Abstract
The persulfide sulfur formed on an active site cysteine residue of pyridoxal 5′-phosphate-dependent cysteine desulfurases is subsequently incorporated into the biosynthetic pathways of a variety of sulfur-containing cofactors and thionucleosides. In molybdenum cofactor biosynthesis, MoeB activates the C terminus of the MoaD subunit of molybdopterin (MPT) synthase to form MoaD-adenylate, which is subsequently converted to a thiocarboxylate for the generation of the dithiolene group of MPT. It has been shown that three cysteine desulfurases (CsdA, SufS, and IscS) of Escherichia coli can transfer sulfur from l-cysteine to the thiocarboxylate of MoaD in vitro. Here, we demonstrate by surface plasmon resonance analyses that IscS, but not CsdA or SufS, interacts with MoeB and MoaD. MoeB and MoaD can stimulate the IscS activity up to 1.6-fold. Analysis of the sulfuration level of MoaD isolated from strains defective in cysteine desulfurases shows a largely decreased sulfuration level of the protein in an iscS deletion strain but not in a csdA/sufS deletion strain. We also show that another iscS deletion strain of E. coli accumulates compound Z, a direct oxidation product of the immediate precursor of MPT, to the same extent as an MPT synthase-deficient strain. In contrast, analysis of the content of compound Z in ΔcsdA and ΔsufS strains revealed no such accumulation. These findings indicate that IscS is the primary physiological sulfur-donating enzyme for the generation of the thiocarboxylate of MPT synthase in MPT biosynthesis.
Keywords: Enzymes, Enzymes/Pyridoxal Phosphate, Metabolism/Amino Acid, Metabolism/Sulfur, Protein/Protein-Protein Interactions, Cysteine Desulfurase, Molybdenum Cofactor, Molybdopterin
Introduction
The presence of sulfur in cofactors and RNA thionucleoside confers significant functionality to these biomolecules because of its unique chemical properties. The sulfur atoms of thiamin, lipoic acid, biotin, iron-sulfur (FeS)2 clusters, and molybdenum cofactors participate in stabilizing reaction intermediates (1, 2), alternating between the reduced dithiol and oxidized disulfide, facilitating electron transfer, or modulating the redox potential of the cofactor (3). The thionucleosides in tRNAs fine-tune the efficiency or accuracy of translation (4, 5), serve as recognition sites for aminoacyl-tRNA synthetase (6), or act as photosensors (7). Despite the diverse chemical environments in which the sulfur atoms reside in cofactors and thionucleosides, it has been found that the incorporation of sulfur into them involves a few enzymes with the persulfide (R-S-SH) groups on the cysteine residues of these enzymes; thus, these enzymes act as the key agents of sulfur transfer (8, 9).
Protein persulfide groups are generated by a family of cysteine desulfurases; these use the cofactor pyridoxal 5′-phosphate to form a persulfide group on an active site cysteine residue by using the free amino acid l-cysteine and generate l-alanine as the second product (10). The terminal sulfur (formally S0) is transferred to the cysteine residues in acceptor proteins to form new persulfide groups that are subsequently used directly in the biosynthesis of cofactors or thionucleosides or transferred to other protein acceptors for the eventual incorporation of sulfur into an end product (8–10).
Escherichia coli contains the following three cysteine desulfurases: CsdA (also termed CSD), SufS (also termed CsdB), and IscS (10, 11). These are classified into 2 groups, I and II, based on differences in four sequence regions and the consensus sequence motif (12). Unlike IscS, which has the consensus group I sequence SSGSACTS around the active site Cys-328, CsdA and SufS possess consensus group II sequences. CsdA is encoded by the csdAE operon, which is presumed to function in the maturation of an FeS protein (13). The activity of CsdA is enhanced by CsdE via sulfur transfer from the persulfide group of the former to the cysteine residue of the latter (13). The sufABCDSE operon containing the gene encoding SufS is specifically adapted to synthesize FeS clusters under conditions of iron starvation or oxidative stress (14, 15). The catalytic activity of SufS is significantly lower than that of CsdA and IscS (16), but it is enhanced by SufE through persulfide sulfur transfer; its catalytic activity is further enhanced by the SufBCD complex (17, 18). IscS, which is the central sulfur-providing enzyme, is involved in the formation of the FeS cluster (19–22), thiamin (23–25), and the thionucleosides 4-thiouridine and 5-methylaminomethyl-2-thiouridine in tRNAs (23, 25–29) via a specific interaction with different proteins. The interaction of IscS with IscU enables the assembly of FeS clusters (21, 22), whereas IscS interacts with ThiI to form ThiS-COSH, which subsequently transfers the sulfur to thiamin (24, 25). The formation of persulfide in ThiI by IscS is a critical event in the biosynthesis of 4-thiouridine in tRNA (25). Unlike the synthesis of 4-thiouridine, the generation of 2-thiouridine requires 5 polypeptides as sulfur carriers between IscS and tRNA-binding MnmA (29). Thus, the elucidation of donor-acceptor pairs and specific structural elements that specify the protein-protein interactions remains a fascinating aspect of sulfur transfer systems.
