Abstract
The ubiquitin-proteasome pathway (UPP) is responsible for most programmed turnover of proteins in eukaryotic cells, and this activity has been known for some time to be involved in transcriptional regulation. More recently, intersections of the UPP and transcription have been discovered that are not proteolytic in nature and appear to revolve around the chaperonin-like activities of the ATPases in the 19 S regulatory subunit of the proteasome. Moreover, monoubiquitylation, which does not signal degradation, has been found to be a key modification of many transcription factors and histones. These various non-proteolytic roles of the UPP in transcription are reviewed here, and plausible mechanistic models are discussed.
Keywords: DNA/Protein Interaction, DNA/Transcription, Proteases/Proteasomes, Proteases/Ubiquitination, Transcription, Transcription/Elongation Factors, Transcription/RNA Polymerase II, Transcription/Yeast
Introduction
Most non-lysosomal protein degradation occurs via the UPP2 in eukaryotic cells (1). In most cases, the protein targeted for degradation is modified with a Lys48-linked poly-Ub chain, a modification catalyzed by ULs, of which there are three types. E1 and E2 ULs are involved in activating the ubiquitin molecule for conjugation, whereas E3 ULs act as matchmakers between the activated Ub-E2 intermediate and substrate proteins. Polyubiquitylated proteins interact with the proteasome, a large multiprotein complex with three proteolytic active sites sequestered inside a barrel-like core 20 S complex (2). The opening to the interior of the barrel is so small that the substrate protein must be unfolded and “fed” through by the action of six AAA class ATPases, now called Rpt1–6. These are part of the 19 S RP that sits immediately atop the 20 S CP. The 19 S RP itself can be dissociated by high salt buffers into a base, which includes the six ATPases, Rpn1, Rpn2, and a “lid” subcomplex (3). All six of the genes that encode these proteins are essential in yeast, indicating that they have non-identical functions. The classical proteolytic activity of the UPP has long been known to regulate transcription indirectly, e.g. by keeping the levels of gene-specific transactivators low under basal conditions. More recently, a more intimate relationship between UPP-mediated turnover of activators, coactivators and other transcription actors has been found to be highly stimulatory for the expression of some genes (4–6). However, this minireview focuses on a rather different intersection of the UPP and eukaryotic transcription, which involves non-proteolytic activities of UPP proteins.
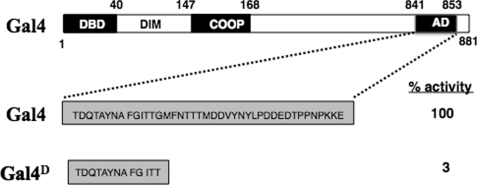
The first hint of such a radical idea was the finding by Johnston and co-workers in 1992 (7) that specific alleles of the Saccharomyces cerevisiae SUG1 and SUG2 genes (sug1-1 and sug2-1), which encode Rpt6 and Rpt4, respectively, suppress the “no growth on galactose” phenotype of gal4D. gal4D encodes a truncated version of the yeast Gal4 activator (8) that lacks about two-thirds of the C-terminal AD (Fig. 1) (7). At the time of this study, it was thought that Sug1 and Sug2 were coactivators whose interaction with Gal4 was weakened by the truncation in Gal4D and reconstituted by a point mutation. When it was discovered soon thereafter that SUG1 and SUG2 encode Rpt6 and Rpt4, respectively (9), most of the community wrote off this result as likely an indirect effect of reducing the rate of proteolysis of a weakly functional Gal4D protein, thus allowing the yeast to limp along. However, other alleles of SUG1 and SUG2 isolated in other genetic screens did not rescue growth of the gal4D strain, despite the fact that these alleles allowed the Gal4D protein to increase to a level higher than that observed in the sug1-1 and sug2-1 backgrounds. Eventually, experiments were published that directly refuted this altered proteolysis model (10), leaving the true mechanism of this effect a mystery.
FIGURE 1.
Schematic representation of the truncation mutant Gal4D. The crude domain structure of the yeast Gal4 protein is shown at the top. The 34-residue core AD is truncated to a 12-amino acid peptide in Gal4D. All other domains of the 881-amino acid protein are unchanged. This protein has only 3% the activity of wild-type Gal4 when expressed at physiological levels. DIM, dimerization domain; COOP, cooperativity domain.
