd:ICR strain [Harlan Sprague-Dawley (HSD), Indianapolis, IA]. After purchase from HSD, eight lines were created, four of these replicate lines have been selectively bred for high voluntary running (HR lines) on days 5 + 6 of a 6-day exposure to wheels (circumference = 1.12 m), while four others have been bred without regard to running as Controls (C). All procedures were approved by and are in accordance with guidelines set forth by the Institutional Animal Care and Use Committee at The University of North Carolina at Chapel Hill.
HR mice in the current experiment originated from one (lab designated #8) of the four HR lines in the 44th generation of artificial selection at the University of California, Riverside (UCR). At a mean age of 170 days (range = 157–174 days), HR males (n = 15) and females (n =15) were shipped from UCR to The Jackson Laboratory (TJL; Bar Harbor, ME) for rederivation. From 12 rederivations, 11 specific pathogen-free litters were successfully weaned and shipped to the University of North Carolina, Chapel Hill (UNC). These mice represented the founders (generation 0) of the UNC breeding colony of HR mice. HR mice for the current experiment originated from generation 2 of the UNC breeding colony.
The second strain was an inbred strain (C57BL/6J). Adult B6 males and females were purchased from TJL for the current experiment. The B6 strain was chosen for several reasons. First, Lightfoot et al. (55) examined variation in wheel running across inbred strains of mice and showed that B6 is a relatively average runner. Although generating a population from HR mice crossed to a poor runner would have maximized phenotypic divergence, it might also have uncovered variation that is not of interest. For example, extremely poor running performance (or no running at all) may not reflect a low predisposition to engage in exercise but instead may reflect a fear of the running wheel or mice that are generally unhealthy, both different traits than the one of current interest (see Ref. 30). Second, B6 is widely employed in many aspects of biomedical research, and its use provides access to a wealth of genomic tools and databases. These genomic tools and databases will be vital in follow-up investigations and future studies of voluntary exercise in the AIL. Third, the B6 strain has proven to be a reliable breeder, which was critical for the production of a large population. Finally, we have previously characterized wheel-running behavior in HR and B6 mice of both sexes and have demonstrated significant differences in running distance, time, average speed, and maximum speed (63).
Breeding design.
At ∼8 wk of age, 44 HR mice (22 males and 22 females) and 44 B6 mice (22 males and 22 females) underwent a reciprocal cross breeding protocol to produce a F1 population. The 44 progenitor HR mice represented brother/sister sibling pairs from 11 different families. In the F1 population, 16 (out of a possible 22) families from one cross-line population (HR♀ × B6♂) and 16 (out of a possible 22) families from a second cross-line population (B6♀ × HR♂) were chosen to propagate the following generation. In both of the reciprocal F1 populations each of the 11 progenitor families was represented at least once.
Once established, the two reciprocal cross-line populations were not mixed. The F2 and F3 generations were carried out as follows. After weaning, mice were housed in sex-specific cages in groups of four or five until 8 wk of age. At ∼8 wk of age, two males and two females were randomly chosen for breeding from within each of the 16 families in each reciprocal cross population. The males and females were bred across families with pairs of brothers and sisters each mated to a different family. In total, 64 mating pairs (32 from each reciprocal cross population) were established each generation. Each unique family was always represented by two breeding pairs to account for possible failure due to infertility of females, low litter size, or loss of pups before weaning. From these mating pairs at least one successful litter per family contributed to the next generation, which yielded no fewer than 16 unique families represented in each reciprocal cross population each generation.
To avoid inbreeding and increase the effective population size, interfamilial matings were assigned each generation utilizing a Latin square design. Pedigree files and genetic relationship matrices were maintained for all generations. All mice were reared in a viral-free facility, maintained at a temperature of 22°C, 30–55% relative humidity, and a light-dark cycle of 12 h:12 h beginning at 0700. Food (Prolab Isopro RMH 3000; calories provided by: protein 26%, fat 14%, carbohydrates 60%) and water were provided ad libitum. During pregnancy and lactation, breeding pairs were provided an enriched diet (Prolab RMH 2000; calories provided by protein 22%, fat 23%, carbohydrates 55%).
A large G4 was produced following the F3 generation. The production of the G4 population was created in a similar fashion to the preceding generations, with the following exceptions. Again, individuals representing the reciprocal cross-line populations (HR♀ × B6♂ and B6♀ × HR♂), established in the F1 generation, were not mixed and were equally represented in the G4. Two males and two females from each of 30 families were chosen from the F3 generation for breeding. The 30 (an additional 30 were created as “spare,” see above) breeding pairs were randomly assigned to groups of five and given the opportunity to mate beginning at ∼8, 9, or 10 wk of age. Mating pairs remained together for the entirety of the breeding process such that each of the 30 original pairs produced three sequential litters. Throughout the breeding process a repeatable synthetic diet (Research Diet D10012G; 20 kcal% protein, 16 kcal% fat, 64 kcal% carbohydrate) and water were provided ad libitum. This staggered mating design, and subsequent availability of adult animals, was done to accommodate space constraints associated with the phenotyping process while reducing any substantial intergroup age differences that may contribute to phenotypic variation.
Prior to and throughout phenotyping, G4 individuals (n = 815) were housed in sex-specific cages in groups of four and provided a repeatable synthetic control diet (Research Diet D10001; 21 kcal% protein, 68 kcal% carbohydrate, 13 kcal% fat) and water ad libitum. G4 individuals were divided into 19 cohorts with ∼45 individuals in each cohort. Cohorts 1–9 (n = 402 in total; 196 females and 206 males) represented one cross-line population (HR♀ × B6♂) established in the F1 generation. Cohorts 10–19 (n = 413 in total; 211 females and 202 males) represented the second cross-line population (B6♀ × HR♂) established in the F1 generation. Cohorts were phenotyped in sequential order spanning a period of 21 wk.
Phenotypic measures.
G4 mice were weighed ( ± 0.1 g) at 4 (mean = 30 days, range 27–32), 6 (mean = 44 days, range 41–46), and 8 (mean = 57 days, range 54–59) wk of age. Immediately following mass measurements, body composition (% fat tissue and % lean tissue) was assessed utilizing an EchoMRI-100 quantitative magnetic resonance whole body composition analyzer (Echo Medical Systems, Houston, TX). Percent body fat (and lean) was calculated as (fat mass/body mass)*100. Following body composition measurements at 8 wk of age, mice were housed individually with access to running wheels (model 80850, circumference = 1.1 m; Lafayette Instruments, Lafayette, IA) for 6 days. A circumference of 1.1 m was chosen to match the conditions under which the HR mice were selectively bred. Daily wheel-running activity was monitored with Running Wheel Activity Software (AWM V9.2, Lafayette Instruments) via Activity Wheel Counters (model 86061, Lafayette Instruments) interfaced with computers. Wheel-running activity was recorded in 1-min intervals for 23–24 h of each of the 6 days of wheel access. From this information, the following daily traits were calculated: total daily revolutions, time spent running (i.e., cumulative 1-min intervals in which at least one revolution was recorded), average speed (total revolutions/time spent running), and maximum speed (highest number of revolutions in any 1-min interval within a 24 h period). Here, we chose to analyze mean values on days 5 and 6 of the 6-day exposure to wheels, as these were the focal days used in the selection experiment that generated the HR line. Finally, food consumption was calculated by weighing (± 0.1 g) food prior to and following the 6 days of wheel access. To minimize any variation in food consumption due to food wasting, bedding was examined following day 6 and any visible pieces of food were accounted for.
