Abstract
The Tat system is used to transport folded proteins across the cytoplasmic membrane in bacteria and archaea and across the thylakoid membrane of plant chloroplasts. Multimers of the integral membrane TatA protein are thought to form the protein-conducting element of the Tat pathway. Nitroxide radicals were introduced at selected positions within the transmembrane helix of Escherichia coli TatA and used to probe the structure of detergent-solubilized TatA complexes by EPR spectroscopy. A comparison of spin label mobilities allowed classification of individual residues as buried within the TatA complex or exposed at the surface and suggested that residues Ile12 and Val14 are involved in interactions between helices. Analysis of inter-spin distances suggested that the transmembrane helices of TatA subunits are arranged as a single-walled ring containing a contact interface between Ile12 on one subunit and Val14 on an adjacent subunit. Experiments in which labeled and unlabeled TatA samples were mixed demonstrate that TatA subunits are exchanged between TatA complexes. This observation is consistent with the TatA dynamic polymerization model for the mechanism of Tat transport.
Keywords: Membrane/Channels, Membrane/Proteins, Methods/Electron Paramagnetic Resonance EPR, Protein/Structure, Protein/Translocation, Transport, Spin Labeling
Introduction
The twin-arginine translocation (Tat)5 transports folded proteins across the cytoplasmic membrane of bacteria and archaea and across the thylakoid membrane of plant chloroplasts (1–4). Proteins are targeted to the Tat pathway by amino-terminal signal peptides bearing a consensus sequence motif that includes consecutive arginine residues (5, 6). Transport through the Tat pathway is driven by the transmembrane proton electrochemical gradient (7). The Tat pathway is vital for many cellular processes, including biogenesis of respiratory and photosynthetic electron transfer chains, formation of the bacterial cell envelope, establishing the nitrogen fixing symbiosis, and bacterial pathogenesis (8).
In the bacterium Escherichia coli the minimal components of the Tat machinery are the integral membrane proteins TatA, TatB, and TatC (9–12). TatA and TatB are sequence-related but functionally distinct proteins. TatA is predicted to be composed of an amino-terminal transmembrane helix (also termed helix-α1) separated by a short hinge region from an amphipathic helix (helix-α2), which is in turn followed by an unstructured, polar, carboxyl-terminal region. The amphipathic helix is proposed to lie along the membrane surface but may undergo a change of topology related to function in which the helix changes location to become membrane-spanning (13). TatA purified in detergent solution forms large, homo-oligomeric complexes of variable size (14–16). Low resolution single particle electron microscopy studies of detergent-solubilized TatA complexes show ring-shaped structures with inner diameters suitable for accommodating typical Tat transported substrates (16, 17). A lid structure is associated with one side of the ring pore. The TatB and TatC proteins form a separate large, hetero-oligomeric complex in detergent solution (17, 18).
Current models for the mechanism of Tat translocation suggest that substrate proteins initially bind to the TatBC complex, which then recruits TatA to form the active translocation site (17, 19–24). TatA is thought to form the protein-conducting element of the system. The task faced by TatA is challenging, because it must allow the transport of large protein substrates of varying sizes while maintaining the membrane permeability barrier to ions and small molecules. There is only limited information about how this is achieved. There is suggestive evidence that TatA may polymerize in response to substrate (21, 22), and it has been suggested that the oligomeric state of TatA may dynamically alter to accommodate substrate proteins of different sizes (16). However, to fully understand how TatA complexes are able to mediate the transmembrane movement of folded proteins structural information at the molecular level of resolution is now required.
In this study we have used site-directed spin labeling to probe the structure and organization of TatA subunits. Site-directed spin labeling is a powerful method to obtain structural information on proteins, such as TatA, that are not easily studied by crystallographic or solution NMR methods (25–28). The room temperature EPR spectra of a covalently attached spin label is sensitive to the dynamics of the label and hence its protein environment (29–31). Thus a set of spin labels along a helix can provide a powerful means of deducing whether a residue is sitting at the protein surface or at a buried site. Unpaired electron spins will also interact, via dipolar coupling, with a nearby spin. The magnitude of this coupling is proportional to 1/rab3, where rab is the inter-spin distance. Measurements of inter-spin coupling can, therefore, be used to estimate the distances between the spins and hence the distances between different regions of spin-labeled proteins (32–34).
In this study we have carried out EPR measurements on purified, detergent-solubilized TatA complexes containing site-directed nitroxide spin labels at consecutive positions within the transmembrane helix. The results suggest a model for the organization of the TatA complex in which a single wall of transmembrane helices interact via residues Ile12 and Val14. We also observed subunit exchange between TatA complexes. This is consistent with models for the mechanism of Tat transport that invoke dynamic TatA polymerization.
MATERIALS AND METHODS
Plasmid Construction
Plasmid pQE80-TatA expresses E. coli TatA with a carboxyl-terminal hexahistidine tag. pQE80-TatA was produced by amplifying the tatA gene from plasmid pUNITATA (35) using the primers TATA5 (11) and TatA_ RS_His(5′-ATACGTGGATCCTTAGTGATGGTGATGGTCATGAGATCTCACCTGCTCTTTATCGTGG-3′), digesting the product with EcoRI and BamHI, and cloning into the same sites in pQE80 (Qiagen). Plasmids expressing single cysteine TatA variants were constructed by using the same strategy as pQE80-TatA but with the appropriate mutant tatA alleles amplified from the pUNITATA-derived plasmids described by Greene et al. (35).