Proteasomal ATPases Stimulate Transcriptional Initiation and Elongation without Engaging in Protein Degradation
With strong evidence in hand that altered proteolysis could not explain the suppression of gal4D by sug1-1 or sug2-1, Ferdous et al. (11) performed experiments designed to investigate whether the proteasomal ATPases might have a direct non-proteolytic role in transcription. An important clue in this regard was an earlier finding that a different allele of SUG1, sug1-20, suppressed a mutation in CDC68 (12), which encodes one of the components of an elongation factor called FACT (13). FACT facilitates elongation through nucleosomes. Furthermore, Rpt6 and FACT co-immunoprecipitate (14).
It was found that a sug1-20 strain was highly sensitive to 6-azauracil, a classic indicator of elongation defects (15), but strains with mutations in 20 S proteasome core component-encoding genes were not (11). Moreover, inactivation of the temperature-sensitive Sug1-20 protein or addition of anti-Rpt6 or anti-Rpt4 antibodies to a transcriptionally active yeast whole cell extract strongly inhibited transcription, whereas inhibition of the proteolytic activity of the proteasome had no effect. Addition of purified 19 S RP completely lacking the 20 S CP to the inactivated extract reconstituted transcriptional activity. These data argued strongly for a direct and non-proteolytic role of the 19 S RP in elongation (11).
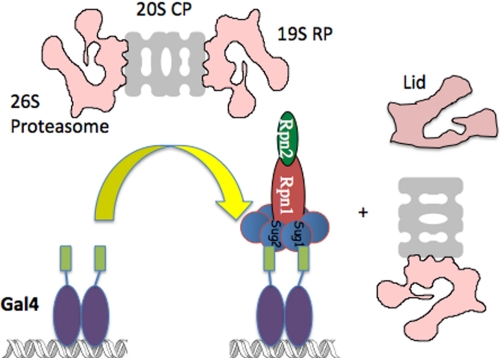
This conclusion was highly controversial at the time. Since this time, however, other studies have appeared that demonstrated non-proteolytic roles for the proteasomal ATPases in driving the expression of other genes (16–18). Further support came from studies of the physical interaction of the Gal4 AD with the 26 S proteasome (19). In vitro, when a GST-Gal4 AD fusion protein was mixed with immunopurified 26 S proteasome, the GST-AD protein retained Rpt1–Rpt6, Rpn1, and Rpn2 but excluded the 20 S core CP and the lid subcomplex of the 19 S RP (Fig. 2), even though the input 26 S proteasome was 100% intact as judged by pulldown with GST-Rad23 protein, which binds the entire proteasome. Additionally, when the association of different proteasomal proteins with GAL or heat shock promoters was analyzed by ChIP, it was found that gene activation resulted in the association of the ATPases, but not the 20 S CP or lid constituents, with the promoter (19, 20). Moreover, strong ChIP signals were observed for the ATPases, but again not the 20 S CP or lid proteins, throughout the gene, consistent with this subcomplex, called APIS, playing an important role in elongation (19). APIS includes the base of the 19 S RP but may also contain additional proteins besides Rpt1–6, Rpn1, and Rpn2.
FIGURE 2.
Gal4 extracts a subcomplex called APIS from the 26 S proteasome. APIS contains at least Rpt1–Rpt6, Rpn1, and Rpn2. See text for details. Sug1 = Rpt6, and Sug2 = Rpt4. The green rectangles represent the ADs of the Gal4 dimer.
The presence of the ATPases at the GAL enhancer also suggests a possible role in initiation of transcription, consistent with the observed interaction of the Rpt proteins with initiation factors (14, 21–23). Indeed, Workman and co-workers (24) have demonstrated that the proteasomal ATPases stimulate the recruitment of the SAGA complex to promoters in yeast.