Following the 6th day of wheel access, mice were weighed (± 0.1 g) and body composition assessed (as described above). Percent change (following wheel access) in percent body fat (and lean) was calculated as [(% following wheel access − % prior to wheel access)/% prior to wheel access]*100. Following decapitation, a trunk blood sample was taken and blood glucose was measured using a portable glucose monitoring system (Contour model; Bayer HealthCare, Tarrytown, NY), and tissues [tail, brain, epididymal (males) and perimetrial (females) fat depots, triceps surae muscles, liver, lungs, spleen] harvested. Wet masses (± 0.001 g) of fat and muscle were recorded on an analytical balance (model XS204; Mettler-Toledo, Columbus, OH). Following dissection (and mass measurements for fat and muscle) tissues were immediately snap frozen in liquid nitrogen.
Statistical analysis.
The Mixed procedure in SAS (version 9.1; SAS Institute, Cary, NC) was used to apply two-way nested analysis of covariance (ANCOVA) models with REML estimation and type 3 tests of fixed effects. The primary grouping factors were parent-of-origin type (whether a G4 individual was descended from a progenitor cross of HR♀ × B6♂ or B6♀ × HR♂, coded as 1 or 0, respectively) and sex (male vs. female), both fixed effects. As each of the 19 phenotyped cohorts contained only one parent-of-origin type, replicate cohort (n = 19 total) was nested within parent-of-origin type, and was always considered a random effect. Degrees of freedom (DF) for testing the parent-of-origin and sex effects were always 1 and 17. The sex effect and the sex × parent-of-origin type interaction were tested relative to the sex × cohort within parent-of-origin type interaction, again with 1 and 17 DF. Family was included as additional random factor nested within cohort. And, because each dam contributed multiple litters, litter was also treated as a random factor and was nested within family. Due to the slight variation in age, it was included as a covariate in all analyses. Additionally, where applicable, body mass, wheel freeness, time of day, and z-transformed (squared) term for time of death were included in the model as covariates. Wheel freeness was calculated as the number of wheel revolutions following acceleration to a given velocity. The z-transformed (squared) term for time of death was included, as it allows for possible nonlinear relationships between time of day and the dependent variable of interest. Traits were transformed as needed to stabilize variances among groups and improve normality of residuals. Statistical significance was judged at P < 0.05, and all P values presented are two-tailed.
RESULTS
Partial Pearson correlations among wheel-running variables and separately among body composition traits revealed several significant pair-wise associations (P < 0.05) (results not shown). Therefore, we first analyzed both groups of related traits with a MANCOVA, utilizing models in SAS Proc Mixed similar to those presented above. Overall contrasts yielded significant results (P < 0.001) for both the wheel running and body composition traits. Therefore, we proceeded by analyzing each trait separately as described in the Statistical analysis section and present these results below.
Since multiple tests were performed on the same set of animals it is appropriate to control the type I error rate. Given the relatively low number of hypotheses tested and the distribution of P values, we adjusted for multiple comparisons using the false discovery rate (FDR) technique in R (v. 2.8.1; R Development Core Team, 2008). Results of the FDR analysis indicated a more conservative alpha level of 0.03 for judging statistical significance (corresponding to an FDR of 5%). Since this correction does not substantially alter the overall pattern of our results we have chosen to present nominal P values for two-tailed statistical tests and emphasize general patterns in the results.
Wheel running.
Among G4 individuals, after adjusting for variation in age and wheel freeness, analysis of wheel-running data revealed that females ran significantly greater distances (revolutions/day, P < 0.0001), ran at higher average speeds (rpm, P = 0.0002), and spent significantly more time running (1-min intervals/day, P < 0.0001) than males (Table 1). With regard to maximum speed (highest number of revolutions in any 1-min interval within a 24 h period), the difference between the sexes was not significant (P = 0.1046, Table 1). As indicated by statistically significant parent-of-origin effects, G4 individuals descended from progenitor (F0) crosses of HR females and C57BL/6J males (HR♀ × B6♂) ran greater distances (P = 0.0004), spent more time running (P = 0.0006), and ran at higher maximum speeds/day (P = 0.0052) than mice descended from the reciprocal B6♀ × HR♂ progenitor crosses (Table 1 and Fig. 1). Additionally, while not statistically significant, mice descended from HR♀ × B6♂ parents tended to run at higher daily average speeds (P = 0.0529, Table 1).
Table 1.
Analysis of mean voluntary running traits from days 5 and 6 of a 6-day exposure to running wheels
| Trait | Transform | n | Sex | Parent-of-Origin | Interaction | Age | Freeness |
|---|---|---|---|---|---|---|---|
| Revolutions/day | ^0.5 | 782 | F1,17 = 81.55 | F1,17 = 19.23 | F1,17 = 3.00 | F1,655 = 2.34 | F1,655 = 1.85 |
| P < 0.0001(+) | P = 0.0004(+) | P = 0.1014 | P = 0.1267(+) | P = 0.1743(+) | |||
| 1-Minute intervals/day* | none | 782 | F1,17 = 106.16 | F1,17 = 17.63 | F1,17 = 0.62 | F1,655 = 8.86 | F1,655 = 11.06 |
| P < 0.0001(+) | P = 0.0006(+) | P = 0.4422 | P = 0.0030(+) | P = 0.0009(+) | |||
| Average speed, rpm | log10 | 782 | F1,17 = 12.99 | F1,17 = 4.33 | F1,17 = 6.93 | F1,655 = 2.68 | F1,655 = 2.99 |
| P = 0.0002(+) | P = 0.0529(+) | P = 0.0175 | P = 0.1021(−) | P = 0.0844(−) | |||
| Maximum speed, rpm | log10 | 782 | F1,17 = 2.94 | F1,17 = 10.27 | F1,17 = 7.23 | F1,655 = 1.02 | F1,655 = 1.18 |
| P = 0.1046(+) | P = 0.0052(+) | P = 0.0155 | P = 0.3140(−) | P = 0.2787(−) |
Data are from nested ANCOVAs and were transformed as necessary to improve normality of residuals. Significance levels (P values: boldface indicates P < 0.05) for the effects of sex (males vs. females), parent-of-origin (HR♀ × C57BL/6J♂ vs. C57BL/6J♀ × HR♂), and the sex by parent-of-origin interaction implemented in SAS PROC MIXED.
1-minute intervals/day: a measure of the amount of time spent running (i.e., cumulative 1-min intervals in which at least 1 revolution was recorded). The following covariates were also included in the analyses: Age, days since birth at the time of initial exposure to running wheel; freeness, number of wheel revolutions following acceleration to a given velocity. Signs following P values indicate direction of effect based on the partial regression from the mixed model: + indicates females > males: parent-of-origin type HR♀ × C57BL/6J♂ > C57BL/6J♀ × HR♂. Running wheel circumference was 1.1 m.
Two of the wheel-running traits [average and maximum speed/day (rpm)] showed significant sex by parent-of-origin interactions (P = 0.0175 and 0.0155, respectively). Thus, for these traits, the parent-of-origin effect was dependent on sex and the phenotypic differences between the sexes were dependent on parent of origin. Inspection of the least-squares means (Fig. 1) along with separate ANCOVAs of male and female mice (Table 2 and Supplemental Table S1)1 indicated the following patterns. For males, there were significant parent-of-origin effects for average (P = 0.0014, Table 2) and maximum (P < 0.0001, Table 2) daily running speed with G4 individuals descended from HR♀ × B6♂ progenitor crosses showing higher values. Conversely, for females, there were no significant parent-of-origin effects for average (P = 0.7414, Table 2) or maximum (P = 0.3016, Table 2) daily running speed. Thus, for running speed (average and maximum), the statistically significant parent-of-origin effects seem to be exclusive to males.
