Abstract
The genetic factors determining the magnitude of the response to exercise training are poorly understood. The aim of this study was to identify quantitative trait loci (QTL) associated with adaptation to exercise training in a cross between FVB/NJ (FVB) and C57BL/6J (B6) mice. Mice completed an exercise performance test before and after a 4-wk treadmill running program, and changes in exercise capacity, expressed as work (kg·m), were calculated. Changes in work in F2 mice averaged 1.51 ± 0.08 kg·m (94.3 ± 7.3%), with a range of −1.67 to +4.55 kg·m. All F2 mice (n = 188) were genotyped at 20-cM intervals with 103 single nucleotide polymorphisms (SNPs), and genomewide linkage scans were performed for pretraining, posttraining, and change in work. Significant QTL for pretraining work were located on chromosomes 14 at 4.0 cM [3.72 logarithm of odds (LOD)] and 19 at 34.4 cM (3.63 LOD). For posttraining work significant QTL were located on chromosomes 3 at 60 cM (4.66 LOD) and 14 at 26 cM (4.99 LOD). Suggestive QTL for changes in work were found on chromosomes 11 at 44.6 cM (2.30 LOD) and 14 at 36 cM (2.25 LOD). When pretraining work was used as a covariate, a potential QTL for change in work was identified on chromosome 6 at 68 cM (3.56 LOD). These data indicate that one or more QTL determine exercise capacity and training responses in mice. Furthermore, these data suggest that the genes that determine pretraining work and training responses may differ.
Keywords: treadmill running, genetic factors
low exercise capacity or cardiorespiratory fitness is comparable to elevated systolic blood pressure, obesity, diabetes, and smoking as a risk factor and predictor of future disease (33, 47). Improving cardiorespiratory fitness through increased physical activity can significantly reduce the risk of all-cause mortality (8), regardless of the level of initial fitness (22). However, there is a high degree of individual variation in the responses to exercise training. For example, there are some individuals who might not show an increase in maximal oxygen consumption (V˙o2max) in response to endurance training (9, 12). Consequently, identifying the genetic factors modulating the adaptations to exercise may provide insight into individual differences in responses to training. However, the genetic factors determining the magnitude of the response to exercise are poorly understood.
Initial studies investigating the genetic basis for exercise capacity and training responses focused on familial resemblance and heritability of performance phenotypes. Results from cross-sectional, twin, and prospective studies indicate that the heritability for training-induced changes in V˙o2max and submaximal power output ranges from 25% to 50% (11). Heritability estimates from family studies also vary between 25% and 50%, depending on sample size (10, 13, 48). In the HERITAGE Family Study, heritability for V˙o2max training responses was estimated to be 47% on the basis of data from 98 two-generation families (9). In subsequent publications from the HERITAGE Family Study, linkage analysis was used to identify chromosomal regions affecting variation in V˙o2max in the sedentary state as well as training responses in V˙o2max (13, 53).
More recently, animal models have been utilized to identify the genetic basis for exercise and exercise training responses. Strain-dependent differences in intrinsic or pretraining exercise capacity measured during a graded treadmill test have been reported for inbred mice and rats (2, 34, 36, 62). In a screen of several inbred mouse strains, Lerman et al. (34) reported that maximal speed achieved during a graded treadmill test was highest in FVB/NJ (FVB) mice and lowest in C57BL/6J (B6) mice. Using a similar protocol, Lightfoot et al. (36) also found that B6 mice had relatively low intrinsic exercise capacity, whereas Balb/cJ mice had high intrinsic exercise capacity. Given these strain-dependent differences, quantitative trait locus (QTL) mapping has been used to identify QTL for intrinsic exercise capacity in rats and mice (37, 65). In contrast, less is known about the change in exercise capacity with training across inbred strains of rodents (30, 42, 63). Large differences were reported for training responses across several inbred strains of rats, and these strain differences persisted whether training was performed at the same relative or absolute workload (30, 63). Using a mouse model of exercise training, Massett and Berk (42) reported that the change in exercise capacity with training varied significantly across inbred (B6, Balb/cJ, and FVB) and hybrid mouse strains. Interestingly, FVB and Balb/cJ mice had similarly high levels of intrinsic exercise capacity but disparate training responses. Estimated broad-sense heritability for posttraining values for distance run in mice ranged from 47% to 65%, suggesting that the variance in adaptation responses to exercise in mice was significantly influenced by genotypic variance. Collectively, these data indicate that variation in both intrinsic exercise capacity and exercise training responses depends, in part, on genotype. Therefore, the aim of this study was to identify QTL affecting changes in exercise capacity, defined as work, after 4 wk of treadmill training in F2 progeny derived from B6 and FVB mice. QTL analysis was also performed to determine loci influencing pretraining and posttraining exercise capacity in mice.
METHODS
Animals.
All procedures adhered to the established National Institutes of Health guidelines for the care and use of laboratory animals and were approved by the University Committee on Animal Resources at the University of Rochester. Standard inbred B6 and FVB mice were obtained from Jackson Laboratories (Bar Harbor, ME). Female FVB mice were mated to male B6 mice to generate F1 offspring. Nineteen F1 breeder pairs were used in an intercross breeding scheme to generate 188 (FVB × B6) F2 mice. This required 22 litters of F2 offspring. All mice were housed in the same room in the vivarium, maintained on a 12:12-h light-dark schedule, and allowed food and water ad libitum. With R/qtlDesign (58), 188 F2 mice are sufficient to detect at least 1 QTL accounting for 10% of the variance in either posttraining work or change in work with a power of 80%.
Exercise performance test.
After 2 days of familiarization with the treadmill, mice completed a graded treadmill run to exhaustion on a motorized rodent treadmill with an electric grid at the rear of the treadmill (Columbus Instruments, Columbus, OH) as described previously (42). Mice performed a 9-min warm-up by walking on the treadmill at 9 m/min and 0° grade. Speed was then increased by 2.5 m/min every 3 min from a starting speed of 10 m/min to a maximum of 40 m/min. The incline progressively increased 5° every 9 min to a maximum of 15° (20, 42). When exhaustion was determined, defined as an inability to maintain running speed despite repeated contact with the electric grid, each mouse was immediately removed from the treadmill and returned to its home cage. Maximal exercise capacity was estimated from each run to exhaustion trial with three parameters: the duration of the run (in min), the distance run (in m), and vertical work performed (in kg·m) (2, 42). In our laboratory, this protocol yields a within-mouse coefficient of variation for work of 10 ± 1% for two tests conducted within 72 h by the same individual (M. P. Massett, unpublished observations).
Exercise training.