Monoubiquitylation Stimulates the Function of Many Activators
While the line of experimentation described above was proceeding, Tansey and co-workers (25) reported a remarkable and unexpected finding that revealed yet another odd connection between the UPP and transcription. Previous reports had already suggested a positive role of E3 ULs in transcriptional activation. For example, a general inverse correlation between the activity of transactivators and their protein levels in cells had been noted. In addition, the knock-out or knockdown of several E3 ULs strongly suppressed the activated transcription of certain genes (26), but the mechanism underlying these observations was unclear. The breakthrough was the realization that this stimulatory effect was due, at least in part, to the monoubiquitylation of transactivators (25) and, as was realized later, coactivators as well (27). Their study was directed toward identification of the E3 UL that operates on the artificial transactivator LexA-VP16 in S. cerevisiae. Using a candidate gene approach, they found that deletion of MET30 strongly suppressed LexA-VP16-mediated transcription. In these cells, the levels of the activator were quite high, but its activity was low. Remarkably, when a monoubiquitin moiety was fused to the N terminus of the artificial activator genetically, the activity of the protein was reconstituted. These data suggested that monoubiquitylation of the transactivator was the critical event mediated by Met30, although the isolation of a bona fide, natively ubiquitylated activator was not accomplished in this study.
This study was soon followed by similar reports of stimulatory monoubiquitylation of other activators, and in some of these cases, the natively monoubiquitylated proteins were observed directly (28–30). Because degradation of a protein generally requires its modification with a Lys48-linked polyubiquitin chain of at least four monomers (31), these studies argued strongly for yet another non-proteolytic effect of the UPP on activated gene transcription.
As is described in other reviews, there is now evidence that in some, but not all, systems, this monoubiquitylation may be coupled inextricably to polyubiquitylation events that result in the subsequent destruction of functional transactivators by the proteasome (5, 26). This coupling between “licensing” of activator function by monoubiquitylation and its subsequent proteolysis provides an elegant mechanism for the cell to keep the transcription of critical activated genes under tight control.
How Does Monoubiquitylation Stimulate Activated Gene Expression?
For quite some time after its initial discovery, the mechanism by which activator monoubiquitylation stimulates its function remained a mystery. The LexA-VP16 study (25) provided some interesting hints. The VP16 AD does not contain any lysine residues, so monoubiquitylation must have occurred on the LexA DBD. Because this is a bacterial protein, one cannot invoke a model in which this modification alters the conformation of the protein in some specific fashion, thus allowing it to interact with some other transcription factor. Instead, it seemed that the ubiquitin moiety itself must interact directly with something that is rate-limiting for transcription. The experiments that led to at least a partial answer to this question emerged from an unlikely direction.
As described above, efforts to understand the suppression of the gal4D mutation by sug1-1 and sug2-1 eventually led to the discovery of a role for the proteasomal ATPases in transcriptional elongation. Ironically, however, the suppressor mutations were cleanly recessive (7), suggesting that they alleviated some inhibitory activity that antagonized the function of the Gal4D protein. This seemed difficult to reconcile with a positive role of Rpt4 and Rpt6 in elongation (11). Another troubling aspect of the findings to that point was that the proteasomal ATPases are recruited to the promoter through direct interactions of Rpt6 and Rpt4 with the ADs of the Gal4 homodimer. In proteolysis, the ATPases function to unfold proteins that interact with the 19 S RP. Why do they not unfold Gal4?
Indeed, when the effect of purified 19 S RP or 26 S proteasome on immobilized Gal4-VP16·DNA complexes was examined, it was found that the activator was dissociated rapidly from the DNA, although not proteolyzed (32). This reaction required ATP hydrolysis and direct binding of the ATPases to the VP16 AD. Because the proteasomal ATPases are highly abundant proteins in the nucleus (33), how do native activators resist this presumably highly inhibitory activity? Possible explanations included a post-translational modification or perhaps the presence of some activator-binding protein that protects it. An experiment designed to probe the former possibility was highly illuminating. When the Gal4-VP16·DNA complex was incubated with HeLa nuclear extract, an essentially quantitative monoubiquitylation of the activator was observed (32). Moreover, this event was found to be dependent on a preceding phosphorylation event at Ser22, located in the same domain (34). Remarkably, a DNA complex containing phosphorylated and monoubiquitylated Gal4-VP16 was completely resistant to disruption by the proteasomal ATPases (32).