Table 2.
Separate-sex analysis of mean voluntary running traits from days 5 and 6 of a 6-day exposure to running wheels
| Trait | Transform | n | Parent-of-Origin | Age | Freeness | |
|---|---|---|---|---|---|---|
| Revolutions/day | ♂ | ^0.5 | 391 | F1, 17 = 33.08 | F1,289 = 1.66 | F1,289 = 5.74 |
| P < 0.0001(+) | P = 0.1985(+) | P = 0.0172(+) | ||||
| ♀ | ^0.5 | 391 | F1,17 = 4.97 | F1,288 = 2.20 | F1,288 = 0.17 | |
| P = 0.0395(+) | P = 0.1393(+) | P = 0.6801(−) | ||||
| 1-Minute intervals/day* | ♂ | none | 391 | F1,17 = 28.88 | F1,289 = 7.05 | F1,289 = 11.86 |
| P < 0.0001(+) | P = 0.0084(+) | P = 0.0007(+) | ||||
| ♀ | none | 391 | F1,17 = 6.59 | F1,288 = 5.11 | F1,288 = 2.97 | |
| P = 0.0200(+) | P = 0.0245(+) | P = 0.0859(+) | ||||
| Average speed, rpm | ♂ | log10 | 391 | F1,17 = 14.57 | F1,289 = 2.28 | F1,289 = 0.01 |
| P = 0.0014(+) | P = 0.1323(−) | P = 0.9191(−) | ||||
| ♀ | log10 | 391 | F1,17 = 0.11 | F1,288 = 0.21 | F1,288 = 5.88 | |
| P = 0.7414(+) | P = 0.6448(−) | P = 0.0159(+) | ||||
| Maximum speed, rpm | ♂ | log10 | 391 | F1,17 = 29.16 | F1,289 = 0.10 | F1,289 = 0.33 |
| P < 0.0001(+) | P = 0.7546(−) | P = 0.5651(+) | ||||
| ♀ | log10 | 391 | F1,17 = 1.13 | F1,288 = 0.22 | F1,288 = 4.83 | |
| P = 0.3016(+) | P = 0.6430(−) | P = 0.0288(−) |
Data are from nested ANCOVAs and were transformed as necessary to improve normality of residuals. Significance levels (P values: boldface indicates P < 0.05) for the effects of parent-of-origin (HR♀ × C57BL/6J♂ vs. C57BL/6J♀ × HR♂) implemented in SAS PROC MIXED.
1-Minute intervals/day: a measure of the amount of time spent running (i.e., cumulative 1 min intervals in which at least 1 revolution was recorded). The following covariates were also included in the analyses: age, days since birth at the time of initial exposure to running wheel; freeness, number of wheel revolutions following acceleration to a given velocity. Signs following P values indicate direction of effect based on the partial regression from the mixed model: + indicates parent-of-origin type HR♀ × C57BL/6J♂ > C57BL/6J♀ × HR♂. Running wheel circumference was 1.1 m.
While parent-of-origin effects on running distance and time spent running did not show statistically significant interactions with sex (P = 0.1014 and 0.4442, respectively; Table 1), they tended to be more pronounced in males than females (especially for running distance; Fig. 1, A and B). G4 males descended from HR♀ × B6♂ progenitor crosses ran 11% more revolutions per day than individuals descended from B6♀ × HR♂ crosses, while females descended from HR♀ × B6♂ ran only 5% more than their counterparts (Table 1 and Fig. 1A). Additionally, separate-sex analyses indicated the parent-of-origin effect for running distance was more robust for males (P < 0.0001) than for females (P = 0.0395) (Table 2). With regard to time spent running, G4 males descended from HR♀ × B6♂ progenitor crosses ran 14% more per day than their counterparts while the same parent-of-origin effect accounted for a 9% increase in females (Table 1 and Fig. 1B). Again, separate-sex analyses (Table 2 and Supplemental Table S1) indicated that the parent-of-origin effect for time spent running was more robust in males (P < 0.0001) than females (P = 0.0200). Hence, while the statistically significant parent-of-origin effects for running distance and time do not appear to be exclusive to males, as seen for running speed (average and maximum), the percent increase and critical values from separate-sex analyses indicate a larger effect in males.
Body composition.
Pre-exercise body composition was assessed at 4, 6, and 8 wk of age (Table 3 and Supplemental Table S2). At 4 and 6 wk of age, female mice weighed significantly less (P < 0.0001) and had a greater percentage of body fat (P < 0.0001), with a lower percentage of lean mass (P ≤ 0.003), compared with males (Table 3). Additionally, mice descended from HR♀ × B6♀ progenitor crosses had significantly lower percentages of fat (P < 0.05) and higher percentages of lean mass (P < 0.05) compared with mice descended from B6♀ × HR♂ (Table 3), although statistical significance was only noted at 6 wk of age after correction for multiple comparisons.
Table 3.
Analysis of body composition traits at 4, 6, and 8 wk of age
| Trait | Transform | n | Sex | Parent-of-Origin | Interaction | Age |
|---|---|---|---|---|---|---|
| ∼4 wk of age | ||||||
| Body mass, g | ^2.5 | 808* | F1,17 = 778.67 | F1,17 = 0.07 | F1,17 = 0.12 | F1,680 = 3.49 |
| P < 0.0001(−) | P = 0.7929(−) | P = 0.73740 | P = 0.0620(+) | |||
| % Fat | log10 | 811* | F1,17 = 208.62 | F1,17 = 4.72 | F1,17 = 0.16 | F1,683 = 0.42 |
| P < 0.0001(+) | P = 0.0442(−) | P = 0.6905 | P = 0.5188(+) | |||
| % Lean | ^2.0 | 811* | F1,17 = 102.23 | F1,17 = 4.56 | F1,17 = 0.01 | F1,682 = 3.79 |
| P < 0.0001(−) | P = 0.0476(+) | P = 0.9287 | P = 0.0518(−) | |||
| ∼6 wk of age | ||||||
| Body mass, g | ^1.5 | 807* | F1,17 = 1583.89 | F1,17 = 0.32 | F1,17 = 0.44 | F1,678 = 8.32 |
| P < 0.0001(−) | P = 0.5792(−) | P = 0.5159 | P = 0.0040(−) | |||
| % Fat | log10 | 808* | F1,17 = 67.67 | F1,17 = 16.00 | F1,17 = 7.56 | F1,679 = 0.00 |
| P < 0.0001(+) | P = 0.0009(−) | P = 0.0137 | P = 0.9760(+) | |||
| % Lean | ^3.0 | 805* | F1,17 = 11.95 | F1,17 = 31.40 | F1,17 = 10.16 | F1,677 = 0.17 |
| P = 0.0030(−) | P < 0.0001(+) | P = 0.0054 | P = 0.6801(+) | |||
| ∼8 wk of age† | ||||||
| Mean body mass, g‡ | none | 810* | F1,17 = 2093.63 | F1,17 = 0.26 | F1,17 = 0.09 | F1,682 = 9.05 |
| P < 0.0001(−) | P = 0.6162(+) | P = 0.7696 | P = 0.0027(−) | |||
| Body mass in, g | none | 810* | F1,17 = 1704.45 | F1,17 = 0.08 | F1,17 = 1.00 | F1,682 = 7.77 |
| P < 0.0001(−) | P = 0.7832(+) | P = 0.3325 | P = 0.0054(−) | |||
| Body mass out, g | none | 810* | F1,17 = 2572.41 | F1,17 = 0.53 | F1,17 = 0.22 | F1,681 = 9.87 |
| P < 0.0001(−) | P = 0.4785(+) | P = 0.6470 | P = 0.0018(−) | |||
| % Change in body mass | none | 811* | F1,17 = 29.33 | F1,17 = 0.82 | F1,17 = 4.97 | F1,683 = 0.13 |
| P < 0.0001(−) | P = 0.3790(−) | P = 0.0396 | P = 0.7208(+) | |||
| % Fat in | ^0.5 | 810* | F1,17 = 23.33 | F1,17 = 19.82 | F1,17 = 14.56 | F1,682 = 0.05 |
| P = 0.0002(+) | P = 0.0003(−) | P = 0.0014 | P = 0.8281(−) | |||
| % Fat out | log10 | 807* | F1,17 = 114.75 | F1,17 = 20.96 | F1,17 = 0.30 | F1,678 = 1.14 |
| P < 0.0001(+) | P = 0.0003(−) | P = 0.5883 | P = 0.2861(−) | |||
| % Change in % fat | none | 804* | F1,17 = 28.32 | F1,17 = 0.71 | F1,17 = 8.70 | F1,676 = 0.27 |
| P < 0.0001(−) | P = 0.4128(−) | P = 0.0090 | P = 0.6026(−) | |||
| % Lean in | ^2.0 | 808* | F1,17 = 1.03 | F1,17 = 26.34 | F1,17 = 11.32 | F1,679 = 0.31 |