Exercise training responses were determined in ∼8-wk-old male (n = 96) and female (n = 92) (FVB × B6) F2 mice as described previously (42). Briefly, male and female mice completed an exercise performance test before and after a 4-wk treadmill running program. The exercise training program consisted of treadmill running 5 days/wk, 60 min/day at a final intensity of 19 m/min up a 10° incline on one of two six-lane rodent treadmills. Treadmill assignment was rotated each day, and lane assignment was randomized for each mouse. Male and female mice from parental B6 (n = 8 male, n = 4 female) and FVB (n = 5 male, n = 6 female) strains were exercise trained concurrently and showed training responses similar to those previously described. A minimal number of sedentary F2 mice were utilized as time controls. These mice were exposed to the treadmill but not made to run and showed no changes in exercise performance.
Genotyping.
Approximately 24 h after the final exercise performance test, heart, soleus, plantaris, and gastrocnemius muscle, and liver samples were harvested from all mice, washed in ice-cold (4°C) saline, frozen in liquid nitrogen, and stored at −80°C for future analyses (42). DNA was extracted from liver samples by the University of Rochester Functional Genomics Center, and genotyping was performed with a competitive allele-specific PCR single nucleotide polymorphism (SNP) genotyping system (KBiosciences, Hoddesdon, UK) (51, 52). All 188 F2 mice were genotyped at 20-cM intervals with SNP markers from a published panel (51, 52).
QTL analysis.
One-dimensional genomewide linkage scans were conducted using R/qtl to identify QTL with main effects (16). Permutation tests (1,000 repetitions) were used to calculate experiment-specific threshold values for logarithm of odds (LOD) scores and determine the significance of linkage between marker genotype and phenotype (17). LOD scores ≥ 3.5 were considered significant (P < 0.05), and those ≥ 2.1 were considered suggestive (P < 0.63). QTL confidence intervals were determined with the 1.5-LOD support interval (15). For each exercise phenotype, potential covariates were determined by stepwise regression analysis (SPSS 16.0 statistical software, Chicago, IL). These covariates were then included as additive and interacting covariates in single genomewide linkage scans. Differences in LOD scores (ΔLOD) ≥ 2.0 between scans that included additive and interacting covariates were considered significant (31). Physiological variables included as covariates for posttraining work were pretraining work (Pearson correlation: r2 = 0.23, P < 0.01) and heart weight in milligrams (r2 = 0.07, P < 0.01), and for change in work soleus muscle weight in milligrams (r2 = 0.07, P < 0.01) was included as a covariate. Because pretraining work varied considerably across F2 animals and was significantly different between parental strains, pretraining work was included as a covariate for single genomewide linkage scans for change in work. The correlation between pretraining work and change in work was relatively small (r2 = 0.03) but significant (P < 0.05). No measured physiological variables were significantly related to pretraining work as determined by stepwise regression analysis. Separate genomewide linkage scans were conducted with sex as an additive and interacting covariate. However, subsequent analyses were not performed on male and female mice separately because there were insufficient progeny per genotype to conduct an analysis with sufficient power (19). To determine the contribution of each QTL and associated covariates to each exercise phenotype, multiple regression analysis was used. All significant and suggestive QTL and covariates were included in the analyses, and individual terms were dropped until all remaining variables were significant (P < 0.05). Separate analyses were run for each exercise phenotype. The percent variance ascribed to each term was determined from this analysis.
Statistical analysis.
Values are expressed as means ± SE. Statistical significance was set at P < 0.05. Comparisons among strains and across genotypes for effect plots were made with analysis of variance followed by Tukey's post hoc comparisons. Pre- vs. posttraining comparisons within a strain were made with paired Student's t-tests. Each exercise phenotype was tested for normal distribution with the Kolmogorov-Smirnov test. Broad-sense heritability was calculated with the following formula: H2 = [VF2 − (1/2VF1 + 1/4VP1 + 1/4VP2)]/VF2, where VP1 and VP2 are the variances of the isogenic parental strains and VF1 and VF2 are the variances of the F1 and F2 offspring (25, 35).
RESULTS
Exercise capacity in parental strains and (FVB × B6) F2 mice.
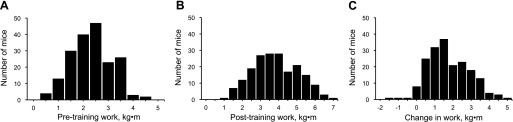
(FVB × B6) F2 mice were an average of 59.0 ± 0.2 days old at the start of the study. Exercise performance phenotypes in F2 mice had a wide range of values and were normally distributed (Fig. 1). Body weight and exercise performance test results are shown in Table 1. There were no significant differences in body weight between B6 and FVB mice. F2 mice (96 male, 92 female) were significantly heavier than B6 and FVB mice before training. After training F2 mice were significantly heavier than B6 mice but not different from FVB mice. Within each group, body weight increased significantly from pretraining to posttraining. However, changes in body weight were not significantly different across groups, although B6 mice gained less weight (0.6 ± 0.2 g) than FVB (1.8 ± 0.6 g) and F2 (1.3 ± 0.1 g) mice. Pretraining, posttraining and change in work varied significantly across groups (Table 1). Pretraining work was significantly different across the three groups, with FVB mice having the highest and B6 mice the lowest values for intrinsic exercise capacity. Four weeks of exercise training significantly increased exercise capacity in all groups. Posttraining work was significantly different across all groups as well. B6 mice had significantly lower posttraining work and FVB mice had significantly higher posttraining work compared with the other groups. Change in work was similar between FVB and F2 mice and significantly lower in B6 mice compared with the other groups. Broad-sense heritability was 0.37 for pretraining work, 0.70 for posttraining work, and 0.53 for change in work. These values are similar to those reported by Lightfoot et al. (38) for voluntary wheel running in an F2 population of mice.
Fig. 1.
Frequency distribution of pretraining work (A), posttraining work (B), and change in work (C) in 188 [FVB/NJ (FVB) × C57BL/6J (B6)] F2 mice. F2 mice performed a graded exercise test to exhaustion before and after 4 wk of exercise training.
Table 1.
Strain comparison of body weight and work before and after exercise training
| Body Weight, g |
Work, kg·m |
|||||
|---|---|---|---|---|---|---|
| Strain | n | Pretraining | Posttraining | Pretraining | Posttraining | Change |
| B6 | 12 | 21.7±0.6 | 22.3±0.5† | 1.31±0.15† | 1.65±0.20† | 0.34±0.12* |
| FVB | 11 | 21.8±0.5 | 23.6±1.1 | 2.88±0.16* | 4.66±0.23* | 1.78±0.17 |
| (FVB×B6)F2 | 188 | 24.6±0.3* | 25.9±0.3 | 2.08±0.06 | 3.59±0.09 | 1.51±0.08 |
Values are means ± SE for n mice. C57BL/6J (B6): n = 8 males, 4 females; FVB/NJ (FVB): n = 5 males, 6 females; F2: n = 96 males, 92 females.
P < 0.05 vs. all other strains;
P < 0.05 vs. F2.
QTL analysis.