Could this stripping activity be the putative inhibitory activity of the proteasomal ATPases suggested by the recessive nature of the sug1-1 and sug2-1 alleles in suppressing gal4D? And was ablation of this stripping activity by the point mutations in sug1-1 and sug2-1 the molecular basis of this suppression? To address this hypothesis, Gal4D function at the level of promoter occupancy was tested. Standard views of how activators work would certainly not anticipate that truncation of part of the AD would compromise DNA-binding activity. Indeed, a Gal4D derivative binds well to promoters in vitro (35). Remarkably though, ChIP experiments showed clearly that the Gal4D protein fails to occupy GAL promoters in otherwise wild-type cells or in strains carrying sug1 or sug2 alleles that do not suppress gal4D. In the sug1-1 strain, Gal4D occupied the promoter ∼60% as well as native Gal4, and the Gal4D-stimulated output of GAL gene expression was ∼60% of that driven by the wild-type activator. This demonstrated that the defect in Gal4D activity was due entirely to reduced promoter occupancy. Moreover, when the proteasome was purified from sug1-1 yeast, this complex showed a sharply reduced ability to strip Gal4 AD constructs from DNA in vitro, although the Sug1-1 protein-containing proteasome remained competent in proteolysis and the ability to bind to ADs. These data (36) argue that Gal4D is indeed hypersensitive to the stripping activity of the wild-type proteasomal ATPases and that the point mutations encoded by sug1-1 and sug2-1 down-regulate this activity without affecting other functions of the ATPases or the proteasome in general.
Part of the Gal4 AD Is Critical for Activator Monoubiquitylation
Is the molecular basis of the hypersensitivity of Gal4D to the stripping activity of the ATPases due to poor monoubiquitylation of the truncated activator? Several lines of evidence support this (36). First, genetic fusion of a monoubiquitin to the N terminus of Gal4D resulted in the rescue of significant GAL gene transcription in a SUG1/SUG2 strain. Second, when a Gal4 derivative containing the entire DNA-binding and activation domains was immobilized on DNA in vitro and then treated with HeLa nuclear extract, it was monoubiquitylated almost quantitatively, and the protein remained bound to DNA. However, when the same experiment was conducted with the Gal4D-like protein (i.e. lacking two-thirds of the Gal4 AD), no ubiquitylation was observed, and the protein was stripped off of the DNA. Perhaps most convincing were ChIP experiments carried out with anti-Gal4 and anti-Ub antibodies in various genetic backgrounds. In wild-type cells, a strong Ub-dependent ChIP signal was observed on the enhancer region of the GAL7 promoter, as was a strong Gal4-dependent signal. Both signals were absent in a gal4D strain. Importantly, the Gal4-dependent ChIP signal, but not the Ub-dependent signal, was observed in a gal4D/sug1-1 strain. This showed that, in the suppressing background, the Gal4D protein is not monoubiquitylated but can occupy the promoter due to the reduced stripping activity of the Sug1-1-containing APIS complex.
These experiments demonstrated that deletion of the C-terminal two-thirds of the Gal4 AD ablates monoubiquitylation of the DBD and thus exposes the activator to the unfolding activity of the proteasomal ATPases. Is this region of the AD the docking site for the E3 UL that targets Gal4 (or the kinase that carries Ser22 phosphorylation, a prerequisite for ubiquitylation)? Consistent with this idea, a soluble peptide representing the region deleted in Gal4D was able to strongly suppress monoubiquitylation of a Gal4 construct in vitro, but control peptides or a peptide corresponding to the N-terminal region of the AD still present in Gal4D had no effect (36). Unfortunately, the E3 UL and kinase that mediate these modifications of Gal4 have not been identified. The E3 UL Dsg1/Mdm30 ubiquitylates Gal4, but this event does not affect Gal4-mediated transcription, so Dsg1 cannot be the E3 UL responsible for stabilizing Gal4-DNA interactions in vivo (37).
It is interesting to note that whether the C-terminal subregion of the Gal4 AD is a kinase- or E3 UL-docking site, it is clearly not a classical AD required for binding to other transcription factors. If Gal4D can hang onto the promoter, then it is a perfectly good activator. Moreover, Gal4-(1–841) is completely devoid of activity, although it can bind to DNA in vivo (7). Thus, these data reveal that the 12-amino acid peptide that is present in Gal4D but absent in Gal4-(1–841) is the core AD of the activator. Remarkably, this short fragment appears to be sufficient to interact with coactivators and APIS. The other 22 residues of the classical Gal4 AD are apparently required only for phosphorylation/ubiquitylation.
How Does Activator Monoubiquitylation Protect against “Stripping?”
