Abstract
Peroxisome proliferator-activated receptor type gamma (PPARγ) is a subgroup of the PPAR transcription factor family. Recent studies indicate that loss of PPARγ is associated with the development of pulmonary hypertension (PH). We hypothesized that the endothelial dysfunction associated with PPARγ inhibition may play an important role in the disease process by altering cellular gene expression and signaling cascades. We utilized microarray analysis to determine if PPARγ inhibition induced changes in gene expression in pulmonary arterial endothelial cells (PAEC). We identified 100 genes and expressed sequence tags (ESTs) that were upregulated by >1.5-fold and 21 genes and ESTs that were downregulated by >1.3-fold (P < 0.05) by PPARγ inhibition. The upregulated genes can be broadly classified into four functional groups: cell cycle, angiogenesis, ubiquitin system, and zinc finger proteins. The genes with the highest fold change in expression: hyaluronan-mediated motility receptor (HMMR), VEGF receptor 2 (Flk-1), endothelial PAS domain protein 1 (EPAS1), basic fibroblast growth factor (FGF-2), and caveolin-1 in PAEC were validated by real time RT-PCR. We further validated the upregulation of HMMR, Flk-1, FGF2, and caveolin-1 by Western blot analysis. In keeping with the microarray results, PPARγ inhibition led to re-entry of cell cycle at G1/S phase and cyclin C upregulation. PPARγ inhibition also exacerbated VEGF-induced endothelial barrier disruption. Finally we confirmed the downregulation of PPARγ and the upregulation of HMMR, Flk-1, FGF2, and Cav-1 proteins in the peripheral lung tissues of an ovine model of PH. In conclusion, we have identified an array of endothelial genes modulated by attenuated PPARγ signaling that may play important roles in the development of PH.
Keywords: microarray, cell signaling, peroxisome proliferator-activated receptor
peroxisome proliferator-activated receptor-γ (PPARγ) was initially identified in adipose tissue and is a key regulator of lipid metabolism and glucose homeostasis (47, 50). Later, it was shown that PPARγ is also expressed in the vasculature including endothelial cells (ECs), vascular smooth muscle cells (VSMCs), and monocytes/macrophages (13, 43). Upon activation by its ligands (fatty acids, arachidonic acid metabolites, and thiazolidinediones, etc), PPARγ forms heterodimers with the retinoid X receptor (RXR), and binds to specific PPAR response elements (PPRE) in the promoter region of its target genes, thereby regulating downstream gene expression (15, 46). PPARγ-mediated gene regulation can also be modulated by its interactions with specific co-activators and co-repressors (15, 26). PPARγ activation has been shown to alleviate atherosclerotic lesion formation and tumor progression (25, 30). The antiproliferative effects on VSMCs and antiangiogenic effects on tumor vasculogenesis by PPARγ activation seem to play important roles in these beneficial effects (9, 31, 53).
Recently, it has been shown that loss of PPARγ is associated with pulmonary hypertension (PH) (35). PPARγ expression has been shown to be significantly reduced in the plexiform lesions of human subjects with PH (2). Reduced PPARγ expression has also been demonstrated in vascular lesions of a rat model of severe PH (2). Hypoxia as well as shear stress has been implicated in reducing PPARγ expression in PH (2). The discovery of an association between PPARγ and the development of PH have brought new perspectives for understanding the etiology of this disease. It is generally believed that derangements in pulmonary endothelium-derived mediators and endothelial dysfunction play pivotal roles in PH. However, our knowledge on the interplay between PPARγ and endothelial dysfunction is limited. It has been shown that decreases in PPARγ expression reduces nitric oxide (NO) production and induces abnormal EC and VSMC growth (4, 35). However, the exact mechanisms underlying PPARγ downregulation and the pathogenesis of PH remain to be defined. We propose that global changes in pulmonary arterial endothelial cell (PAEC) gene expression induced by attenuated PPARγ binding are involved the endothelial dysfunction in pulmonary vasculature. We therefore utilized the powerful microarray techniques to identify target genes regulated by PPARγ in PAEC with the potential to identify novel target genes to develop new therapeutic strategies for the prevention and treatment of PH. We then confirmed a downregulation of PPARγ in our clinically relevant ovine model of congenital heart diseases (CHD) with increased pulmonary blood flow and compared the expression changes discovered in our cell culture studies with those in the lungs of our lambs with PH.
MATERIALS AND METHODS
Reagents.
Cell culture reagents including Dulbecco's modified Eagle's medium (DMEM), M199 medium, antibiotic-antimycotic solution, and fetal bovine serum (FBS) were obtained from Mediatech (Herndon, VA). Nuclear Extraction Kit and PPARγ Transcription Factor Assay Kit were from Cayman Chemical (Ann Arbor, MI). The Bovine Genome Genechips were from Affymetrix (Santa Clara, CA). The RNeasy Mini Kit, QuantiTect Reverse Transcription Kit, QuantiTect SYBR Green PCR kit, Flk-1 siRNA, and HiPerfect Transfection Reagent were from Qiagen (Valencia, CA). GW9662 and propidium iodide were obtained from Sigma (St. Louis, MO). VEGF was from Calbiochem (San Diego, CA). The monoclonal antibody directed against Cav-1 was from BD Transduction Laboratories (San Jose, CA). hyaluronan-mediated motility receptor (HMMR), VEGF receptor 2 (Flk-1), basic fibroblast growth factor (FGF-2), PPARγ, and cyclin C polyclonal antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). The scrambled siRNA and PPARγ siRNA were from Santa Cruz Biotechnology.
Cell culture, PPARγ antagonist treatment, and PPARγ depletion.
Primary cultures of ovine PAEC were isolated as described previously (58). Ovine PAEC or bovine aortic endothelial cells (BAEC) were cultured and maintained in 10% FBS-DMEM until confluence for experimental procedures. Confluent PAEC were treated with vehicle (DMSO) or the PPARγ antagonist GW9662 at 5 μM for 24 h for subsequent analysis. Human pulmonary microvascular endothelial cells (HMVEC) were isolated as previously described (7), cultured in M199 media containing 20% FBS, 100 units heparin/ml, 150 μg endothelial cell growth factor/ml, 1 μg hydrocortisone/ml, 292 mg l-glutamine/l, and 110 mg sodium pyruvate/l. The HMVEC were grown to 70% confluence and transfected with 25 nM scrambled siRNA or PPARγ siRNA, and the cells were utilized for subsequent analyses 24–48 h after transfection.
Measurement of PPARγ transcription factor binding.
PPARγ transcription factor binding was measured by an ELISA method from Cayman according to manufacturer's instructions. Briefly, the assay plate was precoated with a consensus dsDNA sequence of PPRE then the nuclear extracts were added. The plate was then washed to remove unbound reagents, the PPARγ antibody was added, and the plate washed three times. After the addition of an horseradish peroxidase (HRP)-conjugated secondary antibody the plate was again washed three times, the developing substrate was added, and incubated at room temperature for 15–45 min, finally the stop solution was added and the plate was read at 450 nm.
Microarray hybridization procedures.
Our primary cultures of PAEC are of ovine species; however, when this research was conducted, there was no commercially available ovine genechip for microarray studies. We therefore used the bovine genome chip from Affymetrix for a cross-species hybridization. We first carried out a preliminary study and confirmed the validity of this approach by comparing the basal gene expression profile in BAECs (same species hybridization) with that in ovine PAEC (cross-species hybridization). We found that the gene expression pattern was similar between ovine and bovine endothelial cells, as can be seen on the log ratio plot of expression difference (Fig. 1), the log ratio of distribution difference centered on 0; only ∼2.0% of the transcripts differ by >2-fold between the ovine and bovine species (Fig. 1 and Table 1), suggesting that the bovine chips may be used for ovine studies for the vast majority of the genes. For the formal study ovine PAEC were treated with vehicle (DMSO) or the PPARγ antagonist GW9662 (5 μM) for 24 h. Total RNA was then isolated, labeled, and then hybridized to the bovine genome chips. Five genechips were employed in each group in both the preliminary and formal studies.
Fig. 1.
Ovine pulmonary arterial endothelial cells (PAEC) and bovine aortic endothelial cells have similar basal gene expression patterns. The log ratios of gene expression fold change between ovine PAEC and bovine aortic endothelial cells (BAEC) were plotted continuously on the x-/y-axes. The log ratios of gene expression fold change between ovine PAEC and BAEC centered on 0, indicating a similar gene expression pattern between ovine PAEC and BAEC.
Table 1.
Number of differential expressed genes in ovine pulmonary arterial and bovine aortic endothelial cells
| Fold Change Between OV and BA |
|||||
|---|---|---|---|---|---|
| 50–184 | 10–50 | 5–10 | 2–5 | Total | |
| Up in OV | 6 | 47 | 53 | 375 | 481 |
| Down in OV | 7 | 31 | 47 | 384 | 469 |
| Total | 13 | 78 | 100 | 759 | 950 |
Heat map analysis showed that only 2.1% of the ovine genes are upregulated (481/23,000) by >2-fold, and 2.0% of the ovine genes are downregulated (469/23,000) by >2-fold compared with bovine gene expression, suggesting that the bovine chips may be used for ovine studies for the vast majority of the genes.