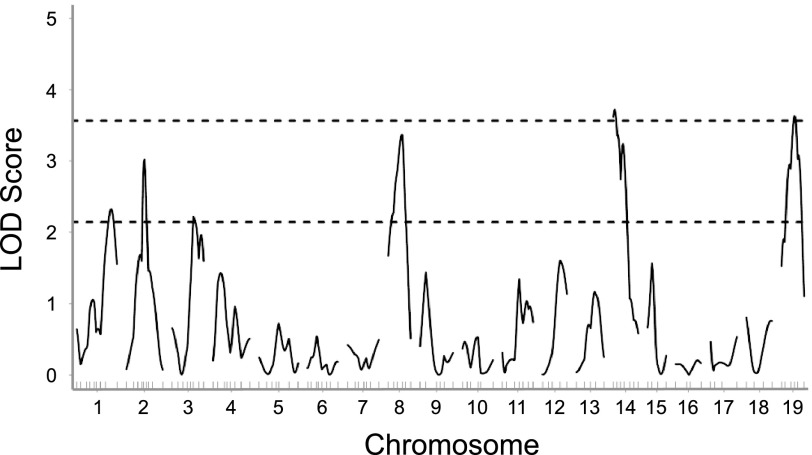
All 188 (FVB × B6) F2 mice were genotyped, and QTL analysis was performed with pretraining, posttraining, and change in work as phenotypes (Figs. 2–4). Significant QTL for pretraining work were identified on chromosomes 14 and 19 (Fig. 2; Table 2). These QTL are designated as endurance exercise QTL 3 and 4 (Eeq3 and Eeq4), respectively, because QTL for a similar phenotype in mice have been designated as Eeq1 and Eeq2 (37). Suggestive QTL were identified on chromosomes 1, 2, 3, and 8, with the QTL on chromosome 8 surpassing a LOD score of 3.0. For the peak marker on chromosome 14, mice homozygous for FVB alleles (FF, 2.4 ± 0.1 kg·m) had significantly greater (P < 0.05) pretraining work compared with mice carrying homozygous B6 alleles (BB, 1.8 ± 0.1 kg·m) and heterozygous (FB, 2.0 ± 0.1 kg·m) mice, suggesting a recessive inheritance. Conversely, homozygous FF mice had significantly lower (P < 0.05) pretraining work (1.7 ± 0.1 kg·m) compared with BB mice (2.2 ± 0.1 kg·m) for the peak marker on chromosome 19; heterozygous FB mice (2.2 ± 0.1 kg·m) were not different from BB mice. This locus showed dominant inheritance, although the increasing allele came from the B6 strain.
Fig. 2.
Genomewide linkage scan for pretraining work in 188 (FVB × B6) F2 progeny. Significant and suggestive logarithm of odds (LOD) thresholds are represented by top (LOD = 3.57) and bottom (LOD = 2.14) horizontal lines, respectively. LOD thresholds were determined by permutation testing using 1,000 permutations.
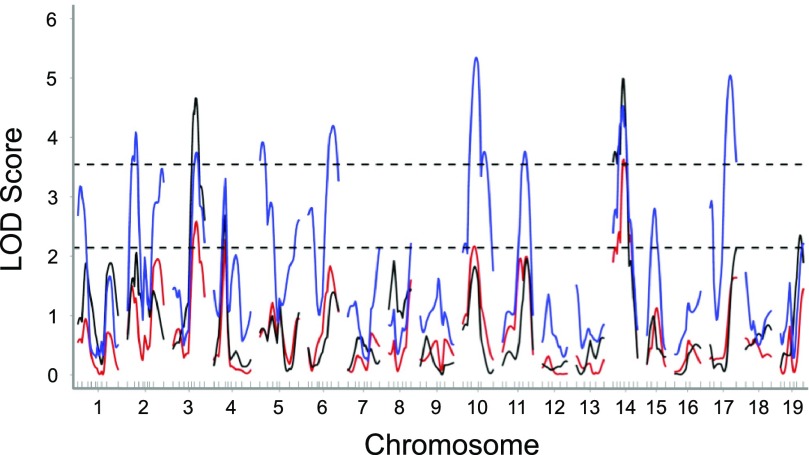
Fig. 3.
Effect of pretraining work and heart weight as covariates on genomewide linkage scans for posttraining work after 4 wk of exercise training in (FVB × B6) F2 mice. Black line, genomewide linkage scan for posttraining work with no covariates; red line, genomewide linkage scan for posttraining work with pretraining work and heart weight as additive covariates; blue line, genomewide linkage scan for posttraining work with pretraining work and heart weight as interacting covariates. Significant and suggestive LOD thresholds are represented by top (LOD = 3.54) and bottom (LOD = 2.14) horizontal lines, respectively. LOD thresholds were determined by permutation testing using 1,000 permutations with no covariate. Differences in LOD scores (ΔLOD) ≥ 2.0 were found on chromosomes 1, 2, 5, 6, 10, and 17 with interacting covariates.
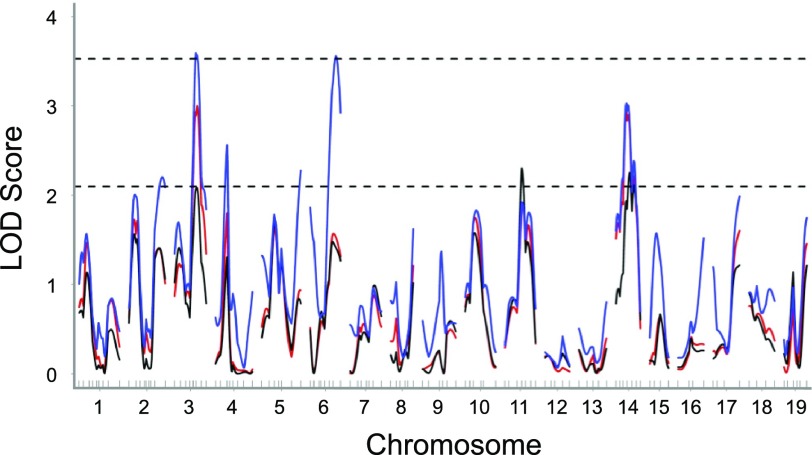
Fig. 4.
Effect of pretraining work as a covariate on genomewide linkage scans for change in work after 4 wk of exercise training in (FVB × B6) F2 mice. Black line, genomewide linkage scan for change in work with no covariates; red line, genomewide linkage scan for change in work with pretraining work as an additive covariate; blue line, genomewide linkage scan for change in work with pretraining work as an interacting covariate. Significant and suggestive LOD thresholds are represented by top (LOD = 3.53) and bottom (LOD = 2.10) horizontal lines, respectively. LOD thresholds were determined by permutation testing using 1,000 permutations. ΔLOD ≥ 2.0 was found on chromosome 6 with pretraining as an interacting covariate.
Table 2.