Protein Purification and Characterization
E. coli strain C43 (36) transformed with the appropriate expression plasmid was cultured aerobically at 37 °C in Luria-Bertani (LB) medium. When the cultures reached an A600 nm of 0.5, expression of the tatA allele was routinely induced with 1 mm final concentration of isopropyl β-d-thiogalactoside, and the growth continued for a further 5 h before harvesting. The TatA Ile11 → Cys variant did not express well under these conditions. Instead, production of this variant was induced by 0.5 mm final concentration isopropyl β-d-thiogalactoside, and the cells were cultured for a further 20 h at 25 °C.
Cells were harvested by centrifugation and resuspended in 20 mm MOPS, pH 7.2 (at 25 °C), 200 mm NaCl (buffer A) containing 2 mm dithiothreitol. Isolation of a crude membrane fraction, solubilization of the membranes with nonapolyoxyethylene dodecyl ether (C12E9, Sigma), and purification of TatA variants from the membrane extract by Ni(II) affinity chromatography were as described earlier (14) with the following modifications. A 5-ml HisTrap Chelating HP column (Amersham Biosciences) was used in the affinity purification step. 2 mm dithiothreitol was included in the buffer at all steps up to and including the Ni(II) affinity column wash step to ensure that the cysteine residue of the TatA variants was in the reduced state. Dithiothreitol was omitted from the affinity column elution step onward to avoid interference with the labeling reaction.
TatA-containing fractions from the Ni(II) affinity column were pooled and incubated for 16 h at 4 °C and then 4 h at room temperature with 1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl (MTSL, Toronto Research Chemicals, Canada). A 10:1 molar excess of MTSL over TatA was employed to produce fully labeled protein, while substoichiometric MTSL:TatA ratios were used for method 1 underlabeling. The sample was then supplemented with Na2EDTA to a final concentration of 20 mm and concentrated to a volume of 0.5 ml using a 100-kDa molecular mass cut-off centrifugal concentrator (Millipore). The concentrated sample was subjected to size-exclusion chromatography on a Superose 6 10/300 GL column (Amersham Biosciences) in buffer A containing 0.1% C12E9. TatA-containing fractions were identified by SDS-PAGE, pooled, dialyzed against buffer A containing 0.1% C12E9, and concentrated to ∼5 mg of protein ml−1 using a 100-kDa molecular mass cut-off microcon centrifugal concentrator (Millipore). Protein concentrations were estimated using the Bio-Rad protein assay with bovine serum albumin as the standard. The degree of labeling was determined in each case by comparing the measured protein concentration to the spin label concentration obtained by integration of the spin intensity of the EPR spectrum. The purified MTSL-labeled TatA variants were analyzed on a Bruker Ultraflex TOF/TOF matrix-assisted laser desorption ionization time-of-flight mass spectrometer, using a sinapinic acid matrix. All variants showed a major peak of mass within 5 Da of that expected for TatA-MTSL. Analysis of mass peak intensities suggested >90% MTSL labeling for all “fully labeled” TatA variants in agreement with the biochemical analysis.
EPR Spectroscopy
Room temperature X-band EPR spectra were acquired using a Bruker EleXsys 500 spectrometer fitted with an ER4123D resonator with samples contained in 0.6-mm inner diameter × 0.84-mm outer diameter quartz tubes (37). A Bruker EleXsys 580 system with an ER4118X-MD resonator was used in the continuous-wave mode to record X-band EPR at 170 K. For this system 3-mm inner diameter × 4-mm outer diameter quartz tubes were used. W-band EPR spectra were acquired using a Bruker, EleXsys 680 spectrometer with samples contained in 0.1-mm inner diameter × 0.5-mm outer diameter quartz tubes.
RESULTS
Preparation of Spin-labeled TatA Complexes and Evidence for Subunit Exchange
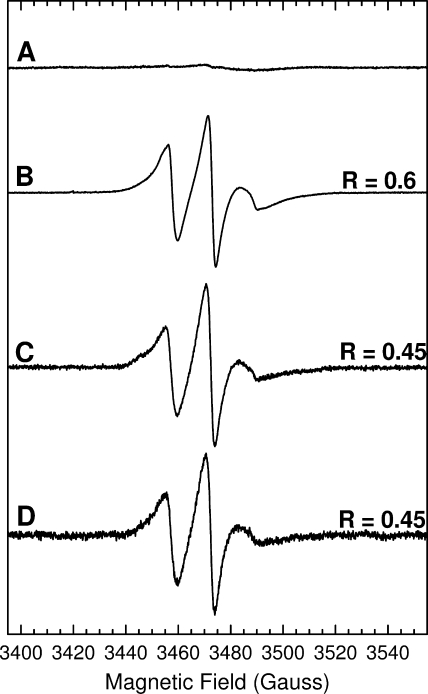
Samples of TatA were prepared with nitroxide radical spin labels at each of positions 9–18. These residues encompass the center and carboxyl-terminal half of the transmembrane helix and correspond to just over 2.5 helical turns. We chose this region of TatA for analysis, because previous studies had shown that this part of TatA forms well defined interactions with other TatA protomers and because cysteine substitutions at the chosen positions do not significantly affect TatA function (35). Appropriate hexahistidine-tagged single cysteine TatA variants were purified in the detergent C12E9 and then labeled with the thiol-specific spin label MTSL (38–40). The purification procedure was essentially identical to the one used previously to obtain low resolution structures of TatA by electron microscopy (16). Thus the structural data from the two studies should be directly comparable. Blue native-PAGE analysis (41) of the purified spin-labeled variants revealed ladders of bands resembling those exhibited by the wild-type TatA protein (15, 16), and spin-labeled protein was eluted in the same fractions in gel filtration as the wild-type TatA (14). To confirm that the spin labels were attached only at the engineered sites, a batch of cysteine-free parental TatA was taken through the same procedure. This resulted in an EPR spectrum devoid of any features from a nitroxide spin label (Fig. 1A).