The microarray protocol was as follows: RNA was isolated using the Qiashredder column and RNeasy Mini kit. All RNA extracted was analyzed for quantity and quality using the Agilent 2100 Bioanalyzer system (Agilent Technologies, Palo Alto, CA). Gene expression profiling was performed using the bovine genome chips. An aliquot of 1 μg of total RNA was converted into double-stranded cDNA (ds-cDNA) by using SuperScript Choice System (GIBCO-BRL Life Technologies, Carlsbad, CA) with an oligo-dT primer containing a T7 RNA polymerase promoter (Genset, San Diego, CA). After second-strand synthesis, the reaction mixture was extracted with phenol-chloroform-isoamyl alcohol, and ds-cDNA was recovered by ethanol precipitation. In vitro transcription was performed on the ds-cDNA using the Enzo RNA transcript Labeling kit. Biotin-labeled cRNA was purified by using an RNeasy affinity column (Qiagen) and fragmented randomly in fragmentation buffer (40 mM Tris-acetate, pH 8.1, 100 mM KOAc, and 30 mM MgOAc) to sizes ranging from 35 to 200 bases by incubating at 94°C for 35 min. The hybridization solutions contained 100 mM MES [2-(N-morpholino) ethanesulfonic acid], 1 M Na+, 20 mM EDTA, and 0.01% Tween 20. The final concentration of fragmented cRNA was 0.05 μg/μl in hybridization solution. A target for hybridization was prepared by combining 40 μl of fragmented transcript with sonicated herring sperm DNA (0.1 mg/ml), bovine serum albumin, and 5 nM control oligonucleotide in a buffer containing 1.0 M NaCl, 10 mM Tris · HCl (pH 7.6), and 0.005% Triton X-100. The target was hybridized for 16 h at 45°C to the array chips (Affymetrix). Arrays were then washed at 50°C with stringent solution and then again at 30°C with nonstringent washes. Arrays were then stained with streptavidin-phycoerythrin (Invitrogen). DNA chips were read at a resolution of 3 μm with a Hewlett-Packard GeneArray Scanner and were analyzed with the GENECHIP software (Affymetrix GCOS 1.1). Both the CEL and DAT files for each hybridization have been uploaded to our server running GeneTraffic v3.2 (Iobion Informatics, La Jolla, CA).
Microarray data analysis.
Microarray data were processed and normalized using RMA (5). Normalized data were subsequently analyzed using the LIMMA (49) package in R. All groups were compared pair wise, and a P value <0.05 was considered significant.
Real-time RT-PCR analysis.
Real-time RT-PCR was employed to verify the regulation of a list of genes of interest. Primers were designed by Primer 3. Table 2 shows all the primer sets utilized. Real time RT-PCR was carried out in two steps. First, total RNA was extracted from cells (or tissues) using the RNeasy kit (Qiagen), and 1 μg total RNA was reverse-transcribed using QuantiTect Reverse Transcription Kit (Qiagen, Hilden) in a total volume of 20 μl. Quantitative real-time PCR was conducted on Mx4000 (Stratagene), using 2 μl of RT product, 12.5 μl of QuantiTect SYBR Green PCR Master Mix (Qiagen, Hilden), and primers (400 nM) in a total volume of 25 μl. The following thermocycling conditions were employed: 95°C for 10 min, followed by 95°C for 30 s, 55°C for 60 s, and 72°C 30 s for 45 cycles. The threshold cycles (Ct) of a serially diluted control sample were plotted to generate a standard curve. Concentration of each sample was calculated by interpolating its Ct on the standard curve and then normalized to β-actin (housekeeping gene) mRNA levels.
Table 2.
Primer pairs for real-time RT-PCR analysis
| Gene Name | Sense Primer | Anti-sense Primer |
|---|---|---|
| Bovine HMMR | 5′-TACAAGCTAGATATT GCCCA-3′ | 5′-CCTCAAGAGACTGCT TAAGG-3′ |
| Ovine Flk-1 | 5′-AGCCCCTGATTACACCACAC-3′ | 5′-AGTCCCGAATCCTCTTCCAT-3′ |
| Bovine EPAS1 | 5′-CCATGCACTGGACTCAGAGA-3′ | 5′-CGAGGGTTGTAGATCACCGT-3′ |
| Bovine FGF2 | 5′-GTGCAAACCGTTACCTTGCT-3′ | 5′-ACTGCCCAGTTCGTTTCAGT-3′ |
| Bovine Cav-1 | 5′-AGCCCAACAACAAGGCTATG-3′ | 5′-GATGCCATCGAAACTGTGTG-3′ |
| Bovine SERPINE1 | 5′-TCATTCTTGCACTGTCTGCC-3′ | 5′-ACACACTGTTTCCTGGGGAG-3′ |
| Bovine STAP2 | 5′-CCGAGATCGGGATTACAAGA-3′ | 5′-GAAGGACCAAGCAGAAGTGG-3′ |
| Bovine β-actin | 5-CTCTTCCAGCCTTCCTTCCT-3′ | 5′-GGGCAGTGATCTCTTTCTGC-3′ |
Western blot analysis.
For the cell culture studies, PAEC or HMVEC were lysed with modified RIPA buffer containing 150 mmol/l NaCl, 50 mmol/l Tris, pH 7.4, 1.0% Nonidet P-40 (NP-40), 0.25% sodium deoxycholate, 1 mmol/l EDTA, and protease inhibitor mixture for 15 min at 4°C and insoluble materials were removed by centrifugation (14,000 g for 15 min). Lung tissues were homogenized in Triton X-100 lysis buffer [20 mM Tris · HCl (pH 7.6), 0.5% Triton X-100, 20% glycerol] supplemented with protease inhibitors (100 μg/ml PMSF, 1 μg/ml leupeptin and aprotinin), clarified by centrifugation at 20,000 g for 20 min at 4°C, and the supernatant was stored at −80°C until use. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Then proteins were run on 4% to 20% gradient SDS-PAGE gel (NuSep), transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA), and blotted with the appropriate primary antibody overnight at 4°C, followed by incubation with the HRP-conjugated secondary antibody (Pierce) for 1 h at room temperature. After the membrane was exposed to Supersignal West Femto Maximum Sensitivity Substrate (Pierce), proteins were detected and quantified on Kodak Image Station 440.
Cell cycle and cell growth analyses.
Ovine PAEC were treated with vehicle or 5 μM GW9662 for 24 h; then the cells were trypsinized, washed with ice-cold PBS, and resuspended in 2 ml of Vindelov's propidium iodide (0.01 M Tris, pH 8.0, 10 mM NaCl, 700 U of RNase, 75 μM propidium iodide, 0.1% NP-40). Cells were incubated at room temperature for 15 min. DNA content was determined using a Becton Dickinson FACS Calibur cell flow cytometer. Cell cycle distribution was analyzed using Cell Quest Pro (BD Bioscience, San Jose, CA). Flow cytometric analyses used 20,000 events/sample, and gating was used to exclude cell aggregates; 40,000 HMVEC were seeded into each well of a 24-well plate. After 24 h the cells were then transfected with 25 nM of a PPARγ-specific siRNA or a scrambled siRNA. The cells were then trypsinized and counted using a hematocytometer at 48-, 72-, and 96-h postplating.
Measurement of transendothelial resistance.
Transendothelial resistance (TER) was measured by electric cell impedance sensing (ECIS) apparatus (Applied Biophysics, Troy, NY) as described (37). Equal number of ovine PAEC or HMVEC were seeded on l-cysteine-coated gold electrode arrays (8W10E). Ovine PAEC were grown to confluence and HMVEC were grown to 70% confluence. Current was applied across the electrodes by 4,000-Hz AC voltage source attached to an amplifier. TER was monitored for 30 min to establish baseline. Ovine PAEC were pretreated with vehicle (DMSO) or GW9662 (5 μM) for 24 h. HMVEC were pretransfected with 25 nM of a PPARγ-specific siRNA or a scrambled siRNA for 48 h. Cells were then exposed or not to vascular endothelial growth factor (VEGF, 100 ng), and ECIS was continuously recorded for 24 h. To confirm the role for PPARγ mediated increases in Flk-1 on VEGF-mediated increased permeability we transiently transfected ovine PAECs with either an Flk-1 siRNA or a scrambled siRNA as a control. After the cells were treated with either DMSO or GW9662 (5 μM) for a further 24 h; then the cells were exposed or not to VEGF (100 ng), and the change in TER was monitored for 24 h.
Ovine model of PH with increased pulmonary blood flow.