A clue as to the mechanism of this protective effect of ubiquitin was the surprising finding that the addition of high levels of free ubiquitin to a test tube containing purified proteasome and a Gal4·DNA complex blocked stripping even though the Gal4 derivative was not monoubiquitylated (32, 38). This strongly suggested either a direct interaction between ubiquitin and the activator, which somehow rendered it insensitive to stripping, or a ubiquitin-APIS interaction, which somehow disrupted the ability of the complex to strip the activator from the DNA. Note that the concentration of free ubiquitin required to observe this effect was quite high (several μm), but the total cellular concentration of ubiquitin is at about this level, leading one to wonder how these data could be physiologically relevant. After all, if untethered ubiquitin can quench the stripping process and if total cellular concentration is at or above the IC50 for this effect, how can stripping be an important cellular process? A resolution to this dilemma was suggested by the further observation that Lys48-linked ubiquitin chains did not exhibit a protecting effect in this assay, arguing that a surface on ubiquitin that is sequestered in the Lys48-linked chains is responsible for the effect (32, 38). In addition, there are many different known ubiquitin-binding proteins in the cell. Thus, the free monoubiquitin concentration in the cell at any one time is likely far less than the low μm concentrations required to see this in trans effect in vitro.
But how is protection achieved? In a series of in vitro experiments that relied, in part, on a novel label transfer technology ideal for probing interactions in multiprotein complexes, Archer et al. (38) demonstrated that monoubiquitin, but not Lys48-linked Ub chains, binds directly to Rpt1 and Rpn1, two components of the APIS complex. Moreover, these Ub-Rpn1/Ub-Rpt1 contacts allosterically dissociate the binding of Rpt4 and Rpt6 to the Gal4 or VP16 AD. In other words, Ub interaction with the APIS complex drives it to release the activator. No interactions whatsoever could be detected between mono-Ub and the 26 S proteasome, arguing that the Ub-binding surfaces of Rpn1 and Rpt1 are occluded until the Gal4 or VP16 AD extracts APIS from the proteasome.
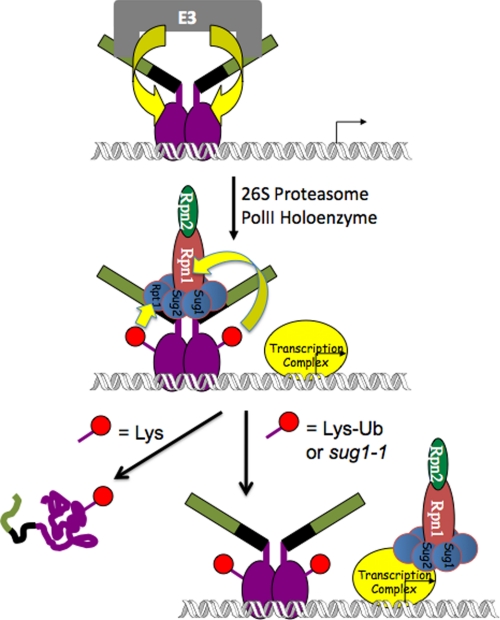
These protein interaction data are therefore consistent with a beautiful “hand-off” model (38) that explains how a monoubiquitylated activator can engage in a dangerous liaison with the APIS complex without becoming a victim to its unfolding activity (Fig. 3). The trick is to hold on only briefly. The first step in recruitment of the APIS complex to an active gene is for the ADs of the activator to extract the ATPases, Rpn1, and Rpn2 (APIS) from the 26 S proteasome. Association of APIS with the AD presumably initiates an unfolding reaction that, left unchecked, would result in processive unfolding of the activator beginning at the C-terminal AD of the protein (39, 40), eventually reaching the DBD and resulting in dissociation from the promoter. When Gal4 is monoubiquitylated, however, the appended ubiquitin molecules (Gal4 is a dimer (41)) bind to Rpt1 and Rpn1, driving APIS to release the activator before unfolding proceeds far enough to disrupt activator function. In the context of the preinitiation complex, Ub-mediated AD-APIS disruption may allow APIS to transfer to the nearby polymerase II holoenzyme, but there are no direct data to support this hypothesis.
FIGURE 3.