Significant and suggestive QTL for pre- and posttraining work and changes in work
| Work, kg·m | Chr | Position, cM | 1.5 LOD, cM | LOD | P Value | Nearest Marker |
|---|---|---|---|---|---|---|
| Pretraining | 1 | 92.0 | 2.32 | 0.501 | rs4222922 | |
| 2 | 48.0 | 3.02 | 0.159 | rs4223268 | ||
| 3 | 56.9 | 2.22 | 0.572 | rs3687177 | ||
| 8 | 36.0 | 3.36 | 0.079 | rs3089148 | ||
| 14 | 4.0 | 0–38 | 3.72* | 0.039 | rs3689508 | |
| 19 | 34.4 | 10–56 | 3.63* | 0.040 | rs3679049 | |
| Posttraining | 3 | 60.0 | 46–76 | 4.66* | 0.002 | rs3687177 |
| 4 | 30.6 | 2.69 | 0.272 | rs3667625 | ||
| 14 | 26.0 | 0–38 | 4.99* | 0.000 | rs3660830 | |
| 17 | 70.3 | 2.14 | 0.626 | rs3023460 | ||
| 19 | 52.0 | 2.35 | 0.454 | rs3023517 | ||
| Change | 11 | 44.6 | 2.30 | 0.480 | rs3023267 | |
| 14 | 36.0 | 2.25 | 0.514 | rs3660830 |
QTL, quantitative trait locus; Chr, chromosome; LOD, peak logarithm of odds score obtained in interval mapping; 1.5 LOD, support interval of 1.5 LOD drop in centimorgans obtained from interval mapping; nearest marker, single nucleotide polymorphism (SNP) marker closest to the LOD peak.
Significant (P < 0.05) QTL.
QTL for posttraining work were found on chromosomes 3, 4, 14, 17, and 19, with those on chromosome 3 (exercise training QTL Etq1) and 14 (Etq2) surpassing the significance threshold (Fig. 3). Genomewide linkage scans for posttraining work with pretraining work and heart weight as covariates identified additional QTL on chromosomes 1, 2, 5, 6, 10, and 17 (ΔLOD ≥ 2.0) (Fig. 3). The genotypes for the SNP markers closest to the peak on chromosome 14 showed dominant inheritance, with mice homozygous for FVB alleles (FF mice; 3.8 ± 0.2 kg·m) having posttraining work values similar to FB mice (3.8 ± 0.1 kg·m) and significantly greater (P < 0.05) than BB mice (2.9 ± 0.2 kg·m). In contrast, for the marker on chromosome 3, the increasing allele came from the B6 strain (4.0 ± 0.2 kg·m) and the inheritance was dominant; posttraining values were similar to those for heterozygous F2 mice (3.7 ± 0.1 kg·m) and significantly greater (P < 0.05) than those for FF mice (2.9 ± 0.2 kg·m).
For change in work, suggestive QTL were identified on chromosomes 11 and 14 (Fig. 4). Soleus weight as a covariate had no influence on any QTL for change in work. When pretraining work was included as an additive covariate, QTL peaks on chromosomes 3 and 14 were increased to LOD scores above 3.0 (Fig. 4). A novel QTL peak on chromosome 6 (ΔLOD ≥ 2.0) was identified when pretraining work was included as an interactive covariate (Fig. 4).
Regression analysis was used to identify the contribution of significant and suggestive QTL and physiological traits to each exercise performance phenotype. For pretraining work, significant QTL on chromosomes 14 and 19 explained 8.5% and 4.6% of total variance, respectively. In the final model, significant and suggestive QTL combined accounted for 29% of the total variance. Multiple regression models for posttraining work and change in work are shown in Table 3. The majority of the loci identified with pretraining work and heart weight as covariates in genomewide scans dropped out in regression analysis, yet the final model explained 38.7% of the total variance (Table 3). Of the QTL included in the final model, the QTL on chromosome 3 contributed the largest variance (9.6%), followed by the QTL on chromosome 14 (6.5%). Pretraining work accounted for the largest percentage of the total variance (22.1%). For change in work, a model including pretraining work as a covariate and significant and suggestive QTL accounted for ∼17% of the total variance (Table 3). Pretraining work accounted for 3.2% of the variance, whereas QTL on chromosomes 3 and 6 contributed 5.4% and 3.5%, respectively, to the total variance. Sex was also used as an additive and interacting covariate for each exercise performance phenotype. The majority of the significant (defined as ΔLOD ≥ 2.0) QTL identified with sex as an additive and interacting covariate were very similar to the QTL identified with other covariates, and most were not included in the final regression model (Supplemental Table S1).1
Table 3.
Multiple regression model for posttraining work and changes in work
| Work, kg·m | Nearest Marker | Covariate | LOD | % Variance | P Value |
|---|---|---|---|---|---|
| Posttraining | Pretraining work, kg·m | 22.1 | 3.35E-07 | ||
| Heart weight, mg | 6.6 | 0.0003 | |||
| rs3687177 | Chr 3 @ 60 cM | 4.66 | 9.6 | 0.002 | |
| rs3667625 | Chr 4 @ 30.6 cM | 2.69 | 3.6 | 0.039 | |
| rs4225021 | Chr 5 @ 6 cM | 3.92* | 1.1 | 0.06 | |
| rs3717445 | Chr 10 @ 41.4 cM | 5.35* | 2.9 | 0.029 | |
| rs3660830 | Chr 14 @ 26 cM | 4.99 | 6.5 | 0.021 | |
| Model | 38.7 | 0.000 | |||
| Change | Pretraining work, kg·m | 3.2 | 0.002 | ||
| rs3687177 | Chr 3 @ 56 cM | 3.59* | 5.4 | 0.001 | |
| rs3727110 | Chr 6 @ 68 cM | 3.56* | 3.5 | 0.016 | |
| rs3023267 | Chr 11 @ 45 cM | 2.30 | 1.9 | 0.05 | |
| rs3660830 | Chr 14 @ 36 cM | 3.03* | 1.3 | 0.04 | |
| Model | 17.2 | 0.0001 |
LOD, peak LOD score obtained in interval mapping; % variance, % of phenotypic variance accounted for by variable; nearest marker, SNP marker closest to the LOD peak.
LOD score is from genomewide scan with covariate(s).
DISCUSSION
There is a high degree of variation in adaptation to exercise training, such that some individuals might not improve their exercise capacity in response to training. Although there is a genetic component influencing the response to training, the genetic factors determining the magnitude of the response to exercise training are poorly understood. With the use of QTL analysis for a mouse model of exercise training, novel significant QTL were identified for posttraining exercise capacity, defined as work, on chromosomes 3 and 14. Suggestive QTL for the change in work with exercise training were identified on chromosomes 11 and 14. These QTL represent novel regions of the mouse genome related to the response to exercise training. Significant and suggestive QTL were also identified for pretraining work on chromosomes 14 and 19. QTL for the three exercise phenotypes were found on chromosome 14, suggesting that genes on this chromosome may be associated with the ability to exercise. There was no overlap among the other QTL for pretraining work (Chr 19), posttraining work (Chr 3), and change in work (Chr 11), implying that the genes that determine pretraining work and training responses might differ.