We utilized an ovine model of CHD with increased pulmonary blood flow as the experimental PH model as we have previously described (40). Briefly, pregnant ewes (137–141 days gestation, term = 145 days) underwent in utero surgery to anastomose an 8.0-mm Gore-tex vascular graft (∼2 mm length; W. L. Gore and Assos., Milpitas, CA) between the ascending aorta and main pulmonary artery of the fetus. The incisions in the uterus and the abdomen were closed, and the sheep were allowed to deliver normally. Two weeks after spontaneous delivery, lambs were anesthetized with ketamine hydrochloride (∼0.3 mg · kg−1 · min−1), diazepam (0.002 mg · kg−1 · min−1), and fentanyl citrate (1.0 μg · kg−1 · h−1), intubated, mechanically ventilated, and a midsternotomy incision was performed. Oxygen saturations were obtained in the aorta, right ventricle, right atrium, and distal pulmonary artery to confirm graft potency and increased pulmonary blood flow. Increase in mean pulmonary arterial pressure was validated. Four biopsies of peripheral lung tissue were harvested from randomly selected lobes, and ∼300 mg of peripheral lung were obtained for each biopsy. At the end of the protocol, all lambs were killed with a lethal injection of pentobarbital sodium followed by bilateral thoracotomy as described in National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. All protocols and procedures were approved by the Committees on Animal Research at the University of California, San Francisco; Medical College of Georgia, Augusta, Georgia; and the German Heart Centre, Munich, Germany.
Statistical analysis.
Statistical calculations were performed using the GraphPad Prism V. 4.01 software. The mean ± SD or SE was calculated for all samples, and significance was determined either by the unpaired t-test or ANOVA. For ANOVA Newman-Keuls post hoc testing was also utilized. A value of P < 0.05 was considered significant.
RESULTS
Overview of gene expression profiles in PPARγ-inhibited PAEC.
Initially we verified that GW9662 (5 μM, 24 h) significantly decreased PPARγ transcription factor binding in ovine PAEC (Fig. 2, P < 0.05 vs. vehicle). Next we identified gene expression changes in GW9662-treated ovine PAEC using bovine genome microarrays. We found the expression levels of 100 genes and expressed sequence tags (ESTs) were upregulated by >1.5-fold and 21 genes and ESTs downregulated by >1.3-fold (P < 0.05) by PPARγ inhibition (Fig. 3). Although the majority of these changes were less than twofold, a number of upregulated genes changed by more than twofold. For example, the HMMR is upregulated by 2.3-fold and Flk-1 (VEGF receptor 2) is upregulated by twofold. A number of other highly upregulated genes are EPAS1 (1.8-fold), FGF2 (1.7-fold), and caveolin-1 (1.4-fold). All the downregulated genes were altered by less than twofold (See microarray data in GEO database). Based on gene annotations, four functional groups were identified in the upregulated genes: cell cycle (Table 3), angiogenesis (Table 4), ubiquitin system (Table 5), and zinc finger proteins (Table 6).
Fig. 2.
GW9662 inhibits peroxisome proliferator-activated receptor (PPAR)γ transcription factor binding in ovine PAEC. Ovine PAEC were treated with vehicle (DMSO) or GW9662 (5 μM) for 24 h, and nuclear extracts were prepared, loaded onto a 96-well plate precoated with dsDNA of PPRE, and detected by the PPARγ antibody. PPARγ transcription factor binding was significantly decreased in GW9662-treated PAEC. Data are means ± SE, n = 4, *P < 0.05 vs. Vehicle.
Fig. 3.
Heat map analysis of the significantly (P < 0.05) regulated genes in GW9662-treated ovine pulmonary arterial endothelial cells. 1, GW9662 vs. vehicle upregulated genes (1.5 fold cut-off); 2, GW9662 vs. vehicle downregulated genes (1.3 fold cut-off).
Table 3.
Cell cycle-related genes
| Probe Set ID | Gene Symbol | Fold Change (GW vs. C) | P Value | FDR | Gene Name |
|---|---|---|---|---|---|
| Bt.29953.2.S1_at | CCNYL1 | 2.48 | 0.001 | 0.134 | cyclin Y-like 1 |
| Bt.16065.1.S1_at | CKAP2 | 2.43 | 0.002 | 0.177 | Cytoskeleton-associated protein 2 |
| Bt.27798.1.A1_at | RBM5 | 2.40 | 0.018 | 0.214 | RNA-binding motif protein 5 |
| Bt.17932.2.S1_a_at | CCNL1 | 1.71 | 0.024 | 0.215 | cyclin L1 |
| Bt.26828.1.S1_at | CNTLN | 1.54 | 0.000 | 0.100 | centlein, centrosomal protein |
| Bt.2551.1.S1_at | CCNA2 | 1.49 | 0.001 | 0.128 | cyclin A2 |
| Bt.19894.1.S1_at | CDC40 | 1.33 | 0.003 | 0.191 | cell division cycle 40 homolog (S. cerevisiae) |
| Bt.5852.1.A1_at | CCNC | 1.31 | 0.018 | 0.214 | cyclin C |
The significantly upregulated cell cycle-related genes are listed. For each gene, the Affymetrix probe set ID, the gene symbol, fold change, P value, the false discovery rate (FDR), and the gene name are given.
Table 4.
Angiogenesis and cell adhesion-related genes
| Probe Set ID | Gene Symbol | Fold Change (GW vs. C) | P Value | FDR | Gene Name |
|---|---|---|---|---|---|
| Bt.27997.1.S1_at | HMMR | 2.24 | 0.000 | 0.051 | hyaluronan-mediated motility receptor (RHAMM) |
| Bt.6125.2.S1_at | JAG1 | 2.20 | 0.001 | 0.128 | jagged 1 (Alagille syndrome) |
| Bt.29836.1.S1_at | FLK-1 | 2.04 | 0.008 | 0.214 | vascular endothelial growth factor receptor 2 |
| Bt.20111.1.S1_at | TNFRSF12A | 1.88 | 0.003 | 0.196 | tumor necrosis factor receptor superfamily, member 12A |
| Bt.22840.1.S1_a_at | CD46 | 1.84 | 0.000 | 0.084 | CD46 molecule, complement regulatory protein |
| Bt.11873.1.A1_at | SSX2IP | 1.81 | 0.007 | 0.214 | synovial sarcoma, X breakpoint 2 interacting protein |
| Bt.4353.1.S1_at | EPAS1 | 1.77 | 0.000 | 0.110 | endothelial PAS domain protein1 |
| Bt.5129.2.A1_at | NNAT | 1.76 | 0.040 | 0.221 | neuronatin |
| Bt.4834.1.S1_at | FGF2 | 1.68 | 0.010 | 0.214 | fibroblast growth factor 2 (basic) (associated with hyaluronan) |
| Bt.27.1.S1_at | YWHAG | 1.60 | 0.009 | 0.214 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
| Bt.28459.1.S1_at | BMP4 | 1.59 | 0.046 | 0.226 | bone morphogenetic protein 4 |
| Bt.25926.1.A1_at | PDE3B | 1.57 | 0.002 | 0.190 | phosphodiesterase 3B, cGMP-inhibited |
| Bt.4781.1.S1_at | ADAM10 | 1.57 | 0.005 | 0.214 | ADAM metallopeptidase domain 10 |
| Bt.25447.1.A1_at | EPHA7 | 1.55 | 0.015 | 0.214 | EPH receptor A7 |
| Bt.28750.1.S1_at | IL13RA1 | 1.53 | 0.003 | 0.250 | interleukin-13 receptor, α-1 |
| Bt.5395.1.S1_a_at | VCAN | 1.47 | 0.000 | 0.118 | versican |
| Bt.8131.1.S1_at | ALCAM | 1.40 | 0.045 | 0.225 | activated leukocyte cell adhesion molecule |
The significantly upregulated angiogenesis and cell adhesion-related genes are listed. For each gene, the Affymetrix probe set ID, the gene symbol, fold change, P value, the FDR, and the gene name are given.
Table 5.
Ubiquitin-related genes
| Probe Set ID | Gene Symbol | Fold Change (GW vs. C) | P Value | FDR | Gene Name |
|---|---|---|---|---|---|
| Bt.14223.3.S1_at | NEDD4 | 1.86 | 0.001 | 0.167 | neural precursor cell expressed, developmentally downregulated 4 |
| Bt.18964.1.A1_at | RWDD4A | 1.85 | 0.001 | 0.167 | RWD domain containing 4A |
| Bt.19024.1.A1_at | NEDD4 | 1.79 | 0.008 | 0.214 | neural precursor cell expressed, developmentally downregulated 4 |
| Bt.18003.1.S1_at | CUL3 | 1.69 | 0.002 | 0.186 | cullin 3 |
| Bt.20830.1.A1_at | CUL5 | 1.49 | 0.042 | 0.223 | cullin 5 |
| Bt.8857.1.S1_at | UBQLN1 | 1.44 | 0.006 | 0.214 | ubiquilin 1 |
| Bt.20364.1.S1_at | UBE2J2 | 1.42 | 0.009 | 0.214 | ubiquitin-conjugating enzyme E2, J2 (UBC6 homolog, yeast) |
| Bt.6350.1.A1_at | UBE2H | 1.42 | 0.019 | 0.214 | ubiquitin-conjugating enzyme E2 H (UBC8 homolog, yeast) |
| Bt.14124.2.S1_at | USP33 | 1.33 | 0.044 | 0.225 | Ubiquitin-specific protease 33 |
The significantly upregulated ubiquitin-related genes are listed. For each gene, the Affymetrix probe set ID, the gene symbol, fold change, P value, the FDR, and the gene name are given.