FIGURE 1.
Comparison of the effects of different spin dilution methods on the room temperature X-band EPR spectra of the spin-labeled TatA Ile15 → Cys variant. A, wild-type TatA at a protein concentration of 1 mm. B, 100% labeled Ile15 → Cys TatA. The protein concentration was 350 μm. C, 10% labeled Ile15 → Cys TatA prepared by sub-stoichiometric addition of MTSL. The protein concentration was 800 μm. D, 10% labeled Ile15 → Cys TatA prepared by dilution of 100% labeled Ile15 → Cys with wild-type TatA. The final protein concentration was 350 μm. Spectral intensities have been normalized for comparison of line shapes. Spectra were recorded under the following conditions: frequency = 9.7 GHz, power = 2.0 milliwatts, modulation amplitude = 1 gauss, temperature = 295 K. R is the ratio of the low field peak height to that of the central peak.
The EPR line shapes recorded at room temperature for the spin-labeled TatA variants contain contributions not only from the motions of the spin label and protein but also from any inter-spin dipolar coupling. Separation of these two effects is necessary to interpret the EPR spectra. Freezing the sample removes the contributions from label and protein motions allowing the analysis of dipolar coupling. Conversely, any contribution from dipolar coupling can be reduced by diluting spins thereby increasing inter-spin distances. In this study two different methods of sub-stoichiometric labeling were used to obtain spin dilution. In method 1 TatA was underlabeled by adding substoichiometric amounts of MTSL to the protein during the labeling reaction. In method 2 fully labeled TatA was mixed with unlabeled wild-type TatA. Identical line shape broadening was seen whichever spin dilution method was employed, and there was no significant difference between 30%, 20, and 10% dilutions. Therefore we assumed that 10% spin dilution is sufficient to remove effects due to dipolar coupling. An example is given in Fig. 1 where the room temperature EPR spectrum of 100% spin-labeled variant Ile-15 → Cys (Fig. 1B) is compared with 10% spin-labeled protein prepared by underlabeling (method 1, Fig. 1C) or prepared by mixing with wild-type protein (method 2, Fig. 1D). Line shape broadening can be characterized by a parameter R, which is the ratio of the height of the low field peak to that of the central resonance line. Both 10% labeled samples have R = 0.45 and, therefore, show identical line shape broadening.
The equivalence of the two spin dilution methods is an important observation, because TatA subunits can only have the same environment after mixing that they do after underlabeling if the subunits equilibrate completely between TatA complexes. If such subunit exchange had not occurred then method 2 would be expected to yield a 9:1 mixture of completely unlabeled and 100% labeled complexes resulting in an EPR spectrum identical to that of a stoichiometrically labeled sample but with a 10-fold reduction in spin intensity. Our data show that subunit exchange was complete by the time the samples had been mixed and an EPR spectrum recorded (about 10 min). Thus the rate of helical exchange is relatively rapid. To investigate the kinetics of exchange in more detail samples were flash-frozen in liquid nitrogen at different times after mixing, and spectral broadening was measured in the low temperature EPR spectrum (data not shown). These experiments showed that subunit exchange was effectively complete at the shortest measurable mixing time (1 min).
Spin Label Mobility
Nitroxide motional dynamics cause spectral broadening in the room temperature EPR of spin-labeled proteins. Various molecular motions contribute to probe mobility, including the flexibility of the spin-labeled side chain relative to the protein, local protein backbone fluctuations, and the overall rotary diffusion of the protein (27). At 9.5 GHz and room temperature the rotational diffusion of oligomeric complexes of TatA will be too slow on the EPR timescale to affect the line shape and can be ignored. By contrast, internal rotations of the spin label around the bonds in its flexible linker, and backbone motions of the protein itself, can both be sufficiently rapid (τ ∼ 1 ns) to influence spectral line shape (26). These motions and their anisotropy will be strongly influenced by packing interactions in the local environment and by the backbone secondary structure. Spectral line shape analysis can, therefore, show whether the spin label is at a buried or surface site and give information about the type of secondary structure in which the spin label is located. Semi-empirical measures of spin label mobility can be derived from the EPR “mobility” parameter ΔH0, measured as the peak-to-peak first derivative width of the central line, and by the spectral breadth parameter, which is represented by the second moment of the spectral breadth, 〈H2〉 (42, 43),
 |
where B is magnetic field, S(B) is the integrated absorbance of the EPR spectrum of the spin label, and the first moment 〈H〉 = ∫B S(B)dB/∫S(B)dB. Using proteins of known structure, correlations between the inverse values of these parameters, ΔH0−1 and 〈H2〉−1, allow distinction between residues that are buried, surface-exposed, in a loop, or at contact sites between secondary structure regions (43).
