Abstract
Galectin-1, a β-galactoside-binding protein highly expressed in the thymus, induces apoptosis of specific thymocyte subsets and activated T cells. Galectin-1 binds to N- and O-glycans on several glycoprotein receptors, including CD7, CD43, and CD45. Here we show that galectin-1 signaling through CD45, which carries both N- and O-glycans, is regulated by CD45 isoform expression, core 2 O-glycan formation and the balance of N-glycan sialylation. Regulation of galectin-1 T cell death by O-glycans is mediated through CD45 phosphatase activity. While galectin-1 signaling in cells expressing low molecular weight isoforms of CD45 requires expression of core 2 O-glycans (high affinity ligands for galectin-1), galectin-1 signaling in cells expressing a high molecular weight isoform of CD45 does not require core 2 O-glycans, suggesting that a larger amount of core 1 O-glycans (low affinity ligands for galectin-1) is sufficient to overcome lack of core 2 O-glycans. Furthermore, regulation of galectin-1 signaling by α2,6-sialylation of N-glycans is not solely dependent on CD45 phosphatase activity and can be modulated by the relative expression of enzymes that attach sialic acid in an α2,6- or α2,3-linkage. Thus, N- and O-glycans modulate galectin-1 T cell death by distinct mechanisms, and different glycosylation events can render thymocytes susceptible or resistant to galectin-1.
Keywords: Apoptosis, Carbohydrate/Glycoprotein, Cell/Apoptosis, Glycoproteins/Carbohydrates, Glycosylation, Phosphorylation/Phosphatases/Tyrosine, Tissue/Organ Systems/Leukocyte/Lymphocyte
Introduction
A functional immune response requires a T cell repertoire that can both recognize pathogens and ignore self. During T cell development in the thymus, thymocytes that either inadequately recognize foreign antigens or are self-reactive are purged by processes termed positive or negative selection, respectively (1). Positive and negative selection are complex processes controlled by a number of pro-apoptotic and anti-apoptotic factors. Both positive and negative thymocyte selection are regulated by galectin-1, an endogenous lectin expressed by thymic epithelial cells (2, 3). Galectin-1 binding to developing thymocytes can influence the strength of T cell receptor (TCR)2 signaling, an important factor in determining if a T cell can properly recognize antigen (3, 4). Galectin-1 can also directly induce apoptosis of specific thymocyte subsets, as well as activated peripheral T cells (5–9). Galectin-1 knock-out mice have aberrant thymocyte selection, leading to an altered T cell repertoire (3), as well as an altered mature T cell response in the periphery (4).
Galectin-1 preferentially binds to lactosamine sequences (Galβ1,4GlcNAc) on both N- and O-glycans, and thus can bind to a wide variety of T cell surface glycoproteins that bear these glycans. In addition, our laboratory has shown that galectin-1 can bind to a glycoprotein receptor, CD43, bearing only core 1 O-glycans and lacking lactosamine sequences; in this case, low affinity/high avidity binding to a highly abundant but less preferred glycan ligand, Galβ1,3GalNAc, is sufficient to induce T cell death (6).
Several T cell surface glycoprotein receptors regulate susceptibility of thymocytes and T cells to galectin-1-induced death, including CD7 (10), CD43 (6), and CD45 (5, 11). Although not absolutely required for susceptibility to galectin-1, CD45 is a major receptor for galectin-1 on T cells, acts as a negative and positive regulator of galectin-1 death, and enhances phagocytic clearance of cells killed by galectin-1 (11–15).
CD45 is a large transmembrane glycoprotein expressed on all nucleated hematopoietic cells. CD45 is estimated to comprise up to 10% of lymphocyte cell surface proteins, and the CD45 intracellular phosphatase domain contributes the majority of tyrosine phosphatase activity in T cells (16–18). Galectin-1 binding to CD45 reduces CD45 phosphatase activity, an effect that appears to be essential for galectin-1 death of CD45-expressing T cells (12–14), although the mechanism by which this occurs is unknown. The extracellular domain of CD45 is variable, with different CD45 isoforms expressed on different lymphocyte subsets at distinct developmental stages. The extracellular domain of CD45 can include 1, 2, or 3 additional domains, termed A, B, and C, which are encoded by exons 4, 5, and 6 in the CD45 gene (16). The A, B, and C domains all contain numerous serine and threonine residues (in mouse, 13, 13, and 16 residues, respectively (16)), so that CD45 isoforms including these domains bear additional O-glycans; the number of O-glycans on CD45 can vary significantly among different isoforms (19). Thymocytes and mature T cells express low molecular weight isoforms of CD45, typically CD45R0 (no additional domains), CD45RA, and CD45RB. Thus T cells have relatively fewer O-glycans compared with B cells, which express full-length CD45RABC with the full complement of O-glycans (20–23). CD45 also bears abundant N-glycans, most of which are found on the membrane proximal region of the molecule, which is common to all CD45 isoforms. The expression of specific CD45 isoforms and N- and O-glycosylation of CD45 are all regulated during T cell development (6, 11, 12, 21, 22, 24–27).
Developing T cells undergo several specific changes in cell surface glycosylation during maturation in the thymus (28, 29). For example, immature cortical thymocytes bear abundant asialo O-glycans that can bind the plant lectin peanut agglutinin (PNA)hi, while expression of α2,3-sialyltransferase I (ST3Gal-I), which creates the SAα2,3Galβ1,3GalNAc sequence on core 1 O-glycans, is up-regulated in mature medullary thymocytes, which are PNAlo (30–32). Additionally, immature cortical thymocytes bear core 2 O-glycans, created by core 2 O-GlcNAc transferase (C2GnT); C2GnT expression is reduced in mature medullary thymocytes, concurrent with up-regulation of ST3Gal-I expression in this subset (2, 32). Similarly, there is higher binding of Sambucus nigra agglutinin (SNA) to mature thymocytes compared with immature thymocytes, indicating increased decoration of glycans with α2,6-linked sialic acid on these cells. Knock-out of either ST3Gal-I or α2,6-sialyltransferase I (ST6Gal-I), both of which preferentially decorate glycans on mature thymocytes, results in aberrant thymocyte development and loss of cells from specific thymocyte subsets (32, 33).
Galectin-1 preferentially kills immature thymocytes, while mature thymocytes are resistant to galectin-1 death (2, 6, 8). Susceptibility to galectin-1 is regulated by the presence or absence of specific N- and O-glycan modifications, and these specific glycans are differentially expressed on thymocyte subsets. Expression of core 2 O-glycans, as found on immature thymocytes, promotes T cell susceptibility to galectin-1-induced death (11, 26, 34, 35). Expression of α2,6-linked sialic acid, as found on mature thymocytes, reduces T cell susceptibility to galectin-1-induced death (9, 12, 36, 37). These modifications can affect O- and N-glycans, respectively, on CD45. However, it is not clear how these precise glycan modifications regulate galectin-1 signaling through CD45 during T cell death, nor how galectin-1 binding to different CD45 isoforms may be affected by glycosylation.
In the present work, we have found that galectin-1 signaling via CD45 is regulated by both the relative abundance and the type of O-glycans on different CD45 isoforms, suggesting a mechanism by which T and B cells are differentially controlled in similar environments. The balance of α2,3- and α2,6-linked sialic acid on N-glycans influences galectin-1 binding to T cells and thymocytes, and removal of α2,6-linked sialic acids from N-glycans on mature thymocytes is sufficient to render the cells susceptible to galectin-1 death. Thus, thymocyte and T cell susceptibility to galectin-1 is controlled both by developmentally regulated glycosylation as well as by expression of specific glycoforms of CD45.
EXPERIMENTAL PROCEDURES
Cell Lines
Murine cell lines PhaR2.1 (gift of Dr. M. Pierce), BW5147 (gift of Dr. R. Hyman), T200RABC and T200C817S (gift of Dr. P. Johnson) were maintained in DMEM (Invitrogen) supplemented with 10 mm GlutaMAX (Invitrogen), 10% fetal bovine serum (Hyclone), and 100 units/ml penicillin and 0.1 mg/ml streptomycin (Biowhittaker). BW5147-C2GnT and vector control cells (34), PhaR ST3 and vector control cells, T200-C2GnT and vector control cells, and Rev-C2GnT and vector control cells were maintained in the above supplemented DMEM with 0.2 mg/ml Zeocin (Invitrogen). PhaR ST6 and vector control cells (12) were maintained in the above supplemented DMEM with 800 μg/ml G418 (Gemini). T200 cells doubly transfected with C2GnT and CD45RABC or CD45E613R and corresponding controls were maintained in DMEM with 0.2 mg/ml Zeocin (Invitrogen) and 0.8 mg/ml G418 (Gemini). All murine cell lines were maintained in 10% CO2 in a humidified atmosphere.