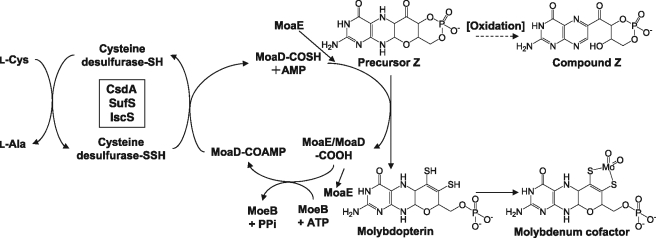
Molybdenum cofactors (Moco) also contain a sulfur atom as a functionally essential component. These cofactors are required for the activity of a variety of molybdoenzymes, including nitrate reductase, sulfite oxidase, and xanthine dehydrogenase (30, 31). A molybdenum cofactor consists of a mononuclear molybdenum covalently bound to the dithiolene moiety of a tricyclic pterin referred to as molybdopterin (MPT) (Fig. 1). In E. coli, MPT is generated from precursor Z, which is the first intermediate formed from GTP (32). Under experimental conditions, precursor Z is rapidly oxidized to 6-alkyl pterin, which is termed compound Z, by air or iodine (33). The dithiolene moiety is inserted into precursor Z by MPT synthase, a tetrameric protein comprising 2 small MoaD subunits and 2 large MoaE subunits (34). Each of the MPT sulfurs is derived from one of the 2 MoaD subunits that carry a C-terminal thiocarboxylate (35). MoaD with the C-terminal Gly-Gly motif is adenylated by MoeB via formation of the MoaD/MoeB complex (36). The next step in the C-terminal thiocarboxylation of MoaD has not been completely established. In a previous study, it is suggested that l-cysteine can be used as a sulfur source, and cysteine desulfurases in E. coli serve as a sulfur-donating enzyme for the synthesis of MPT in the in vitro system (36). However, the contributions of three cysteine desulfurases to MPT biosynthesis in vivo are unknown. Recent findings that IscS plays a role in sulfur transfer via protein-protein interaction with a certain partner (8, 9) led us to reconsider cysteine desulfurases involved in in vivo MPT synthesis.
FIGURE 1.
Proposed pathway for the biosynthesis of molybdenum cofactor in E. coli. The structures of precursor Z, MPT, molybdenum cofactor, and the oxidation product of precursor Z, i.e. compound Z, are shown. The incorporation of sulfur atoms into precursor Z is catalyzed by MPT synthase (MoaE/MoaD). Three cysteine desulfurases (CsdA, SufS, and IscS) transfer the sulfur atom from l-cysteine to the C-terminal thiocarboxylate of MoaD in vitro (36, 41).
In the present study, we have investigated the functional and physical interactions of three cysteine desulfurases with MoeB and MoaD. Each of three cysteine desulfurases could catalyze the transfer of 35S from [35S]cysteine to MoaD in the presence of MoeB and Mg-ATP. However, only IscS was activated by MoeB. MoaD further enhanced the IscS activity in the presence of MoeB. Surface plasmon resonance studies further revealed that IscS specifically binds to MoeB and MoaD. In contrast, neither CsdA nor SufS interacted with MoeB or MoaD. We also show that the sulfuration level of MoaD isolated from strains defective in IscS exhibits a large decrease in contrast to that of MoaD expressed in a csdA/sufS deletion strain. In accord with this result, a genetic study exhibits that an ΔiscS strain, but not the ΔcsdA and ΔsufS strains, accumulates compound Z. Based on the results presented here, we propose that IscS is the primary physiological sulfur-donating enzyme in the conversion of precursor Z to MPT.
EXPERIMENTAL PROCEDURES
Bacteria, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli cells were grown aerobically at 37 °C in a Luria-Bertani (LB) medium unless otherwise noted.
TABLE 1.
E. coli strains and plasmids used in this study
| Strains | Genotype | Source |
|---|---|---|
| BW25113 | lacIq, rrnBT14, ΔlacZWJ16, hsdR514, ΔaraBADAH33, ΔrhaBADLD78 | NBRPa collection |
| ΔcsdA | BW25113 ΔcsdA | NBRPa collection |
| ΔsufS | BW25113 ΔsufS | NBRPa collection |
| ΔiscS | BW25113 ΔiscS | NBRPa collection |
| ΔmoaD | BW25113 ΔmoaD | NBRPa collection |
| MC1061 | F'araD139 Δ(ara leu)7696 Δ(lacY74) galU galK hsdr hsdM+ strA | (23) |
| CL100 | MC1061 ΔiscS | (23) |
| CL102 | MC1061 ΔcsdA/ΔsufS | (26) |
| Plasmids | Property | Source |
| pUC118 | lac promoter-based expression vector | Takara Shuzo |
| pET14b | T7 RNA polymerase-based expression vector | Novagen |
| pCSD1 | csdA gene cloned into EcoRI and PstI sites of pUC118 | (12) |
| pCSDB | sufS gene cloned into EcoRI and HindIII sites of pUC118 | (16) |
| pEF1 | iscS gene cloned into NdeI and HindIII sites of pET21a | (11) |
| pIscS | An XbaI-HindIII iscS fragment of pEF1 was ligated into pUC118 | This work |
| pMoaD | moaD gene cloned into NdeI and BamHI sites of pET14b | This work |
| pMoeB | moeB gene cloned into NdeI and BamHI sites of pET14b | This work |
| pMW15aD | moaD gene cloned into NcoI and BamHI sites of pET15b | (35) |
a National BioResource Project (NIG, Japan):E.coli.