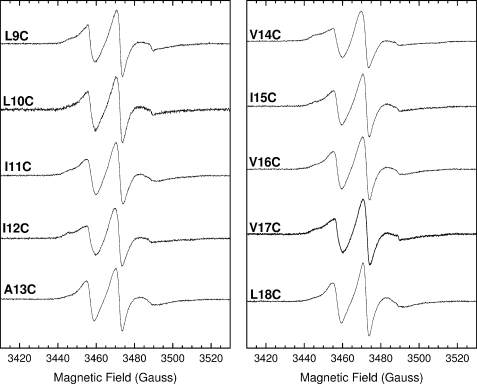
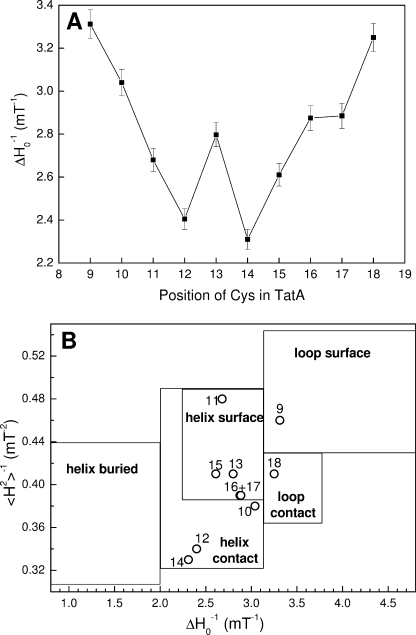
Room temperature X-band EPR spectra were collected for the TatA variants successively spin-labeled from residue Leu-9 to Leu-18 (Fig. 2), and spin mobility parameters were calculated (Fig. 3). In each case the protein was 10% spin-labeled by dilution of 100% labeled TatA with wild-type TatA. As discussed in the previous section this removes any detectable broadening from interspin dipolar coupling and hence the EPR line shapes reflect only the motional dynamics of each spin label. Fig. 3A shows the variation of side-chain mobility, represented by ΔH0−1 values, along the transmembrane helix. Positions 12 and 14 have the most restricted mobility, whereas positions 9, 10, and 18 are significantly more mobile than the average value of 2.8 millitesla−1. This suggests two sites of significant inter-protein contact at the center of the region probed, namely residues 12 and 14. By plotting 〈H2〉−1 against ΔH0−1 for each sequence position residues can be assigned to regions of secondary structure and of inter-protein contact (Fig. 3B). This plot suggests that residues 12 and 14 are at contact interfaces, whereas residues 11, 13, and 15–17 are exposed at a helical surface.
FIGURE 2.
Room temperature X-band EPR spectra of 10% spin-labeled TatA complexes. Each TatA variant was spin-labeled to 10% by dilution of 100% labeled TatA with wild-type TatA. The final spin label concentrations ranged from 35 to 80 μm. Spectral intensities have been normalized to aid comparison of line shapes. Spectra were recorded under the following conditions: frequency = 9.7 GHz, power = 2.0 milliwatts, modulation amplitude = 1 gauss, temperature = 295 K.
FIGURE 3.
Mobility of spin labels attached to the transmembrane helix of TatA. A, variation of the mobility parameter ΔH0−1 along the transmembrane helix of TatA. The ΔH0−1 values were calculated from the room temperature EPR spectra given in Fig. 2. Error bars are calculated from the results of duplicate sample preparations. B, correlation of the mobility parameters 〈H2〉−1 (inverse second moment) and ΔH0−1 (inverse of the central line width) for spin labels at positions 9–18 within TatA. Mobility parameters were calculated from the room temperature X-band EPR spectra shown in Fig. 2. Assignment of structural environments is based on the compilation of Bordignon and Steinhoff (43).
Estimating Inter-helical Distances
In frozen solution the motions of the spin labels are quenched, and any broadening in EPR line shape is due to dipolar coupling between labels. The strength of the dipolar coupling depends on the separation between the interacting labels and thus gives information on the distance between the labeled positions. If the interacting spin separation is ∼5 Å or less the dipolar coupling may become large in comparison with the energy of the microwave photon, rendering the interacting spins EPR silent.
X-band EPR spectra were collected for the ten spin-labeled TatA variants at 170 K for both 10 and 100% labeled samples. Fig. 4A shows the spectra for a representative variant. The changes in line shape on dilution are characteristic of those observed in the presence of weak magnetic dipolar interactions between nitroxide spin labels (32). The broadening in the 100% labeled spectrum is similar for all label positions between 9 and 18. Thus the distance between the same positions in adjacent protomers is approximately the same for all positions analyzed. Crucially, comparison of the integrated EPR signal intensities of the fully labeled and underlabeled samples shows no loss of spin concentration with increased labeling. Thus there is no evidence for loss of signal intensity that would be indicative of very close interactions between labels. We, therefore, conclude that no residue in the transmembrane region of TatA is in close proximity to the identical residue in an adjacent TatA protomer.
FIGURE 4.
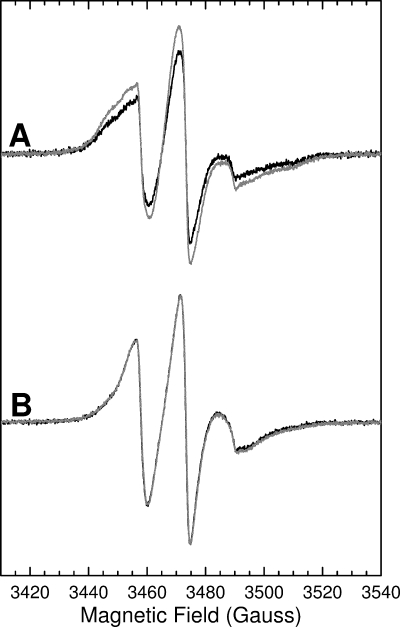
Analysis of inter-spin distances between nearest-neighbor spin labels in the transmembrane helix of TatA. In A, black line, 100% labeled Ile15 → Cys TatA. The protein concentration was 350 μm. Gray line, 10% labeled Ile15 → Cys TatA prepared by sub-stoichiometric addition of MTSL. The total protein concentration was 800 μm (gray line). The spectral intensities have been normalized to allow comparison of line shapes. Spectra were recorded under the following conditions: frequency = 9.65 GHz, power = 0.006 milliwatt, modulation amplitude = 1 gauss, temperature = 170 K. In B, black line, 100% labeled Ile15 → Cys TatA as in A. Gray line, simulation of the spectrum of 100% labeled Ile15 → Cys TatA using a fitting routine that takes account of dipolar broadening.