Table 6.
Zinc finger genes
| Probe Set ID | Gene Symbol | Fold Change (GW vs. C) | P Value | FDR | Gene Name |
|---|---|---|---|---|---|
| Bt.18309.1.A1_at | SLC39A10 | 1.64 | 0.020 | 0.214 | solute carrier family 39 (zinc transporter), member 10 |
| Bt.13849.1.S1_at | SLC30A1 | 1.62 | 0.000 | 0.065 | solute carrier family 30 (zinc transporter), member 1 |
| Bt.16125.3.S1_at | RNFT1 | 1.55 | 0.005 | 0.214 | ring finger protein, transmembrane 1 |
| Bt.27414.1.S1_at | RNF168 | 1.47 | 0.046 | 0.226 | ring finger protein 168 |
| Bt.19940.1.S1_at | LNX2 | 1.44 | 0.004 | 0.205 | ligand of numb-protein X2 |
| Bt.23611.2.S1_at | TIPARP | 1.43 | 0.009 | 0.214 | TCDD-inducible poly (ADP-RIBOSE) polymerase |
| Bt.13708.2.S1_a_at | ATMIN | 1.42 | 0.022 | 0.214 | ATM interactor |
| Bt.27247.1.A1_at | WHSC1L1 | 1.39 | 0.039 | 0.221 | Wolf-Hirschhorn syndrome candidate 1-like1 |
| Bt.25649.1.A1_at | CAST ERAP2 | 1.39 | 0.045 | 0.225 | calpastatin endoplasmic reticulum aminopeptidase 2 |
| Bt.25310.1.A1_at | ZNF181 | 1.38 | 0.030 | 0.217 | zinc finger protein 181 |
| Bt.13990.1.S1_at | ZCCHC8 | 1.38 | 0.018 | 0.214 | zinc finger, CCHC domain containing 8 |
| Bt.20064.1.S1_at | TSHZ1 | 1.37 | 0.007 | 0.214 | teashirt zinc finger homeobox1 |
| Bt.20726.1.S1_at | LOC507724 | 1.34 | 0.029 | 0.216 | odorant response abnormal 4 |
| Bt.26803.1.A1_at | LOC789240 | 1.32 | 0.001 | 0.142 | similar to testis derived transcript |
| Bt.7676.2.S1_a_at | CALR | 1.32 | 0.016 | 0.214 | calreticulin |
| Bt.15872.1.S1_at | SLU7 | 1.32 | 0.003 | 0.201 | SLU7 splicing factor homolog (S. cerevisiae) |
| Bt.22605.1.A1_at | ZCWPW1 | 1.34 | 0.010 | 0.214 | zinc finger, CW type with pwwp domain1 |
| Bt.10397.2.S1_at | RNF20 | 1.31 | 0.013 | 0.214 | ring finger protein 20 |
The significantly upregulated zinc finger-related genes are listed. For each gene, the Affymetrix probe set ID, the gene symbol, fold change, P value, the FDR, and the gene name are given.
Validation of candidate genes by quantitative real-time RT-PCR in GW9662-treated ovine PAEC and PPARγ-depleted HMVEC.
Among the most upregulated genes, HMMR is involved in cell migration (45, 48); Flk-1 is actively involved in angiogenesis and endothelial barrier function (32); EPAS1, also known as HIF-2 alpha, is an important transcription factor in hypoxia response and is implicated in the development of lung vessels (14); FGF-2 is critical in angiogenesis (33, 52); and caveolin-1 has been documented as a negative regulator of endothelial nitric oxide synthase (eNOS) activity (59) and thus may inhibit vascular relaxation. As these genes are all strong candidates for mediating endothelial dysfunction in PH, we verified the regulation of these genes by real-time RT-PCR. Treatment of PAEC with the pharmacological inhibitor GW9662 (5 μM) led to the significant upregulation of HMMR mRNA (1.9-fold, Fig. 4A), Flk-1 mRNA (4.4-fold, Fig. 4B), EPAS1 mRNA (1.3-fold, Fig. 4C), FGF2 mRNA (1.4-fold, Fig. 4D), and caveolin-1 mRNA (2.1-fold, Fig. 4E). We also confirmed the downregulation of SERPINE1 mRNA (1.4-fold, Fig. 4F) and STAP2 mRNA (2.2-fold, Fig. 4G) by real-time RT-PCR. To further verify the regulations of these genes by PPARγ in a more clinically relevant system, we utilized PPARγ siRNA to deplete PPARγ expression (Fig. 4H) and PPARγ transcription factor binding (Fig. 4I) in HMVEC and again tested the regulation of these genes. We found, in PPARγ-depleted HLMVEC, there was a significant upregulation of HMMR mRNA (3-fold, Fig. 4J), Flk-1 mRNA (1.8-fold, Fig. 4K), EPAS1 mRNA (1.9-fold, Fig. 4L), FGF2 mRNA (1.8-fold, Fig. 4M), and caveolin-1 mRNA (1.4-fold, Fig. 4N). Thus, we confirmed, using both a pharmacological inhibitor and an siRNA depletion technique, that PPARγ inhibition altered the regulation of these genes in two different EC lines, corroborating the microarray data.
Fig. 4.
PPARγ inhibition or depletion increases hyaluronan-mediated motility receptor (HMMR),VEGF receptor 2 (Flk-1), endothelial PAS domain protein 1 (EPAS1), basic fibroblast growth factor (FGF-2), and caveolin-1 mRNA levels in ovine and human PAEC. To pharmacologically inhibit PPARγ signaling, ovine PAEC were treated with vehicle (DMSO) or GW9662 (5 μM) for 24 h. Total RNA was isolated and the levels of HMMR (A), FLK-1 (B), EPAS1 (C), FGF2 (D), and caveolin-1 (E), SERPINE1 (F), and STAP2 (G) were quantified by SYBR Green real-time RT-PCR analyses. HMMR mRNA was upregulated by 1.9-fold (A); Flk-1 mRNA was upregulated by 4.4-fold (B); EPAS1 mRNA was upregulated by 1.3-fold (C); FGF2 mRNA was upregulated by 1.4-fold (D); Cav-1 mRNA was upregulated by 2.3-fold (E), SERPINE1 mRNA was downregulated by 1.4-fold (F), and STAP2 mRNA was downregulated by 2.2-fold (G). To deplete PPARγ expression, HMVEC were transfected with a PPARγ siRNA or a scrambled siRNA (Scra siRNA) for 48 h, and Western blot analysis (H) and transcription factor binding assays (I) were then utilized to confirm the decrease in PPARγ expression and binding respectively. Data are means ± SE, n = 6 for the Western blot assays and N + 4 for the transcription assays, *P < 0.05 vs. Scra siRNA. In the Western blot assays, protein loading was normalized by reprobing the blot with β-actin. Total RNA was the isolated from the PPARγ-depleted cells, and the levels of HMMR (J), FLK-1 (K), EPAS1 (L), FGF2 (M), and caveolin-1 (N) quantified by SYBR Green real-time RT-PCR analyses. HMMR mRNA was upregulated by 3-fold (J); Flk-1 mRNA was upregulated by 1.8-fold (K); EPAS1 mRNA was upregulated by 1.9-fold (L); FGF2 mRNA was upregulated by 1.8-fold (M); and Cav-1 mRNA was upregulated by 1.4-fold (N). Data are means ± SE, n = 6, *P < 0.05 vs. Scra siRNA. All mRNA data was normalized to β-actin mRNA levels.
Validation of candidate genes at the protein level in GW9662-treated ovine PAEC and PPARγ-depleted HMVEC.
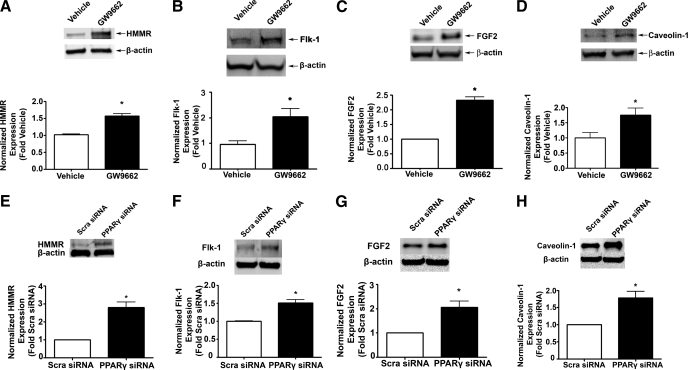
We next validated the upregulation of HMMR, Flk-1, EPAS1, FGF2, and caveolin-1 in GW9662-treated PAEC and PPARγ-depleted HMVEC at the protein level. Except for EPAS1, for which we were unable to detect a band, we found that GW9662 treatment significantly increased the protein level of HMMR (1.6-fold, Fig. 5A), Flk-1 (2-fold, Fig. 5B), FGF2 (2.3-fold, Fig. 5C), and caveolin-1 (1.8-fold, Fig. 5D). Depleting PPARγ by siRNA in HMVEC also significantly increased the protein level of HMMR (2.8-fold, Fig. 5E), Flk-1 (1.5-fold, Fig. 5F), FGF2 (2.1-fold, Fig. 5G), and caveolin-1 (1.8-fold, Fig. 5H). Thus, the expression changes of these genes identified by the microarray studies are not only confirmed at mRNA level but also related to changes in protein expression with functional implications. We next wished to determine if the gene expression changes induced functional consequences in the cells.