ST3Gal-III Messenger RNA Isolation and RT-PCR
Total RNA from PhaR2.1 T cells was isolated using TRIzol reagent according to the manufacturer's protocol. Murine ST3N (ST3Gal3, EC 2.4.99.6, GenBankTM NM009176) cDNA was obtained using a one-step RT-PCR kit (Invitrogen). Forward primer sequence containing the EcoR1 digestion site was: aagaattcgccgccaccatgggactcttggtatttg; reverse primer sequence containing XbaI digestion site was: acctctagaatttcagataccgctgcttaagtc. The final PCR product was cloned into TOPO-TA-PCR vector (Invitrogen).
Construction of ST3Gal-III Expression Vectors
cDNA was cut from the TOPO-TA-PCR vector containing the sequence of ST3N with digestion enzymes EcoR I and XbaI (New England Biolabs). Digested cDNA was purified and ligated into pcDNA3.1/Zeo(+) (Invitrogen) linearized by the same two restriction enzymes. The ligated products were transformed into chemically competent Escherichia coli DH5α (Novagen) to amplify the expression vectors. Plasmid was isolated, and the insert was verified by restriction enzyme analysis and DNA sequencing.
Construction of CD45-E613R Mutant
cDNA of full-length murine CD45RABC (20) in pBCMGS-neo (gift of Dr. P. Johnson) was mutated with a site-directed mutagenesis kit (Stratagene). Primers used, which encode the E613R mutation, were: caaaaggaagattgccgatcggggcagactgttcctggctg and the reverse complement. Mutagenesis was verified by sequencing (UC Davis Sequencing).
Transfection
pcDNA3.1/Zeo(+)-ST3N plasmid or vector alone were transfected into PhaR2.1 cells. Following selection in culture containing 0.3 mg/ml Zeocin, positive PhaR2.1 clones were identified by Maackia amurensis lectin II (MAA) flow cytometry.
T200− and Rev1.1 cell lines were transfected with C2GnT in the pcDNA3.1 vector (Invitrogen) as done previously (34). T200-C2GnT and vector control cells were then cotransfected with CD45RABC or CD45E613R in the pBCMGS-neo vector (38, 39). All cells were selected in 0.3 mg/ml Zeocin (Invitrogen) and/or 900 μg/ml G418 (Gemini). CD45 transfectants were cloned and C2GnT +/− clones were paired for matched levels of CD45 expression by flow cytometry.
Thymocytes
Thymocytes from C57/BL6 mice were isolated as described (6). Thymocytes were resuspended in RPMI medium supplemented with 10% fetal bovine serum, 100 mm HEPES (Invitrogen), 10 mm GlutaMAX, 2.5 mg/liter glucose, 0.1% 2-mercaptoethanol (Fisher Scientific), and 100 units/ml penicillin and 0.l mg/ml streptomycin and cultured at 5% CO2.
Flow Cytometry
All flow cytometry was done at the UCLA Johnsson Comprehensive Cancer Center Flow Cytometry Core Facility on the BD FACScan or BD LSR I.
Death Assays
Galectin-1 death assays were performed essentially as described (40). Briefly, 2 × 105 cells were resuspended in complete DMEM with galectin-1 (20 μm unless otherwise noted) in 1.2 mm dithiothreitol (DTT, Fisher Scientific) or buffer control for 4.5 h. Cells were disaggregated with 0.1 m β-lactose (Agros), washed in annexin-V binding buffer, and stained with annexin-V FITC or annexin-V PE (Molecular Probes) and 7-aminoactinomycin D (7-AAD, Invitrogen). T200RABC and T200C817S cells were stained with annexin-V PE (Invitrogen) and 7-AAD. Percent cell death was calculated by normalizing the % of cells that are annexin-V+ in treated samples to % of cells that are annexin-V+ in buffer control and 100%.
In galectin-1 death assays with bpV(phen) phosphatase inhibitor (Calbiochem) (11), 2 × 105 PhaR2.1, PhaR ST6, or PhaR ST3 cells were resuspended in complete DMEM with 20 μm bpV(phen) and incubated at 37 °C and 10% CO2 for 3 h. Then galectin-1 or buffer control was added, and cells were incubated and assayed as above.
Thymocytes were assayed for death with bpV(phen) essentially as described for cell lines above, with the following exceptions. 5 × 105 cells were resuspended in 10 μm bpV(phen) for 3 h, followed by incubation with DTT or galectin-1. Thymocytes were stained with CD4-FITC (Biolegend), CD8a-PE (Biolegend), and 7-AAD, and 5 μl CountBright Absolute Counting beads (Invitrogen) were added to each sample. 750 beads were collected per sample. Thymocytes were gated into DN, DP, CD4, and CD8 subsets by CD4-FITC and CD8a-PE staining. Percent cell loss was calculated by finding the percent reduction in live cell counts per cell type per sample in galectin-1-treated versus buffer-treated control samples.
For neuraminidase assays, thymocytes were resuspended at 3 × 106 cells/ml in 100 units/ml Clostridium perfringens or Salmonella typhimurium LT2 neuraminidase (New England Biolabs), or buffer control (50 mm sodium citrate, 100 mm NaCl, 100 μg/ml BSA, pH 6.0) for 1 h at 37 °C. Sialic acid removal was verified by flow cytometric analysis with SNA or MAA lectin (see below). Thymocytes were then resuspended in RPMI at 2.5 × 106/ml with galectin-1 or buffer control for 4.5 h. Galectin-1 or buffer control-treated thymocytes were then stained with CD4-FITC, CD8a-PE, 7-AAD, and CountBright Beads as above, and assayed for cell loss.
Surface Staining
For phenotyping of cell lines, 2 × 105 cells were blocked with 1% bovine serum albumin (BSA, Sigma) and incubated with 0.2 μg of antibody for 1 h. Antibodies used were: 30-F11-FITC (BD Biosciences) for CD45, Rat IgG2b/κ-FITC (Biolegend), CD43 1B11-PE (Biolegend) for core 2 O-glycans, and rat IgG2a/κ-PE (Biolegend). For T200RABC or T200C817S cells, 30-F11-biotin (BD Biosciences) and Rat IgG2b/κ-biotin (BD Biosciences) were used to assay CD45 expression. Biotinylated 30-F11 or isotype control stains were bound by streptavidin-PerCP (BD Biosciences).
For lectin phenotyping of cell lines, 2 × 105 cells were blocked with 1% BSA and then stained with 1 μg/106 cells BSA-biotin, PNA-biotin, MAA-biotin, or 0.3 μg/106 cells SNA-biotin (Vector Labs). Cells were washed and bound lectin was detected with 5 μg/ml streptavidin-FITC (Jackson IR).
For lectin phenotyping of thymocytes, 1 × 106 thymocytes were stained with lectins as above, except that biotinylated lectins were detected with 5 μg/ml streptavidin-PerCP (BD Biosciences). Thymocytes were co-stained with CD4-FITC (Biolegend) and CD8a-APC (BD Biosciences) to allow gating of DN, DP, CD4, and CD8 populations.
For galectin-1 binding, 5 × 105 cells were suspended in PBS with the indicated amount of galectin-1-biotin for 10 min with or without 50 mm β-lactose. Cells were washed in PBS and then stained in 5 μg/ml streptavidin-FITC. Mean fluorescence intensity was corrected for autofluorescence for each cell type.
Phosphatase Activity Assays
1 × 106 cells were resuspended in 250 μl of complete DMEM and 20 μm galectin-1 and 1.2 mm DTT or buffer control (in triplicate). Cells were incubated in round bottom 96-well plates for 15 min or the indicated times, washed twice with 1× TBS, then lysed in Nonidet P-40 lysis buffer (50 mm Tris, 150 mm NaCl, 1% Nonidet P-40 (Igepal CA-630, Sigma), 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin (Sigma), 10 μg/ml aprotinin (Sigma)). Lysate was cleared by centrifugation for 20 min. 1 part lysate was mixed with 9 parts phosphatase assay buffer (150 mm NaCl, 100 mm HEPES, 1 mm DTT, 1 mm EDTA, 50 nm okadaic acid, and 10 mm pNpp) with or without 40 μm bpV(phen) and incubated at room temperature for 2 h. Equal volume 1 m NaOH (Sigma) was then added to stop the pNpp reaction. Samples were plated in duplicate and were read on a colorimetric spectrophotometer (Bio-Rad Benchmark Plus) at A415. A415 readings were normalized to blank and background activity readings for each cell type (except T200C817S in supplemental Fig. S1, which was normalized to background activity of T200RABC control).
Western Blots and Immunoprecipitations
106 cells were washed in PBS and lysed in Nonidet P-40 lysis buffer and cleared by centrifugation for 30 min at 4 °C. Lysates were assayed for protein concentration by Bradford Assay (Bio-Rad), and diluted to 1 mg/ml protein. 100 μg of protein lysate was mixed with 10 μl of protein G beads (Pierce) and 1 μg of goat anti-mouse CD45 polyclonal antibody (R&D Systems) or 1 μg of goat IgG (Zymed Laboratories Inc./Invitrogen) overnight at 4 °C. Beads were washed with lysis buffer and denatured in NuPAGE sample buffer and reducing agent (Invitrogen). Immunoprecipitate was run on NuPAGE Novex 3–8% Tris-acetate gels (Invitrogen) per the manufacturer's protocols. Gels were transferred to polyvinylidene difluoride membrane (Bio-Rad) and blocked in 3% BSA and 0.01% Tween.