Model for the interaction of the Gal4 transactivator with the proteins in the UPP. It is proposed that the piece of the Gal4 AD missing in the truncated Gal4D protein (green rectangles) interacts with an (uncharacterized) E3 UL that mediates monoubiquitylation (red circle) of the Gal4 DBD (purple ovals). Upon interaction with the proteasome, Gal4 extracts the APIS complex (see Fig. 2) through interactions between the 12 amino acids remaining in the Gal4D AD (black rectangles). This initiates an unfolding reaction that, if not terminated quickly, would result in the unfolding of the Gal4 DBD and disruption of the protein·DNA complex (left side). This occurs if Gal4 is not ubiquitylated. However, the appended Ub moieties bind to Rpt1 and Rpn1 after the APIS complex has been extracted from the proteasome. These interactions mediate dissolution of the binding of the Gal4 AD to Rpt6 and Rpt4 (Sug1 and Sug2, respectively). It is proposed, but not proven, that this allows the APIS complex to move into the polymerase II (PolII) complex, where it is important for stimulating elongation, perhaps by helping to manipulate chromatin structure on the template.
Remaining Questions and Challenges
The work on the yeast Gal4 system has provided a satisfying and relatively detailed picture of how this particular activator interacts with the proteasome to recruit the ATPases and other associated proteins to stimulate transcription via non-proteolytic mechanisms. To the best of my knowledge, the models presented in this minireview are consistent with all of the available biochemical and genetic data for the GAL system. There is some evidence that these conclusions will be fairly general. Genome-wide ChIP-chip studies have shown that the proteasomal ATPases, but not the 20 S core complex, are resident on hundreds of different yeast genes (42, 43), consistent with the idea that the non-proteolytic stimulation of promoter escape and elongation seen in detailed studies of the GAL and heat shock (20) systems is likely operative on many other genes in yeast. However, it is important to point out that the same genomic data show clearly that hundreds of genes are transcribed efficiently without the involvement of APIS. A much smaller amount of data suggests that this may also be true in mammalian cells. What distinguishes APIS-dependent from APIS-independent genes is completely unclear.
A major unresolved issue is to determine exactly how APIS stimulates the elongation process. Ezhkova and Tansey (44) have reported a link between histone ubiquitylation, Rpt6, and histone methylation. Moreover, as mentioned above, we demonstrated that the proteasomal ATPases co-immunoprecipitate with the histone chaperone FACT (14), consistent with the known genetic interaction between these factors (12). Thus, an attractive model is that the unfolding activity of the proteasomal ATPases acts to transiently disrupt chromatin structure in the path of an elongating RNA polymerase II complex in collaboration with FACT. But this model remains to be tested seriously.
The effects of APIS recruitment and transcription factor monoubiquitylation could be broader than those uncovered in the studies reviewed above. The remarkable connection between monoubiquitylation and the APIS-mediated stripping reaction would appear to be sufficient to explain all of the observations in the GAL system. However, p53 has long been known to be a target for several different monoubiquitylation events, some of which are stimulatory (29, 30) and some inhibitory (45–47) to the transcription of its target genes, so clearly context effects will be important in many cases. Even for the clearly stimulatory events, monoubiquitylation may mediate events downstream of promoter occupancy. Kurosu and Peterlin (48) have provided evidence that Ub and the VP16 AD can collaborate to allow LexA-VP16 to more efficiently recruit the elongation factor P-TEFb to promoters. Indeed, it is not clear if the protection-from-stripping model applies to the artificial activator LexA-VP16 because, in Δmet30 cells, the presumably non-ubiquitylated form of LexA-VP16 was found to be associated with its target promoter by ChIP (25) but nonetheless activated transcription only weakly, again arguing for an important role of monoubiquitylation downstream of promoter occupancy. It should be noted that the LexA DBD·operator complex is an unusually tight protein·DNA complex and thus may be less vulnerable to APIS-mediated stripping than physiologically relevant eukaryotic activators.
Conclusion
In summary, it is now clear that non-proteolytic activities of the UPP play an important and integral role in the transcription of many genes. There was a great deal of resistance to this idea at first, given the prevailing view at the beginning of this century that the only role of the 19 S RP in the cell was as a piece of the 26 S proteasome, devoted entirely to proteolysis. This is clearly not the case. Ironically, a direct precedent for this kind of idea was available from the prokaryotic world. ClpX, an ATPase that plays a 19 S RP-like role in bacterial proteolysis, operates independently of the ClpP protease in phage Mu transposition and functions to actively disrupt otherwise very stable protein·DNA complexes non-proteolytically (49). More generally, I would argue that unfoldases such as the APIS complex will be generally required for many, if not all, cellular processes driven by multiprotein complexes that must alter their composition and quaternary structure over the course of the catalytic cycle. The cell cannot rely on equilibrium processes to drive these transitions. Thus, we anticipate that the discoveries described here probably represent the tip of the iceberg in learning how ATP-dependent unfoldases participate non-proteolytically in nucleic acid metabolism.