Fig. 5.
PPARγ inhibition or depletion increases HMMR, FLK-1, EPAS1, FGF2, and caveolin-1 protein levels in ovine and human PAEC. To pharmacologically inhibit PPARγ signaling, ovine PAEC were treated with vehicle (DMSO) or GW9662 (5 μM) for 24 h. To deplete PPARγ expression, HMVEC were transfected with a PPARγ siRNA or a Scra siRNA for 48 h. In each case, whole cell extracts (20 μg) were then subjected to Western blot analysis to determine changes in HMMR, FLK-1, FGF2, and caveolin-1 protein levels. A representative image is shown for each Western blot. In ovine PAEC, HMMR protein was upregulated by 1.6-fold (A); Flk-1 protein was upregulated by 1.9-fold (B); FGF2 protein was upregulated by 2.3-fold (C); and caveolin-1 protein was upregulated by 1.8-fold (D). In HMVEC, HMMR protein was upregulated by 2.8-fold (E); Flk-1 protein was upregulated by 1.5-fold (F); FGF2 protein was upregulated by 2.1-fold (G); and caveolin-1 protein was upregulated by 1.8-fold (H). Data are means ± SE, n = 6, *P < 0.05 vs. vehicle. All protein levels were normalized by reprobing with β-actin.
PPARγ inhibition induces cell cycle re-entry in PAEC and cell proliferation in HMVEC.
In the microarray studies we found a group of cell cycle-related genes were upregulated by PPARγ inhibition (Table 3). We therefore examined the effects of PPARγ inhibition on EC growth in both GW9662-treated PAEC and PPARγ-depleted HMVEC. We did not observe any gross difference in cell growth between vehicle- and GW9662-treated PAEC. However, using flow cytometry analysis we found that PPARγ inhibition more than doubled the percentage of S-phase cells (Fig. 6A), although PPARγ inhibition did not significantly change the percentage of G0/G1-phase cells (Fig. 6B) or the percentage of G2/M-phase cells (Fig. 6C). Based on the microarray result that PPARγ inhibition significantly upregulated cyclin C expression, and cyclin C may promote exit at G0/G1 and reentry into S phase (44), we speculated that cyclin C may be involved in the increase in S-phase cells. We therefore carried out Western blot analysis to examine the cyclin C protein levels in vehicle- and GW9662-treated PAEC. Our data indicate that GW9662 significantly upregulated cyclin C (3.3-fold, Fig. 6D). These results indicate that PPARγ inhibition induces cell cycle reentry at G1/S in PAEC and that increased levels of cyclin C may underlie this process. We also found that depleting PPARγ using siRNA in HMVEC led to significant increase in both cell proliferation (Fig. 6E) and also significant increases in cyclin C protein levels (Fig. 6F).
Fig. 6.
PPARγ inhibition increases the number of cells in S phase and cyclin C expression in ovine PAEC, while PPARγ depletion increases cell proliferation in human PAEC. Ovine PAEC were treated with vehicle (DMSO) or GW9662 (5 μm) for 24 h. The cells were then harvested and labeled with propidium iodide (75 μM). Cell flow cytometry was then used to identify changes in cell cycle progression. PPARγ inhibition significantly increased the percentage of cells in S phase (A). However, PPARγ inhibition did not alter the percentage of cells in the G0/G1 phase (B) or the G2/M phase (C); n = 6, *P < 0.05 vs. vehicle-treated PAEC. In addition, Western blot analysis identified that PPARγ inhibition significantly increased cyclin C expression in ovine PAEC (D). Shown is a representative image with protein loading normalized by reprobing with β-actin. Data are means ± SE, n = 3, *P < 0.05 vs. vehicle. Furthermore, PPARγ depletion with a PPARγ-specific siRNA significantly increased human pulmonary microvascular endothelial cell (HMVEC) proliferation (E). Data are means ± SD, n = 4, *P < 0.05 vs. Scra siRNA, and this correlated with increased cyclin C protein levels in HMVEC (F). HMVEC were transiently transfected with a PPARγ siRNA or Scra siRNA as a control for 48 h, and the proteins were quantified as described above; a representative image is shown. Data are means ± SE, n = 4, *P < 0.05 vs. Scra siRNA.
PPARγ inhibition exacerbates VEGF-induced barrier disruption in PAEC and HMVEC.
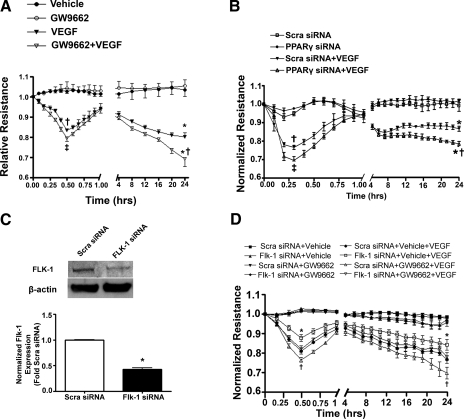
The GW9662-mediated increase in FLK-1 (VEGF receptor 2) suggested that PPARγ inhibition could potentiate VEGF-induced endothelial barrier disruption. To test this, we monitored barrier function using ECIS to measure changes in TER continuously for 24 h in GW9662-treated PAEC and PPARγ-depleted HMVEC. Our data indicate that PPARγ inhibition (Fig. 7A) or depletion (Fig. 7B) both acutely (0.25–0.5 h) and chronically (24 h) significantly decreased TER following VEGF treatment, indicating an exacerbation in VEGF-induced barrier disruption. We then attenuated the expression of Flk-1 in PAECs using an siRNA technique (Fig. 7C), and demonstrated that the exacerbating effect of GW9662 on VEGF-induced barrier disruption was abolished (Fig. 7D), confirming the key role played by Flk-1 in the effect of PPARγ inhibition on VEGF-mediated barrier disruption.
Fig. 7.
PPARγ inhibition or depletion exacerbates VEGF-induced endothelial barrier disruption in ovine PAEC and HMVEC. A: ovine PAEC were plated on gold microelectrodes and grown until there was a plateau of impedance for at least 6 h. PAEC were then treated with either vehicle (DMSO) or GW9662 (5 μM) for 24 h, after which cells were exposed or not to VEGF (100 ng), and transendothelial resistance (TER) was monitored for 24 h. Alone GW9662 did not alter basal TER; however, it significantly decreased TER acutely (at 0.5 h) and chronically (24 h) in the presence of VEGF. Data are means ± SD, n = 6, †VEGF vs. vehicle at 0.5 h; ‡GW9662 + VEGF vs. VEGF at 0.5 h; *VEGF vs. vehicle at 24 h; *†GW9662 + VEGF vs. VEGF at 24 h, †, ‡, *, *† indicate P < 0.05. B: HMVEC were plated on gold microelectrodes and grown until 70% confluence then transfected with a PPARγ siRNA or a Scra siRNA. After 48 h the cells were exposed or not to VEGF (100 ng), and TER was monitored for 24 h. Alone PPARγ knock-down did not alter basal TER; however, it significantly decreased TER acutely (at 0.3 h) and chronically (24 h) in the presence of VEGF. Data are means ± SD, n = 6. †Scra siRNA + VEGF vs. Scra siRNA at 0.3 h; ‡PPARγ siRNA + VEGF vs. Scra siRNA + VEGF at 0.3 h; *Scra siRNA + VEGF vs. Scra siRNA at 24 h; *†PPARγ siRNA +VEGF vs. Scra siRNA + VEGF at 24 h, †, ‡, *, *† indicate P < 0.05. C: ovine PAECs were transiently transfected with a Flk-1 siRNA or Scra siRNA as a control for 48 h, and the proteins were harvested for Western blot analysis. The Flk-1 siRNA significantly reduces Flk-1 protein levels (a representative image is shown). Data are means ± SE, n = 4. *P < 0.05. D: ovine PAECs were transiently transfected with a Flk-1 siRNA or Scra siRNA as a control for 24 h, followed by treatment with either DMSO or GW9662 (5 μM) for a further 24 h. The cells were then exposed or not to VEGF (100 ng), and the TER was monitored over 24 h. The Flk-1 siRNA significantly attenuates the effect of GW9662 on VEGF-induced barrier disruption (both acutely and chronically). *Flk-1 siRNA + Vehicle + VEGF vs. Scra siRNA + Vehicle + VEGF; †Scra siRNA + GW9662 + VEGF vs. Scra siRNA + Vehicle + VEGF.
Confirmation of protein changes in an ovine model of PH.
