Abstract
Phosphorylation-dependent ubiquitination and ensuing down-regulation and lysosomal degradation of the interferon α/β receptor chain 1 (IFNAR1) of the receptor for Type I interferons play important roles in limiting the cellular responses to these cytokines. These events could be stimulated either by the ligands (in a Janus kinase-dependent manner) or by unfolded protein response (UPR) inducers including viral infection (in a manner dependent on the activity of pancreatic endoplasmic reticulum kinase). Both ligand-dependent and -independent pathways converge on phosphorylation of Ser535 within the IFNAR1 degron leading to recruitment of β-Trcp E3 ubiquitin ligase and concomitant ubiquitination and degradation. Casein kinase 1α (CK1α) was shown to directly phosphorylate Ser535 within the ligand-independent pathway. Yet given the constitutive activity of CK1α, it remained unclear how this pathway is stimulated by UPR. Here we report that induction of UPR promotes the phosphorylation of a proximal residue, Ser532, in a pancreatic endoplasmic reticulum kinase-dependent manner. This serine serves as a priming site that promotes subsequent phosphorylation of IFNAR1 within its degron by CK1α. These events play an important role in regulating ubiquitination and degradation of IFNAR1 as well as the extent of Type I interferon signaling.
Keywords: Cytokines/Interferons, Phosphorylation/Kinases/Serine/Threonine, Phosphorylation/Serine/Threonine, Proteases/Ubiquitination, Receptors/Cytokine, Receptors/Regulation, Receptors/Threonine-Serine Kinases, IFNAR1
Introduction
Ligand-induced down-regulation of cell surface receptors represents a major mode of actions for the branch of signaling that leads to its elimination (1). For example, Type I interferons (including IFNα and IFNβ),2 the cytokines that play a paramount role in anti-viral defense (2) and elicit potent antiproliferative effects (3), stimulate down-regulation of the cell surface levels of their receptor (4, 5). This receptor consists of IFNAR1 and IFNAR2 chains and functions via activation of associated Janus kinase (Tyk2 and Jak1) leading to activating tyrosine phosphorylation of the signal transducers and activators of transcription proteins (STAT1 and STAT2). In turn, these STAT proteins govern transcription of IFN-stimulated genes whose products mediate anti-viral, anti-proliferative, and immunomodulatory functions (reviewed in Refs. 6 and 7). Early studies have reported that IFNAR1 is rapidly down-regulated and degraded upon internalization in response to IFNα (8, 9).
Mechanisms of ligand-induced degradation of IFNAR1 rely on IFNα/β-stimulated and Tyk2 catalytic activity-dependent phosphorylation of this receptor chain on Ser535/539 within a specific phospho-degron (10–12). This phosphorylation enables the recognition of IFNAR1 by the β-Trcp2/HOS F-box protein, followed by the recruitment of the SCFβ-Trcp E3 ubiquitin ligase (10, 11). This ligase facilitates polyubiquitination of IFNAR1 on a specific cluster of lysines. Through a yet to be identified mechanism, this site-specific ubiquitination results in an exposure of a previously masked linear endocytic motif that enables the recruitment of the AP2 complex and ensuing internalization of IFNAR1 and of entire Type I IFN receptor (13, 14). Ubiquitination of IFNAR1 was also shown to stimulate post-internalization sorting of this chain to the lysosomes for efficient proteolysis (13).