Supplementary Material
Acknowledgments
I thank Prof. Stephen A. Johnston and my co-workers for many stimulating discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM087283. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- UPP
- ubiquitin-proteasome pathway
- Ub
- ubiquitin
- UL
- ubiquitin ligase
- RP
- regulatory particle
- CP
- core particle
- AD
- activation domain
- GST
- glutathione S-transferase
- ChIP
- chromatin immunoprecipitation
- DBD
- DNA-binding domain.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Coux O., Tanaka K., Goldberg A. L. (1996) Annu. Rev. Biochem. 65, 801–847 [DOI] [PubMed] [Google Scholar]
- 3.Saeki Y., Toh-e A., Yokosawa H. (2000) Biochem. Biophys. Res. Commun. 273, 509–515 [DOI] [PubMed] [Google Scholar]
- 4.Muratani M., Tansey W. P. (2003) Nat. Rev. Mol. Cell Biol. 4, 192–201 [DOI] [PubMed] [Google Scholar]
- 5.Kodadek T., Sikder D., Nalley K. (2006) Cell 127, 261–264 [DOI] [PubMed] [Google Scholar]
- 6.Collins G. A., Lipford J. R., Deshaies R. J., Tansey W. P. (2009) Nature 461, E7; Discussion E8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swaffield J. C., Bromberg J. F., Johnston S. A. (1992) Nature 357, 698–700 [DOI] [PubMed] [Google Scholar]
- 8.Lohr D., Venkov P., Zlatanova J. (1995) FASEB J. 9, 777–787 [DOI] [PubMed] [Google Scholar]
- 9.Rubin D. M., Coux O., Wefes I., Hengartner C., Young R. A., Goldberg A. L., Finley D. (1996) Nature 379, 655–657 [DOI] [PubMed] [Google Scholar]
- 10.Russell S. J., Johnston S. A. (2001) J. Biol. Chem. 276, 9825–9831 [DOI] [PubMed] [Google Scholar]
- 11.Ferdous A., Gonzalez F., Sun L., Kodadek T., Johnston S. A. (2001) Mol. Cell 7, 981–991 [DOI] [PubMed] [Google Scholar]
- 12.Xu Q., Singer R. A., Johnston G. C. (1995) Mol. Cell. Biol. 15, 6025–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orphanides G., LeRoy G., Chang C. H., Luse D. S., Reinberg D. (1998) Cell 92, 105–116 [DOI] [PubMed] [Google Scholar]
- 14.Sun L., Johnston S. A., Kodadek T. (2002) Biochem. Biophys. Res. Commun. 296, 991–999 [DOI] [PubMed] [Google Scholar]
- 15.Powell W., Reines D. (1996) J. Biol. Chem. 271, 6866–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhat K. P., Turner J. D., Myers S. E., Cape A. D., Ting J. P., Greer S. F. (2008) Mol. Immunol. 45, 2214–2224 [DOI] [PubMed] [Google Scholar]
- 17.Lassot I., Latreille D., Rousset E., Sourisseau M., Linares L. K., Chable-Bessia C., Coux O., Benkirane M., Kiernan R. E. (2007) Mol. Cell 25, 369–383 [DOI] [PubMed] [Google Scholar]
- 18.Morris M. C., Kaiser P., Rudyak S., Baskerville C., Watson M. H., Reed S. I. (2003) Nature 423, 1009–1013 [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez F., Delahodde A., Kodadek T., Johnston S. A. (2002) Science 296, 548–550 [DOI] [PubMed] [Google Scholar]
- 20.Sulahian R., Sikder D., Johnston S. A., Kodadek T. (2006) Nucleic Acids Res. 34, 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makino Y., Yoshida T., Yogosawa S., Tanaka K., Muramatsu M., Tamura T. A. (1999) Genes Cells 4, 529–539 [DOI] [PubMed] [Google Scholar]
- 22.Weeda G., Rossingnol M., Fraser R. A., Winkler G. S., Vermeulen W., van 't Veer L. J., Ma L., Hoeijmakers J. H., Egly J. M. (1997) Nucleic Acids Res. 25, 2274–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swaffield J. C., Melcher K., Johnston S. A. (1995) Nature 374, 88–91 [DOI] [PubMed] [Google Scholar]