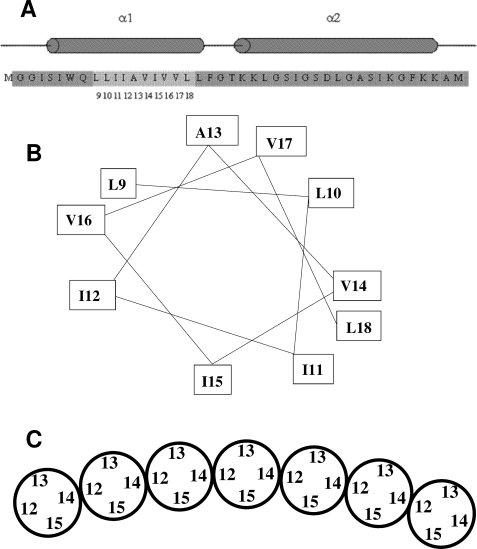
The mobility analysis described above identified Ile12 and Val14 as likely contact points between TatA protomers. A helical wheel representation of the TatA transmembrane region (Fig. 5B) shows that these residues lie on opposite sides of the helix. Thus, the TatA helices in the complex must be interacting either in an arrangement that brings residue Ile12 into contact with Val14 (Ile12-Val14), or in an arrangement involving Ile12 to Ile12 and Val14 to Val14 contacts (Ile12-Ile12). The Ile12-Ile12 model, in contrast to the Ile12-Val14 model, predicts spin quenching for TatA variants that are 100% labeled at either position 12 or 14. Because such quenching is not observed, we infer that TatA protomers form the Ile12-Val14 arrangement with contacts between Ile12 and Val14 (Fig. 5C).
FIGURE 5.
Model for the arrangement of the transmembrane helices in the TatA complex. A, sequence and predicted secondary structure of the amino-terminal portion of E. coli TatA. Residues 1–43 of the 89 amino acid E. coli TatA protein are shown. The predicted α-helical regions of the protein are indicated by cylinders above the sequence. The numbered, lighter gray region indicates the sites where residues were substituted with cysteines and spin-labeled in this study. B, helical wheel representation of the sites within the TatA transmembrane helix to which spin labels were attached in this work. C, model for the arrangement of TatA transmembrane helices within the TatA complex derived from the spin labeling data presented in this work. The helices are shown schematically and end-on. Only a section of a TatA oligomer is shown. It is envisaged that the ends of the oligomer are extended by further subunits to form a ring. Note that the depicted organization with residue 15 facing the interior of the ring and residue 13 the outside of the ring is arbitrary. The opposite orientation is equally plausible.
To directly test whether residues Ile12 and Val14 are in contact we mixed TatA that had been 100% labeled at position 12 with an equal quantity of TatA 100% labeled at position 14. The integrated intensity of the resulting room temperature X-band EPR spectrum decreased by ∼30% compared with the normalized sum of the integrated intensities of the samples prior to mixing (Fig. 6A). The spin quenching confirms that positions 12 and 14 are in sufficiently close proximity to produce a loss of signal intensity due to dipolar coupling. By contrast, mixing of variants 100% spin-labeled at positions 13 and 15 gave exactly the same integrated EPR intensity as that of the normalized sum of the two individual components (Fig. 6B), showing that these two spin labels do not approach closely.
FIGURE 6.
Room temperature X-band EPR spectra of mixtures of spin-labeled TatA variants. A, 100% spin-labeled Ile12 → Cys was mixed with an equimolar amount of 100% spin-labeled Val14 → Cys. B, 100% spin-labeled Ala13 → Cys was mixed with an equimolar amount of 100% spin-labeled Ile15 → Cys. Black lines, experimental spectra; gray lines, spectra calculated by summing each component of the mixture and dividing by two. Spectra were recorded under the following conditions: frequency = 9.7 GHz, power = 2.0 milliwatts, modulation amplitude = 1 gauss, temperature = 295 K.
In summary, these data show that the helical faces containing residues 12 and 14 are in Ile12-Val14 contact, but that the faces containing residues 13 and 15 do not have close contacts with each other. The most straightforward interpretation of these data is that the TatA transmembrane helices are arranged in a single line with each helix oriented in the same way as the preceding helix (Fig. 5C). We have simulated the dipolar broadening data in the light of this model and used this fit to estimate spin-spin separations. In the model each spin label in a 100% labeled sample has two nearest neighbors at the same distance. Hence we have fitted these data to an exchange-coupled trimer with equidistant spin labels. Because the dipolar coupling dies away as 1/rab3 the contributions to broadening from the second nearest neighbors can be ignored. Full details of the fitting method and assumptions are given in the supplemental material. Fig. 4B shows one example of the observed broadening at 170 K together with the simulated spectrum. The spin-spin distances estimated on the basis of this model for each of position 9–18 all lie between ∼14 and 15.5 Å with a mean of ∼14.7 ± 1.0 Å. This distance equates well to the van der Waals diameters of typical helices, which vary between 13 and 16 Å depending on the nature of the side chains and the tightness of packing (44, 45). The fitted distances are, thus, in good agreement with our inferred model for the arrangement of the TatA transmembrane helices in which the spin labels are predicted to be positioned at a width of one helix apart (Fig. 5C).