Intriguingly, the mechanisms of ligand-induced receptor down-regulation could be also utilized by other unrelated stimuli. As a result, a cell might be rendered refractory to a particular ligand even before this ligand has had a chance to initiate signaling. For example, we have recently described a basal ligand- and JAK-independent mechanism of Ser535 phosphorylation, ubiquitination, and degradation of IFNAR1. This relatively low efficacy mechanism contributes to an ability of cells to avoid the anti-proliferative effects of high levels of IFNAR1 expression (15). Interestingly, this mechanism could be robustly induced by some ligand-independent stimuli including the products of tobacco smoking (16) and the inducers of unfolded protein responses (UPRs) such as treatment with thapsigargin (TG) or forced overexpression of the receptor (15, 17). Importantly, UPR is known to be induced by a rapid synthesis of viral proteins during infection with diverse viruses (18, 19). Indeed, we have recently demonstrated that infection with vesicular stomatitis virus (VSV) or hepatitis C virus (HCV) promotes Ser535 phosphorylation-dependent ubiquitination and down-regulation of IFNAR1 in a manner that does not require JAK activity but relies on activation of pancreatic endoplasmic reticulum kinase (PERK) by UPR. Serendipitously for these viruses, these events enabled them to decrease the efficacy of IFNα/β signaling and to use this mechanism (along with numerous virus-specific means previously described in Refs. 20 and 21) to avoid the IFN-induced anti-viral defenses (17). Given that IFNα is used for treatment of chronic viral infections including hepatitis C (22, 23), identification of signaling pathways that mediate UPR-stimulated IFNAR1 degradation is of obvious importance.
Casein kinase 1α (CK1α) was purified and characterized as a bona fide Ser535 IFNAR1 kinase that functioned within the ligand-independent pathway but was dispensable for IFNα-induced Ser535 phosphorylation (24). Although TG- or virus-induced Ser535 phosphorylation and down-regulation of IFNAR1 required CK1α activity, this kinase activity per se was not increased by UPR (24), suggesting that UPR signaling abets CK1α via another mechanism. Here we report that UPR stimulates phosphorylation of yet another serine residue proximal to the phospho-degron of IFNAR1 that increases the efficacy of IFNAR1 degron phosphorylation by CK1α. We further demonstrate that this priming site plays an important role in ligand- and JAK-independent regulation of IFNAR1 ubiquitination and degradation as well as in the regulation of the extent of cellular responses to IFNα/β.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
TG, cycloheximide, and methylamine HCl were purchased from Sigma. Human pCDNA3- FLAG-IFNAR1 mammalian expression construct and retroviral pBABE-puro-based construct for expression of FLAG-tagged mouse IFNAR1 as well as GST-IFNAR1 bacterial expression vector were described previously (10). Mutants lacking the priming sites (Ser532 in human IFNAR1 and Ser523 in mouse IFNAR1) were generated by site-directed mutagenesis. The sequence of mutants was confirmed by dideoxy sequencing. Constructs for expression of human Myc-tagged CK1α (a kind gift from J. Wade Harper, Harvard University, Cambridge, MA) was described previously (25). Vector for expression of FLAG-STAT1 was kindly provided by J. Darnell (Rockefeller University, New York, NY). HA-tagged Leishmania CK1 (L-CK1) pEF-BOS-based expression vector (wild type or kinase dead K40R mutant) was described elsewhere (24). pLKO.1-puro (Sigma) vector-based small hairpin RNA constructs targeted against PERK or irrelevant control were described previously (17). Construct for bacterial expression of GST-CK1α (described in Ref. 26) was a kind gift from Jiandong Chen (H. Lee Moffitt Cancer Center, Tampa, FL). Construct for bacterial expression of constitutively active PERK (ΔN-PERK described in Ref. 27), as well as for mammalian expression of wild type or catalytically inactive PERK (K618R) (27), were previously described. Human IFNα (Roche Applied Science) and murine IFNβ (PBL) were purchased.