To further confirm that the gene expression changes caused by PPARγ inhibition may indeed play a role in PH, we examined the expression of PPARγ and the expression of identified upregulated genes HMMR, Flk-1, FGF2, and caveolin-1 by Western blot analysis in the peripheral lung tissues of our ovine model of PH associated with increased pulmonary blood flow (Shunt). We first demonstrated that PPARγ is significantly downregulated in the peripheral lung tissues of Shunt lambs (Fig. 8A). We also found that HMMR (2.8-fold, Fig. 8B), FLK-1 (3-fold, Fig. 8C), FGF2 (2.1-fold, Fig. 8D), and caveolin-1 (4.6-fold, Fig. 8E) are all significantly upregulated in the peripheral lung tissues of Shunt lambs. These in vivo results are consistent with our in vitro data, strongly suggesting that PPARγ inhibition-induced regulation of these genes may play important roles in the development of PH.
Fig. 8.
Expression of PPARγ, HMMR, Flk-1, FGF2, and caveolin-1 in a lamb model of pulmonary hypertension. PPARγ (A), HMMR (B), Flk-1 (C), FGF2 (D), and caveolin-1 (E) protein levels were determined by Western blot analysis in extracts prepared from peripheral lung tissue of 2-wk old Shunt and control lambs. A representative image is shown for each Western blot. PPARγ protein is significantly decreased in the Shunt lamb (2.7-fold, A), while the protein levels of HHMR (2.8-fold, B), Flk-1 (3-fold, C), FGF2 (2.1-fold, D), and caveolin-1 (4.6-fold, E) were all significantly increased. Data are means ± SE, n = 4, *P < 0.05 vs. control. All protein levels were normalized by reprobing with β-actin.
DISCUSSION
In this study, we utilized the powerful microarray technique to investigate gene expression changes following PPARγ inhibition in PAEC and assessed their potential roles in PH. Based on previous findings that loss of PPARγ is associated with PH (2) and PPARγ activation with PPARγ agonists significantly reduces the PH phenotype in a variety of rodent models (18, 34, 35), we hypothesized that the global gene expression changes caused by PPARγ inhibition underlie the pulmonary vascular remodeling in PH. We found 100 genes and ESTs were upregulated by >1.5-fold and 21 genes and ESTs were downregulated by >1.3-fold in PPARγ-inhibited PAEC (P < 0.05). Compared with the usual cutting point using a fold change at 2.0, our array results exhibited small fold changes. We believe this is partly due to the moderate inhibition of PPARγ transcription factor binding by GW9662 treatment. However, this was chosen to more accurately mimic the clinical situation where PPARγ signaling is attenuated but not completely ablated. However, it is also possible that the attenuation of hybridization signals inherent in cross-species hybridization (3) may also play a role in the limiting the identified fold change. Furthermore, it is possible that due to cross-species differences in sequence our studies may not detect all genes that are dysregulated by loss of PPARγ binding activity. To alleviate this concern we also determined the false discovery rate for each gene that had a P < 0.05 and found that the range was between 5 and 25% with the majority being <20%.
The upregulated genes can be broadly classified into four categories: cell cycle-related genes, angiogenesis-related genes, ubiquitin-related genes, and zinc finger proteins. FLK-1, HMMR, FGF2, and caveolin-1 are among the top upregulated genes, and their regulations were confirmed at both mRNA and protein levels in PAEC exposed to the PPARγ pharmacological inhibitor GW9662, and these results were reproduced in PPARγ-depleted HMVEC. We further demonstrated PPARγ inhibition led to re-entry of cell cycle at G1/S phase, cell proliferation, and exacerbated VEGF-induced endothelial barrier disruption. Finally we identified the downregulation of PPARγ and the upregulation of FLK-1, HMMR, FGF2, and caveolin-1 in the peripheral lung tissues of an ovine model of PH with increased pulmonary blood flow, thus establishing a link between PPARγ-inhibition mediated changes in PAEC gene expression in vitro and the development of the PH phenotype.
PH is the most common manifestation of pulmonary vascular diseases caused by diverse clinical conditions including chronic obstructive pulmonary diseases, congenital heart defects, autoimmune diseases, etc (42). The decrease in NO and prostacyclin and the increase in ET-1, NADPH oxidase, and Rho/Rho kinase activity have been demonstrated to be important mediators of pulmonary vasoconstriction and thus play important roles in PH (35, 39). However, our understanding of the pathogenesis of PH is far from complete as little is known about the onset of pulmonary vascular remodeling in this disease process. In addition, despite the current therapeutic measures, which include inhaled NO and phosphodiesterase-5 inhibitor (sildenafil) to target the NO/cGMP axis, prostacyclin analogs to target the prostacyclin axis, and endothelin receptor blockers to target the endothelin axis, PH remains a great source of morbidity and mortality. The inadequacy of these measures to regress pulmonary vascular remodeling may be one reason for the suboptimal therapeutic efficiency. Thus, it is imperative to identify the underlying mechanisms for the initiation of pulmonary vascular remodeling and develop new therapeutic strategies.
Plexiform lesions and concentric-obliterative muscularization lesions can both occur in PH, in which plexiform lesions occur as solitary lesions, whereas concentric-obliterative lesions appear to be only associated with, and proximal to, plexiform structures (11). The cells in the plexiform lesions have an endothelial origin (51). One characteristic of ECs is that when they reach confluence, contact inhibition prevents them from further dividing and they become quiescent maintaining a monolayer. However, rather than growing in a typical monolayer that defines a patent vessel, the ECs in the plexiform lesions exhibit a proproliferative and apoptosis-resistant phenotype in which the “law of the monolayer” is lost (51). It is still not clear what produces the “proliferative” phenotype of these ECs. Previous studies showed that the ECs in the plexiform lesion stained negative for the cyclin-kinase inhibitor marker p27Kip1 (11). Here we demonstrate that PPARγ inhibition increases gene expression of a list of cell cycle-related genes, including the G1/S-associated cyclin C (44) and G2/M-associated cyclin A2 (55). We also demonstrated that PPARγ inhibition significantly increased the percentage of S-phase cells. Moreover, we have directly shown that depleting PPARγ promotes HMVEC proliferation. These findings are consistent with previous reports that PPARγ agonists block events for re-entry of quiescent VSMCs into cell cycle, specifically G0/G1 to S phase, inhibiting VSMCs proliferation (23, 56). The arrest on VSMCs cell cycle progression has been proposed to be associated with the antiatherogenic utilities of PPARγ agonists on systematic vascular diseases (12, 23). In VSMCs, PPARγ agonists seem to induce cell cycle arrest by inhibiting retinoblastoma protein phosphorylation and upregulating the CDK inhibitor p27Kip1 (6, 22). However, in our studies, we found cyclin C protein expression was increased, which may play an important role in expediting G1/S progression. We were not able to detect a significant increase in G2/M-phase cells, nor can we confirm an increase in cyclin A2 at protein level by Western blot analysis. However, our data highlight a potential role for PPARγ signaling in regulating cell proliferation and normal cell cycle progression through the G1/S phase. The re-entry of cell cycle at G1/S in PPARγ-inhibited PAEC may be a critical event in the plexiform lesion formation in PH. However, further studies will be required to test this hypothesis.
Although our results demonstrate that PPARγ inhibition overdrives PAEC cell cycle progression at G1/S, which most likely gives them a proliferative phenotype, these ECs may also need an amenable environment for their outgrowth. In fact, the plexiform lesions are not end-stage lesions, they are dynamic angiogenic processes (10, 54). The executive summary from the world symposium Primary Pulmonary Hypertension 1998 states that the plexiform lesion may represent “endothelial cells that are involved prominently in angiogenesis, perhaps akin to a neoplastic process” (19). In keeping with this, we discovered that PPARγ inhibition upregulated mRNA and protein expression of a list of angiogenesis-related genes. For example, the VEGF type-2 receptor Flk-1 was upregulated as was FGF-2, another important angiogenic factor (33, 52). In addition to upregulating VEGF receptor 2/Flk-1, which has an angiogenic implication, we also found that PPARγ inhibition both acutely and chronically exacerbated VEGF-induced disruption of the endothelial barrier function in PAEC as well as in PPARγ depleted HMVEC. In vivo, this loss of barrier function would potentially expose the underlying cells to circulating mitogens (51) and therefore may promote VSMCs proliferation and aggravate vascular remodeling in PH. We also found that HMMR was upregulated by PPARγ inhibition in PAEC and HMVEC. HMMR has been shown to play an important role in EC migration (45, 48). We further verified the upregulation of these angiogenic proteins in the peripheral lung tissues of an ovine model of PH with increased pulmonary blood flow. Thus, we conclude that PPARγ inhibition not only confers a proliferative phenotype on PAEC by regulating cell-cycle progression but also provides an angiogenic milieu by increasing gene expression of angiogenic factors. Indeed, previous studies have shown that PPARγ activation caused antiangiogenic effects in ECs by downregulating VEGF and FGF-2 expression (29, 30). These findings are consistent with our findings that PPARγ inhibition increased VEGF-2 receptor and FGF2 expression both in vitro and in vivo. We also demonstrated that PPARγ inhibition upregulated caveolin-1 expression at both mRNA and protein levels in PAEC and HMVEC. Furthermore, caveolin-1 was found to be upregulated in the peripheral lung tissues of our ovine model of PH with increased pulmonary blood flow. Decreases in NO production are well established in PH (35), although these decreases are not necessarily accompanied by a decrease in eNOS expression (35, 38). Caveolin-1 is a well-recognized negative regulator of eNOS activity through its ability to limit calcium-calmodulin binding (59). Thus, we propose that the upregulation of caveolin-1 may play an important role in the sequential decrease in NO production we have observed in our ovine model of PH (36). In addition, since NO is known to inhibit VSMC proliferation (1), the upregulation of caveolin-1 may also indirectly promote pulmonary VSMC proliferation and vascular muscularization by reducing NO production. However, it is worth noting that due to the limitations of the peripheral lung tissue used in our in vivo studies we cannot definitively ascribe the protein changes to the endothelium.