Pretraining work.
Previous studies in mice, rats, and humans demonstrated that intrinsic exercise capacity is determined, in part, by genetic factors (13, 37, 53, 65). In the present study, (FVB × B6) F2 mice completed a graded exercise test to exhaustion before exercise training, and significant QTL for pretraining work were identified on chromosomes 14 and 19. Lightfoot et al. (37) used a similar exercise protocol to screen several strains of mice for endurance exercise performance. This group identified significant QTL on chromosomes 8 and X in an F2 cross between Balb/cJ and DBA/2J mouse strains. The confidence interval for the QTL on chromosome 8 was relatively large but overlaps with a suggestive QTL for pretraining work identified in the present study (Table 2). Ways et al. (65) performed a genomewide linkage scan for intrinsic aerobic exercise capacity in an F2 population derived from Copenhagen and DA rat strains. A significant QTL was identified on chromosome 16 (D16Rat17), and two suggestive QTL localized to chromosomes 3 (D3Rat56) and 16 (D16Rat55). The two QTL on rat chromosome 16 are homologous to regions of mouse chromosome 8, overlapping with QTL reported in the present study (pretraining work) and by Lightfoot et al. (37) (Supplemental Fig. S1). Several linkage markers for V˙o2max in the sedentary state in humans (13, 53) also fall within the 1.5 LOD intervals for pretraining work (Supplemental Fig. S1), suggesting potential concordance among mouse, rat, and human QTL.
Training responses.
Although intrinsic exercise capacity has been measured in a wide range of inbred strains, little data is available for training responses across mouse strains. Previously, we reported (42) significant differences in posttraining work and change in work among B6, Balb/cJ, and FVB mice after 4 wk of exercise training on a treadmill. On the basis of our previous finding and the common use of B6 and FVB strains in generating genetically modified mice, these strains were chosen for QTL analysis of exercise training responses. In the present study, (FVB × B6) F2 mice completed a similar exercise training program and showed a wide variation in posttraining work and change in work (Table 1; Fig. 1). Linkage analysis revealed two novel significant QTL for posttraining work on chromosomes 3 and 14. In the final regression model, these loci explained ∼9.6% and 6.5% of the variance in posttraining work (Table 3). Genomewide scans for change in work identified suggestive QTL on chromosomes 11 and 14. These QTL contributed <2% each of the total variance in the response to exercise training in the final multiple regression model. However, QTL peaks on chromosomes 3, 6, and 14 were identified with the addition of pretraining work as a covariate. The QTL on chromosomes 3 and 14 overlap with Etq1 and Etq2, whereas the locus on chromosome 6 was not previously identified. Several peak markers for QTL related to changes in V˙o2max or mean power output (MPO) identified in the HERITAGE Family Study fall within the 1.5 LOD intervals for QTL for posttraining work (13, 53) (Supplemental Fig. S1). Unfortunately, only peak markers are reported, so the degree of overlap between mouse and human exercise training-related QTL cannot be determined from the present data. However, the finding that several of these markers for QTL for training responses in V˙o2max and MPO in humans map to locations homologous to QTL regions identified in the present study suggests potential concordance between mouse and human exercise training QTL.
Earlier reports suggested that the genes influencing variation in pretraining exercise capacity are different from those determining the responses to training (9, 13, 53). A comparison of significant and suggestive QTL for pretraining work and change in work identified in the present study support this concept. There is minimal overlap between QTL for pretraining work and change in work with or without covariates (Figs. 2 and 3A; Tables 2 and 3). In contrast, there is some overlap between pre- and posttraining and considerable overlap between posttraining and change in work QTL. This is not surprising given the strong relationship between posttraining work and either pretraining work (r = 0.47, P < 0.01) or the change in work (r = 0.78, P < 0.01). However, the significant QTL for pretraining work on chromosome 19 was not present in posttraining work, and one new QTL on chromosome 3 was identified in the scan for posttraining work, suggesting that the effect of some gene or genes on exercise capacity is modified (increased or decreased) by exercise training.
Exercise performance is a polygenic trait determined by multiple genes each having a small effect on the phenotype. Each of the QTL detected in this study explains ≤10% of the variation in the exercise performance phenotypes. The percent variances attributed to the QTL in the present study are comparable to those for blood pressure (21, 61), circadian behaviors (59), and blood parameters (49) in similarly sized F2 populations. Although larger F2 sample sizes can improve the power to detect QTL of small to medium effects (4), some balance is required between phenotyping large populations and QTL discovery (38). In addition, the finding that there is some overlap of QTL for intrinsic exercise capacity in independent crosses (present study and Ref. 37) as well as the agreement across mouse, rat, and human studies for exercise and exercise training QTL (Supplemental Fig. S1) support the validity of the QTL identified in the present study (1).
Candidate genes.
Association studies in humans have demonstrated to varying degrees that polymorphisms in genes for endurance phenotypes [i.e., angiotensin-converting enzyme (ACE), actinin, α3 (ACTN3), creatine kinase, muscle (CKM), hypoxia-inducible factor 1, α (HIF1A), nitric oxide synthase 3 (NOS3), peroxisome proliferator-activated receptor, γ, coactivator 1, α (PPARGC1A)] influence endurance performance or training responses (14). Transgenic or knockout mice have been generated to alter the expression level of genes already known to be relevant to acute or chronic exercise. In most cases, deletion or overexpression of genes globally or in heart or skeletal muscle results in marked changes in exercise performance or responses to training (23, 26, 29, 41, 45, 46, 64). Yet, based on searches of NCBI and MGI databases, few, if any of these well-known genes associated with exercise or exercise training were located in the QTL regions for Etq1 or Etq2. However, a number of genes are found on chromosomes 3 and 14 that are related to cardiac or skeletal muscle contraction or signaling. These genes include calsequestrin 2 (Casq2) (54), epidermal growth factor (Egf) (27), and gap junction membrane channel protein α5 (Gja5 or Cx40) (3) on chromosome 3 and myosin, heavy polypeptide (Myh6) (28), annexin A7 (Anxa7) (56), and bone morphogenic protein 4 (Bmp4) (24, 39) on chromosome 14. All but one of these genes (Bmp4) contain multiple SNPs that are polymorphic between B6 and FVB strains, and two, Casq2 and Anxa7, contain SNPs that are classified as coding nonsynonymous SNPs. Abnormalities in Casq2 genes are associated with exercise-induced ventricular arrhythmias (55). Anxa7-deficient mice exhibit disturbances in electrical conduction in the heart and increased incidence of ventricular tachycardia (56). Anxa7 has also been linked to excitation-contraction coupling in skeletal muscle and is redistributed from the plasma membrane to the cytoplasm in muscular dystrophy (6).