Blots were probed with 0.1 μg/ml goat anti-mouse CD45 polyclonal antibody (R&D Systems) and donkey anti-goat horseradish peroxidase (HRP) secondary (Novus) for CD45 blots. Lectin blots were probed with 0.1 μg/ml SNA-biotin (Vector Labs) and streptavidin-HRP secondary (Zymed Laboratories Inc./Invitrogen).
Glycan Microarray
200 μg/ml biotinylated galectin-1 (5) were assayed at Core H at the Consortium for Functional Glycomics on the mammalian version 4 slide glycan microarray. Data shown in this publication are from glycans (A) 320: [Neu5Aca2–3Galb1–4GlcNAcb1–2Mana1–3(Neu5Aca2–3Galb1–4GlcNAcb1–2Mana1–6)Manb1–4GlcNAcb1– 4GlcNAcb-Sp12], (B) 313: [Neu5Aca2–3Galb1–4GlcNAcb1– 2Mana1–3(Neu5Aca2–6Galb1–4GlcNAcb1–2Mana1– 6)Manb1–4GlcNAcb1–4GlcNAcb-Sp12], (C) 321: [Neu5Aca2– 6Galb1–4GlcNAcb1–2Mana1–3(Neu5Aca2–3Galb1–4GlcNAcb1–2Mana1–6)Manb1-4GlcNAcb1–4GlcNAcb-Sp12],and (D) 53: [Neu5Aca2–6Galb1–4GlcNAcb1–2Mana1–3(Neu5Aca2–6Galb1–4GlcNAcb1–2Mana1–6)Manb1– 4GlcNAcb1–4GlcNAcb-Sp12]. Data shown are relative fluorescence units (RFUs) ± the S.E. of six replicate assays.
RESULTS
Differential Sialylation of Thymocytes at Distinct Developmental Stages
During development, thymocytes undergo several changes in N- and O-glycosylation of cell surface glycoproteins. Earlier studies of glycosylation patterns in the thymus focused on differences between cortical (immature) and medullary (mature) thymocytes, or used only a single marker, such as CD3, to distinguish thymocyte subsets at different points of maturation (2, 29, 30, 41, 42). However, multiparameter flow cytometry has allowed separation of thymocytes into four distinct developmental subsets; these include the most immature cells that express neither CD4 nor CD8 (double negative, DN), immature cells that are undergoing TCR rearrangement and express both CD4 and CD8 (double positive, DP), and mature cells that express a functional TCR and either CD4 or CD8 (single positive, SP) (1). Few studies have analyzed differences in glycosylation among these well-defined thymocyte subsets (33, 43).
To more precisely characterize the sialylation of both N- and O-glycans on thymocyte subsets, we used three plant lectins: PNA, which binds asialo core 1 O-glycan structures, SNA, which binds α2,6-linked sialic acid primarily on N-glycans, and MAA, which binds α2,3-linked sialic acid primarily on N-glycans. Total murine thymocytes were stained with CD4, CD8, and the indicated lectin. Using flow cytometry, cells were separated into DN, DP, CD4SP, and CD8SP subsets (Fig. 1A), and reactivity with the indicated lectin for each subset was determined.
FIGURE 1.
Differential glycosylation of murine thymocyte subsets. A, identification of thymocyte subsets. Thymocytes were stained with CD4-FITC and CD8-PE. Representative gating to identify DN (CD4- CD8-), DP (CD4+ CD8+), CD4 (CD4+ CD8-), and CD8 (CD4- CD8+) cells is shown. B, DP thymocytes bear asialo O-glycans. Thymocytes were stained with CD4-FITC, CD8-PE, and PNA-biotin. Lectin binding was detected with streptavidin-PerCP. Thymocytes were gated on DN, DP, CD4, and CD8 subpopulations and PerCP MFI of each subset determined. C, SP thymocytes bear abundant α2,6-linked sialic acid. Cells were stained and gated as above, except that SNA-biotin binding was detected. D, SP thymocytes bear abundant α2,3-linked sialic acid. Cells were stained and gated as above, except that MAA-biotin binding was detected. In B–D, data are mean MFI ± S.E. of cells from four animals.
As shown in Fig. 1B, exposure of asialo core 1 O-glycans increases at the DN-DP transition; we observed strong reactivity of DP cells with PNA, while DN cells had minimal binding. While previous studies have described increased PNA binding to “immature” versus “mature” thymocytes, the present analysis identifies a clear difference between DN and DP cells with regard to PNA staining. In contrast, both CD4-SP and CD8-SP subsets lose PNA reactivity, compared with DP cells; previous studies had found reduced PNA binding to medullary compared with cortical thymocytes (2, 29). Work from our laboratory and others has shown that sialylation of core 1 O-glycans antagonizes susceptibility to galectin-1-induced cell death (6, 11, 44). Thus, the PNA profile of these four thymocyte subsets is consistent with our observation that DP cells are highly susceptible to galectin-1-induced cell death, while DN and SP thymocytes, which have reduced levels of asialo O-glycans, are resistant to galectin-1 death.
Fig. 1, C and D, show that there is also an increase in both α2,6- and α2,3-linked sialic acids (detected by SNA and MAA, respectively) on thymocytes at the DP-SP transition, especially on CD4 cells; the former is consistent with previous reports that SNA preferentially binds to mature thymocytes (33, 41). As mentioned above, CD4 and CD8-SP thymocytes are resistant to galectin-1 (8); as overexpression of ST6Gal-I enzyme, which adds α2,6-linked sialic acid to N-glycans, blocks galectin-1 death, and peripheral CD4-Th2 cells, which are resistant to galectin-1, express ST6Gal-I and bind SNA (9, 12), this indicates that addition of α2,6-linked sialic acids to SP thymocytes also protects these cells from galectin-1 death.
O-Glycans on CD45 Regulate Galectin-1 Binding, Phosphatase Activity, and Cell Death
As mentioned above, CD45 is one of the most abundant glycoproteins on the T cell surface, and glycosylation of CD45 is regulated during thymocyte development. While not essential for galectin-1 T cell death, CD45 is a major cell surface receptor for galectin-1, and galectin-1 binding to T cells results in a reduction in the intracellular tyrosine phosphatase activity of CD45 (12–14, 44). In T cells and T cell lines expressing CD45, CD45 must be appropriately glycosylated for galectin-1 death to occur. Cells that express CD45 but not core 2 O-glycans are not susceptible to galectin-1 death, while expression of C2GnT adds core 2 O-glycans to CD45 and renders cells susceptible to galectin-1 (11, 26, 34). While core 2 O-glycans can also be found on CD43, another major cell surface receptor for galectin-1, addition of core 2 O-glycans specifically to CD43 is not essential for galectin-1 death (6). Thus, we asked if addition of core 2 O-glycans was essential for the galectin-1-induced reduction in CD45 phosphatase activity that is necessary for galectin-1 T cell death.
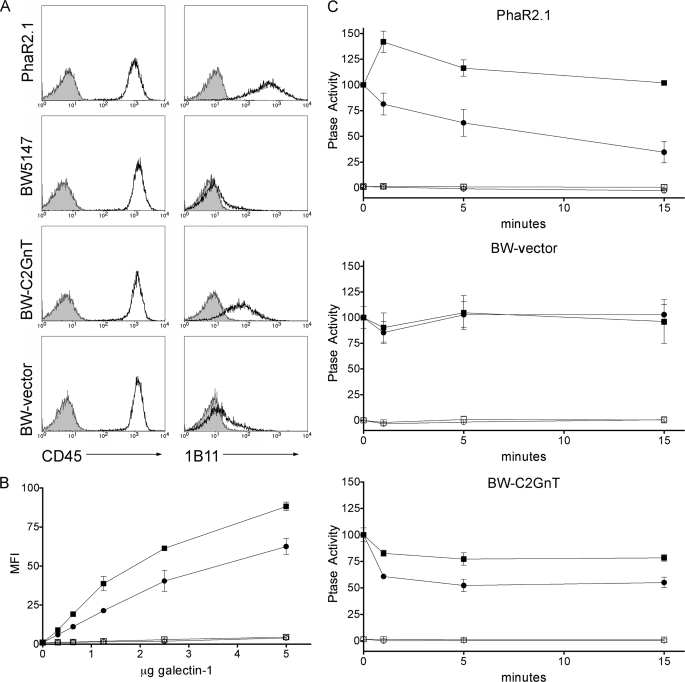
We expressed C2GnT in the CD45RB+ BW5147 thymoma cell line (11, 34); all cell lines constructed for this study are described in Fig. 2. As a positive control, we used the PhaR2.1 cell line, a phytohemagglutinin resistant mutant of BW5147, which expresses C2GnT and CD45RB and is highly susceptible to galectin-1 (11). C2GnT expression in BW5147 cells resulted in addition of core 2 O-glycans to cell surface glycoproteins detected by the 1B11 mAb (45), while all cell lines expressed comparable amounts of CD45 (Fig. 3A). Expression of C2GnT in BW5147 cells resulted in increased galectin-1 binding to the cell surface (Fig. 3B).