| P = 0.3233(+) | P < 0.0001(+) | P = 0.0037 | P = 0.5753(+) | |||
| % Lean out | ^3.0 | 810* | F1,17 = 0.70 | F1,17 = 36.07 | F1,17 = 2.37 | F1,682 = 1.19 |
| P = 0.4140(+) | P < 0.0001(+) | P = 0.1420 | P = 0.2763(+) | |||
| % Change in % lean | none | 808* | F1,17 = 0.18 | F1,17 = 1.81 | F1,17 = 5.57 | F1,680 = 0.14 |
| P = 0.6792(−) | P = 0.1965(−) | P = 0.0305 | P = 0.7090(+) | |||
Data were from nested ANCOVAs and transformed as necessary to improve normality of residuals. Significance levels (P values: bold indicates P < 0.05) for the effects of sex (males versus females), parent-of-origin (HR♀ × C57BL/6J♂ vs. C57BL/6J♀ × HR♂), and the sex by parent-of-origin interaction implemented in SAS PROC MIXED. Signs following P values indicate direction of effect based on the partial regression from the mixed model: +females > males: parent-of-origin type HR♀ × C57BL/6J♂ > C57BL/6J♀ × HR♂.
At ∼8 wk of age body composition measures were taken immediately prior to (in) and following (out) 6 days of wheel access. Percent body fat (and lean) was calculated as (fat mass/body mass)*100. Percent change variables were calculated as [(out − in)/in]*100.
Mean body mass was calculated as (body mass in + body mass out)/2.
At 8 wk of age, all body composition measurements were made immediately prior to and following 6 days of wheel access. Females were significantly smaller than males (P < 0.0001) prior to and following wheel access, with no parent-of-origin effect (Table 3). Wheel access reduced body mass in both males and females, with males having a larger percent change (P < 0.0001). Prior to and following wheel access, females had percent fat values that were significantly larger than males (P < 0.0001) with no difference in percent lean mass (P > 0.3). After 6 days of wheel access, males lost a significantly larger percentage of fat compared with females (on a relative basis, P < 0.0001), with no difference in percent change in percent lean mass (P = 0.6792). With regard to parent-of-origin effects, mice descended from HR♀ × B6♂ progenitor crosses had significantly lower percentages of fat (P = 0.0003) and significantly higher percentages of lean mass (P < 0.0001), both pre- and postwheel access (Table 3).
Significant interactions between sex and parent-of-origin were observed at 6 wk of age for percent fat (P = 0.0137) and lean mass (P = 0.0054). Examination of least-squares means (Supplemental Table S2) and assessment of the sexes utilizing separate ANCOVAs (results not shown) revealed that parent-of-origin effects were more pronounced in females than males. That is, while both males and females descended from HR♀ × B6♂ progenitor crosses had significantly less fat and more lean mass (compared to B6♀ × HR♂) the quantitative effect was greater in females. At 8 wk of age significant interactions between sex and parent-of-origin type were detected for percent fat (P = 0.0014) and lean mass (P = 0.0037) prior to exercise (Table 3). These significant interaction terms, and the patterns revealed by them, were similar to results at 4 wk of age. Namely, males and females descended from HR♀ × B6♂ progenitor crosses were significantly leaner, but the quantitative effect was greater in females.
Following 6 days of wheel access, there was a marginally significant sex by parent-of-origin interaction for percent change in body mass (P = 0.0396, Table 3). The mean values (± SE) for the four subgroups were as follows: −3.7% ± 0.5, −5.0% ± 0.5, −6.4% ± 0.5, and −6.2% ± 0.5 for F4 females descended from HR♀ × B6♂, females descended from B6♀ × HR♂, males descended from HR♀ × B6♂, and males descended from B6♀ × HR♂, respectively (Supplemental Table S2). Thus, the parent-of-origin effect appears to be greater in females than in males, with female progeny from HR♀ × B6♂ parental origin having the smallest percent change in body mass following 6 days of wheel access. Additionally, significant interaction terms were observed for relative changes in percent fat (P = 0.0090) and percent lean mass (P = 0.0305) following the 6-day wheel trial (Table 3). Males descended from the two progenitor crosses (HR♀ × B6♂ and B6♀ × HR♂) had similar percent fat losses resulting from wheel exposure (−37% ± 2 and −35% ± 2, respectively), while female percent fat losses were more varied between the parent-of-origin types (−26% ± 2 and −32% ± 2, respectively; Supplemental Table S2). Results for relative change in percent lean mass were similar.
DISCUSSION
Parent-of-origin effects.
Along with generally similar results from an F1 cross between the same replicate of the HR selection lines and a control line (Hannon RM, Kelly SA, Keeney BK, Malisch JL, Garland T Jr., unpublished data), the current findings represent the first evidence directly implicating parent-of-origin effects (and in some cases their dependence on sex) in the regulation of voluntary activity levels. Since our data are from an advanced G4 intercross, we discuss for the first time that such effects may be inherited via X-linked or mtDNA genes or may be persisting across several generations through epigenetic effects. Here, we discuss some of the potential mechanisms through which parent-of-origin effects may be modulating voluntary activity and body composition in mice in a transgenerational manner.
One potential explanation for the transgenerational parent-of-origin effect on exercise and body composition is dosage effects of X-linked QTL (see Ref. 78 for examples of X-linked gene regulation). With regard to body composition, multiple QTL have been previously mapped to the X chromosome for fatness, body weight, and weight gain (see Fig. 1 in Ref. 66 and references therein). However, among human and mouse studies that have identified specific genetic markers associated with voluntary activity levels, none have implicated the X chromosome thus far (13, 22, 51, 56, 72). It is important to note, however, that only a minority of these studies (51, 56) performed analyses with sex-linked markers, whereas the others either did not or could not examine X chromosome inheritance. In the current experiment, G4 males descended from HR♀ × C57BL/6J♂ progenitor crosses have an approximate 62% probability of inheriting an HR X allele while males descended from C57BL/6J♀ × HR♂ have only a 38% probability, while G4 females have a 69 and 31% chance of inheriting a HR X allele, respectively. Given these percentages and the strong parent-of-origin effects observed for many of the G4 phenotypes, an X-linked QTL would need to have extremely large allelic effects to be a primary regulator of the phenotypic differences observed between the HR♀ × B6♂ and B6♀ × HR♂ progenitor crosses.