We also found that PPARγ inhibition led to the upregulation of a number of ubiquitin-proteasome system (UPS)-related genes. These include the ubiquitin-protein ligase NEDD4, Cullin3, Cullin5, and ubiquitin-conjugating enzyme E2, J2, and H. The UPS is the major pathway for mediating nonlysosomal proteolysis of intracellular proteins (57). It plays important roles in a variety of fundamental cellular processes such as proliferation, cell cycle progression, DNA damage repair, and cell death (57). Interactions between PPARγ and the UPS could thus affect multiple cellular pathways (16). PPARγ signaling can alter protein levels and downstream pathways not only by altering their expression but also by modulating the activity of the UPS in target-specific manner (16). Conversely, the UPS could also play a role in regulating the level and activity of PPARγ as well as its co-activators and co-repressors. Interestingly, alterations in UPS are frequently found in cancer (17). Oncogenes and tumor suppressor proteins are often the targets of UPS alterations (8), and during the oncogenic process their susceptibility to proteasomal-dependent degradation can be altered (41). However, at the moment little is known about the relationship between the UPS and PH, and further studies will be required to identify the role, if any, of UPS in the development of PH.
Finally, we also found that PPARγ inhibition upregulated a number of zinc finger genes. Zinc finger proteins have diverse functions. Some of them are transcription factors and are involved in regulating gene expression (27). Other zinc finger proteins are known to modulate PPARγ transcription factor binding. For example, the DNA-binding domain (DBD) of PPARγ has two sets of zinc fingers (20); the specificity and polarity of PPAR-DNA binding seem to be at least in part due to features in the zinc finger domains of PPARγ (21). In addition, the DNA-binding partner of PPARγ, RXR, also has a DBD with two zinc fingers (28). Thus, we predict that some of the upregulated zinc finger proteins, including the zinc finger protein 181 and zinc finger-CCHC domain containing 8, may serve either as transcription factors for downstream genes or as co-activators or co-repressors for PPARγ signaling. Another distinct family of upregulated zinc finger proteins belongs to the ring finger protein family, including the ring finger protein-transmembrane 1, the ring finger protein 168, and the ring finger protein 20. Ring finger proteins can be involved in mediating ubiquitin ligase activity (24), suggesting a link to the UPS system we have also found to be altered by PPARγ inhibition. We speculate that because of the diverse functions of the zinc finger proteins, they may play diverse roles in PH.
In conclusion, we have identified a number of gene families that are regulated by the loss of PPARγ signaling. We found that PPARγ inhibition promoted cell-cycle progression and cell proliferation and upregulated angiogenesis-related genes in PAEC and HMVEC. From this we speculate that antiproliferation and antiangiogenic therapeutic measures could be utilized in addition to the current vasodilator therapies for the treatment of PH. The inhibition of PPARγ signaling also led to upregulation of ubiquitin-related proteins and zinc finger proteins; however, their implications in PH remain to be defined but potentially open new avenues for innovative research.
GRANTS
This research was supported in part by National Institutes of Health (NIH) Grants HL-60190 (to S. M. Black), HL-67841 (to S. M. Black), HL-72123 (to S. M. Black), HL-70061 (to S. M. Black), HL-084739 (to S. M. Black), R21HD-057406 (to S. M. Black), and HL-61284 (to J. R. Fineman); an award from the American Heart Association (AHA) Mountain States Affiliates (to S. M. Black); and by a grant from the Fondation Leducq (to S. M. Black, J. R. Fineman, S. Fratz, J. Hess, and A. Göerlach). A. Smith was supported in part by NIH training Grant 5T32HL-06699, and S. Aggarwal was supported in part by a predoctoral fellowship from the AHA Southeast Affiliates. This work was also supported in part by Programmatic Development (to S. M. Black, JDC, and ADV) and Seed (to S. Kumar) awards from the Cardiovascular Discovery Institute of the Medical College of Georgia.
DISCLOSURES
No conflicts of interest are declared by the author(s).
REFERENCES
- 1.Ahanchi SS, Varu VN, Tsihlis ND, Martinez J, Pearce CG, Kapadia MR, Jiang Q, Saavedra JE, Keefer LK, Hrabie JA, Kibbe MR. Heightened efficacy of nitric oxide-based therapies in type II diabetes mellitus and metabolic syndrome. Am J Physiol Heart Circ Physiol 295: H2388–H2398, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ameshima S, Golpon H, Cool CD, Chan D, Vandivier RW, Gardai SJ, Wick M, Nemenoff RA, Geraci MW, Voelkel NF. Peroxisome proliferator-activated receptor gamma (PPARgamma) expression is decreased in pulmonary hypertension and affects endothelial cell growth. Circ Res 92: 1162–1169, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Bar-Or C, Czosnek H, Koltai H. Cross-species microarray hybridizations: a developing tool for studying species diversity. Trends Genet 23: 200–207, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Bishop-Bailey D, Hla T. Endothelial cell apoptosis induced by the peroxisome proliferator-activated receptor (PPAR) ligand 15-deoxy-Delta12, 14-prostaglandin J2. J Biol Chem 274: 17042–17048, 1999 [DOI] [PubMed] [Google Scholar]
- 5.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Bruemmer D, Blaschke F, Law RE. New targets for PPARgamma in the vessel wall: implications for restenosis. Int J Obes (Lond) 29, Suppl 1: S26–S30, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Burg J, Krump-Konvalinkova V, Bittinger F, Kirkpatrick CJ. GM-CSF expression by human lung microvascular endothelial cells: in vitro and in vivo findings. Am J Physiol Lung Cell Mol Physiol 283: L460–L467, 2002 [DOI] [PubMed] [Google Scholar]
- 8.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1: 193–199, 1999 [DOI] [PubMed] [Google Scholar]
- 9.Collins AR. Pleiotropic vascular effects of PPARgamma ligands. Drug News Perspect 16: 197–204, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Cool CD, Kennedy D, Voelkel NF, Tuder RM. Pathogenesis and evolution of plexiform lesions in pulmonary hypertension associated with scleroderma and human immunodeficiency virus infection. Hum Pathol 28: 434–442, 1997 [DOI] [PubMed] [Google Scholar]
- 11.Cool CD, Stewart JS, Werahera P, Miller GJ, Williams RL, Voelkel NF, Tuder RM. Three-dimensional reconstruction of pulmonary arteries in plexiform pulmonary hypertension using cell-specific markers. Evidence for a dynamic and heterogeneous process of pulmonary endothelial cell growth. Am J Pathol 155: 411–419, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Nigris F, Lerman LO, Napoli C. New insights in the transcriptional activity and coregulator molecules in the arterial wall. Int J Cardiol 86: 153–168, 2002 [DOI] [PubMed] [Google Scholar]
- 13.Duan SZ, Usher MG, Mortensen RM. Peroxisome proliferator-activated receptor-gamma-mediated effects in the vasculature. Circ Res 102: 283–294, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Favier J, Kempf H, Corvol P, Gasc JM. Coexpression of endothelial PAS protein 1 with essential angiogenic factors suggests its involvement in human vascular development. Dev Dyn 222: 377–388, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Forman BM, Chen J, Evans RM. The peroxisome proliferator-activated receptors: ligands and activators. Ann NY Acad Sci 804: 266–275, 1996 [DOI] [PubMed] [Google Scholar]
- 16.Genini D, Carbone GM, Catapano CV. Multiple interactions between peroxisome proliferators-activated receptors and the ubiquitin-proteasome system and implications for cancer pathogenesis. PPAR Res 2008: 195065, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 100: 57–70, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Hansmann G, Wagner RA, Schellong S, Perez VA, Urashima T, Wang L, Sheikh AY, Suen RS, Stewart DJ, Rabinovitch M. Pulmonary arterial hypertension is linked to insulin resistance and reversed by peroxisome proliferator-activated receptor-gamma activation. Circulation 115: 1275–1284, 2007. [DOI] [PubMed] [Google Scholar]