FIGURE 2.
Derivation of cell lines used in this study. The PhaR2.1 cell line is a derivative of BW5147 that has spontaneously re-expressed C2GnT. The T200− cell line is a derivative of BW5147 that lacks CD45. The Rev1.1 cell line is a derivative of the T200− cell line that has spontaneously re-expressed the extracellular and transmembrane regions of CD45.
FIGURE 3.
Expression of core 2 O-glycans increases galectin-1 binding and is required for galectin-1 inhibition of CD45 tyrosine phosphatase activity in cells expressing CD45RB. A, expression of C2GnT adds core 2 O-glycans to T cell surface glycoproteins. PhaR2.1, BW5147, BW-C2GnT, or BW-vector cells were stained with CD45-FITC or 1B11-PE (open) or isotype control (filled). 1B11 binding to BW-C2GnT cells demonstrates core 2 O-glycan addition. B, BW-C2GnT (squares) or BW-vector (circles) cells were bound by biotinylated galectin-1 for 10 min with (open) or without (filled) 50 mm lactose. Bound galectin-1 was detected with streptavidin-FITC, and MFI was determined. Data are mean fluorescence, corrected for autofluorescence, ± S.D. of duplicate samples. C, 1 × 106 PhaR2.1, BW-vector, or BW-C2GnT cells were incubated with 20 μm galectin-1 (circles) or buffer control (squares) for the shown times. Cells were then washed and lysed, and phosphatase activity was measured with (open) or without (filled) bpV(phen). Data are mean ± S.D. of triplicate samples.
As shown in Fig. 3C, addition of galectin-1 to PhaR2.1 cells resulted in a substantial reduction in tyrosine phosphatase activity, while no reduction of phosphatase activity was seen in BW5147 cells that do not express C2GnT. In contrast, galectin-1 binding to BW5147 cells expressing C2GnT resulted in a significant and sustained decrease, ∼40%, in tyrosine phosphatase activity in these cells. While 1B11 reactivity of C2GnT-expressing BW5147 cells was not as high as that for PhaR2.1 cells (Fig. 3A), the degree of core 2 O-glycosylation on C2GnT-expressing BW5147 cells was clearly adequate to permit galectin-1 binding to modulate CD45 phosphatase activity. Thus, addition of core 2 O-glycans to CD45RB, the isoform expressed by thymocytes and the BW5147 thymoma cell line, was necessary and sufficient to restore galectin-1 signaling through CD45.
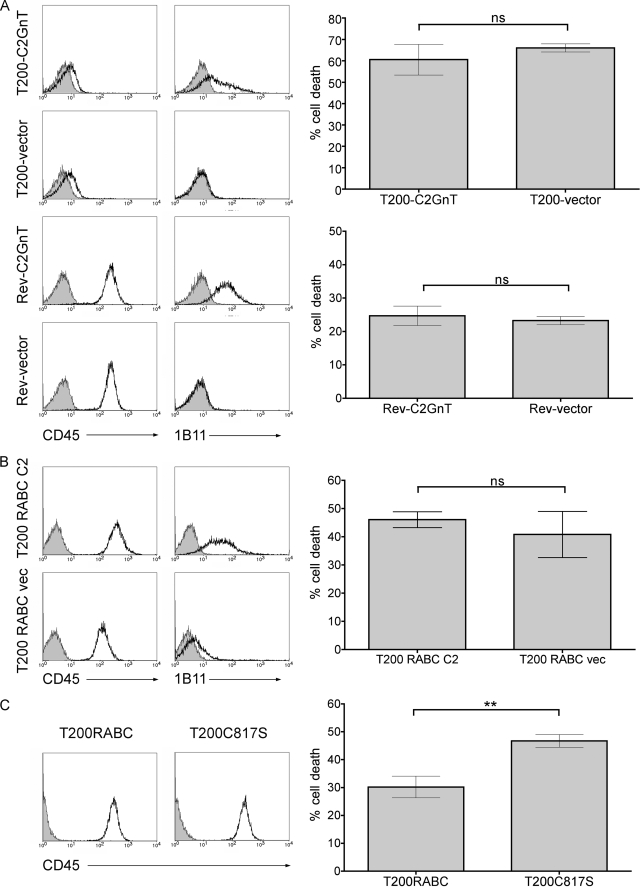
As mentioned above, core 2 O-glycan expression is only required for galectin-1 T cell death if the cells express CD45 (11). While core 2 O-glycan expression clearly affected CD45 intracellular phosphatase activity (Fig. 3C), core 2 O-glycan modification of the CD45 extracellular domain could also enhance galectin-1 binding and cell death by increasing the concentration of galectin-1 on the cell surface and making more galectin-1 locally available to other, smaller glycoprotein receptors, such as CD7, that are required for cell death (6, 46). To determine whether dependence on core 2 O-glycans for galectin-1 cell death is mediated through the CD45 extracellular domain, we expressed C2GnT in the T200− cell line, a derivative of BW5147 that lacks CD45, and the Rev1.1 cell line, a derivative of T200− that bears a truncated form of CD45 expressing only the transmembrane and extracellular domains. We have previously shown that both these cell lines are susceptible to galectin-1 death, even in the absence of core 2 O-glycans (11). Following C2GnT transfection, expression of core 2 O-glycans was demonstrated by increased binding of the 1B11 mAb (Fig. 4A). However, the presence or absence of core 2 O-glycans had no effect on the susceptibility of either the T200− cells, or the Rev1.1 cells (Fig. 4A) to galectin-1 cell death, as we observed equivalent susceptibility to galectin-1 in T200− or Rev1.1 cells with or without core 2 O-glycans. These data demonstrate that the primary effect of core 2 O-glycan modification of CD45 is to allow galectin-1 regulation of CD45 intracellular tyrosine phosphatase activity.
FIGURE 4.
CD45 regulation of galectin-1 susceptibility requires the intracellular phosphatase domain, and CD45 isoform usage determines the requirement for core 2 O-glycans. A, T cells lacking the intracellular domain of CD45 do not require C2GnT expression for galectin-1 susceptibility. Left, T200− or Rev1.1 cells were transfected with cDNA encoding C2GnT or vector alone, and the presence of core 2 O-glycans determined by staining with 1B11-PE (open) or isotype control (filled). CD45 expression was determined with CD45-FITC (open) or isotype control (filled). Histograms are of representative clones. Right, T200-C2GnT, T200-vector, Rev-C2GnT, or Rev-vector cells were treated with galectin-1 or buffer control, and cell death was analyzed by staining with annexin-V-FITC and 7-AAD. T200− and Rev1.1 cells were susceptible to galectin-1 death, and cells with or without core 2 O-glycans had comparable levels of cell death. Data are mean ± S.D. of at least duplicate samples, and representative of at least four experiments. B, expression of core 2 O-glycans does not enhance galectin-1 susceptibility in T cells expressing CD45RABC. Left, T200− cells were transfected with CD45RABC and C2GnT (T200 RABC C2) or CD45RABC and vector control (T200 RABC vec), and core 2 O-glycan expression was determined as in A. Histograms are of representative clones. Right, cell death assays were performed as in A. Data are mean ± S.D. of triplicate samples. C, loss of CD45 phosphatase activity enhances T cell susceptibility to galectin-1. Left, T200− cells transfected with CD45RABC (T200RABC) or the inactive phosphatase mutant CD45RABC-C817S (T200C817S) were stained for CD45 as above. Right, while cells expressing CD45RABC were sensitive to galectin-1, complete loss of CD45 phosphatase activity in the C817S mutant enhanced susceptibility to galectin-1. Cell death assays were performed as above. Data are mean ± S.E. of triplicate samples from three independent experiments. Significance was determined by Student's t test.
CD45RB, the isoform of CD45 found on BW5147 cells and on most thymocytes, bears fewer O-glycans than the CD45RABC isoform; the CD45RABC isoform, which is typically expressed on B cells, includes all three alternatively spliced regions in the extracellular domain, which have abundant serine/threonine residues. As discussed above, core 2 O-glycan modification of cells expressing CD45RB is required for galectin-1 death. To determine if an increase in total core 1 (low affinity galectin-1 ligand) O-glycans could compensate for lack of core 2 (high affinity galectin-1 ligand) O-glycans, we transfected full-length murine CD45RABC into T200− cells with and without C2GnT. We compared cells expressing comparable levels of CD45, and the presence of core 2 O-glycans was confirmed by 1B11 binding (Fig. 4B). Surprisingly, there was no significant difference in susceptibility to galectin-1 cell death between cells co-transfected with CD45RABC and C2GnT over cells transfected with CD45RABC alone. Furthermore, T200− cells transfected with CD45RABC, but not with C2GnT, showed a 20% reduction in phosphatase activity after galectin-1 binding (supplemental Fig. S1). These data indicate that that the greater abundance of low affinity core 1 O-glycan ligands present on CD45RABC versus CD45RB is sufficient to allow galectin-1 binding and signaling, even without core 2 O-glycan elongation.