In the final multiple regression model for posttraining work heart weight was included as a covariate. Heart weight accounted for ∼7% of the variance in posttraining work. We reported previously (42) that exercise-trained FVB mice exhibited higher heart weights and higher ratios of heart weight to body weight than sedentary control mice. This cardiac hypertrophy was not observed in exercise-trained B6 mice compared with their sedentary controls (42). Therefore, genes related to cardiac structure and function are logical candidates for posttraining work QTL. However, despite the common themes among a number of genes found in the intervals for Etq1 and Etq2, it is premature to identify any as true candidate genes because of the large number of genes contained in these QTL regions.
In contrast to posttraining work, the suggestive QTL for change in work contain genes more closely associated with exercise and exercise training. The QTL on mouse chromosome 11 contains angiotensin I-converting enzyme (Ace), sarcoglycan, δ (Sgcd) (18), and nitric oxide synthase 2 (Nos2). The potential QTL interval on chromosome 6 includes several genes associated with maintenance of blood glucose levels and obesity. Among these genes are ankyrin repeat domain 26 (Ankrd26) (5), calcium channel, voltage-dependent, L type, α1C subunit (Cacna1c) (57, 60), and adiponectin receptor 2 (Adipor2) (7, 66), which have all been linked to regulation of insulin and blood glucose levels. Obesity-related genes include peroxisome proliferator-activated receptor γ (Pparg) (32, 43), arachidonate 5-lipoxygenase (Alox5) (44), and histamine receptor H1 (Hrh1) (40), which may regulate fat mass. Polymorphic SNPs have been identified in most of these genes for the parental FVB and B6 strains, except Nos2 and Ace. The suggestive QTL for the change in work identified in the present study contained 500 or more genes per interval, which is expected for an initial genomewide linkage scan (50). Therefore, additional efforts to narrow these regions are required before true candidate genes can be identified.
GRANTS
This work was supported by funds from National Heart, Lung, and Blood Institute (NHLBI) Grant HL-085918, an American Heart Association Scientist Development Grant, an American Physiological Society Research Career Enhancement Award to M. P. Massett, and NHLBI Grant HL-62826 to B. C. Berk.
DISCLOSURES
No conflicts of interest are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Amy Mohan, Sarah McCarty, and Dr. Slava Korshunov for help with the animal experiments and Hong Liu for help with DNA extraction.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Complex Trait Consortium The nature and identification of quantitative trait loci: a community's view. Nat Rev Genet 4: 911–916, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbato JC, Koch LG, Darvish A, Cicila GT, Metting PJ, Britton SL. Spectrum of aerobic endurance running performance in eleven inbred strains of rats. J Appl Physiol 85: 530–536, 1998. [DOI] [PubMed] [Google Scholar]
- 3. Bastide B, Neyses L, Ganten D, Paul M, Willecke K, Traub O. Gap junction protein connexin40 is preferentially expressed in vascular endothelium and conductive bundles of rat myocardium and is increased under hypertensive conditions. Circ Res 73: 1138–1149, 1993. [DOI] [PubMed] [Google Scholar]
- 4. Beavis WD. The power and deceit of QTL experiments: lessons from comparative QTL studies. In: Corn and Sorghum Research Conference. Washington, DC: American Seed Trace Association, 1994, p. 252–268. [Google Scholar]
- 5. Bera TK, Liu XF, Yamada M, Gavrilova O, Mezey E, Tessarollo L, Anver M, Hahn Y, Lee B, Pastan I. A model for obesity and gigantism due to disruption of the Ankrd26 gene. Proc Natl Acad Sci USA 105: 270–275, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berchtold MW, Brinkmeier H, Muntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiol Rev 80: 1215–1265, 2000. [DOI] [PubMed] [Google Scholar]
- 7. Bjursell M, Ahnmark A, Bohlooly YM, William-Olsson L, Rhedin M, Peng XR, Ploj K, Gerdin AK, Arnerup G, Elmgren A, Berg AL, Oscarsson J, Linden D. Opposing effects of adiponectin receptors 1 and 2 on energy metabolism. Diabetes 56: 583–593, 2007. [DOI] [PubMed] [Google Scholar]
- 8. Blair SN, Kohl HW, 3rd, Barlow CE, Paffenbarger RS, Jr, Gibbons LW, Macera CA. Changes in physical fitness and all-cause mortality. A prospective study of healthy and unhealthy men. JAMA 273: 1093–1098, 1995. [PubMed] [Google Scholar]
- 9. Bouchard C, An P, Rice T, Skinner JS, Wilmore JH, Gagnon J, Perusse L, Leon AS, Rao DC. Familial aggregation of V˙o2max response to exercise training: results from the HERITAGE Family Study. J Appl Physiol 87: 1003–1008, 1999. [DOI] [PubMed] [Google Scholar]
- 10. Bouchard C, Daw EW, Rice T, Perusse L, Gagnon J, Province MA, Leon AS, Rao DC, Skinner JS, Wilmore JH. Familial resemblance for V˙o2max in the sedentary state: the HERITAGE family study. Med Sci Sports Exerc 30: 252–258, 1998. [DOI] [PubMed] [Google Scholar]
- 11. Bouchard C, Dionne FT, Simoneau JA, Boulay MR. Genetics of aerobic and anaerobic performances. Exerc Sport Sci Rev 20: 27–58, 1992. [PubMed] [Google Scholar]
- 12. Bouchard C, Rankinen T. Individual differences in response to regular physical activity. Med Sci Sports Exerc 33: S446–S453, 2001. [DOI] [PubMed] [Google Scholar]
- 13. Bouchard C, Rankinen T, Chagnon YC, Rice T, Perusse L, Gagnon J, Borecki I, An P, Leon AS, Skinner JS, Wilmore JH, Province M, Rao DC. Genomic scan for maximal oxygen uptake and its response to training in the HERITAGE Family Study. J Appl Physiol 88: 551–559, 2000. [DOI] [PubMed] [Google Scholar]
- 14. Bray MS, Hagberg JM, Perusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc 41: 35–73, 2009. [DOI] [PubMed] [Google Scholar]
- 15. Broman KW. Review of statistical methods for QTL mapping in experimental crosses. Lab Anim (NY) 30: 44–52, 2001. [PubMed] [Google Scholar]
- 16. Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890, 2003. [DOI] [PubMed] [Google Scholar]