Construction of Plasmids
A DNA fragment containing the iscS gene and a Shine-Dalgarno sequence was cleaved from the XbaI and HindIII sites of pEF1 (11) and inserted into the XbaI and HindIII sites of pUC118 to yield pIscS. The DNA fragment containing moaD was PCR-amplified from the chromosomal DNA of E. coli K-12 MG1655 with specific primers (5′-GGGAATTCCATATGATTAAAGTTCTTTTTTTCGC-3′ and 5′-CGCGGATCCTTAACCTCCGGTTACCGGCG-5′) and cloned into the NdeI and BamHI sites of pET14b to give a plasmid, pMOAD. For the cloning of moeB, a PCR product obtained with primers 5′-GGGAATTCCATATGGCGGAACTCAGCGATCAGG-3′ and 5′-CGCGGATCCTTACTGCCCACACACCTCAC-3′ was ligated into pET14b to yield a plasmid, pMOEB.
Expression and Purification of Proteins
pCSD1 (12) and pCSDB (16) were introduced into E. coli JM109 to give CsdA- and SufS-overexpressing strains, respectively. E. coli BL21(DE3)/pLysS carrying pEF1 and E. coli BL21(DE3) carrying pMOAD and pMOEB were used for the expression of IscS, MoaD, and MoeB, respectively. Cells were cultured in LB with 100 mg/liter of ampicillin at 26 °C for 5 h. After induction by 1 mm isopropyl-β-d-thiogalactopyranoside for 6 h, the cells were harvested by centrifugation, disrupted by sonication, and purified by column chromatography as detailed in supplemental Experimental Procedures. For the quantification of thiocarboxylated MoaD in different E. coli strains, MoaD was expressed from pMW15aD (35) in the DE3 lysogens of E. coli strains MC1061 (23), CL100 (iscS deletion mutant) (23), and CL102 (csdA/sufS deletion double mutant) (26). Strains were grown in an LB medium supplemented with 100 μm isopropyl-1-thio-β-d-galactopyranoside and 150 mg/liter ampicillin at 30 °C to an A600 = 0.8 and harvested by centrifugation at 5,000 × g. MoaD was purified as described previously (34).
35S Transfer Assay
All the experiments were performed as described by Loiseau et al. (13) with modifications. Under anaerobic conditions, purified proteins were pretreated with 10 mm dithiothreitol and then repurified by gel filtration before use. 20 μm cysteine desulfurase was incubated with 10 μm PLP and 200 μm 2 mCi/mmol l-[35S]cysteine (1,075 Ci/mmol, American Radiolabeled Chemicals, St. Louis, MO, diluted by nonradioactive l-cysteine) at 37 °C for 30 min. The excess of cysteine was removed by desalting the protein over a Micro Bio-spin 6 column to give a persulfide form of cysteine desulfurase. A standard reaction mixture contains 10 μm persulfide-form cysteine desulfurase, 10 μm MoeB, 10 μm MoaD, 0.575 mm ATP, and 5.75 mm MgCl2 in 20 mm Tris-HCl, pH 7.5 in a final volume of 12 μl. After incubation at 37 °C for 30 min, the solution was mixed with an SDS-PAGE loading buffer without a reductant. Samples were immediately analyzed by gel electrophoresis using a 15% polyacrylamide gel containing 0.1% SDS but in the absence of an added reducing agent. 35S was visualized by exposure on a phosphor screen and analyzed using Typhoon 9210 (GE Healthcare).
Enzyme Assays
The cysteine desulfurase activities of CsdA, SufS, and IscS were quantitated by assaying the amount of sulfide formed from cysteine as described previously (22). A reaction mixture containing 1 mm l-cysteine, 10 μm PLP, 10 mm MgCl2, 1 mm ATP, 5 mm dithiothreitol, and the enzyme in a 50 mm Tris-HCl buffer, pH 8.0, was incubated at 30 °C.
Surface Plasmon Resonance Analysis
Surface plasmon resonance analysis was carried out with BIACORE X (Biacore AB, Uppsala, Sweden). MoaD or MoeB in a 10 mm acetate buffer (pH 6.0) was immobilized on a sensor chip NTA (Biacore AB). The chip was blocked with a purified N-terminal His6-tagged protein 2,4-diaminopentanoate dehydrogenase (37) from Thermoanaerobacter ethanolicus. The flow rate was 20 μl/min, and the sensor chip was regenerated with 350 mm EDTA and 500 mm NiCl2.
Analysis of the Sulfuration Level of MoaD from Different E. coli Strains
75 μm purified MoaD from different E. coli strains, MC1061, CL100, and CL102, 75 μm MoaE (34), and precursor Z (38) in excess amounts were incubated for 30 min in 400 μl of 100 mm Tris, pH 7.2. The MPT produced was oxidized to Form A and quantified following published procedures (36, 39, 40).
Preparation of Acid Extracts of E. coli Cells
E. coli cells were cultured in an LB medium for 18 h at 37 °C with vigorous shaking and harvested by centrifugation. The cell pellets were washed twice with 30 ml of water (H2O), suspended in 3 ml of H2O/g wet weight, and frozen at −30 °C. Acid extracts of the cells were prepared by essentially following the procedure proposed by Wuebbens and Rajagopalan (38). The extracts were concentrated to 900 μl by rotary evaporation and centrifuged to remove the precipitated protein. A portion of the supernatant (12.5–25 μl) was analyzed using high-performance liquid chromatography (HPLC) following air oxidation, which enabled the conversion of precursor Z to compound Z.
HPLC Analysis
All HPLC analyses were performed at 25 °C using a Capcell Pak C18 SG120 column (4.6 × 250 mm, Shiseido, Japan) and a Shimadzu HPLC system (Shimadzu, Japan). The column was equilibrated with 5 mm ammonium acetate (pH 5.0) with a flow rate of 1.0 ml/min. Eluents were monitored for absorption at 300 nm.