Although G4 mice descended from HR♀ × B6♂ are phenotypically different than mice descended from B6♀ × HR♂, in some cases, the parent-of-origin effects are dependent on sex and the phenotypic differences between the sexes are dependent on parent-of-origin. If genes located on the X chromosome are at least partially responsible for the variation in activity and body composition between reciprocal crosses, then the source of the significant statistical interactions between parent-of-origin and sex may be somewhat explained through mechanisms associated with X chromosome inactivation (see Ref. 23 and references therein, also see Ref. 74). For females, X chromosome alleles specific to HR or B6 may be escaping inactivation (see Ref. 23 and references therein), and this could potentially dilute any parent-of-origin effect observed in females. Alternatively, sex-specific differential regulation of wheel-running activity has been hypothesized to be mediated through estrogen/testosterone pathways (54). It is possible that genes that regulate sex-specific hormones and their pathways may be colocalized on sex chromosomes with sites related to activity levels.
Mitochondrial DNA (mtDNA) variation between HR and C57BL/6J is another potential source contributing to the parent-of-origin effects observed in the current study. In mice, mtDNA variations have been previously linked to experimental autoimmune encephalomyelitis, corticosterone and neurotransmitter response to psychological stress, and anxiety-like behavior (32, 89). Currently, we do not know if HR and C57BL/6J differ in their mtDNA sequence, but if such variation did exist then it could influence exercise and body composition traits, both of which rely heavily on energy efficiency, and effects would persist transgenerationally. In response to selective breeding, HR mice exhibit several phenotypic characteristics [e.g., high maximal oxygen consumption (69); alterations in muscle metabolic capacities after wheel running, (46)] that may be explained by sequence variation in the mitochondrial genome.
We acknowledge that founder effects could potentially account for the phenotypic differences observed between parent-of-origin types in the G4 population. However, given that the 44 progenitor HR mice represented brother/sister sibling pairs from 11 different families (see materials and methods) we believe it is unlikely that founder effects play a significant role in explaining reciprocal cross phenotypic differences.
In addition to direct genetic effects, the observed parent-of-origin effects may be modulated via such epigenetic mechanisms as genomic imprinting (68). Approximately 100 imprinted genes have been identified (3) and have been shown to be important for a variety of complex traits, including development and growth (3, 12, 15, 53, 62). Furthermore, it has been demonstrated that imprinting effects can be sex dependent (37). In the current experiment, it is important to note that for imprinting to contribute the parent-of-origin effects, HR and B6 mice would have to exhibit differential imprinting (maternal or paternal). Given the hypothesized function of imprinting in evolutionary processes we cannot rule out this possibility, but view imprinting as less likely relative to the other proposed mechanisms (see Refs. 8, 64).
While direct genetic/epigenetic influences may be the most likely mechanism underlying the parent-of-origin effects, environmental influences during pregnancy and development have also been shown to be important in shaping behavior and body composition (5, 17, 18, 21, 39, 83). This is especially paramount given recent examples of the potential for parent-of-origin effects to be modulated through transgenerationally inherited epigenetic mechanisms (1, 16, 27, 43).
In the current study, we had no direct measures of differences in maternal (or paternal) behavior (e.g., activity, food consumption), or physiology (e.g., metabolic rate) in the progenitors of each generation, and thus we cannot directly evaluate whether parent-of-origin effects on activity and body composition in the G4 mice were related to early developmental perturbations. In fact, previous studies on maternal care behavior in HR lines at earlier generations have revealed only minimal differences compared with controls (34). However, Malisch et al. (57, 58) demonstrated that female and male HR mice exhibit an ∼200% increase in home-cage activity (in the absence of a wheel) compared with control lines. In recent generations, home-cage activity during pregnancy and/or lactation and maternal care and its effects on offspring phenotype have not been quantified or evaluated in HR lines (but see Ref. 34) but may now become a priority.
The absence of F1 phenotypic data in the current experiment may lead to alternative interpretations of the observed parent-of-origin effects, especially in regard to their transgenerational nature. While the F1 generation of the cross utilized in this experiment was not phenotyped, data from another reciprocal cross involving the same HR line and a control line have produced similar results (Hannon RM, Kelly SA, Keeney BK, Malisch JL, Garland T Jr., unpublished data). Wheel running phenotypes of this F1 generation revealed that F1 mice from HR dams had higher running values (revolutions/day, time, average speed, and maximum speed), mirroring results from the current G4 generation. Furthermore, the effects were generally stronger for males than for females, with significance noted for revolutions/day and time spent running. Despite the difference between the two reciprocal crosses with regard to the strain that the HR mice were crossed with and the laboratory where phenotyping was conducted, the similarity and sex-specific magnitude of the results are striking and allow us to infer that the F1 from the current experiment would have yielded similar results. Therefore, given the assumption that F1 results from HR × ICR represent F1 results from HR × B6 this significantly increases confidence in the veracity of our findings that parent-of-origin effects on voluntary exercise levels and body composition in mice are transgenerational.
As outlined in materials and methods (Statistical analysis), due to logistical constraints, cohorts were tested in sequential order and thus cohort was confounded with parent of origin. We adopted several approaches to ensure that a sequence of testing effect was not involved in the parent-of-origin effects discussed above. First, to account for any cohort-to-cohort variation we included cohort as a nested factor in all statistical models. Second, we examined the LS means and standard errors from models that replaced parent of origin with cohort as a primary grouping factor. We did not observe a discernable pattern between cohorts or among parent-of-origin types (cohorts 1–9 vs. cohorts 10–19). Lastly, since any temporal variation in sequence order is likely to be minimized between closely tested cohorts, we grouped cohorts 7–12, separated by only a 6 wk period of time, together and treated them as one large population. With this subset of data, the parent-of-origin effects remained significant.
In this study we chose to evaluate mean running values on days 5 and 6 of the 6-day exposure to wheels. We acknowledge that mice may take longer periods (up to 2 wk) to acclimate to running wheels and reach a plateau with regard to running distance. A number of factors may contribute to the acclimation process causing day-to-day variation among individuals in the trajectory of initial wheel-running behavior. We chose the current paradigm to reflect as accurately as possible the conditions under which the HR mice were selectively bred (75), to measure the same phenotype selected for and minimize any interpretational issues associated with measuring a different phenotype. While initial “learning curves” may be present in some strains of mice, this does not seem to be the case for HR mice compared with ICR control mice (see Fig. 5 in Ref. 31). However, we point out that our results are relevant to the current methods, and caution should be taken when extrapolating these findings to other related measures of voluntary exercise.
Sex effects.
Females ran significantly more, for longer periods of time, and at higher average speeds than males. These results are generally consistent with previous investigations comparing the differences in activity in female vs. male mice of a variety of strains, including the two strains used in the current study and the other replicates of HR selection lines as well as their control lines (55, 75). These sex-specific differences in voluntary activity levels have been hypothesized to be mediated by estrogen and testosterone pathways (reviewed in Ref. 54). As outlined by Lightfoot, mechanisms involved in the estrogen-α pathway are currently the most detailed and may reflect differential release of dopamine as a possible mechanism leading to the increased physical activity in females (see Fig. 4 in Ref. 54). However, while plausible mechanisms are beginning to be proposed, a plethora of alternative hypotheses regarding the mechanistic regulation of exercise via sex hormones are still largely in their infancy.