DISCUSSION
Single molecule electron microscopy of detergent-solubilized TatA oligomers reveals ring-shaped structures with variable diameters that have been interpreted as capped transmembrane pores (16). The average outer ring diameters were between 60 and 100 Å with a constant wall thickness of between 25 and 30 Å. Assuming the ring has a transmembrane orientation in the membrane, it can be inferred that the transmembrane helix of TatA is located in the ring wall. To what extent the amphipathic helix and C-tail of TatA also contribute to the wall structure rather than to the cap structure is unclear. This electron microscopy model raises a number of questions about the ways in which the TatA transmembrane helices might pack together to form a variable diameter ring. Is the repeat packing unit a single helix or higher aggregates such as dimers, trimers, or tetramers? Which residues lie at the inter-helical binding surfaces, and how many binding interfaces are there around the helix?
Here we have demonstrated that X-band EPR spectra of a good signal-to-noise ratio can be obtained from samples of detergent-solubilized TatA oligomers labeled with the nitroxide radical MTSL covalently attached to cysteine residues at every position from 9 to 18 along the transmembrane helix. Blue native PAGE analysis of the spin-labeled samples showed a ladder of oligomers similar to those seen with wild-type TatA confirming that the spin-labeled samples form oligomers of similar size and repeat unit. In interpreting our EPR data we have made the reasonable assumption that any structure built of multiple copies of identical helices will have a repeating pattern of high regularity with each helix having identical inter-helical binding regions. Our data indicate that the transmembrane helices are positioned side by side with residues Ile12 on one side of each helix and Val14 on the other forming the closest helical contacts, resulting in an Ile12-Val14 arrangement of helices around the ring (Fig. 5C). No other helical contact regions have been detected within this set of spin-labeled residues. Hence we conclude that only a single line of transmembrane helices form the pore wall. Our data leave open the possibility that other portions of the TatA molecule, most likely the amphipathic helix, could be positioned in the ring on either side of the wall of transmembrane helices. Analysis of the size differences between TatA complexes by blue native PAGE (15, 16) or by fluorescence labeling (21) have suggested a minimum difference in size of 3–4 subunits. On this basis it has been suggested that TatA complexes are built up from small bundles of TatA proteins. However, there is no evidence from our data to suggest there are more than two helical binding interfaces. In addition the equivalence of the EPR spectra obtained by spin dilution by underlabeling and by mixing with unlabeled protein is only readily explained if single subunit exchange is taking place. The TatA complex model we propose based on interactions between transmembrane helices provides no obvious constraints on the ring size or its incremental growth by the addition of single subunits.
Analysis of the room temperature spin label motions of 10% spin-labeled TatA identifies the steric constraints on residue motions caused by the presence of tertiary protein contacts. The mobility parameter ΔH0−1 can be normalized to give a “scaled mobility” factor, Ms, that allows comparison of ΔH0−1 values between different proteins (26, 27),
where ΔHi−1 and ΔHm−1 are typical widths for strongly immobilized and highly mobile site-directed protein spin labels, respectively, and are assigned values of 8.4 and 2.1 gauss (26, 27). The mean mobility values, 〈Ms〉, of spin labels have been measured in a variety of transmembrane proteins with different helical packing densities. Lac Permease and bacteriorhodopsin possess helices with 〈Ms〉 values of 0.12 and 0.25, respectively, as a consequence of their tight helical packing (27). By contrast, spin labels attached to helices in the channel-forming domain of colicin E1 have a higher 〈Ms〉 value of 0.48. Spin labels attached to the transmembrane helices in TatA give a 〈Ms〉 value of 0.45, indicating a relatively low packing density comparable with that of the colicin E1 channel (27).
The spin label-derived mobilities of individual TatA residues 9–18 (Fig. 3A) do not show the ∼3.8-residue repeating pattern that would be expected for a pair of identical helices lying parallel to one another with similar interactions along their length. On the contrary there are two residues of low mobility lying at the center of the helix, 12 and 14, with the other residues of rising mobility toward both ends of the helix. These observations suggest that the transmembrane helices are tilted relative to each other such that they only interact strongly in the central region. Consistent with this interpretation a recent solid state NMR study of a truncated Bacillus subtilis TatA protein in a bicelle environment suggested that the long axis of the transmembrane helix lies at an angle of 17° to the bilayer normal (46). A similar pattern of spin mobilities along a helix has been found in studies of the leucine zipper GCN4 with a clear helical contact pattern in the helical binding region of the zipper and less restricted mobilities observed at one end where the pair of helices open out to embrace DNA (27).
A key observation of this study is that spin dilution obtained by mixing labeled and unlabeled proteins gives the same EPR spectra as spin dilution by sub-stoichiometric labeling. This shows that transmembrane helices of TatA freely exchange between TatA complexes in detergent solution. Helix exchange on mixing is complete in less than a minute and is, thus, sufficiently rapid to be of physiological relevance. It has been suggested that TatA undergoes dynamic polymerization in response to substrate (21–24) and possibly also to adapt the size of the TatA ring to the size of the substrate (16). Our observation of facile exchange of helices between complexes is consistent with, and provides additional support for, such models of TatA mechanism.