- 19.Haworth SG. The pathology of pulmonary hypertension. Primary Pulmonary Hypertension: Executive Summary from the 2nd World Symposium Primary Pulmonary Hypertension Évian-les-Bains, France, 1998 [Google Scholar]
- 20.Hihi AK, Michalik L, Wahli W. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell Mol Life Sci 59: 790–798, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu MH, Palmer CN, Song W, Griffin KJ, Johnson EF. A carboxyl-terminal extension of the zinc finger domain contributes to the specificity and polarity of peroxisome proliferator-activated receptor DNA binding. J Biol Chem 273: 27988–27997, 1998 [DOI] [PubMed] [Google Scholar]
- 22.Hsueh WA, Jackson S, Law RE. Control of vascular cell proliferation and migration by PPAR-gamma: a new approach to the macrovascular complications of diabetes. Diabetes Care 24: 392–397, 2001 [DOI] [PubMed] [Google Scholar]
- 23.Hsueh WA, Law RE. PPARgamma and atherosclerosis: effects on cell growth and movement. Arterioscler Thromb Vasc Biol 21: 1891–1895, 2001 [DOI] [PubMed] [Google Scholar]
- 24.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102: 549–552, 2000 [DOI] [PubMed] [Google Scholar]
- 25.Kim EH, Surh YJ. The role of 15-deoxy-delta(12,14)-prostaglandin J(2), an endogenous ligand of peroxisome proliferator-activated receptor gamma, in tumor angiogenesis. Biochem Pharmacol 76: 1544–1553, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Kliewer SA, Willson TM. The nuclear receptor PPARgamma - bigger than fat. Curr Opin Genet Dev 8: 576–581, 1998 [DOI] [PubMed] [Google Scholar]
- 27.Klug A. Zinc finger peptides for the regulation of gene expression. J Mol Biol 293: 215–218, 1999 [DOI] [PubMed] [Google Scholar]
- 28.Lee MS, Kliewer SA, Provencal J, Wright PE, Evans RM. Structure of the retinoid X receptor alpha DNA binding domain: a helix required for homodimeric DNA binding. Science 260: 1117–1121, 1993 [DOI] [PubMed] [Google Scholar]
- 29.Margeli A, Kouraklis G, Theocharis S. Peroxisome proliferator activated receptor-gamma (PPAR-gamma) ligands and angiogenesis. Angiogenesis 6: 165–169, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Marx N, Duez H, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors and atherogenesis: regulators of gene expression in vascular cells. Circ Res 94: 1168–1178, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Marx N, Walcher D. Vascular effects of PPARgamma activators - from bench to bedside. Prog Lipid Res 46: 283–296, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Mirzapoiazova T, Kolosova I, Usatyuk PV, Natarajan V, Verin AD. Diverse effects of vascular endothelial growth factor on human pulmonary endothelial barrier and migration. Am J Physiol Lung Cell Mol Physiol 291: L718–L724, 2006. [DOI] [PubMed] [Google Scholar]
- 33.Nicoli S, De Sena G, Presta M. Fibroblast growth factor 2-induced angiogenesis in zebrafish: the zebrafish yolk membrane (ZFYM) angiogenesis assay. J Cell Mol Med 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nisbet RE, Bland JM, Kleinhenz DJ, Mitchell PO, Walp ER, Sutliff RL, Hart CM. Rosiglitazone attenuates chronic hypoxia-induced pulmonary hypertension in a mouse model. Am J Respir Cell Mol Biol 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nisbet RE, Sutliff RL, Hart CM. The role of peroxisome proliferator-activated receptors in pulmonary vascular disease. PPAR Res 2007: 18797, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oishi PE, Wiseman DA, Sharma S, Kumar S, Hou Y, Datar SA, Azakie A, Johengen MJ, Harmon C, Fratz S, Fineman JR, Black SM. Progressive dysfunction of nitric oxide synthase in a lamb model of chronically increased pulmonary blood flow: a role for oxidative stress. Am J Physiol Lung Cell Mol Physiol 295: L756–L766, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearse DB, Shimoda LA, Verin AD, Bogatcheva N, Moon C, Ronnett GV, Welsh LE, Becker PM. Effect of cGMP on lung microvascular endothelial barrier dysfunction following hydrogen peroxide. Endothelium 10: 309–317, 2003 [DOI] [PubMed] [Google Scholar]
- 38.Polikandriotis JA, Mazzella LJ, Rupnow HL, Hart CM. Peroxisome proliferator-activated receptor gamma ligands stimulate endothelial nitric oxide production through distinct peroxisome proliferator-activated receptor gamma-dependent mechanisms. Arterioscler Thromb Vasc Biol 25: 1810–1816, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Rabinovitch M. Molecular pathogenesis of pulmonary arterial hypertension. J Clin Invest 118: 2372–2379, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy VM, Meyrick B, Wong J, Khoor A, Liddicoat JR, Hanley FL, Fineman JR. In utero placement of aortopulmonary shunts. A model of postnatal pulmonary hypertension with increased pulmonary blood flow in lambs. Circulation 92: 606–613, 1995 [DOI] [PubMed] [Google Scholar]
- 41.Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann Intern Med 145: 676–684, 2006 [DOI] [PubMed] [Google Scholar]
- 42.Rich S, Rabinovitch M. Diagnosis and treatment of secondary (non-category 1) pulmonary hypertension. Circulation 118: 2190–2199, 2008 [DOI] [PubMed] [Google Scholar]
- 43.Rios-Vazquez R, Marzoa-Rivas R, Gil-Ortega I, Kaski JC. Peroxisome proliferator-activated receptor-gamma agonists for management and prevention of vascular disease in patients with and without diabetes mellitus. Am J Cardiovasc Drugs 6: 231–242, 2006 [DOI] [PubMed] [Google Scholar]
- 44.Sage J. Cyclin C makes an entry into the cell cycle. Dev Cell 6: 607–608, 2004 [DOI] [PubMed] [Google Scholar]
- 45.Savani RC, Cao G, Pooler PM, Zaman A, Zhou Z, DeLisser HM. Differential involvement of the hyaluronan (HA) receptors CD44 and receptor for HA-mediated motility in endothelial cell function and angiogenesis. J Biol Chem 276: 36770–36778, 2001 [DOI] [PubMed] [Google Scholar]
- 46.Schoonjans K, Martin G, Staels B, Auwerx J. Peroxisome proliferator-activated receptors, orphans with ligands and functions. Curr Opin Lipidol 8: 159–166, 1997 [DOI] [PubMed] [Google Scholar]
- 47.Schoonjans K, Staels B, Auwerx J. The peroxisome proliferator activated receptors (PPARS) and their effects on lipid metabolism and adipocyte differentiation. Biochim Biophys Acta 1302: 93–109, 1996 [DOI] [PubMed] [Google Scholar]
- 48.Slevin M, Krupinski J, Gaffney J, Matou S, West D, Delisser H, Savani RC, Kumar S. Hyaluronan-mediated angiogenesis in vascular disease: uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biol 26: 58–68, 2007. [DOI] [PubMed] [Google Scholar]
- 49.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004 [DOI] [PubMed] [Google Scholar]
- 50.Spiegelman BM. Peroxisome proliferator-activated receptor gamma: A key regulator of adipogenesis and systemic insulin sensitivity. Eur J Med Res 2: 457–464, 1997 [PubMed] [Google Scholar]
- 51.Stevens T. Molecular and cellular determinants of lung endothelial cell heterogeneity. Chest 128: 558S–564S, 2005 [DOI] [PubMed] [Google Scholar]
- 52.Tomanek RJ, Hansen HK, Christensen LP. Temporally expressed PDGF and FGF-2 regulate embryonic coronary artery formation and growth. Arterioscler Thromb Vasc Biol 28: 1237–1243, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Touyz RM, Schiffrin EL. Peroxisome proliferator-activated receptors in vascular biology-molecular mechanisms and clinical implications. Vascul Pharmacol 45: 19–28, 2006 [DOI] [PubMed] [Google Scholar]
- 54.Tuder RM, Groves B, Badesch DB, Voelkel NF. Exuberant endothelial cell growth and elements of inflammation are present in plexiform lesions of pulmonary hypertension. Am J Pathol 144: 275–285, 1994 [PMC free article] [PubMed] [Google Scholar]
- 55.Viallard JF, Lacombe F, Belloc F, Pellegrin JL, Reiffers J. [Molecular mechanisms controlling the cell cycle: fundamental aspects and implications for oncology]. Cancer Radiother 5: 109–129, 2001 [DOI] [PubMed] [Google Scholar]
- 56.Wakino S, Law RE, Hsueh WA. Vascular protective effects by activation of nuclear receptor PPARgamma. J Diabetes Complications 16: 46–49, 2002 [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol 3: 255–261, 2006 [PubMed] [Google Scholar]
- 58.Wedgwood S, Mitchell CJ, Fineman JR, Black SM. Developmental differences in the shear stress-induced expression of endothelial NO synthase: changing role of AP-1. Am J Physiol Lung Cell Mol Physiol 284: L650–L662, 2003 [DOI] [PubMed] [Google Scholar]
- 59.Williams TM, Lisanti MP. The Caveolin genes: from cell biology to medicine. Ann Med 36: 584–595, 2004 [DOI] [PubMed] [Google Scholar]