Cell Culture, Treatment, and Viral Infection
All of the cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum (Hyclone) and various selection antibiotics when indicated. Human HeLa and 293T cells were obtained from ATCC. Mouse embryo fibroblasts from IFNAR1−/− mice and their wild type counterparts were kindly provided by S. Hemmi (Institute for Molecular Biology, Zürich, Switzerland). To obtain reconstituted cells expressing wild type or mutant IFNAR1, these cells were transduced by pBabe-Puro-based mIFNAR1 constructs and selected in puromycin for 2 weeks before analysis. 11,1-Tyk2-null cells reconstituted with catalytically inactive Tyk2 (KR cells) (12) were a generous gift of S. Pellegrini (Pasteur Institute, Paris, France). PERK-deficient mouse embryo fibroblasts were generous gift from D. Ron (New York University, New York, NY). Huh7 and derivative cells that express a complete HCV genome were a kind gift from R. Aldalbe (University of Navarra, Pamplona, Spain). These cells (described in detail in Refs. 17 and 28) were cultured in the presence of 500 μg/ml of G418. Transfection of 293T cells, HeLa cells, and Huh7 cells was carried out with Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's recommendations. VSV (Indiana serotype, a gift from R. Harty, University of Pennsylvania, Philadelphia, PA) was propagated in HeLa cells. For infection, the cells were inoculated with a multiplicity of infection 0.1–0.2 of VSV for 1 h, washed, and incubated with fresh medium as indicated.
Antibodies and Immunotechniques
Commercially available antibodies against pSTAT1, p-eIF2a, STAT1 (Cell Signaling), eIF2a (Biosources), hIFNAR1, (Santa Cruz), FLAG (M2), β-actin (Sigma), mouse IFNAR1 (Leinco), and ubiquitin (clone FK2;Biomol) were purchased. Monoclonal antibodies against human IFNAR1 that were used for immunoprecipitation (EA12) or immunoblotting (GB8) were described in detail elsewhere (29). Monoclonal 23H12 antibody against the M protein of VSV (VSV-M) was a generous gift from D. S. Lyles (Wake Forest University School of Medicine, Winston-Salem, NC). Antibodies against IFNAR1 phosphorylated on Ser535 (11) and against PERK (17) were described previously. Polyclonal antibody against IFNAR1 phosphorylated on Ser532 (Ser523 in the mouse receptor) was raised in rabbits using synthetic monophosphopeptide EDHKKYSSQTpSQDSGNYSNEDE in collaboration with PhosphoSolutions Inc. (Golden, CO). Antibody was further affinity-purified using monophosphopeptide affinity columns and tested for specificity by immunoblotting. Immunoprecipitations, immunoblotting, in vivo ubiquitination assay using denaturing immunoprecipitation, and assessment of the kinetics of degradation of IFNAR1 by cycloheximide chase were carried out as described previously (10, 11, 13, 15, 17).
In Vitro Kinase Assay
Kinase assays were carried out as described in detail elsewhere (24). Briefly, 2 μg of substrates (bacterially expressed and purified GST-IFNAR1, wild type, or S532A mutant) were incubated with 4 μg of lysate (from untreated or thapsigargin-treated cells) that were cleared of CK1α (by immunodepletion) and 0.25 μg of bacterially produced GST-CK1α (where indicated) in kinase buffer (25 mm Tris HCl, pH 7.4, 10 mm MgCl2, 1 mm NaF, 1 mm NaVO3) and ATP (1 mm). Where indicated, 100 μg of bacterially produced ΔN-PERK or undepleted lysates from 293T cells were used as a source of kinase activity. Radiolabel was provided as [γ-32P]ATP (1 μCi; Amersham Biosciences). The reactions were carried out at 30 °C for 30 min shaking at 600 rpm on the tabletop incubator. The products were analyzed either by immunoblotting with phospho-specific antibodies or by autoradiography.