This study, like previous electron microscopy studies (16, 17), was carried out on purified detergent-solubilized TatA complexes and the possibility that detergent extraction may be influencing the aggregation state of TatA should be borne in mind. Specifically, there is evidence that TatA polymerization in membranes requires a substrate-activated TatBC complex and the transmembrane proton electrochemical gradient (21, 22, 24, 47), yet in the absence of both of these factors detergent-solubilized TatA is found to be in the form of large complexes (14–17, 19). The size distribution of TatA complexes in membranes and detergent solution are, however, similar, and there is evidence from diffusion behavior that TatA complexes in membranes, like detergent-solubilized TatA, have a ring morphology (16, 21). It has been argued that detergent extraction pushes the TatA polymerization equilibrium toward the fully assembled state (21).
In an earlier study we determined the propensity of the TatA transmembrane helix cysteine variants to form disulfide bonds with the same cysteine residue in other TatA molecules within the native membrane environment (35). That study found that cross-links could be formed at positions on all sides of the transmembrane helix and that the Ile12 → Cys variant showed the highest degree of cross-linking. At first sight these observations appear inconsistent with the single wall model for TatA derived here (Fig. 5C) in which Ile12 in one helix is close to Ile14, but distant from Ile12 in the adjacent helix. However, as discussed above, it is possible that the two studies are analyzing TatA in different oligomeric states with the cross-linking study, being in de-energized membranes, probing the disassembled state, whereas the EPR analyses in detergent solution are characterizing the polymerized state. Further, it is not established whether the disulfide contacts detected in the cross-linking study correspond to interactions within assembled TatA multimers, or whether they represent collisional contacts between TatA monomers (or complexes) and, therefore, correlate with the surface exposure of the residue under analysis. The observation that cross-links can be formed between positions on all sides of the helix strongly suggests either a dynamic structure or the lack of a single structure. Overall the evidence supports a model of a dynamic structure of TatA helices. Resolving the question of how the structure of TatA in membranes differs from the structure adopted in detergent solution will require further work, including spin-labeling studies of TatA in a membrane environment. However, such experiments will have to await development of a method for functional reconstitution of purified TatA into membranes.
Supplementary Material
This work was supported by Grants BBS/B/03637, BB/D012074/1, and BB/D004578/1 from the Biotechnology and Biological Sciences Research Council, and Grant GR/S57402/01 from the Engineering and Physical Sciences Research Council, by a National Research Council Canada Fellowship (to S. M. S.), and by an equipment grant from the Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Equations S1–S3, and Fig. S1.
- Tat
- twin-arginine translocation
- MOPS
- 4-morpholinepropanesulfonic acid
- MTSL
- 1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl
- C12E9
- nonapolyoxyethylene dodecyl ether.
REFERENCES
- 1.Berks B. C., Palmer T., Sargent F. (2003) Adv. Microb. Physiol. 47, 187–254 [DOI] [PubMed] [Google Scholar]
- 2.Lee P. A., Tullman-Ercek D., Georgiou G. (2006) Annu. Rev. Microbiol. 60, 373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cline K., Theg S. M. (2007) in The Enzymes, Molecular Machines Involved in Protein Transport across Cellular Membranes, Elsevier, San Diego, CA, Vol. XXV, pp. 455–485 [Google Scholar]
- 4.Natale P., Brüser T., Driessen A. J. (2008) Biochim. Biophys. Acta 1778, 1735–1756 [DOI] [PubMed] [Google Scholar]
- 5.Chaddock A. M., Mant A., Karnauchov I., Brink S., Herrmann R. G., Klösgen R. B., Robinson C. (1995) EMBO J. 14, 2715–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berks B. C. (1996) Mol. Microbiol. 22, 393–404 [DOI] [PubMed] [Google Scholar]
- 7.Mould R. M., Robinson C. (1991) J. Biol. Chem. 266, 12189–12193 [PubMed] [Google Scholar]
- 8.Berks B. C., Palmer T., Sargent F. (2005) Curr. Opin. Microbiol. 8, 174–181 [DOI] [PubMed] [Google Scholar]
- 9.Weiner J. H., Bilous P. T., Shaw G. M., Lubitz S. P., Frost L., Thomas G. H., Cole J. A., Turner R. J. (1998) Cell 93, 93–101 [DOI] [PubMed] [Google Scholar]
- 10.Bogsch E. G., Sargent F., Stanley N. R., Berks B. C., Robinson C., Palmer T. (1998) J. Biol. Chem. 273, 18003–18006 [DOI] [PubMed] [Google Scholar]
- 11.Sargent F., Bogsch E. G., Stanley N. R., Wexler M., Robinson C., Berks B. C., Palmer T. (1998) EMBO J. 17, 3640–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sargent F., Stanley N. R., Berks B. C., Palmer T. (1999) J. Biol. Chem. 274, 36073–36082 [DOI] [PubMed] [Google Scholar]