- 17. Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coral-Vazquez R, Cohn RD, Moore SA, Hill JA, Weiss RM, Davisson RL, Straub V, Barresi R, Bansal D, Hrstka RF, Williamson R, Campbell KP. Disruption of the sarcoglycan-sarcospan complex in vascular smooth muscle: a novel mechanism for cardiomyopathy and muscular dystrophy. Cell 98: 465–474, 1999. [DOI] [PubMed] [Google Scholar]
- 19. Curtsinger JW. Sex specificity, life-span QTLs, and statistical power. J Gerontol A Biol Sci Med Sci 57: B409–B414, 2002. [DOI] [PubMed] [Google Scholar]
- 20. Desai KH, Sato R, Schauble E, Barsh GS, Kobilka BK, Bernstein D. Cardiovascular indexes in the mouse at rest and with exercise: new tools to study models of cardiac disease. Am J Physiol Heart Circ Physiol 272: H1053–H1061, 1997. [DOI] [PubMed] [Google Scholar]
- 21. DiPetrillo K, Tsaih SW, Sheehan S, Johns C, Kelmenson P, Gavras H, Churchill GA, Paigen B. Genetic analysis of blood pressure in C3H/HeJ and SWR/J mice. Physiol Genomics 17: 215–220, 2004. [DOI] [PubMed] [Google Scholar]
- 22. Erikssen G, Liestol K, Bjornholt J, Thaulow E, Sandvik L, Erikssen J. Changes in physical fitness and changes in mortality. Lancet 352: 759–762, 1998. [DOI] [PubMed] [Google Scholar]
- 23. Fewell JG, Osinska H, Klevitsky R, Ng W, Sfyris G, Bahrehmand F, Robbins J. A treadmill exercise regimen for identifying cardiovascular phenotypes in transgenic mice. Am J Physiol Heart Circ Physiol 273: H1595–H1605, 1997. [DOI] [PubMed] [Google Scholar]
- 24. Frank DB, Abtahi A, Yamaguchi DJ, Manning S, Shyr Y, Pozzi A, Baldwin HS, Johnson JE, de Caestecker MP. Bone morphogenetic protein 4 promotes pulmonary vascular remodeling in hypoxic pulmonary hypertension. Circ Res 97: 496–504, 2005. [DOI] [PubMed] [Google Scholar]
- 25. Gershenfeld HK, Paul SM. Mapping quantitative trait loci for fear-like behaviors in mice. Genomics 46: 1–8, 1997. [DOI] [PubMed] [Google Scholar]
- 26. Haubold KW, Allen DL, Capetanaki Y, Leinwand LA. Loss of desmin leads to impaired voluntary wheel running and treadmill exercise performance. J Appl Physiol 95: 1617–1622, 2003. [DOI] [PubMed] [Google Scholar]
- 27. Iwamoto R, Yamazaki S, Asakura M, Takashima S, Hasuwa H, Miyado K, Adachi S, Kitakaze M, Hashimoto K, Raab G, Nanba D, Higashiyama S, Hori M, Klagsbrun M, Mekada E. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc Natl Acad Sci USA 100: 3221–3226, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones WK, Grupp IL, Doetschman T, Grupp G, Osinska H, Hewett TE, Boivin G, Gulick J, Ng WA, Robbins J. Ablation of the murine alpha myosin heavy chain gene leads to dosage effects and functional deficits in the heart. J Clin Invest 98: 1906–1917, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kinugawa S, Wang Z, Kaminski PM, Wolin MS, Edwards JG, Kaley G, Hintze TH. Limited exercise capacity in heterozygous manganese superoxide dismutase gene-knockout mice: roles of superoxide anion and nitric oxide. Circulation 111: 1480–1486, 2005. [DOI] [PubMed] [Google Scholar]
- 30. Koch LG, Green CL, Lee AD, Hornyak JE, Cicila GT, Britton SL. Test of the principle of initial value in rat genetic models of exercise capacity. Am J Physiol Regul Integr Comp Physiol 288: R466–R472, 2005. [DOI] [PubMed] [Google Scholar]
- 31. Korstanje R, Li R, Howard T, Kelmenson P, Marshall J, Paigen B, Churchill G. Influence of sex and diet on quantitative trait loci for HDL cholesterol levels in an SM/J by NZB/BlNJ intercross population. J Lipid Res 45: 881–888, 2004. [DOI] [PubMed] [Google Scholar]
- 32. Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, Eto K, Tsubamoto Y, Okuno A, Murakami K, Sekihara H, Hasegawa G, Naito M, Toyoshima Y, Tanaka S, Shiota K, Kitamura T, Fujita T, Ezaki O, Aizawa S, Kadowaki T. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell 4: 597–609, 1999. [DOI] [PubMed] [Google Scholar]
- 33. Laukkanen JA, Lakka TA, Rauramaa R, Kuhanen R, Venalainen JM, Salonen R, Salonen JT. Cardiovascular fitness as a predictor of mortality in men. Arch Intern Med 161: 825–831, 2001. [DOI] [PubMed] [Google Scholar]
- 34. Lerman I, Harrison BC, Freeman K, Hewett TE, Allen DL, Robbins J, Leinwand LA. Genetic variability in forced and voluntary endurance exercise performance in seven inbred mouse strains. J Appl Physiol 92: 2245–2255, 2002. [DOI] [PubMed] [Google Scholar]
- 35. Li X, Quigg RJ, Zhou J, Xu S, Maninde G, Mohan S, Baylink DJ. A critical evaluation of the effect of population size and phenotypic measurement on QTL detection and localization using a large F2 murine mapping population. Genet Mol Biol 29: 166–173, 2006. [Google Scholar]
- 36. Lightfoot JT, Turner MJ, Debate KA, Kleeberger SR. Interstrain variation in murine aerobic capacity. Med Sci Sports Exerc 33: 2053–2057, 2001. [DOI] [PubMed] [Google Scholar]
- 37. Lightfoot JT, Turner MJ, Knab AK, Jedlicka AE, Oshimura T, Marzec J, Gladwell W, Leamy LJ, Kleeberger SR. Quantitative trait loci associated with maximal exercise endurance in mice. J Appl Physiol 103: 105–110, 2007. [DOI] [PubMed] [Google Scholar]
- 38. Lightfoot JT, Turner MJ, Pomp D, Kleeberger SR, Leamy LJ. Quantitative trait loci for physical activity traits in mice. Physiol Genomics 32: 401–408, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu W, Selever J, Wang D, Lu MF, Moses KA, Schwartz RJ, Martin JF. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc Natl Acad Sci USA 101: 4489–4494, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Masaki T, Chiba S, Yasuda T, Noguchi H, Kakuma T, Watanabe T, Sakata T, Yoshimatsu H. Involvement of hypothalamic histamine H1 receptor in the regulation of feeding rhythm and obesity. Diabetes 53: 2250–2260, 2004. [DOI] [PubMed] [Google Scholar]
- 41. Mason SD, Howlett RA, Kim MJ, Olfert IM, Hogan MC, McNulty W, Hickey RP, Wagner PD, Kahn CR, Giordano FJ, Johnson RS. Loss of skeletal muscle HIF-1alpha results in altered exercise endurance. PLoS Biol 2: e288, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Massett MP, Berk BC. Strain-dependent differences in responses to exercise training in inbred and hybrid mice. Am J Physiol Regul Integr Comp Physiol 288: R1006–R1013, 2005. [DOI] [PubMed] [Google Scholar]