Finally, to determine the requirement for CD45 phosphatase activity in galectin-1 death, we used T200− cells transfected with either CD45RABC or a CD45RABC mutant C817S in the intracellular domain. This mutation completely abrogates CD45 phosphatase activity, which is essentially all the tyrosine phosphatase activity in these cells (supplemental Fig. S1) (47). Again, we compared cells expressing equivalent levels of CD45 (Fig. 4C). As we saw in Fig. 3B, cells expressing the CD45RABC isoform (T200RABC) were susceptible to galectin-1. Cells expressing the phosphatase-dead CD45RABC mutant (T200C817S) were also susceptible to galectin-1 and actually demonstrated a modest but statistically significant increase in cell death compared with cells expressing wild-type T200RABC (Fig. 4C). As galectin-1 binding to wild-type CD45 only inhibits phosphatase activity by 20–50% (12–14) (Fig. 3), this suggests that complete inhibition of CD45 phosphatase activity further enhances susceptibility of cells to galectin-1.
Sialic Acid Addition to N-Glycans Regulates Galectin-1 Binding, CD45 Tyrosine Phosphatase Activity, and Galectin-1 Death
Prior work in our laboratory has shown that galectin-1 binding to N-glycans in general and to N-glycans on CD45 in particular is required for galectin-1 signaling and T cell death (8, 12). The factors that regulate galectin-1 binding to N-glycans are not entirely known. However, galectin-1 binding to soluble or immobilized lactosamine-containing glycans terminated with α2,6-linked sialic acid is reduced, compared with binding to glycans with lactosamine sequences capped with α2,3-linked sialic acid (37, 46, 48). Overexpression of ST6Gal-I, which adds α2,6-linked sialic acid to N-glycans, also reduces galectin-1 binding to and signaling in T cell lines (12). As shown in Fig. 1, sialic acid is found in both α2,3- and α2,6-linkages on cell surface N-glycans on different thymocyte subsets. We therefore wished to investigate how relative levels of α2,3- and α2,6-linked sialic acid on cell surface N-glycans would affect galectin-1 binding, signaling, and T cell death.
We expressed α2,3-sialyltransferase III (ST3Gal-III), a sialyltransferase that preferentially adds α2,3-linked sialic acid to N-glycans, in PhaR2.1 cells to create PhaR ST3 cells, as we had done with the ST6Gal-I to create PhaR ST6 cells (12). We compared SNA and MAA lectin staining of parental PhaR2.1 cells with PhaR ST6 and Phar ST3 cells (Fig. 5A). Cells with high levels of cell surface α2,6-linked sialic acid (detected by SNA binding) had reduced levels of cell surface α2,3-linked sialic acid (detected by MAA binding), and vice versa, suggesting that ST6Gal-I and ST3Gal-III compete for glycoprotein substrates, and that changes in the expression level of one of these enzymes alters the relative proportion of both linkages. CD45 was specifically modified by both enzymes, as CD45 from PhaR ST6 cells had increased abundance of α2,6-linked sialic acid, and CD45 from PhaR ST3 cells had less α2,6-linked sialic acid, compared with control cells (Fig. 5B). This indicates that the changes in lectin binding observed on whole cells, detected by flow cytometry, occurs specifically on CD45, a major galectin-1 receptor on T cells.
FIGURE 5.
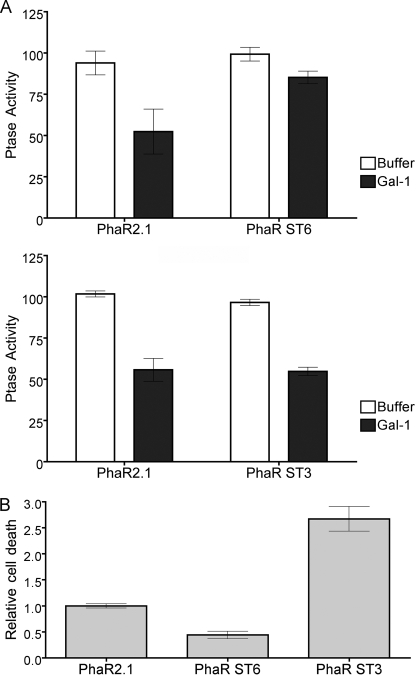
ST6Gal-I and ST3Gal-III expression modulates the balance of sialic acid linkages on T cells to regulate galectin-1 binding. A, ST6Gal-I (PhaR ST6) and ST3Gal-III (PhaR ST3) were expressed in PhaR2.1 cells and cell surface sialylation was determined by reactivity with SNA-biotin or MAA-biotin, detected by streptavidin-FITC. Cells expressing ST6Gal-I had increased SNA binding and reduced MAA binding, and cells expressing ST3Gal-III had increased MAA binding and reduced SNA binding, indicating that the enzymes compete for acceptor substrates. Data are mean fluorescence intensity ± 95% CI for representative samples of three independent assays. B, overexpression of ST6Gal-I increased α2,6-linked sialic acid on CD45, while expression of ST3Gal-III reduced α2,6-linked sialic acid on CD45. Lysates of PhaR2.1, PhaR ST6, and PhaR ST3 cells were immunoprecipitated with anti-CD45 (45) or control IgG (C). Precipitates were probed with SNA-biotin, stripped, and reprobed with anti-CD45 to confirm equal loading. C, galectin-1 binding to bi-antennary N-glycans is regulated by addition of α2,6-linked sialic acid. Galectin-1 binding to a glycan microarray was performed, and binding to bi-antennary complex N-glycans decorated with different terminal sialic acid linkages was determined. Relative binding to a bi-antennary complex N-glycan with two α2,3 (column A), one α2,3 and one α2,6 (columns B and C), or two α2,6 (column D)-linked sialic acids. Data are mean relative fluorescence units (RFU) ± S.E. of six replicate determinations. Statistical significance was measured by one-way analysis of variance with Bonferroni's multiple comparison post-test. D, alteration of sialic acid balance on the cell surface results in change in galectin-1 binding. PhaR2.1 (■, □), PhaR ST6 (▾, ▿), or PhaR ST3 (●, ○) were incubated with indicated concentration of biotinylated galectin with (open) or without (filled) 50 mm lactose for 10 min, washed, and stained with streptavidin-FITC. Data are mean fluorescence intensity (corrected for autofluorescence) ± S.D. of duplicate samples.
Because a multi-antennary N-glycan can bear a sialic acid residue at the terminus of each branch, we investigated the effect of different sialic acid linkages on galectin-1 binding to a biantennary glycan. We analyzed the binding efficiency of galectin-1 to biantennary N-glycans with (column A) two α2,3-, (columns B and C) one α2,3- and one α2,6-, or (column D) two α2,6-linked sialic acids on a glycan microarray (Fig. 5C). The greatest galectin-1 binding was observed for the glycan with only α2,3-linked sialic acid. Galectin-1 binding was reduced by about 40% when the biantennary glycan had one sialic acid in an α2,6-linkage and one sialic acid in an α2,3-linkage, although there was no preference for one branch over another. However, galectin-1 binding was dramatically reduced (greater than 10,000-fold) when both branches were terminated with α2,6-linked sialic acid.
We observed a similar effect of the ratio of α2,6- to α2,3-linked sialic acids on galectin-1 binding to cells. Indeed, while PhaR2.1 cells bearing glycans terminated with an intermediate number of α2,6-linked sialic acids bound galectin-1, small changes in α2,6-α2,3-linkage ratio resulted in a dramatic shift in galectin-1 binding; galectin-1 binding to PhaR ST3 cells was increased, and galectin-1 binding to PhaR ST6 cells was reduced, relative to parental PhaR2.1 cells (Fig. 5D). Thus, modifying the ratio of α2,6-linked to α2,3-linked sialic acids on cell surface glycans can significantly modulate the binding and signaling capability of galectin-1 for the cell.
As mentioned above, overexpression of ST6Gal-I in T cells inhibited the reduction in tyrosine phosphatase activity that occurs after galectin-1 binding (12). As shown in Fig. 6A, this effect is specific for the α2,6-linkage; PhaR ST6 cells demonstrated no significant decrease in tyrosine phosphatase activity after galectin-1 binding compared with parental PhaR2.1 cells, while PhaR ST3 cells had a similar reduction in tyrosine phosphatase activity compared with parental PhaR2.1 cells. Additionally, while PhaR ST6 cells had a 60% reduction in galectin-1 T cell death compared with PhaR2.1 cells, PhaR ST3 cells were >2-fold more susceptible to galectin-1 than PhaR2.1 cells. This suggests that galectin-1 T cell death is exquisitely sensitive to the relative balance of α2,3- and α2,6-linked sialic acids on cell surface glycans, and that small changes in the expression level of either responsible enzyme may alter the sialic acid linkage ratio and thus galectin-1 signaling.