RESULTS AND DISCUSSION
35S Transfer from l-Cysteine to the MoaD Thiocarboxylate Catalyzed by Cysteine Desulfurases
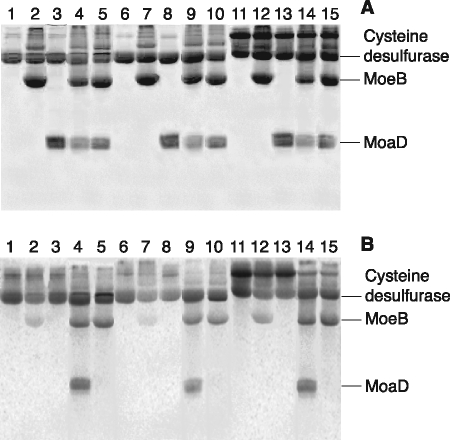
We examined the relative abilities of CsdA, SufS, and IscS to transfer sulfur from l-cysteine to the C-terminal group of MoaD. To visualize sulfur flow in the reaction, persulfide forms of the cysteine desulfurases were incubated with MoeB and MoaD under anaerobic conditions and subjected to SDS-PAGE analysis without a reducing agent (Fig. 2). The tracer assay of 35S radioactivity visualized by the phosphorimager analysis shows that sulfur was transferred from each of three cysteine desulfurases to MoaD in the presence of MoeB and Mg-ATP (Fig. 2, lanes 4, 9, and 14). No such transfer to MoaD was seen in the absence of either MoeB or Mg-ATP (Fig. 2, lanes 3, 5, 8, 10, 13, and 15). These results showed that all three of the cysteine desulfurases, CsdA, SufS, and IscS, could similarly transfer the persulfide sulfur to the C-terminal group of MoaD in the defined in vitro reaction and are consistent with the previous observation (36). MoeB was slightly labeled by 35S in the presence of either of the cysteine desulfurases alone (Fig. 2, lanes 2, 7, and 12). Multiple sequence alignment analysis of MoeBs from various organisms showed that there were two highly conserved cysteine residues (Cys-142 and Cys-187) (supplemental Fig. S2). We performed the sulfur transfer experiment to MoaD using a C142A/C187A double mutant of MoeB. The result showed that the mutation had no effect on the sulfur transfer to MoaD, and even the C142A/C187A mutant MoeB was also labeled by 35S (supplemental Fig. S3). Thus, the 35S-labeling of MoeB is most probably caused by nonspecific transfer of persulfide of cysteine desulfurases to one or more of the cysteine residues exposed on the surface of MoeB. This is in accordance with previous study suggesting that MoeB itself is not a carrier of sulfur for the sulfur transfer reaction to active MPT synthase (41).
FIGURE 2.
Analysis of the sulfur transfer from cysteine desulfurases to MoaD by 35S-labeled l-cysteine. Lane 1, CsdA; lane 2, CsdA and MoeB; lane 3, CsdA and MoaD; lane 4, CsdA, MoeB, and MoaD; lane 5, CsdA, MoeB, and MoaD (without ATP); lane 6, SufS; lane 7, SufS and MoeB; lane 8, SufS and MoaD; lane 9, SufS, MoeB, and MoaD; lane 10, SufS, MoeB, and MoaD (without ATP); lane 11, IscS; lane 12, IscS and MoeB; lane 13, IscS and MoaD; lane 14, IscS, MoeB, and MoaD; lane 15, IscS, MoeB, and MoaD (without ATP). The gel was stained with Coomassie Brilliant Blue (A), and the 35S radioactivity was visualized on a phosphorimaging plate (B).
Activation of IscS by MoeB and MoaD
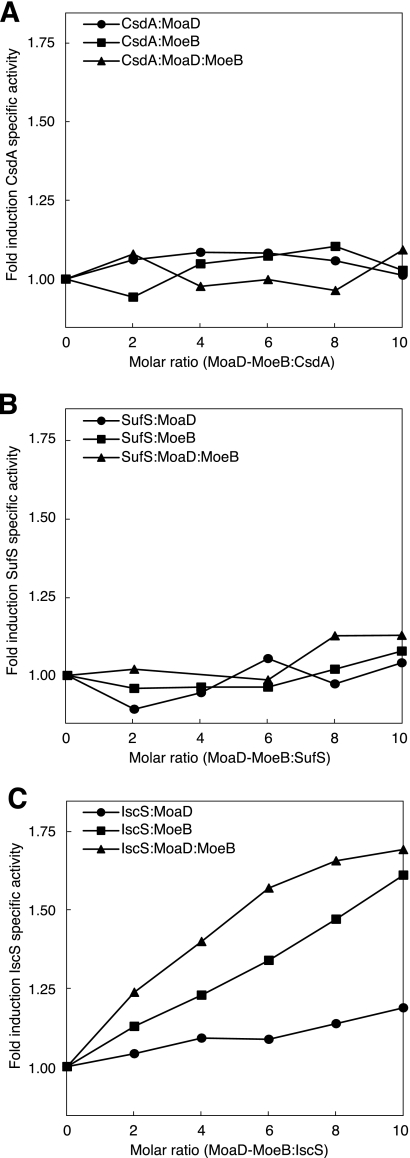
Previous studies have shown that the activity of cysteine desulfurases is enhanced by their physiological interacting partners, such as CsdE for CsdA (13), SufE-SufBCD for SufS (17, 18), and IscU (42), ThiI (43), and TusA (29) for IscS. The mechanism of the activation is not completely understood, but it is proposed that the binding by partner proteins probably induces a conformational change of the flexible loop carrying the persulfide-forming cysteine residue in cysteine desulfurase to facilitate persulfide formation and/or its subsequent cleavage (44, 45). Therefore, it seemed possible that MoeB and/or MoaD could alter the enzyme activity of a physiologically relevant cysteine desulfurase. To determine whether MoeB and MoaD affect the activity of three cysteine desulfurases, we measured the activity of CsdA, SufS, and IscS in the presence of MoeB and MoaD. The specific activities of purified CsdA, SufS, and IscS were 0.2 units/mg, 0.01 units/mg, and 0.24 units/mg, respectively. The addition of MoeB increased the desulfurase activity of IscS (1.6-fold in the presence