RESULTS
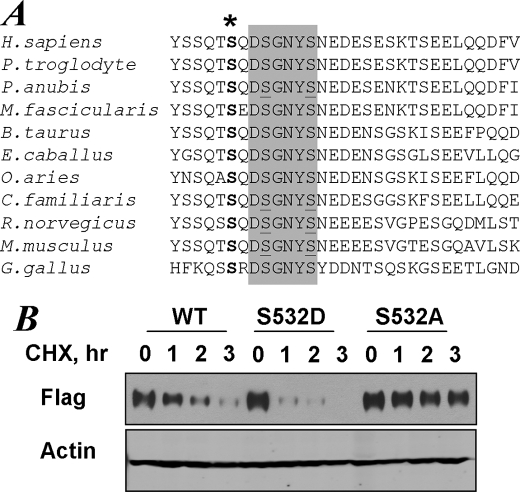
We sought to investigate how inducers of UPR promote phosphorylation-dependent ubiquitination and degradation of IFNAR1. Previous studies demonstrated that these signals feed into the ligand-independent pathway (15, 17) that utilizes CK1α, which directly phosphorylates Ser535 within the degron of IFNAR1 (24). Given that constitutively high activity of CK1α was not further stimulated in cells treated with UPR inducers yet lysates from these cells augmented the ability of CK1α to phosphorylate Ser535 in vitro (24), we proposed that UPR signaling may lead to additional post-translational modification of IFNAR1 that improves its phosphorylation by CK1α on Ser535. Indeed, a large body of literature suggests that priming phosphorylation of a substrate at a Ser/Thr residue in the n − 3 position may greatly increase its phosphorylation by various casein kinase 1 species (30–37). Analysis of primary sequences of IFNAR1 showed that a highly conserved Ser residue (Ser532 in humans and Ser523 in mice) is located at this position and may act as a priming phosphorylation site (Fig. 1A).
FIGURE 1.
The conserved priming site within IFNAR1 regulates the intrinsic stability of the protein. A, alignment of primary sequences of IFNAR1 from indicated species including Homo sapiens, Pan troglodyte, Papia anubis, Macaca fascicularis, Bos taurus, Equus caballus, Ovis aries, Canis familiaris, Rattus norvegicus, Mus musculus, and Gallus gallus. The phospho-degron sequences are shaded, and serine residues within the degron are underlined. The conserved putative priming site (Ser532 in human IFNAR1) is denoted by an asterisk. B, degradation of FLAG-IFNAR1 (wild type (WT) or S532A mutant) overexpressed in 293T cells was analyzed by cycloheximide (CHX, 2 mm) chase for the indicated times followed by immunoblotting using anti-FLAG antibody. The levels of β-actin were also analyzed as a loading control.
Ligand-independent IFNAR1 phosphorylation, ubiquitination, and degradation are readily observed in cells that overexpress this receptor (15, 17). We compared the stability of wild type FLAG-IFNAR1 expressed in 293T cells with its mutant counterpart that lacks Ser532 using cycloheximide chase assay. In this assay, the levels of protein become indicative of its proteolytic turnover because they are assessed under conditions when protein synthesis in cells is inhibited for various times. Replacement of Ser at the putative priming site within IFNAR1 with Ala yielded a receptor chain that displayed a noticeably longer half-life (Fig. 1B). Furthermore, a substitution of this serine residue with a phospho-mimicking Asp produced IFNAR1 mutant protein that underwent a more robust turnover that of its wild type counterpart (Fig. 1B). This result indicates that priming phosphorylation might be important for regulating the rate of IFNAR1 proteolytic turnover.
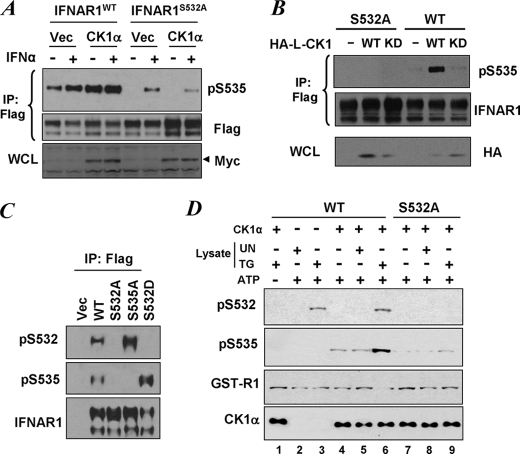
We next determined whether the priming site contributes to CK1-mediated phosphorylation of the IFNAR1 degron on Ser535. In line with our previous observations (15, 17, 24), forced expression of wild type FLAG-IFNAR1 in 293T cells allowed us to observe the basal level of Ser535 phosphorylation, and co-expression of Myc-tagged CK1α further increased this phosphorylation. Under these conditions, Ser535 phosphorylation was not found in mutant IFNAR1S532A (Fig. 2A), although phosphorylation of another proximal serine mutant, S529A, remained unaffected (data not shown). This result could be explained neither by differences in the levels of Myc-CK1α expression (Fig. 2A, bottom panel) nor by the possibility that mutation in Ser532 might alter the recognition of the Ser(P)535-specific epitope by the antibody, because Ser535 phosphorylation of IFNAR1S532A mutant was still observed in the cells treated with IFNα. These observations suggest that the priming site is indispensable for ligand-independent IFNAR1 degron phosphorylation but not when phosphorylation is induced by IFNα.