- 24.Lee D., Ezhkova E., Li B., Pattenden S. G., Tansey W. P., Workman J. L. (2005) Cell 123, 423–436 [DOI] [PubMed] [Google Scholar]
- 25.Salghetti S. E., Caudy A. A., Chenoweth J. G., Tansey W. P. (2001) Science 293, 1651–1653 [DOI] [PubMed] [Google Scholar]
- 26.Collins G. A., Tansey W. P. (2006) Curr. Opin. Genet. Dev. 16, 197–202 [DOI] [PubMed] [Google Scholar]
- 27.Wu R. C., Feng Q., Lonard D. M., O'Malley B. W. (2007) Cell 129, 1125–1140 [DOI] [PubMed] [Google Scholar]
- 28.Greer S. F., Zika E., Conti B., Zhu X. S., Ting J. P. (2003) Nat. Immunol. 4, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 29.Rajendra R., Malegaonkar D., Pungaliya P., Marshall H., Rasheed Z., Brownell J., Liu L. F., Lutzker S., Saleem A., Rubin E. H. (2004) J. Biol. Chem. 279, 36440–36444 [DOI] [PubMed] [Google Scholar]
- 30.Le Cam L., Linares L. K., Paul C., Julien E., Lacroix M., Hatchi E., Triboulet R., Bossis G., Shmueli A., Rodriguez M. S., Coux O., Sardet C. (2006) Cell 127, 775–788 [DOI] [PubMed] [Google Scholar]
- 31.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferdous A., Sikder D., Gillette T. G., Nalley K., Kodadek T., Johnston S. A. (2007) Genes Dev. 20, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell S. J., Steger K. A., Johnston S. A. (1999) J. Biol. Chem. 274, 21943–21952 [DOI] [PubMed] [Google Scholar]
- 34.Ferdous A., O'Neal M., Nalley K., Sikder D., Kodadek T., Johnston S. A. (2008) Mol. BioSyst. 4, 1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y., Reece R. J., Ptashne M. (1996) EMBO J. 15, 3951–3963 [PMC free article] [PubMed] [Google Scholar]
- 36.Archer C. T., Delahodde A., Gonzalez F., Johnston S. A., Kodadek T. (2008) J. Biol. Chem. 283, 12614–12623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muratani M., Kung C., Shokat K. M., Tansey W. P. (2005) Cell 120, 887–899 [DOI] [PubMed] [Google Scholar]
- 38.Archer C. T., Burdine L., Liu B., Ferdous A., Johnston S. A., Kodadek T. (2008) J. Biol. Chem. 283, 21789–21798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeuchi J., Chen H., Coffino P. (2007) EMBO J. 26, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M., MacDonald A. I., Hoyt M. A., Coffino P. (2004) J. Biol. Chem. 279, 20959–20965 [DOI] [PubMed] [Google Scholar]
- 41.Carey M., Kakidani H., Leatherwood J., Mostashari F., Ptashne M. (1989) J. Mol. Biol. 209, 423–432 [DOI] [PubMed] [Google Scholar]
- 42.Auld K. L., Brown C. R., Casolari J. M., Komili S., Silver P. A. (2006) Mol. Cell 21, 861–871 [DOI] [PubMed] [Google Scholar]
- 43.Sikder D., Johnston S. A., Kodadek T. (2006) J. Biol. Chem. 281, 27346–27355 [DOI] [PubMed] [Google Scholar]
- 44.Ezhkova E., Tansey W. P. (2004) Mol. Cell 13, 435–442 [DOI] [PubMed] [Google Scholar]
- 45.Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. (2003) Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 46.Brooks C. L., Li M., Gu W. (2004) Cell Cycle 3, 436–438 [PubMed] [Google Scholar]
- 47.Brooks C. L., Li M., Gu W. (2007) J. Biol. Chem. 282, 22804–22815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurosu T., Peterlin B. M. (2004) Curr. Biol. 14, 1112–1116 [DOI] [PubMed] [Google Scholar]
- 49.Levchenko I., Luo L., Baker T. A. (1995) Genes Dev. 9, 2399–2408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.