of 10-fold molar excess of MoeB) but had no significant effect on the activity of CsdA and SufS (Fig. 3). The addition of MoaD to CsdA, SufS, and IscS had little effect on the desulfurase activity. However, the addition of MoaD to a mixture of IscS and MoeB detectably enhanced the desulfurase activity to levels much higher than those observed for the addition of MoeB alone to IscS (Fig. 3C). The enhancement of desulfurase activity by MoaD in the presence of MoeB is specific for IscS, as the addition of MoaD together with MoeB to CsdA and SufS had only a slight effect on their activities (Fig. 3, A and B). The activation of IscS by MoeB and further enhancement of the activity by MoaD resembles the activation of SufS by SufE and the additional enhancement of the SufS-SufE complex by SufBCD (18). In the activation of SufS by SufE-SufBCD, the persulfide sulfur of SufS is transferred to SufE and subsequently to SufBCD via protein-protein interactions (18, 46). However, the mechanism of activation of IscS by MoeB and MoaD may be different from that of SufS activation by SufE-SufBCD because specific sulfuration of MoeB was not observed (Fig. 3) (41).
FIGURE 3.
Activation of cysteine desulfurase activity by MoaD or MoeB. Formations of sulfide from l-cysteine were measured after 30 min for CsdA (A) and IscS (C) and 90 min for SufS (B).
Evidence for the Specific Interaction of IscS with MoeB and MoaD
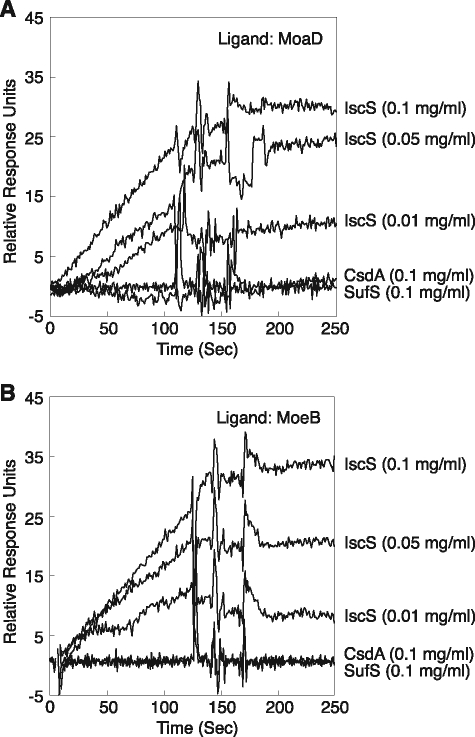
An intrinsic function of a protein in vivo is highly dependent on its specific interactions with other proteins. In fact, the activation of a cysteine desulfurase by a partner protein is coincident with the specific interaction between them (13, 18, 42, 46, 47). To investigate whether the observed activation of IscS by MoeB and MoaD involves specific physical protein-protein interactions, we conducted surface plasmon resonance measurements. MoeB or MoaD was immobilized on a sensor chip, while CsdA, SufS, or IscS was added to a solution as the analyte. For both MoeB and MoaD, the response unit was increased by increasing the concentration of IscS (Fig. 4). Binding of IscS to MoaD and MoeB showed KD values of 21 and 75 nm, respectively. In contrast, no measurable interaction was observed when SufS or CsdA was used as an analyte (Fig. 4). These data imply that the MoeB-MoaD complex presumably formed in vivo (41, 48) preferentially binds to IscS during the thiocarboxylation of MoaD.
FIGURE 4.
Surface plasmon resonance analysis to examine the interaction between MoaD (A) or MoeB (B) and three cysteine desulfurases of E. coli.
Analysis of the Sulfuration Level of MoaD Isolated from Strains Defective in Cysteine Desulfurases
Because the MoaD-thiocarboxylate group is the direct sulfur donor for the conversion of precursor Z to MPT, it was of interest to analyze the sulfuration level of MoaD isolated from cysteine desulfurase mutants. The purified protein MoaD from the MC1061, CL100, and CL102 strains was incubated with purified MoaE and precursor Z. Produced MPT was quantified by a conversion to Form A. As shown in Fig. 5, a largely reduced level of MPT formation was detected (quantified as Form A) when MoaD was purified from the iscS-deficient strain CL100, whereas MoaD isolated from the csdA/sufS deletion mutant strain CL102 showed the same activity as the corresponding wild-type strain. The amount of about 1% of activity of MoaD purified from the CL100 strain is consistent with previous results, which show that the CL100 strain contained only a level of 10% Moco of wild-type strain (36). This result strongly suggests that IscS is involved in the sulfuration of MoaD. Because no effect on the sulfuration level of MoaD was obtained using the CL102 strain, CsdA and SufS are not involved in Moco biosynthesis in vivo.
FIGURE 5.