FIGURE 6.
Overexpression of ST6Gal-I, but not ST3Gal-III, inhibits galectin-1-induced reduction of CD45 phosphatase and death. A, PhaR2.1, PhaR ST6, and PhaR ST3 cells were incubated with 20 μm galectin-1 or buffer control for 15 min prior to determination of tyrosine phosphatase activity of cell lysates. Data are mean ± S.D. of triplicate samples. B, PhaR2.1, PhaR ST6, or PhaR ST3 cells were incubated with 20 μm galectin-1 or buffer control. % cell death was normalized to buffer control, and then PhaR ST6 and PhaR ST3 death were normalized relative to PhaR2.1 cell death. Data are mean ± S.E. of triplicate samples from seven independent experiments.
Resistance of SP Thymocytes to Galectin-1 Is Dependent upon the Presence of Sialic Acid, Rather than Phosphatase Activity
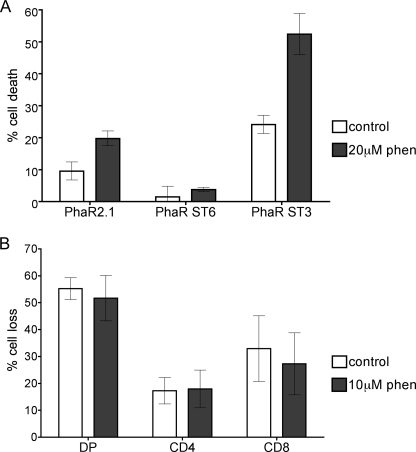
As shown in Fig. 3, addition of core 2 O-glycans to CD45 is necessary for reduction of tyrosine phosphatase activity after galectin-1 binding, as well as for susceptibility to galectin-1 death. Resistance to galectin-1 can be overcome in T cells that express CD45 but lack core 2 O-glycans with a pharmacological tyrosine phosphatase inhibitor, bpV(phen) (11). To ask if the resistance to galectin-1 death that we observed in cells overexpressing ST6Gal-I was also dependent on CD45 phosphatase activity, we asked if bpV(phen) would enhance death of PhaR ST6 cells (Fig. 7A). Surprisingly, bpV(phen) increased the sensitivity of both PhaR2.1 and Phar ST3 cells to galectin-1, but had no effect on the susceptibility of Phar ST6 cells to galectin-1. This indicates that the mechanism by which increased cell surface α2,6-linked sialic acid inhibits galectin-1 binding, signaling and T cell death is not solely due to blocking the ability of galectin-1 to decrease CD45 tyrosine phosphatase activity, as we observed for core 2 O-glycans (Fig. 3).
FIGURE 7.
Inhibition of tyrosine phosphatase activity cannot overcome the block to galectin-1 death in T cells expressing high levels of α2,6-linked sialic acid. A, tyrosine phosphatase inhibitor bpV(phen) increases galectin-1 death of PhaR2.1 and PhaR ST3 cells, but does not affect PhaR ST6 cells. Phar2.1, PhaR ST6, and PhaR ST3 cells were treated with 20 μm bpV(phen) or buffer control for 3 h, then incubated with 20 μm galectin-1 or buffer control for an additional 4.5 h. Data are mean ± S.D. of triplicate samples. B, treatment with bpV(phen) does not increase galectin-1-induced death in SP thymocytes. Total thymocytes were incubated with 10 μm bpV(phen) and followed by 20 μm galectin-1 as above. Cells were stained with 7-AAD, anti-CD4, and anti-CD8, and gated for DP, CD4, and CD8 populations. Data are mean ± S.D. of triplicate samples from two independent experiments.
As shown in Fig. 1, sialylation of both N- and O-glycans is increased in SP thymocytes, which are resistant to galectin-1 death (8). In T cell lines, while resistance to galectin-1 death because of lack of core 2 O-glycans on CD45 can be overcome by inhibiting CD45 phosphatase activity with bpV(phen) (11), resistance to galectin-1 death due to cell surface α2,6-linked sialic acid was not (Fig. 7A). To ask if primary SP thymocyte resistance to galectin-1 is also due to a phosphatase-independent mechanism, we treated thymocytes with galectin-1 with or without bpV(phen) (Fig. 7B). As we saw with PhaR ST6 cells, the resistance of CD4 and CD8 SP thymocytes to galectin-1 was unaltered by the addition of bpV(phen), suggesting that the primary reason that SP thymocytes are resistant to galectin-1 death is the presence of α2,6-linked sialic acid on N-glycans rather than the lack of core 2 O-glycans.
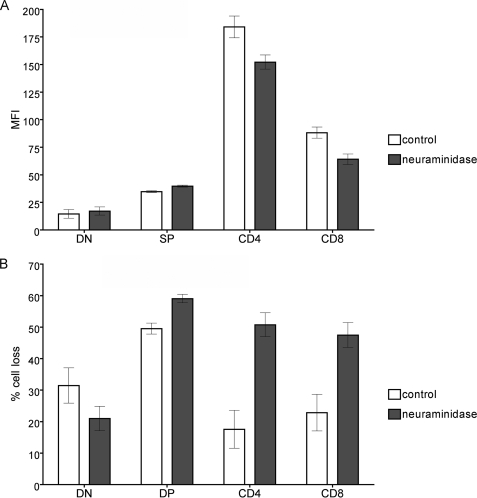
To determine if removal of α2,6-linked sialic acids from cell surface glycans would restore susceptibility to galectin-1, we treated thymocytes with C. perfringens neuraminidase, which primarily removes sialic acid in α2,6- and α2,3-linkages from underlying glycans, and confirmed that the enzyme removed α2,6-linked sialic acid by flow cytometry with SNA (Fig. 8A). Neuraminidase treatment had no effect on SNA binding to DN and DP thymocytes, but reduced SNA binding to CD4 and CD8 SP thymocytes. We then tested neuraminidase-treated and control-treated thymocytes for sensitivity to galectin-1 death. Neuraminidase treatment dramatically enhanced the susceptibility of CD4 and CD8 SP thymocytes to galectin-1; the galectin-1 death of SP cells was equivalent to that of DP thymocytes (Fig. 8B). Importantly, a relatively modest decrease in the amount of α2,6-linked sialic acid on the cell surface (Fig. 7A) was sufficient to dramatically increase galectin-1 sensitivity. In contrast, when thymocytes were treated with S. typhimurium neuraminidase that selectively removes α2,3-linked sialic acid, we observed no change in susceptibility to galectin-1 cell death (data not shown). These results demonstrate that, as we have previously found for peripheral CD4 Th2 cells (9), SP thymocyte resistance to galectin-1 death results from an increase in α2,6-linked sialic acid on cell surface glycans; however, small changes, such as a modest decrease in ST6Gal-I activity or increase in ST3Gal-III activity, may be sufficient to render these cells susceptible to galectin-1 binding, signaling, and death.
FIGURE 8.
Removal of sialic acid from thymocytes renders SP thymocytes sensitive to galectin-1. A, thymocytes were treated with 100 units/ml C. perfringens neuraminidase or buffer control for 1 h. Cells were stained with CD4-FITC and CD8-PE, and SNA-biotin and streptavidin PerCP, and gated for DN, DP, CD4, and CD8 subpopulations. SNA binding is reduced on CD4 and CD8 cells after neuraminidase treatment. Data are mean fluorescence intensity ± S.D. of cells from two animals. B, loss of α2,6-linked sialic acid enhances galectin-1 susceptibility of CD4 and CD8 SP thymocytes. Cells were treated with neuraminidase or buffer control as above, followed by 20 μm galectin-1 or buffer control for 4.5 h. Cells were stained with 7-AAD, anti-CD4, and anti-CD8, and gated for DN, DP, CD4, and CD8 subpopulations. Cell loss from each subpopulation was determined. Data are mean ± S.D. of triplicate samples.
DISCUSSION
The present work demonstrates that there are at least two independent glycan-dependent mechanisms that regulate T cell susceptibility to galectin-1-induced cell death. First, the presence or absence of core 2 branches on O-glycans regulates the binding of galectin-1 to CD45RB and the ability of galectin-1 to reduce CD45 tyrosine phosphatase activity (Fig. 3), and lack of core 2 O-glycans can be overcome by a pharmacological tyrosine phosphatase inhibitor (11). Second, α2,6-linked sialic acids on N-glycans block galectin-1 binding to T cells; while this mechanism also inhibits galectin-1-induced reduction in CD45 phosphatase activity, it cannot be overcome by a tyrosine phosphatase inhibitor (Figs. 5–7). Both mechanisms likely function in vivo to regulate susceptibility of thymocyte subsets to galectin-1 T cell death. The decoration of O-glycans with core 2 branches and N-glycans with terminal α2,6-linked sialic acid are tightly regulated during T cell development and activation, so that both glycosylation events can alter the threshold for galectin-1 binding and signaling.