The results of sex effects on body composition are generally consistent with existing human literature examining weight loss patterns via body fat loss or oxidation in men and women as a result of exercise training (42, 44), suggesting that these sex differences in weight loss patterns may result from greater precision in energy balance among women than among men. That is, when participating in exercise-training programs, women often eat more to compensate for the negative energy balance resulting from aerobic activity while men generally do not (Ref. 42, cf. Ref. 77 on sex differences in the maintenance of body mass in the HR mice). Sex differences in weight loss patterns resulting from exercise training may be attributable to changes in circulating hormones, differences in primary substrate utilization, or a combination of these two factors along with others. Several studies (29, 45, 61) have demonstrated that females derive a relatively larger contribution from lipids to oxidative metabolism during exercise, although others (11, 60) have observed similar contributions of lipids and carbohydrates. Future analyses will focus on whether percent change differences (among individuals and between sexes) in body mass, percent fat, and percent lean varied in accordance with running distance, duration, average speed, or maximum speed.
The results of the current investigation are an important step toward uncovering the factors contributing to the variation associated with the predisposition of voluntary activity levels. The G4 advanced intercross line we have created is one of the largest and potentially most powerful experimental populations of mice created for the purpose of dissecting and understanding the genetic architecture controlling voluntary exercise. Here, we have described strong, transgenerational parent-of-origin and sex effects on exercise and outlined several possible mechanisms that may be modulating these observations. We hypothesize that several of these factors are likely making a joint contribution to the parent-of-origin effects, as opposed to a single major cause. Forthcoming genotypic and linkage analyses will be useful in determining the role of X-linked loci and genomic imprinted QTL. Further studies will be needed to elucidate the role of the mitochondrial genome and maternal environmental effects.
GRANTS
This work was supported by National Institutes of Health (NIH) Grant DK-076050 to D. Pomp. S. A. Kelly was supported through a National Institute of Mental Health-funded (5T32MH-075854-04) Interdisciplinary Obesity Training program. Phenotypes were collected using the Animal Metabolism Phenotyping core facility within UNC's Clinical Nutrition Research Center (funded by NIH Grant DK-056350).
DISCLOSURES
No conflicts of interest are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Z. Yun for assistance with animal care and data collection. We thank Chris Wiesen at UNC's Odum Institute for Research in Social Science for statistical consultation.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Abbott A. Obesity linked to grandparental diet [online]. Nature News. Nature. http://www.nature.com/news/2008/081120/full/news.2008.1240.html [ 2008].
- 2. Barrick CJ, Dong A, Waikel R, Corn D, Yang F, Threadgill DW, Smyth SS. Parent-of-origin effects on cardiac response to pressure overload in mice. Am J Physiol Heart Circ Physiol 297: H1003– H1009, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beechey CV, Cattanach BM, Blake A, Peters J. Mouse Imprinting Data and References (online). Oxfordshire, UK: MRC Harwell, http://www.har.mrc.ac.uk/research/genomic_imprinting/ ( 2008). [Google Scholar]
- 4. Belter JG, Carey HV, Garland T., Jr Effects of voluntary exercise and genetic selection for high activity levels on HSP70 expression in house mice. J Appl Physiol 96: 1270– 1276, 2004. [DOI] [PubMed] [Google Scholar]
- 5. Bick-Sander A, Steiner B, Wolf SA, Babu H, Kempermann G. Running during pregnancy transiently increases postnatal hippocampal neurogenesis in the offspring. Proc Natl Acad Sci USA 103: 3852– 3857, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blair SN, LaMonte MJ, Nichman MZ. The evolution of physical activity recommendations: how much is enough? Am J Clin Nutr 79: 913S– 920S, 2004. [DOI] [PubMed] [Google Scholar]
- 7. Booth FW, Chakravarthy MV, Gordon SE, Spangenburg EE. Waging war on physical inactivity: using modern molecular ammunition against an ancient enemy. J Appl Physiol 93: 3– 30, 2002. [DOI] [PubMed] [Google Scholar]
- 8. Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecol Lett 11: 106– 115, 2008. [DOI] [PubMed] [Google Scholar]
- 9. Bray MS, Hagberg JM, Perusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc 41: 35– 73, 2009. [DOI] [PubMed] [Google Scholar]
- 10. Bronikowski AM, Carter PA, Swallow JS, Girard I, Rhodes JS, Garland T., Jr Open-field behavior of house mice selectively bred for high voluntary wheel-running. Behav Genet 31: 309– 316, 2001. [DOI] [PubMed] [Google Scholar]
- 11. Burguera B, Proctor D, Dietz N, Guo Z, Joyner M, Jensen MD. Leg free fatty acid kinetics during exercise in men and women. Am J Physiol Endocrinol Metab 278: E113– E117, 2000. [DOI] [PubMed] [Google Scholar]
- 12. Burt A, Trivers R. Genes in Conflict. Cambridge, MA: Harvard University Press, 2006. [Google Scholar]
- 13. Cai G, Cole SA, Butte N, Bacino C, Diego V, Tan K, Goring HH, O'Rahilly S, Farooqi IS, Commuzzie AG. A quantitative trait locus on chromosome 18q for physical activity and dietary intake in Hispanic children. Obes Res 14: 1596– 1604, 2006. [DOI] [PubMed] [Google Scholar]
- 14. Chakravarthy M, Booth F. Eating, exercise, and “thrift” genotypes: connecting the dots toward an evolutionary understanding of modern chronic diseases. J Appl Physiol 96: 3– 10, 2004. [DOI] [PubMed] [Google Scholar]
- 15. Cheverud JM, Hager R, Roseman C, Fawcett G, Wang B, Wolf JB. Genomic imprinting effects on adult body composition in mice. Proc Natl Acad Sci USA 105: 4253– 4258, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chong S, Youngson NA, Whitelaw E. Heritable germline epimutation is not the same as transgenerational epigenetic inheritance. Nat Genet 39: 574– 575, 2007. [DOI] [PubMed] [Google Scholar]
- 17. Clapp JF. Exercise during pregnancy. A clinical update. Clin Sports Med 19: 273– 286, 2001. [DOI] [PubMed] [Google Scholar]
- 18. Clapp JF. Influence of endurance exercise and diet on human placental development and fetal growth. Placenta 27: 527– 34, 2006. [DOI] [PubMed] [Google Scholar]
- 19. Constancia M, Kelsey G, Reik W. Resourceful imprinting. Nature 432: 53– 57, 2004. [DOI] [PubMed] [Google Scholar]
- 20. Cook RD, Weisberg S. Applied Regression Including Computing and Graphics. New York: Wiley Press, 1999. [Google Scholar]
- 21. Curley JP, Champagne FA, Bateson P, Keverne EB. Transgenerational effects of impaired maternal care on behaviour of offspring and grandoffspring. Animal Behav 75: 1551– 1561, 2008. [Google Scholar]