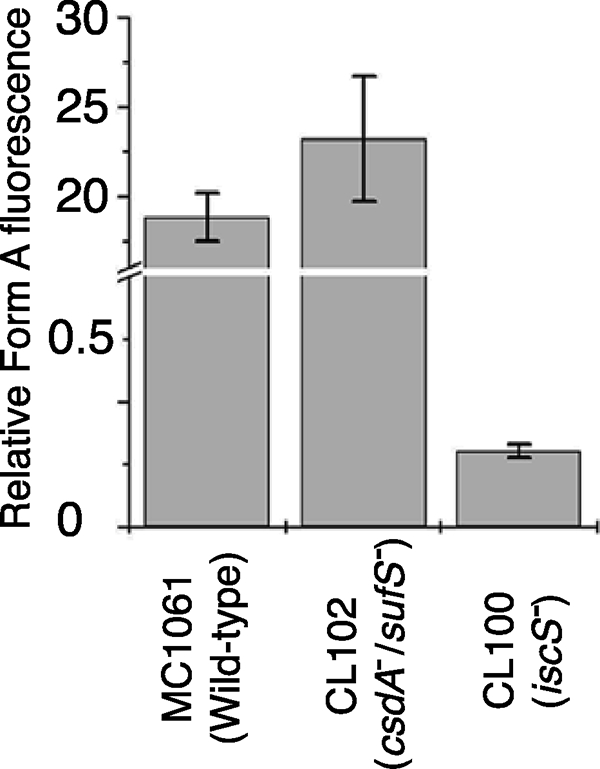
Detection of the sulfuration level of MoaD after expression in different E. coli mutant strains. The sulfuration level of MoaD was determined by the amount of MPT formed after incubation of purified MoaD, MoaE, and precursor Z in excess. The amount of MPT formed during the reaction was quantified as Form A using the peak height.
Accumulation of Compound Z in the ΔiscS Strain
Previously, Leimkühler and Rajagopalan (36) showed that precursor Z is not accumulated in the iscS− strain; however, the crude extract of the iscS− strain was able to convert externally added precursor Z to MPT to the same level as the parental strain. Accordingly, they suggested that the low level of MPT in an iscS− strain is due to the impaired activity of the Fe-S protein MoaA involved in the synthesis of precursor Z, and, therefore, it could not be clearly determined whether IscS is required for the in vivo sulfuration of MPT synthase. Because the amount of Moco in the iscS mutant was determined to be 10% of that in the wild-type strain, it was concluded that CsdA, SufS, or other sulfur transferases in the cells are able to compensate to some extent for the function of IscS in the Fe-S cluster formation of MoaA and the conversion of precursor Z to MPT. Therefore, the amount of precursor Z in an iscS− strain seemed to differ depending on the balance of CsdA, SufS, or other sulfur transferases available for Fe-S cluster synthesis and for the sulfuration of MoaD.
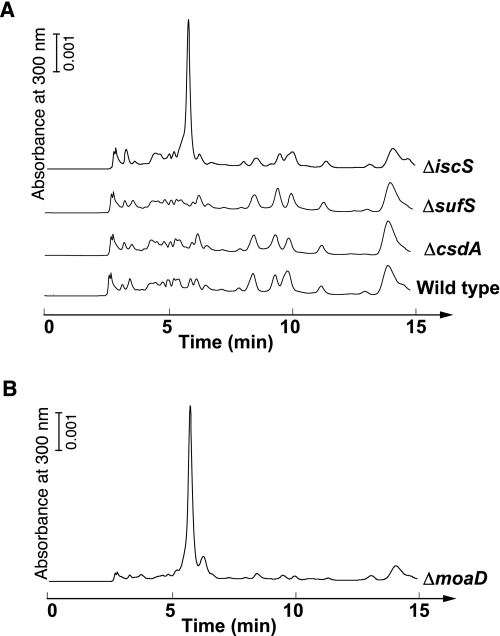
To test this possibility, we examined whether compound Z is accumulated in E. coli strains deficient in csdA, sufS, or iscS. An earlier study demonstrated that the accumulated compound Z could be released by direct acidification of whole-cell suspensions of appropriate E. coli MPT-deficient mutants, such as the ΔmoaD and ΔmoeB strains (33). HPLC analysis of the acid extracts of E. coli cells revealed that the peak absorbing at 300 nm, which eluted at 5.75 min, was observed in the ΔiscS strain (Fig. 6A); the ΔmoaD strain also accumulated compounds absorbing at 300 nm with the same retention time as the ΔiscS strain (Fig. 6B). In contrast, such accumulation was not observed in the ΔcsdA and ΔsufS strains as well as in the wild-type strain (Fig. 6A). HPLC/ESI-MS, ESI-MS/MS, and UV-Vis absorption analyses clearly identified the accumulated 300 nm absorbing material to be compound Z (supplemental Fig. S2, S3, and S4). A genetic complementation analysis of the ΔiscS strain by the expression of IscS further confirmed that the accumulation of compound Z is specifically caused by the deletion of the iscS gene (supplemental Fig. S5). These results indicate that MoaA is active in the strain. CsdA, SufS, or other sulfur transferases presumably support the assembly of the FeS cluster in MoaA present in the ΔiscS strain. However, the contributions of the three cysteine desulfurases for it are still unclear and will be interesting to study in the future.
FIGURE 6.
Analysis of the acid extracts of E. coli cells. The acid extracts of the wild-type, ΔcsdA, ΔsufS, ΔiscS, and ΔmoaD strains were analyzed using HPLC with a Capcell Pak C18 SG120 column after air oxidation. The volumes of the analyzed samples were 12.5 μl for the ΔmoaD strain and 25 μl for the other strains.