- 13.Gouffi K., Gérard F., Santini C. L., Wu L. F. (2004) J. Biol. Chem. 279, 11608–11615 [DOI] [PubMed] [Google Scholar]
- 14.Porcelli I., de Leeuw E., Wallis R., van den Brink-van der Laan E., de Kruijff B., Wallace B. A., Palmer T., Berks B. C. (2002) Biochemistry 41, 13690–13697 [DOI] [PubMed] [Google Scholar]
- 15.Oates J., Barrett C. M., Barnett J. P., Byrne K. G., Bolhuis A., Robinson C. (2005) J. Mol. Biol. 346, 295–305 [DOI] [PubMed] [Google Scholar]
- 16.Gohlke U., Pullan L., McDevitt C. A., Porcelli I., de Leeuw E., Palmer T., Saibil H. R., Berks B. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10482–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarry M. J., Schäfer E., Chen S., Buchanan G., Greene N. P., Lea S. M., Palmer T., Saibil H. R., Berks B. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13284–13289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolhuis A., Mathers J. E., Thomas J. D., Barrett C. M., Robinson C. (2001) J. Biol. Chem. 276, 20213–20219 [DOI] [PubMed] [Google Scholar]
- 19.Cline K., Mori H. (2001) J. Cell Biol. 154, 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alami M., Lüke I., Deitermann S., Eisner G., Koch H. G., Brunner J., Müller M. (2003) Mol. Cell 12, 937–946 [DOI] [PubMed] [Google Scholar]
- 21.Leake M. C., Greene N. P., Godun R. M., Granjon T., Buchanan G., Chen S., Berry R. M., Palmer T., Berks B. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15376–15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dabney-Smith C., Cline K. (2009) Mol. Biol. Cell 20, 2060–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori H., Cline K. (2002) J. Cell Biol. 157, 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dabney-Smith C., Mori H., Cline K. (2006) J. Biol. Chem. 281, 5476–5483 [DOI] [PubMed] [Google Scholar]
- 25.Hubbell W. L., Gross A., Langen R., Lietzow M. A. (1998) Curr. Opin. Struct. Biol. 8, 649–656 [DOI] [PubMed] [Google Scholar]
- 26.Hubbell W. L., Cafiso D. S., Altenbach C. (2000) Nat. Struct. Biol. 7, 735–739 [DOI] [PubMed] [Google Scholar]
- 27.Columbus L., Hubbell W. L. (2002) Trends Biochem. Sci. 27, 288–295 [DOI] [PubMed] [Google Scholar]
- 28.Perozo E., Cortes D. M., Sompornisut P., Kloda K., Martinac B. (2002) Nature 418, 942–948 [DOI] [PubMed] [Google Scholar]
- 29.Hubbell W. L., Altenbach C. (1994) Curr. Opin. Struct. Biol. 4, 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbell W. L., Mchaourab H. S., Altenbach C., Lietzow M. A. (1996) Structure 4, 779–793 [DOI] [PubMed] [Google Scholar]
- 31.Borbat P. P., Costa-Filho A. J., Earle K. A., Moscicki J. K., Freed J. H. (2001) Science 291, 266–269 [DOI] [PubMed] [Google Scholar]
- 32.Steinhoff H. J., Radzwill N., Thevis W., Lenz V., Brandenburg D., Anston A., Dodson G., Wollmer A. (1997) Biophys. J. 73, 3287–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persson M., Harbridge J. R., Hammarström P., Mitri R., Mårtensson L. G., Carlsson U., Eaton G. R., Eaton S. S. (2001) Biophys. J. 80, 2886–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radzwill N., Gerwert K., Steinhoff H. J. (2001) Biophys. J. 80, 2856–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greene N. P., Porcelli I., Buchanan G., Hicks M. G., Schermann S. M., Palmer T., Berks B. C. (2007) J. Biol. Chem. 282, 23937–23945 [DOI] [PubMed] [Google Scholar]
- 36.Miroux B., Walker J. E. (1996) J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 37.Carl P. (2003) The ER4123D CW-Resonator: Dedicated to Spin Labels, in Bruker BioSpin Application Note, Bruker BioSpin, Coventry, UK [Google Scholar]
- 38.Berliner L. J. (1983) Ann. N. Y. Acad. Sci. 414, 153–161 [DOI] [PubMed] [Google Scholar]
- 39.Hubbell W. L., Altenbach C. (1994) in Membrane Protein Structure: Experimental Approaches (White S. H. ed) pp. 224–248, Oxford University Press, London, UK [Google Scholar]
- 40.Feix J. B., Klug C. S. (1998) in Spin Labeling: The Next Millennium, Biological Magnetic Resonance (Berliner L. J. ed) Vol. 14, pp. 251–281, Plenum Press, New York [Google Scholar]
- 41.Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 42.Mchaourab H. S., Lietzow M. A., Hideg K., Hubbell W. L. (1996) Biochemistry 35, 7692–7704 [DOI] [PubMed] [Google Scholar]
- 43.Bordignon E., Steinhoff H.-J. (2007) in Membrane Protein Structure and Dynamics (Hemminga M. A., Berliner L. J. eds) pp. 129–164, Springer, New York [Google Scholar]
- 44.Barton L. L. (2005) Structural and Functional Relationships in Prokaryotes, pp. 45–52, Springer, New York [Google Scholar]
- 45.Miles A. J., Sansom C. E., Wallace B. A. (2005) in Essays in Bioinformatics (Moss D. S., Jelaska S., Pongor S. eds) Vol. 368, pp. 96–150, IOS Press, Amsterdam, The Netherlands [Google Scholar]
- 46.Müller S. D., De Angelis A. A., Walther T. H., Grage S. L., Lange C., Opella S. J., Ulrich A. S. (2007) Biochim. Biophys. Acta 1768, 3071–3079 [DOI] [PubMed] [Google Scholar]
- 47.Ridder A. N., de Jong E. J., Jongbloed J. D., Kuipers O. P. (2009) J. Bacteriol. 191, 4410–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.