CD45 is highly N- and O-glycosylated and is a major cell surface receptor for galectin-1 on T cells. While not absolutely required for galectin-1 death, all nucleated hematopoietic cells express CD45, so CD45 would be an important regulator of thymocyte and T cell susceptibility to galectin-1 in vivo. CD45 can both positively and negatively regulate susceptibility to galectin-1; appropriate glycosylation of CD45 with core 2 O-glycans permits galectin-1 binding to decrease CD45 tyrosine phosphatase activity, an essential step in the galectin-1 pathway. The absolute decrease in CD45 tyrosine phosphatase activity after galectin-1 binding does not appear to depend on the expression level of CD45; indeed, it has been noted that very high levels of expression of CD45 blunt signaling strength compared with lower levels of CD45 expression3 (17, 22, 49). Thus, changes in glycosylation of CD45, rather than changes in the amount of CD45 that is expressed, may be the primary factor that regulates susceptibility to galectin-1 death during thymocyte development.
CD45 phosphatase activity is not required for phosphatidylserine externalization and membrane permeabilization during galectin-1 death; indeed, we observed enhanced death of T cells expressing an enzymatically inactive CD45 mutant compared with wild-type CD45 (Fig. 4). In contrast, we have recently shown that membrane blebbing and cytoskeletal degradation during galectin-1 death require the CD45 phosphatase domain (15). To ask if constitutively active CD45 phosphatase would oppose galectin-1 death, we used a CD45RABC mutant reported to have a constitutively active phosphatase domain, the CD45 E613R “wedge” mutant (50). However, we found that galectin-1 binding reduced the phosphatase activity of the CD45 E613R mutant to a level comparable to that observed for wild-type CD45; moreover, T200− cells expressing the E613R mutant were susceptible to galectin-1 death (supplemental Fig. S2). Thus, we could not use this mutant to determine the role of the CD45 phosphatase activity in opposing galectin-1 signaling and death.
T cells typically express lower molecular weight isoforms of CD45, such as CD45RB or CD45R0 (22, 51). These lower molecular weight isoforms carry fewer O-glycans compared with CD45RABC, which is expressed primarily by B cells (19). We found that T cells expressing CD45RB were dependent on expression of C2GnT for susceptibility to galectin-1, while cells expressing CD45RABC did not require C2GnT expression. Interestingly, mice overexpressing C2GnT have impaired T cell immune responses and humoral immune defects because of impaired T cell stimulation of B cells (52). Similarly, mice lacking ST3Gal-I, which inhibits core 2 elongation on O-glycans, have increased apoptosis of CD8+ T cells (32). Thus, an increase in core 2 O-glycans, either by overexpression of C2GnT or deletion of ST3Gal-I, results in impaired T cell function and loss of T cells. This suggests that the balance of core 1 and core 2 structures on O-glycans is more important in T cell regulation than in B cell regulation, likely because of expression of smaller CD45 isoforms by T cells. Indeed, aberrant CD45 isoform expression on T cells has been noted in autoimmune and immunodeficiency diseases (53–55).
N-Glycosylation is also integral to T cell development and function. A recent report has shown that, in mice lacking ST6Gal-I, there is a significant defect in thymocyte cellularity, starting at the DN stage (33). As the presence of α2,6-linked sialic acid protects thymocytes from galectin-1 death (Fig. 8), there may be increased loss of thymocytes in these mice because of endogenous galectin-1.
While galectin-1 resistance caused by the lack of core 2 O-glycans could be overcome by a tyrosine phosphatase inhibitor, we found that inhibition of galectin-1 binding and signaling by α2,6-linked sialic acid on N-glycans could not be overcome by a phosphatase inhibitor. This suggests that, in the absence of core 2 O-glycans, galectin-1 can bind to other receptors involved in galectin-1 death, such as CD43 and CD7; core 2 O-glycans are only required for inhibition of CD45 phosphatase activity. In contrast, an increase in α2,6-linked sialic acid on N-glycans may reduce binding of galectin-1 to other receptors, such as CD7, a small glycoprotein that is exclusively N-glycosylated and is absolutely required for galectin-1 death of human T cells (10, 34, 56, 57). Although a homologue of CD7 is not known to be expressed on murine T cells, a similar N-glycosylated protein may act as a pro-apoptotic receptor for galectin-1 in the mouse, and α2,6-linked sialic acid on N-glycans may affect galectin-1 binding to this receptor.
As mentioned above, aberrant expression of glycosyltransferases, including C2GnT, ST3Gal-I, and ST6Gal-I, leads to defects or abnormalities in T cell development, activation, and regulation (32, 33, 52). Indeed, the presence of large, highly glycosylated structures on receptors like CD45 and CD43 are necessary for TCR signaling and proper thymocyte positive and negative selection (58). Specific CD45 isoforms contribute to the intensity of TCR signals (55, 59). Additionally, galectin-1 regulates the threshold of TCR signaling and tunes thymocyte selection (4, 60). Together, these findings demonstrate that the presentation of specific N- and O-glycans by CD45 on thymocytes, influenced both by CD45 isoform usage and regulated glycosyltransferase expression, is critical for proper thymocyte development and selection involving galectin-1.
Supplementary Material
Acknowledgments
We thank Mary Clark and Dr. Omai Garner for helpful suggestions and critical reading of the manuscript. We also thank Dr. David Smith and Jamie Heinberg-Molinaro of the Glycan-Protein Interaction Core (Core H) of The Consortium for Functional Glycomics (funded by NIGMS grant number GM62116) at Emory University School of Medicine, Atlanta, GA, for the glycan array analysis. We also thank Dr. Pauline Johnson of the University of British Columbia for the T200RABC and T200C817S cell lines. We thank the UCLA Jonsson Comprehensive Cancer Center and UCLA AIDS Institute Shared Flow Cytometry Resource (NIH CA16042 and AI28697).
This work was supported, in whole or in part, by National Institutes of Health Grant GM63281 (to L. G. B.) and National Institutes of Health Training Grant T32 AI52031 (to L. A. E.). This work was also supported by a grant from the University of California, Cancer Research Coordinating Committee (to L. G. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
L. A. Earl and L. G. Baum, unpublished observations.
- TCR
- T cell receptor
- PNA
- peanut agglutinin
- ST3Gal-I
- α2,3-sialyltransferase I
- C2GnT
- core 2 O-GlcNAc transferase
- SNA
- S. nigra agglutinin
- ST6Gal-I
- α2,6-sialyltransferase I
- MAA
- Maackia amurensis lectin II
- 7-AAD
- 7-aminoactinomycin D
- DN
- double negative
- DP
- double positive
- SP
- single positive
- MFI
- mean fluorescence intensity
- ST3Gal-III
- α2,3-sialyltransferase III
- FITC
- fluorescein isothiocyanate
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- DTT
- dithiothreitol
- DMEM
- Dulbecco's modified Eagle's medium.