The studies presented here identify the protein involved in the sulfur transfer for the formation of the dithiolene moiety of Moco in E. coli. Earlier work demonstrated that any of the three NifS-like sulfur transferases, namely IscS, CsdA, or SufS, is capable of mobilizing and transferring sulfur from l-cysteine to MoaD in vitro (36). The minimal requirements for the activation of inactive MPT synthase in vitro were shown to be MoeB, Mg-ATP, l-cysteine, and a sulfur transferase. Thus, the specific protein involved in sulfur transfer to MoaD remained unknown in E. coli. Using different approaches, we show the IscS is the specific cysteine desulfurase transferring the sulfur to the MoaD protein.
It was recently shown that, in humans, the cysteine desulfurase Nfs1 acts as direct sulfur donor for MOCS3 in the cytosol (49). Thus, in humans and E. coli, the same protein components participate in the sulfur transfer pathway to MPT synthase during Moco biosynthesis: l-cysteine is the direct sulfur donor for a cysteine desulfurase, which transfers the sulfur to the adenylated small subunit of MPT synthase. In humans, this sulfur transfer occurs with the aid of the C-terminal rhodanese-like domain of MOCS3. For this additional sulfur transfer step, there must have been an evolutionary advantage for eukaryotes, which, likely, made this reaction more specific.
Supplementary Material
Acknowledgments
We thank Dr. Wolfgang Bückel (Max Planck Institute for Terrestrial Microbiology, Marburg), Dr. Rudolf K. Thauer (Max Planck Institute for Terrestrial Microbiology, Marburg), Dr. Roland Lill (Philipps University Marburg, Marburg), and Dr. Antonio J. Pierik (Philipps University Marburg, Marburg) for kind suggestions concerning this study. We thank Charles Lauhon (University of Wisconsin) for providing strains CL100 and CL102. The other E. coli mutant strains were kindly provided by the National BioResource Project (NIG, Japan):E.coli.
This work was supported in part by a Grant-in-aid for Scientific Research on Priority Areas (B) 17370037 (to N. E.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by Grant-in-aid for Encouragement of Young Scientists 18780073 (to H. M.) from the Japan Society for the Promotion of Science, by the National Project on Protein Structural and Functional Analyses, by a Grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (21st Century COE on Kyoto University Alliance for Chemistry), and by Deutsche Forschungsgemeinschaft Grant LE1171/5, and the Fonds der Chemischen Industrie (to S. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental materials and Figs. S1–S7.
- FeS
- iron-sulfur
- MPT
- molybdopterin
- Moco
- molybdenum cofactors
- PLP
- pyridoxal 5′-phosphate
- HPLC
- high performance liquid chromatography
- ESI-MS
- electrospray ionization mass spectrometry.
REFERENCES
- 1.Kluger R. (1987) Chem. Rev. 87, 863–876 [Google Scholar]
- 2.Knowles J. R. (1989) Annu. Rev. Biochem. 58, 195–221 [DOI] [PubMed] [Google Scholar]
- 3.Hille R., Rétey J., Bartlewski-Hof U., Reichenbecher W. (1998) FEMS Microbiol. Rev. 22, 489–501 [DOI] [PubMed] [Google Scholar]
- 4.Sen G. C., Ghosh H. P. (1976) Nucleic Acids Res. 3, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama S., Watanabe T., Murao K., Ishikura H., Yamaizumi Z., Nishimura S., Miyazawa T. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4905–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers K. C., Crescenzo A. T., Söll D. (1995) Biochimie 77, 66–74 [DOI] [PubMed] [Google Scholar]
- 7.Ramabhadran T. V., Fossum T., Jagger J. (1976) J. Bacteriol. 128, 671–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessler D. (2006) FEMS Microbiol. Rev. 30, 825–840 [DOI] [PubMed] [Google Scholar]
- 9.Mueller E. G. (2006) Nat. Chem. Biol. 2, 185–194 [DOI] [PubMed] [Google Scholar]
- 10.Mihara H., Esaki N. (2002) Appl. Microbiol. Biotechnol. 60, 12–23 [DOI] [PubMed] [Google Scholar]
- 11.Mihara H., Kurihara T., Yoshimura T., Esaki N. (2000) J. Biochem. 127, 559–567 [DOI] [PubMed] [Google Scholar]
- 12.Mihara H., Kurihara T., Yoshimura T., Soda K., Esaki N. (1997) J. Biol. Chem. 272, 22417–22424 [DOI] [PubMed] [Google Scholar]
- 13.Loiseau L., Ollagnier-de Choudens S., Lascoux D., Forest E., Fontecave M., Barras F. (2005) J. Biol. Chem. 280, 26760–26769 [DOI] [PubMed] [Google Scholar]
- 14.Takahashi Y., Tokumoto U. (2002) J. Biol. Chem. 277, 28380–28383 [DOI] [PubMed] [Google Scholar]
- 15.Outten F. W., Djaman O., Storz G. (2004) Mol. Microbiol. 52, 861–872 [DOI] [PubMed] [Google Scholar]