FIGURE 2.
Priming phosphorylation is required for the ligand-independent phosphorylation of IFNAR1 degron. A, degron phosphorylation of FLAG-IFNAR1 (wild type (WT) or S532A mutant) co-expressed in 293T cells with Myc-tagged human CK1α or empty vector (Vec) and treated or not with IFNα (1000 IU/ml for 30 min as indicated) was analyzed by FLAG immunoprecipitation (IP) followed by immunoblotting using the indicated antibodies. The levels of Myc-CK1α in whole cell lysates were also determined. B, FLAG-IFNAR1 (wild type or S532A mutant) was co-expressed in 293T cells with HA-tagged Leishmania CK1 (HA-L-CK1, wild type, or kinase dead (KD)) and purified by FLAG immunoprecipitation. Phosphorylation of the IFNAR1 degron and levels of IFNAR1 were analyzed by immunoblotting using the indicated antibodies. The levels of HA-L-CK1 in whole cell lysates (WCL) were also determined. C, characterization of anti-Ser(P)532 antibody. FLAG-IFNAR1 proteins (wild type, S535A, or S532A mutants) were expressed in 293T cells, immunopurified, and analyzed using the indicated antibodies. Vec, reactions from cells transfected with empty vector (pCDNA3). D, 293T cells were left untreated (UN) or were treated with TG (1 μm for 30 min) and harvested. Lysates from these cells were twice immunodepleted with antibodies against CK1α, and the CK1α-free supernatants (4 μg) were used alone (lanes 2 and 3) or together with 0.5 μg of bacterially produced recombinant GST-CK1α (lanes 1 and 4–9) for in vitro phosphorylation of GST-IFNAR1 (wild type, lanes 1–6, or S532A mutant, lanes 7–9) in the presence of ATP (except in lane 1) at 30 °C for 30 min as indicated. Phosphorylation of GST-IFNAR1 on Ser532 and Ser535, levels of GST-IFNAR1 (using anti-GST antibody), and levels of CK1α were analyzed by immunoblotting.
Similar to human CK1α, the Leishmania L-CK1 was also shown to promote phosphorylation of IFNAR1 on Ser535 upon expression in human or mouse cells (24). In line with this report, expression of wild type HA-tagged L-CK1, but not of a kinase dead mutant of L-CK1, stimulated Ser535 phosphorylation of co-expressed FLAG-IFNAR1WT (Fig. 2B). However, phosphorylation of Ser535 in the S532A mutant was not observed under these conditions (Fig. 2B). Together these data further indicate that priming phosphorylation might be required for ligand-independent CK1-mediated phosphorylation of the IFNAR1 degron.
To determine whether the putative priming site is phosphorylated in cells, we generated a polyclonal anti-Ser(P)532 antibody (see “Experimental Procedures”). FLAG-IFNAR1 proteins expressed in 293T cells were immunopurified and analyzed by immunoblotting using this antibody as well as the previously characterized anti-Ser(P)535 antibody (11). The latter antibody recognized wild type receptor but neither the S535A mutant nor the S532A mutant, whereas S532D mutant exhibited an increased phosphorylation on Ser535 (Fig. 2C). This result is consistent with data shown in Fig. 2A. Importantly, the anti-Ser(P)532 antibody recognized both wild type and the S535A mutant (but not the S532A or S532D priming site mutants; Fig. 2C), indicating that overexpressed IFNAR1 undergoes phosphorylation on the putative priming site in cells.