- 22. DeMoor MHM, Posthuma D, Hottenga JJ, Willemsen G, Boomsma D, De Geus EJC. Genome-wide linkage scan for exercise participation in Dutch sibling pairs. Eur J Hum Genet 15: 1252– 1259, 2007. [DOI] [PubMed] [Google Scholar]
- 23. Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol 23: 297– 307, 2007. [DOI] [PubMed] [Google Scholar]
- 24. Dong C, Li WD, Geller F, Lei L, Li D, Gorlova OY, Hebebrand J, Amos CI, Nicholls RD, Price RA. Possible genomic imprinting of three human obesity-related genetic loci. Am J Hum Genet 76: 427– 437, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dumke CL, Rhodes JS, Garland T, Jr, Maslowski E, Swallow JG, Wetter AC, Cartee GD. Genetic selection of mice for high voluntary wheel-running: effect on skeletal muscle glucose uptake. J Appl Physiol 91: 1289– 1297, 2001. [DOI] [PubMed] [Google Scholar]
- 26. Duselis AR, Wiley CD, O'Neill MO, Vrana PB. Genetic evidence for a maternal effect locus controlling genomic imprinting and growth. Genesis 43: 155– 165, 2005. [DOI] [PubMed] [Google Scholar]
- 27. Francis D, Diorio J, Liu D, Meaney MJ. Nongenomic transmission across generations of maternal behavior and stress response in the rat. Science 286: 1155– 1158, 1999. [DOI] [PubMed] [Google Scholar]
- 28. Friedenreich CM, Orenstein MR. Physical activity and cancer prevention: Etiologic evidence and biological mechanisms. J Nutr 132: 3456S– 3464S, 2002. [DOI] [PubMed] [Google Scholar]
- 29. Friedlander AL, Casazza GA, Horning MA, Huie MJ, Piacentini MF, Trimmer JK, Brooks GA. Training-induced alterations of carbohydrate metabolism in women: women respond differently from men. J Appl Physiol 85: 1175– 1186, 1998. [DOI] [PubMed] [Google Scholar]
- 30. Garland T., Jr Selection experiments: an under-utilized tool in biomechanics and organismal biology. In: Vertebrate Biomechanics and Evolution, edited by Bels VL, Gasc JP, Casinos A. Oxford: BIOS Scientific, 2003. [Google Scholar]
- 31. Garland T, Jr, Kelly SA. Phenotypic plasticity and experimental evolution. J Exp Biol 209: 2344– 2361, 2006. [DOI] [PubMed] [Google Scholar]
- 32. Gimsa U, Kanitz E, Otten W, Ibrahim SM. Behavior and stress reactivity in mouse strains with mitochondrial DNA variations. Ann NY Acad Sci 1153: 131– 138, 2009. [DOI] [PubMed] [Google Scholar]
- 33. Girard I, Rezende EL, Garland T., Jr Leptin levels and body composition of mice selectively bred for high voluntary activity. Physiol Biochem Zool 80: 568– 579, 2007. [DOI] [PubMed] [Google Scholar]
- 34. Girard I, Swallow JG, Carter PA, Koteja P, Rhodes JS, Garland T., Jr Maternal-care behavior and life-history traits in house mice (Mus domesticus) artificially selected for high voluntary wheel-running activity. Behav Processes 57: 37– 50, 2002. [DOI] [PubMed] [Google Scholar]
- 35. Goos LM, Ezzatian P, Schachar R. Parent-of-origin effects in attention-deficit hyperactivity disorder. Psychiatr Res 149: 1– 7, 2007. [DOI] [PubMed] [Google Scholar]
- 36. Guo Y, Shen H, Liu Y, Wang W, Xiong D, Xiao P, Liu Y, Zhao L, Recker RR, Deng H. Assessment of genetic linkage and parent-of-origin effects on obesity. J Clin Endocrinol Metab 91: 4001– 4005, 2006. [DOI] [PubMed] [Google Scholar]
- 37. Hager R, Cheverud JM, Leamy LJ, Wolf JB. Sex dependent imprinting effects on complex traits in mice. BMC Evol Biol 8: 303, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hager R, Cheverud JM, Wolf JB. Maternal effects as the cause of parent-of-origin effects that mimic genomic imprinting. Genetics 178: 1755– 1762, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hager R, Cheverud JM, Wolf JB. Change in maternal environment induced by cross-fostering alters genetic and epigenetic effects on complex traits in mice. Proc R Soc B 276: 2949– 2954, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hager R, Johnstone RA. The genetic basis of family conflict resolution in mice. Nature 421: 533– 535, 2003. [DOI] [PubMed] [Google Scholar]
- 41. Hager R, Johnstone RA. The influence of phenotypic and genetic effects on maternal provisioning and offspring weight gain in mice. Biol Lett 2: 81– 84, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hagobian TA, Sharoff CG, Stephens BR, Wade GN, Silva JE, Chipkin SR, Braun B. Effects of exercise on energy-regulating hormones and appetite in men and women. Am J Physiol Regul Integr Comp Physiol 296: R233– R242, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hanson MA, Gluckman PD. Developmental origins of health and disease: new insights. Basic Clin Pharmacol Toxicol 2: 90– 93, 2008. [DOI] [PubMed] [Google Scholar]
- 44. Henderson GC, Fattor JA, Horning MA, Faghihnia N, Johnson ML, Mau TL, Luke-Zeitoun M, Brooks GA. Lipolysis and fatty acid metabolism in men and women during the postexercise recovery period. J Physiol 584: 963– 981, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horton TJ, Pagliassotti MJ, Hobbs K, Hill JO. Fuel metabolism in men and women during and after long-duration exercise. J Appl Physiol 85: 1823– 1832, 1998. [DOI] [PubMed] [Google Scholar]
- 46. Houle-Leroy P, Garland T, Jr, Swallow JG, Guderley H. Effects of voluntary activity and genetic selection on muscle metabolic capacities in house mice Mus domesticus. J Appl Physiol 89: 1608– 1616, 2000. [DOI] [PubMed] [Google Scholar]
- 47. Inouel M, Yamamoto S, Kurahashi N, Iwasaki M, Sasazuki S, Tsugane S. Daily total physical activity level and total cancer risk in men and women: results from a large-scale population-based cohort study in Japan. Am J Epidemiol 168: 391– 403, 2008. [DOI] [PubMed] [Google Scholar]
- 48. Isles AR, Wilkinson S. Imprinted genes, cognition and behaviour. Trends Cogn Sci 4: 309– 318, 2000. [DOI] [PubMed] [Google Scholar]
- 49. Kas MJH, de Mooij-van Malsen JG, de Krom M, van Gassen KLI, van Lith HA, Olivier B, Oppelaar H, Hendriks J, de Wit M, Groot Koerkamp MJA, Holstege FCP, van Oost BA, de Graan PNE. High-resolution genetic mapping of mammalian motor activity levels in mice. Genes Brain Behav 8: 13– 22, 2009. [DOI] [PubMed] [Google Scholar]
- 50. Keverne EB, Curley JP. Epigenetics, brain evolution and behaviour. Front Neuroendocrinol 29: 398– 412, 2008. [DOI] [PubMed] [Google Scholar]
- 51. Leamy LJ, Pomp D, Lightfoot JT. An epistatic genetic basis for physical activity traits in mice. J Hered 99: 639– 646, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lerman I, Harrison BC, Freeman K, Hewett TE, Allen DL, Robbins J, Leinwand LA. Genetic variability in forced and voluntary endurance exercise performance in seven inbred mouse strains. J Appl Physiol 92: 2245– 2255, 2002. [DOI] [PubMed] [Google Scholar]
- 53. Li LL, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA. Regulation of maternal behaviour and offspring growth by paternally expressed Peg3. Science 284: 330– 333, 1999. [DOI] [PubMed] [Google Scholar]
- 54. Lightfoot JT. Sex hormones' regulation of rodent physical activity: a review. Int J Biol Sci 4: 126– 132, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lightfoot JT, Turner MJ, Daves M, Vordermark A, Kleeberger SR. Genetic influence on daily wheel running activity level. Physiol Genomics 19: 270– 276, 2004. [DOI] [PubMed] [Google Scholar]