- 43. Medina-Gomez G, Virtue S, Lelliott C, Boiani R, Campbell M, Christodoulides C, Perrin C, Jimenez-Linan M, Blount M, Dixon J, Zahn D, Thresher RR, Aparicio S, Carlton M, Colledge WH, Kettunen MI, Seppanen-Laakso T, Sethi JK, O'Rahilly S, Brindle K, Cinti S, Oresic M, Burcelin R, Vidal-Puig A. The link between nutritional status and insulin sensitivity is dependent on the adipocyte-specific peroxisome proliferator-activated receptor-gamma2 isoform. Diabetes 54: 1706–1716, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mehrabian M, Allayee H, Stockton J, Lum PY, Drake TA, Castellani LW, Suh M, Armour C, Edwards S, Lamb J, Lusis AJ, Schadt EE. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat Genet 37: 1224–1233, 2005. [DOI] [PubMed] [Google Scholar]
- 45. Momken I, Lechene P, Koulmann N, Fortin D, Mateo P, Doan BT, Hoerter J, Bigard X, Veksler V, Ventura-Clapier R. Impaired voluntary running capacity of creatine kinase-deficient mice. J Physiol 565: 951–964, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Momken I, Lechene P, Ventura-Clapier R, Veksler V. Voluntary physical activity alterations in endothelial nitric oxide synthase knockout mice. Am J Physiol Heart Circ Physiol 287: H914–H920, 2004. [DOI] [PubMed] [Google Scholar]
- 47. Myers J, Prakash M, Froelicher V, Do D, Partington S, Atwood JE. Exercise capacity and mortality among men referred for exercise testing. N Engl J Med 346: 793–801, 2002. [DOI] [PubMed] [Google Scholar]
- 48. Perusse L, Gagnon J, Province MA, Rao DC, Wilmore JH, Leon AS, Bouchard C, Skinner JS. Familial aggregation of submaximal aerobic performance in the HERITAGE Family study. Med Sci Sports Exerc 33: 597–604, 2001. [DOI] [PubMed] [Google Scholar]
- 49. Peters LL, Lambert AJ, Zhang W, Churchill GA, Brugnara C, Platt OS. Quantitative trait loci for baseline erythroid traits. Mamm Genome 17: 298–309, 2006. [DOI] [PubMed] [Google Scholar]
- 50. Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet 8: 58–69, 2007. [DOI] [PubMed] [Google Scholar]
- 51. Petkov PM, Cassell MA, Sargent EE, Donnelly CJ, Robinson P, Crew V, Asquith S, Haar RV, Wiles MV. Development of a SNP genotyping panel for genetic monitoring of the laboratory mouse. Genomics 83: 902–911, 2004. [DOI] [PubMed] [Google Scholar]
- 52. Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res 14: 1806–1811, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rico-Sanz J, Rankinen T, Rice T, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Quantitative trait loci for maximal exercise capacity phenotypes and their responses to training in the HERITAGE Family Study. Physiol Genomics 16: 256–260, 2004. [DOI] [PubMed] [Google Scholar]
- 54. Sato Y, Ferguson DG, Sako H, Dorn GW, 2nd, Kadambi VJ, Yatani A, Hoit BD, Walsh RA, Kranias EG. Cardiac-specific overexpression of mouse cardiac calsequestrin is associated with depressed cardiovascular function and hypertrophy in transgenic mice. J Biol Chem 273: 28470–28477, 1998. [DOI] [PubMed] [Google Scholar]
- 55. Scheinman MM, Lam J. Exercise-induced ventricular arrhythmias in patients with no structural cardiac disease. Annu Rev Med 57: 473–484, 2006. [DOI] [PubMed] [Google Scholar]
- 56. Schrickel JW, Brixius K, Herr C, Clemen CS, Sasse P, Reetz K, Grohe C, Meyer R, Tiemann K, Schroder R, Bloch W, Nickenig G, Fleischmann BK, Noegel AA, Schwinger RH, Lewalter T. Enhanced heterogeneity of myocardial conduction and severe cardiac electrical instability in annexin A7-deficient mice. Cardiovasc Res 76: 257–268, 2007. [DOI] [PubMed] [Google Scholar]
- 57. Schulla V, Renstrom E, Feil R, Feil S, Franklin I, Gjinovci A, Jing XJ, Laux D, Lundquist I, Magnuson MA, Obermuller S, Olofsson CS, Salehi A, Wendt A, Klugbauer N, Wollheim CB, Rorsman P, Hofmann F. Impaired insulin secretion and glucose tolerance in beta cell-selective Cav1.2 Ca2+ channel null mice. EMBO J 22: 3844–3854, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sen S, Satagopan JM, Churchill GA. Quantitative trait locus study design from an information perspective. Genetics 170: 447–464, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shimomura K, Low-Zeddies SS, King DP, Steeves TD, Whiteley A, Kushla J, Zemenides PD, Lin A, Vitaterna MH, Churchill GA, Takahashi JS. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res 11: 959–980, 2001. [DOI] [PubMed] [Google Scholar]
- 60. Sinnegger-Brauns MJ, Hetzenauer A, Huber IG, Renstrom E, Wietzorrek G, Berjukov S, Cavalli M, Walter D, Koschak A, Waldschutz R, Hering S, Bova S, Rorsman P, Pongs O, Singewald N, Striessnig JJ. Isoform-specific regulation of mood behavior and pancreatic beta cell and cardiovascular function by L-type Ca2+ channels. J Clin Invest 113: 1430–1439, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sugiyama F, Churchill GA, Li R, Libby LJ, Carver T, Yagami K, John SW, Paigen B. QTL associated with blood pressure, heart rate, and heart weight in CBA/CaJ and BALB/cJ mice. Physiol Genomics 10: 5–12, 2002. [DOI] [PubMed] [Google Scholar]
- 62. Totsuka Y, Nagao Y, Horii T, Yonekawa H, Imai H, Hatta H, Izaike Y, Tokunaga T, Atomi Y. Physical performance and soleus muscle fiber composition in wild-derived and laboratory inbred mouse strains. J Appl Physiol 95: 720–727, 2003. [DOI] [PubMed] [Google Scholar]
- 63. Troxell ML, Britton SL, Koch LG. Selected contribution: variation and heritability for the adaptational response to exercise in genetically heterogeneous rats. J Appl Physiol 94: 1674–1681, 2003. [DOI] [PubMed] [Google Scholar]
- 64. Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol 2: e294, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ways JA, Cicila GT, Garrett MR, Koch LG. A genome scan for loci associated with aerobic running capacity in rats. Genomics 80: 13–20, 2002. [DOI] [PubMed] [Google Scholar]
- 66. Yamauchi T, Nio Y, Maki T, Kobayashi M, Takazawa T, Iwabu M, Okada-Iwabu M, Kawamoto S, Kubota N, Kubota T, Ito Y, Kamon J, Tsuchida A, Kumagai K, Kozono H, Hada Y, Ogata H, Tokuyama K, Tsunoda M, Ide T, Murakami K, Awazawa M, Takamoto I, Froguel P, Hara K, Tobe K, Nagai R, Ueki K, Kadowaki T. Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat Med 13: 332–339, 2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.