We next tested whether this priming phosphorylation is directly mediated by CK1α or by another kinase that is induced by UPR. Incubation of recombinant CK1α with wild type GST-IFNAR1 substrate and ATP in vitro resulted in phosphorylation of Ser535 but not of Ser532 (Fig. 2D, lane 4). This result confirms the previously published suggestion that CK1α is a direct kinase for the IFNAR1 degron residue Ser535 (24) but also indicates that phosphorylation of the putative priming site might be mediated by another kinase. Indeed phosphorylation of Ser532 was detected in CK1α-depleted lysates from cells treated with TG, an inducer of UPR. Moreover, the extent of this phosphorylation was not changed when recombinant CK1α was added to this reaction (Fig. 2D, compare lanes 3 and 6).
Importantly, a combination of CK1α and lysates from TG-treated cells increased the efficacy of phosphorylation of Ser535 in a manner that depended on the integrity of Ser532 as seen from the reaction using the GST-IFNAR1S532A mutant (lane 6 versus lane 9). These results suggest that TG treatment induces activity of an unknown (yet different from CK1α) protein kinase that phosphorylates IFNAR1 on Ser532. Furthermore, this phosphorylation increases the efficacy of CK1α-mediated phosphorylation of Ser535 within the degron of IFNAR1, suggesting that Ser532 represents a bona fide priming site.
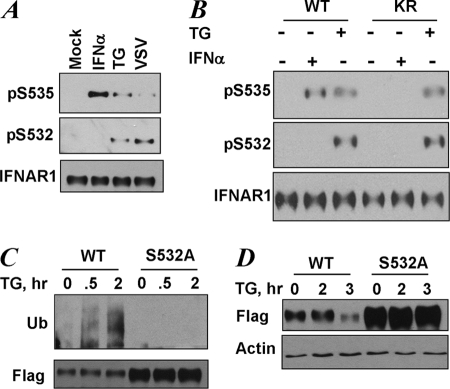
We next sought to investigate whether phosphorylation of the priming site may occur within the context of endogenous IFNAR1 in cells where UPR is induced. Treatment of HeLa cells with TG or infection of these cells with VSV led to phosphorylation of endogenous IFNAR1 on both Ser532 and Ser535 (Fig. 3A). In line with previously published results (11–13), treatment of cells with IFNα stimulated Ser535 phosphorylation. However, priming phosphorylation on Ser532 in response to the ligand was not efficient (Fig. 3A). This result, together with ligand-induced Ser535 phosphorylation of the IFNAR1S532A mutant (Fig. 2A), suggests that IFNα-induced signaling is capable of promoting IFNAR1 degron phosphorylation in a manner that does not require priming phosphorylation. Furthermore, in human KR cells (which harbor catalytically inactive Tyk2 and were shown not to support IFNα-induced IFNAR1 phosphorylation, ubiquitination, and degradation) (12, 15), the phosphorylation of the priming Ser532 site and of the degron Ser535 in response to TG was also detected (Fig. 3B). Collectively, these results suggest that phosphorylation of the priming site occurs in a ligand- and Tyk2-independent manner and is dispensable for the ligand-induced pathway.
FIGURE 3.