REFERENCES
- 1.Hogquist K. A., Baldwin T. A., Jameson S. C. (2005) Nat. Rev. Immunol. 5, 772–782 [DOI] [PubMed] [Google Scholar]
- 2.Baum L. G., Pang M., Perillo N. L., Wu T., Delegeane A., Uittenbogaart C. H., Fukuda M., Seilhamer J. J. (1995) J. Exp. Med. 181, 877–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu S. D., Whiting C. C., Tomassian T., Pang M., Bissel S. J., Baum L. G., Mossine V. V., Poirier F., Huflejt M. E., Miceli M. C. (2008) Blood 112, 120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu S. D., Tomassian T., Bruhn K. W., Miller J. F., Poirier F., Miceli M. C. (2009) J. Immunol. 182, 5283–5295 [DOI] [PubMed] [Google Scholar]
- 5.Perillo N. L., Pace K. E., Seilhamer J. J., Baum L. G. (1995) Nature 378, 736–739 [DOI] [PubMed] [Google Scholar]
- 6.Hernandez J. D., Nguyen J. T., He J., Wang W., Ardman B., Green J. M., Fukuda M., Baum L. G. (2006) J. Immunol. 177, 5328–5336 [DOI] [PubMed] [Google Scholar]
- 7.Perillo N. L., Uittenbogaart C. H., Nguyen J. T., Baum L. G. (1997) J. Exp. Med. 185, 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi S., Earl L. A., Jacobs L., Baum L. G. (2008) J. Biol. Chem. 283, 12248–12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toscano M. A., Bianco G. A., Ilarregui J. M., Croci D. O., Correale J., Hernandez J. D., Zwirner N. W., Poirier F., Riley E. M., Baum L. G., Rabinovich G. A. (2007) Nat. Immunol. 8, 825–834 [DOI] [PubMed] [Google Scholar]
- 10.Pace K. E., Hahn H. P., Pang M., Nguyen J. T., Baum L. G. (2000) J. Immunol. 165, 2331–2334 [DOI] [PubMed] [Google Scholar]
- 11.Nguyen J. T., Evans D. P., Galvan M., Pace K. E., Leitenberg D., Bui T. N., Baum L. G. (2001) J. Immunol. 167, 5697–5707 [DOI] [PubMed] [Google Scholar]
- 12.Amano M., Galvan M., He J., Baum L. G. (2003) J. Biol. Chem. 278, 7469–7475 [DOI] [PubMed] [Google Scholar]
- 13.Fouillit M., Joubert-Caron R., Poirier F., Bourin P., Monostori E., Levi-Strauss M., Raphael M., Bladier D., Caron M. (2000) Glycobiology 10, 413–419 [DOI] [PubMed] [Google Scholar]
- 14.Walzel H., Schulz U., Neels P., Brock J. (1999) Immunol. Lett. 67, 193–202 [DOI] [PubMed] [Google Scholar]
- 15.Pang M., He J., Johnson P., Baum L. G. (2009) J. Immunol. 182, 7001–7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas M. L. (1989) Annu. Rev. Immunol. 7, 339–369 [DOI] [PubMed] [Google Scholar]
- 17.McNeill L., Salmond R. J., Cooper J. C., Carret C. K., Cassady-Cain R. L., Roche-Molina M., Tandon P., Holmes N., Alexander D. R. (2007) Immunity 27, 425–437 [DOI] [PubMed] [Google Scholar]
- 18.Alexander D. R. (2000) Semin Immunol. 12, 349–359 [DOI] [PubMed] [Google Scholar]
- 19.Furukawa K., Funakoshi Y., Autero M., Horejsi V., Kobata A., Gahmberg C. G. (1998) Eur. J. Biochem. 251, 288–294 [DOI] [PubMed] [Google Scholar]
- 20.Thomas M. L., Reynolds P. J., Chain A., Ben-Neriah Y., Trowbridge I. S. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5360–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNeill L., Cassady R. L., Sarkardei S., Cooper J. C., Morgan G., Alexander D. R. (2004) Immunol. Lett. 92, 125–134 [DOI] [PubMed] [Google Scholar]
- 22.Hermiston M. L., Xu Z., Weiss A. (2003) Annu. Rev. Immunol. 21, 107–137 [DOI] [PubMed] [Google Scholar]
- 23.Earl L. A., Baum L. G. (2008) Immunol. Cell Biol. 86, 608–615 [DOI] [PubMed] [Google Scholar]
- 24.Daniels M. A., Hogquist K. A., Jameson S. C. (2002) Nat. Immunol. 3, 903–910 [DOI] [PubMed] [Google Scholar]
- 25.Trowbridge I. S., Thomas M. L. (1994) Annu. Rev. Immunol. 12, 85–116 [DOI] [PubMed] [Google Scholar]
- 26.Galvan M., Tsuboi S., Fukuda M., Baum L. G. (2000) J. Biol. Chem. 275, 16730–16737 [DOI] [PubMed] [Google Scholar]
- 27.Hernandez J. D., Klein J., Van Dyken S. J., Marth J. D., Baum L. G. (2007) Int. Immunol. 19, 847–856 [DOI] [PubMed] [Google Scholar]
- 28.Hernandez J. D., Baum L. G. (2002) Glycobiology 12, 127R–136R [DOI] [PubMed] [Google Scholar]
- 29.Reisner Y., Linker-Israeli M., Sharon N. (1976) Cell. Immunol. 25, 129–134 [DOI] [PubMed] [Google Scholar]
- 30.Gillespie W., Paulson J. C., Kelm S., Pang M., Baum L. G. (1993) J. Biol. Chem. 268, 3801–3804 [PubMed] [Google Scholar]
- 31.Wu W., Harley P. H., Punt J. A., Sharrow S. O., Kearse K. P. (1996) J. Exp. Med. 184, 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Priatel J. J., Chui D., Hiraoka N., Simmons C. J., Richardson K. B., Page D. M., Fukuda M., Varki N. M., Marth J. D. (2000) Immunity 12, 273–283 [DOI] [PubMed] [Google Scholar]
- 33.Marino J. H., Tan C., Davis B., Han E. S., Hickey M., Naukam R., Taylor A., Miller K. S., Van De Wiele C. J., Teague T. K. (2008) Glycobiology 18, 719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabrera P. V., Amano M., Mitoma J., Chan J., Said J., Fukuda M., Baum L. G. (2006) Blood 108, 2399–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motran C. C., Molinder K. M., Liu S. D., Poirier F., Miceli M. C. (2008) Eur. J. Immunol. 38, 3015–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stowell S. R., Arthur C. M., Mehta P., Slanina K. A., Blixt O., Leffler H., Smith D. F., Cummings R. D. (2008) J. Biol. Chem. 283, 10109–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leppänen A., Stowell S., Blixt O., Cummings R. D. (2005) J. Biol. Chem. 280, 5549–5562 [DOI] [PubMed] [Google Scholar]
- 38.Wang Y., Johnson P. (2005) J. Biol. Chem. 280, 14318–14324 [DOI] [PubMed] [Google Scholar]
- 39.Karasuyama H., Kudo A., Melchers F. (1990) J. Exp. Med. 172, 969–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pace K. E., Hahn H. P., Baum L. G. (2003) Methods Enzymol. 363, 499–518 [DOI] [PubMed] [Google Scholar]
- 41.Baum L. G., Derbin K., Perillo N. L., Wu T., Pang M., Uittenbogaart C. (1996) J. Biol. Chem. 271, 10793–10799 [DOI] [PubMed] [Google Scholar]
- 42.Holladay S., Blaylock B., Smith B., Luster M. (1993) Immunol. Invest. 22, 517–529 [DOI] [PubMed] [Google Scholar]
- 43.Jones A. T., Federsppiel B., Ellies L. G., Williams M. J., Burgener R., Duronio V., Smith C. A., Takei F., Ziltener H. J. (1994) J. Immunol. 153, 3426–3439 [PubMed] [Google Scholar]
- 44.Lantéri M., Giordanengo V., Hiraoka N., Fuzibet J. G., Auberger P., Fukuda M., Baum L. G., Lefebvre J. C. (2003) Glycobiology 13, 909–918 [DOI] [PubMed] [Google Scholar]
- 45.Barran P., Fellinger W., Warren C. E., Dennis J. W., Ziltener H. J. (1997) Glycobiology 7, 129–136 [DOI] [PubMed] [Google Scholar]
- 46.Pace K. E., Lee C., Stewart P. L., Baum L. G. (1999) J. Immunol. 163, 3801–3811 [PubMed] [Google Scholar]
- 47.Fortin M., Steff A. M., Felberg J., Ding I., Schraven B., Johnson P., Hugo P. (2002) J. Immunol. 168, 6084–6089 [DOI] [PubMed] [Google Scholar]
- 48.Di Virgilio S., Glushka J., Moremen K., Pierce M. (1999) Glycobiology 9, 353–364 [DOI] [PubMed] [Google Scholar]
- 49.Zamoyska R. (2007) Immunity 27, 421–423 [DOI] [PubMed] [Google Scholar]
- 50.Majeti R., Xu Z., Parslow T. G., Olson J. L., Daikh D. I., Killeen N., Weiss A. (2000) Cell 103, 1059–1070 [DOI] [PubMed] [Google Scholar]
- 51.Hathcock K. S., Laszlo G., Dickler H. B., Sharrow S. O., Johnson P., Trowbridge I. S., Hodes R. J. (1992) J. Immunol. 148, 19–28 [PubMed] [Google Scholar]
- 52.Tsuboi S., Fukuda M. (1998) J. Biol. Chem. 273, 30680–30687 [DOI] [PubMed] [Google Scholar]
- 53.Tackenberg B., Nitschke M., Willcox N., Ziegler A., Nessler S., Schumm F., Oertel W. H., Hemmer B., Sommer N. (2003) Autoimmunity 36, 117–121 [DOI] [PubMed] [Google Scholar]
- 54.Dawes R., Petrova S., Liu Z., Wraith D., Beverley P. C. L., Tchilian E. Z. (2006) J. Immunol. 176, 3417–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Windhagen A., Sönmez D., Hornig-Do H. T., Kalinowsky A., Schwinzer R. (2007) Clin. Exp. Immunol. 150, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sempowski G. D., Lee D. M., Kaufman R. E., Haynes B. F. (1999) Crit. Rev. Immunol. 19, 331–348 [PubMed] [Google Scholar]
- 57.Roberts A. A., Amano M., Felten C., Galvan M., Sulur G., Pinter-Brown L., Dobbeling U., Burg G., Said J., Baum L. G. (2003) Mod. Pathol. 16, 543–551 [DOI] [PubMed] [Google Scholar]
- 58.Irles C., Symons A., Michel F., Bakker T. R., van der Merwe P. A., Acuto O. (2003) Nat. Immunol. 4, 189–197 [DOI] [PubMed] [Google Scholar]
- 59.Xu Z., Weiss A. (2002) Nat. Immunol. 3, 764–771 [DOI] [PubMed] [Google Scholar]
- 60.Vespa G. N., Lewis L. A., Kozak K. R., Moran M., Nguyen J. T., Baum L. G., Miceli M. C. (1999) J. Immunol. 162, 799–806 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.