- 16.Mihara H., Maeda M., Fujii T., Kurihara T., Hata Y., Esaki N. (1999) J. Biol. Chem. 274, 14768–14772 [DOI] [PubMed] [Google Scholar]
- 17.Ollagnier-de-Choudens S., Lascoux D., Loiseau L., Barras F., Forest E., Fontecave M. (2003) FEBS Lett. 555, 263–267 [DOI] [PubMed] [Google Scholar]
- 18.Outten F. W., Wood M. J., Munoz F. M., Storz G. (2003) J. Biol. Chem. 278, 45713–45719 [DOI] [PubMed] [Google Scholar]
- 19.Schwartz C. J., Djaman O., Imlay J. A., Kiley P. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9009–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokumoto U., Takahashi Y. (2001) J. Biochem. 130, 63–71 [DOI] [PubMed] [Google Scholar]
- 21.Smith A. D., Agar J. N., Johnson K. A., Frazzon J., Amster I. J., Dean D. R., Johnson M. K. (2001) J. Am. Chem. Soc. 123, 11103–11104 [DOI] [PubMed] [Google Scholar]
- 22.Urbina H. D., Silberg J. J., Hoff K. G., Vickery L. E. (2001) J. Biol. Chem. 276, 44521–44526 [DOI] [PubMed] [Google Scholar]
- 23.Lauhon C. T., Kambampati R. (2000) J. Biol. Chem. 275, 20096–20103 [DOI] [PubMed] [Google Scholar]
- 24.Taylor S. V., Kelleher N. L., Kinsland C., Chiu H. J., Costello C. A., Backstrom A. D., McLafferty F. W., Begley T. P. (1998) J. Biol. Chem. 273, 16555–16560 [DOI] [PubMed] [Google Scholar]
- 25.Kambampati R., Lauhon C. T. (2000) J. Biol. Chem. 275, 10727–10730 [DOI] [PubMed] [Google Scholar]
- 26.Lauhon C. T. (2002) J. Bacteriol. 184, 6820–6829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mihara H., Kato S., Lacourciere G. M., Stadtman T. C., Kennedy R. A., Kurihara T., Tokumoto U., Takahashi Y., Esaki N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6679–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson K., Lundgren H. K., Hagervall T. G., Björk G. R. (2002) J. Bacteriol. 184, 6830–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeuchi Y., Shigi N., Kato J., Nishimura A., Suzuki T. (2006) Mol. Cell 21, 97–108 [DOI] [PubMed] [Google Scholar]
- 30.Hille R. (2002) Trends Biochem. Sci. 27, 360–367 [DOI] [PubMed] [Google Scholar]
- 31.Mendel R. R., Bittner F. (2006) Biochim. Biophys. Acta 1763, 621–635 [DOI] [PubMed] [Google Scholar]
- 32.Schwarz G., Mendel R. R. (2006) Annu. Rev. Plant Biol. 57, 623–647 [DOI] [PubMed] [Google Scholar]
- 33.Johnson J. L., Wuebbens M. M., Rajagopalan K. V. (1989) J. Biol. Chem. 264, 13440–13447 [PubMed] [Google Scholar]
- 34.Rudolph M. J., Wuebbens M. M., Rajagopalan K. V., Schindelin H. (2001) Nat. Struct. Biol. 8, 42–46 [DOI] [PubMed] [Google Scholar]
- 35.Wuebbens M. M., Rajagopalan K. V. (2003) J. Biol. Chem. 278, 14523–14532 [DOI] [PubMed] [Google Scholar]
- 36.Leimkühler S., Rajagopalan K. V. (2001) J. Biol. Chem. 276, 22024–22031 [DOI] [PubMed] [Google Scholar]
- 37.Fonknechten N., Perret A., Perchat N., Tricot S., Lechaplais C., Vallenet D., Vergne C., Zaparucha A., Le Paslier D., Weissenbach J., Salanoubat M. (2009) J. Bacteriol. 191, 3162–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wuebbens M. M., Rajagopalan K. V. (1993) J. Biol. Chem. 268, 13493–13498 [PubMed] [Google Scholar]
- 39.Johnson J. L., Hainline B. E., Rajagopalan K. V., Arison B. H. (1984) J. Biol. Chem. 259, 5414–5422 [PubMed] [Google Scholar]
- 40.Leimkühler S., Rajagopalan K. V. (2001) J. Biol. Chem. 276, 1837–1844 [DOI] [PubMed] [Google Scholar]
- 41.Leimkühler S., Wuebbens M. M., Rajagopalan K. V. (2001) J. Biol. Chem. 276, 34695–34701 [DOI] [PubMed] [Google Scholar]
- 42.Kato S., Mihara H., Kurihara T., Takahashi Y., Tokumoto U., Yoshimura T., Esaki N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5948–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kambampati R., Lauhon C. T. (1999) Biochemistry 38, 16561–16568 [DOI] [PubMed] [Google Scholar]
- 44.Loiseau L., Ollagnier-de-Choudens S., Nachin L., Fontecave M., Barras F. (2003) J. Biol. Chem. 278, 38352–38359 [DOI] [PubMed] [Google Scholar]
- 45.Behshad E., Parkin S. E., Bollinger J. M., Jr. (2004) Biochemistry 43, 12220–12226 [DOI] [PubMed] [Google Scholar]
- 46.Layer G., Gaddam S. A., Ayala-Castro C. N., Ollagnier-de Choudens S., Lascoux D., Fontecave M., Outten F. W. (2007) J. Biol. Chem. 282, 13342–13350 [DOI] [PubMed] [Google Scholar]
- 47.Kurihara T., Mihara H., Kato S., Yoshimura T., Esaki N. (2003) Biochim. Biophys. Acta 1647, 303–309 [DOI] [PubMed] [Google Scholar]
- 48.Lake M. W., Wuebbens M. M., Rajagopalan K. V., Schindelin H. (2001) Nature 414, 325–329 [DOI] [PubMed] [Google Scholar]
- 49.Marelja Z., Stöcklein W., Nimtz M., Leimkühler S. (2008) J. Biol. Chem. 283, 25178–25185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.