UPR induces phosphorylation of the priming site that is required for increased ubiquitination and degradation of IFNAR1. A, endogenous IFNAR1 proteins immunopurified from HeLa cells that were untreated (Mock) or were treated with TG (1 μm for 30 min), IFNα (6000 IU/ml for 30 min), or infected with VSV (0.1 multiplicity of infection for 1 h followed by an additional 10 h of incubation in virus-free medium) as indicated, were analyzed by immunoblotting using the indicated antibodies. B, endogenous IFNAR1 proteins immunopurified from 11.1-Tyk2-null derivative cell lines reconstituted with wild type Tyk2 (WT) or catalytically deficient Tyk2 (KR) were treated with TG or IFNα (in doses indicated in A) for 30 min. C, FLAG-IFNAR1 proteins (wild type or S532A mutant) were expressed in 293T cells. The cells were pretreated with a lysosomal inhibitor (methylamine HCl, 10 mm) for 1 h to prevent degradation of ubiquitinated receptors. Then the cells were treated with TG (1 μm for the indicated times), and FLAG-IFNAR1 proteins were immunopurified under denaturing conditions and analyzed by immunoblotting using antibodies against ubiquitin (Ub, upper panel) and FLAG (lower panel). D, down-regulation of the levels of FLAG-IFNAR1 (wild type or S532A mutant) expressed in 293T cells treated with TG (1 μm for indicated times) was analyzed by immunoblotting using FLAG antibody. Comparable loading was verified by anti-β-actin immunoblot (lower panel).
We have previously reported that induction of UPR promotes ubiquitination and degradation of endogenous or exogenously expressed wild type IFNAR1 in human cells (17). Here we sought to determine the role of phosphorylation of the priming site in UPR-induced ubiquitination of IFNAR1. Treatment of cells with TG noticeably increased the extent of ubiquitination of wild type IFNAR1 but not of the S532A mutant (Fig. 3C). Furthermore, this mutant was less sensitive to a decrease in the levels of IFNAR1 induced by TG (Fig. 3D). These data suggest that the priming phosphorylation of IFNAR1 plays an important role in the ubiquitination and down-regulation of IFNAR1 in response to UPR induction.
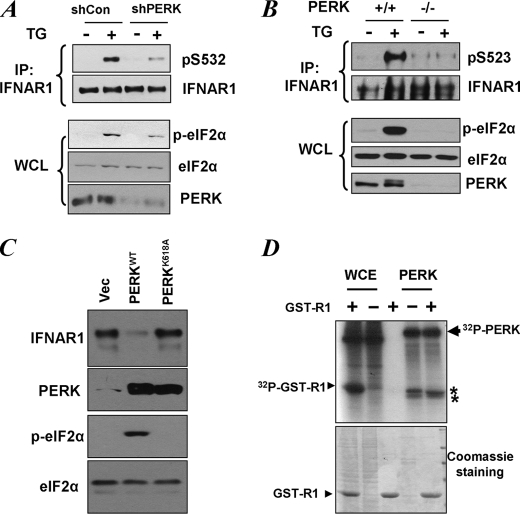
UPR stimulates Ser535 phosphorylation of IFNAR1 and accelerates ubiquitination and degradation of this receptor in a manner that relies on PERK activity (17). We next sought to investigate whether PERK is required for phosphorylation of the priming site within IFNAR1. Transfection of HeLa cells with small hairpin RNA targeted against PERK led to a partial knockdown of this kinase as evident from its decreased level and decreased phosphorylation of its known substrate eIF2α in cells treated with TG (Fig. 4A, lower set of panels). Under these conditions, the efficacy of TG-induced phosphorylation of IFNAR1 on the priming Ser532 was also decreased (Fig. 4A, upper set of panels). This result suggests that PERK is required for UPR-induced priming phosphorylation. Consistent with this suggestion, TG-induced phosphorylation of mouse IFNAR1 on Ser523 (analogous to Ser532 in human receptor) was not observed in mouse embryo fibroblasts from PERK knock-out animals (Fig. 4B). Given that PERK plays an important role in UPR-induced ubiquitination and degradation (17) and these events also depend on the priming site of IFNAR1 (Fig. 3, C and D), the findings that PERK regulates Ser532 phosphorylation also indicate that this kinase might function upstream of the phosphorylation of the priming site.
FIGURE 4.