- 56. Lightfoot JT, Turner MJ, Pomp D, Kleeberger SR, Leamy LJ. Quantitative trait loci (QTL) for physical activity traits in mice. Physiol Genomics 32: 401– 408, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Malisch JL, Breuner CW, Gomes FR, Chappell MA, Garland T., Jr Circadian pattern of total and free corticosterone concentrations, corticosteroid-bonding globulin, and physical activity in mice selectively bred for high voluntary wheel-running behavior. Gen Comp Endocrinol 156: 210– 217, 2008. [DOI] [PubMed] [Google Scholar]
- 58. Malisch JL, Breuner CW, Kolb EM, Wada H, Hannon RM, Chappell MA, Middleton KM, Garland T., Jr Behavioral despair and home-cage activity in mice with chronically elevated baseline corticosterone concentrations. Behav Genet 39: 192– 201, 2009. [DOI] [PubMed] [Google Scholar]
- 59. Manson JE, Skerrett PJ, Greenland P, VanItalie B. The escalating pandemics of obesity and sedentary lifestyle. A call to action for clinicians. Arch Intern Med 164: 249– 258, 2004. [DOI] [PubMed] [Google Scholar]
- 60. Marliss EB, Kreisman SH, Manzon A, Halter JB, Vranic M, Nessim SJ. Gender differences in glucoregulatory responses to intense exercise. J Appl Physiol 88: 457– 466, 2000. [DOI] [PubMed] [Google Scholar]
- 61. McKenzie S, Phillips SM, Carter SL, Lowther S, Gibala MJ, Tarnopolsky MA. Endurance exercise training attenuates leucine oxidation and BCOAD activation during exercise in humans. Am J Physiol Endocrinol Metab 278: E580– E587, 2000. [DOI] [PubMed] [Google Scholar]
- 62. Morison IM, Ramsay RP, Spencer HG. A census of mammalian imprinting. Trends Genet 21: 457– 465, 2005. [DOI] [PubMed] [Google Scholar]
- 63. Nehrenberg DL, Hua K, Estrada-Smith D, Garland T, Jr, Pomp D. Voluntary exercise and its effects on body composition depend on genetic selection history. Obesity 17: 1402– 1409, 2009. [DOI] [PubMed] [Google Scholar]
- 64. Pardo-Manuel de Villena F, de la Casa Esperon E, Sapienza C. Natural selection and the function of genome imprinting: beyond the silenced minority. Trends Genet 16: 573– 579, 2000. [DOI] [PubMed] [Google Scholar]
- 65. Perusse L, Tremblay A, LeBlanc C, Bouchard C. Genetic and environmental influences on level of habitual physical activity and exercise participation. Am J Epidemiol 129: 1012– 1022, 1989. [DOI] [PubMed] [Google Scholar]
- 66. Pomp D, Allan MF, Wesolowski SR. Quantitative genomics: Exploring the genetic architecture of complex trait predisposition. J Anim Sci 82: E300– E312, 2004. [DOI] [PubMed] [Google Scholar]
- 67. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; (//www.R-project.org) ( 2008). [Google Scholar]
- 68. Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet 2: 21– 32, 2001. [DOI] [PubMed] [Google Scholar]
- 69. Rezende EL, Gomes FR, Malisch JL, Chappell MA, Garland T., Jr Maximal oxygen consumption in relation to subordinate traits in lines of house mice selectively bred for high voluntary wheel running. J Appl Physiol 101: 477– 485, 2006. [DOI] [PubMed] [Google Scholar]
- 70. Seabra AF, Mendonc DM, Goring HH, Thomis MA, Maia JA. Genetic and environmental factors in familial clustering in physical activity. Eur J Epidemiol 23: 205– 211, 2008. [DOI] [PubMed] [Google Scholar]
- 71. Shimomura K, Low-Zeddies SS, King DP, Steeves TDL, Whiteley A, Kushla J, Zemenides PD, Lin A, Vitaterna MH, Churchill GA, Takahashi JS. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res 11: 959– 980, 2001. [DOI] [PubMed] [Google Scholar]
- 72. Simonen RL, Rankinen T, Perusse L, Rice T, Rao DC, Chagnon Y, Bouchard C. Genome-wide linkage scan for physical activity levels in the Quebec family study. Med Sci Sports Exerc 35: 1355– 1359, 2003. [DOI] [PubMed] [Google Scholar]
- 73. Spencer HG. Effects of genomic imprinting on quantitative traits. Genetica 136: 285– 293, 2009. [DOI] [PubMed] [Google Scholar]
- 74. Starmer J, Magnuson T. A new model for random X chromosome inactivation. Development 136: 1– 10, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Swallow JG, Carter PA, Garland T., Jr Artificial selection for increased wheel-running behavior in house mice. Behav Genet 28: 227– 237, 1998. [DOI] [PubMed] [Google Scholar]
- 76. Swallow JG, Garland T, Jr, Koteja P, Carter PA. Food consumption and body composition in mice selected for high wheel running activity. J Comp Physiol B 171: 651– 659, 2001. [DOI] [PubMed] [Google Scholar]
- 77. Swallow JG, Koteja P, Carter PA, Garland T., Jr Artificial selection for increased wheel-running activity in house mice results in decreased body mass at maturity. J Exp Biol 202: 2513– 2520, 1999. [DOI] [PubMed] [Google Scholar]
- 78. Thorvaldsen JL, Verona RI, Bartolomei MS. X-tra! X-tra! News from the mouse X chromosome. Dev Biol 298: 344– 353, 2006. [DOI] [PubMed] [Google Scholar]
- 79. Umemori J, Nishi A, Lionikas A, Sakaguchi T, Kuriki S, Blizard DA, Koide T. QTL analyses of temporal and intensity components of home-cage activity in KJR and C57BL/6J strains. BMC Genetics 29: 10– 40, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. US Department of Health and Human Services. Physical Activity and Health: a Report of the Surgeon General. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, 1996. [Google Scholar]
- 81. Vaanholt LM, Meerlo P, Garland T, Jr, Visser GH, van Dijk G. Plasma adiponectin is increased in mice selectively bred for high wheel-running activity, but not by wheel running per se. Horm Metab Res 39: 377– 383, 2007. [DOI] [PubMed] [Google Scholar]
- 82. Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 27: 363– 388, 2007. [DOI] [PubMed] [Google Scholar]
- 83. Weaver ICG, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl J, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci 7: 847– 854, 2004. [DOI] [PubMed] [Google Scholar]
- 84. Westerterp KR. Physical activity as determinant of daily energy expenditure. Physiol Behav 93: 1039– 1043, 2008. [DOI] [PubMed] [Google Scholar]
- 85. Westerterp KR, Speakman JR. Physical activity energy expenditure has not declined since the 1980s and matches energy expenditures of wild mammals. Int J Obes 32: 1256– 1263, 2008. [DOI] [PubMed] [Google Scholar]
- 86. Whitelaw NC, Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr Opin Genet Dev 18: 273– 279, 2008. [DOI] [PubMed] [Google Scholar]
- 87. Wittkopp PJ, Haerum BK, Clark AG. Parent-of-origin effects on mRNA expression in Drosophila melanogaster not caused by genomic imprinting. Genetics 173: 1817– 1821, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yang HeS, Vitaterna MH, Laposky AD, Shimomura K, Turek FW. Genetic analysis of daily physical activity using a mouse chromosome substitution strain. Physiol Genomics 39: 47– 55, 2009. [DOI] [PubMed] [Google Scholar]
- 89. Yu X, Gimsa U, Wester-Rosenlof L, Kanitz E, Otten W, Kunz M, Ibrahim SM. Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res 19: 159– 165, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.