Role of PERK in UPR-induced phosphorylation of priming site of IFNAR1. A, HeLa cells transfected with small hairpin RNA against PERK or against GFP (shCon) were treated with TG (1 μm for 30 min), and endogenous IFNAR1 proteins were immunopurified (IP) and analyzed for phosphorylation on the priming site and for total levels by immunoblotting using the indicated antibodies. Phosphorylation of a known PERK substrate eIF2α (as well as its total levels) and the levels of PERK itself were also determined in whole cell lysates (WCL). B, mouse embryo fibroblasts from wild type or PERK knock-out animals were treated with TG as indicated. The levels and priming phosphorylation of endogenous murine IFNAR1 on Ser523 (analogue of human Ser532) were analyzed by immunoblotting using indicated antibodies. The phosphorylation and levels of eIF2α and the levels of PERK in whole cell lysates were also determined. C, levels of endogenous IFNAR1 in 293T cells transfected with wild type or the catalytically deficient mutant (K618A) of PERK were analyzed by immunoprecipitation followed by immunoblotting using an anti-IFNAR1 antibody. The levels of PERK, phosphorylated PERK, and levels of eIF2α were also examined. D, whole cell extracts (WCE) from 293T cells or recombinant bacterially produced constitutively active ΔN-PERK were incubated alone or with GST-IFNAR1 in the presence of radiolabeled [γ-32P]ATP as indicated. Resulting phosphorylation of GST-IFNAR1 or contaminants and autophosphorylation of PERK was determined by SDS-PAGE followed by Coomassie staining and autoradiography. The positions of PERK, GST-IFNAR1, and some irrelevant contaminants (denoted by asterisks) are indicated. Vec, empty vector.
Expression of wild type but not catalytically inactive PERK mutant led to a noticeable down-regulation of endogenous IFNAR1 (Fig. 4C), suggesting that kinase activity of PERK is required for ligand-independent IFNAR1 degradation. We next sought to determine whether PERK may serve as a direct kinase for the priming site. Incubation of recombinant active PERK with GST-IFNAR1 and ATP in an in vitro kinase assay similar to the one shown in Fig. 2D did not yield any phosphorylation of the substrate on Ser532 that would be detectable by immunoblotting using anti-Ser(P)532 antibody (data not shown). Furthermore, when this reaction was carried out in the presence of radiolabeled ATP, we could not detect the incorporation of phosphate into GST-IFNAR1 (Fig. 4D). Having excluded the possibilities that the integrity of the substrate might be somehow compromised (by analyzing protein load using Coomassie staining and demonstrating that this very substrate was efficiently phosphorylated by the whole cell lysate) or that the kinase was inactive (given efficient autophosphorylation and phosphorylation of contaminants denoted by asterisks in Fig. 4D), we conclude that PERK is not capable of directly phosphorylating IFNAR1. This result together with data from Fig. 2D demonstrating induction of a Ser532 kinase in cells treated with TG also suggests that UPR stimulates a PERK-dependent activation of another serine kinase that functions as a direct kinase for priming phosphorylation. Alternatively, PERK activity might negatively regulate a hypothetical Ser532 phosphatase.
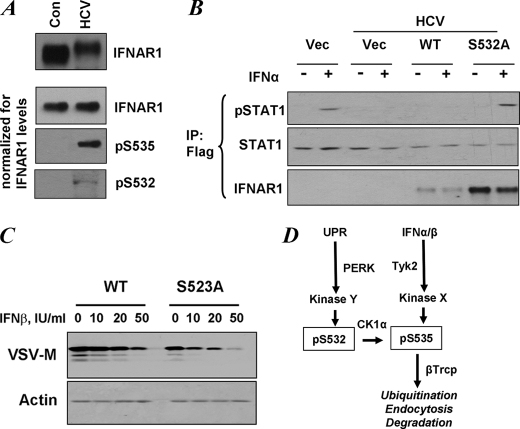
UPR induced by some viruses including VSV and HCV was shown not only to down-regulate IFNAR1 but also to inhibit the extent of IFNα/β signaling, providing these viruses with the means to evade the control from the Type I IFN system (17). We next sought to determine whether phosphorylation of the priming site is important for attenuation of cellular responses to IFN. In line with previously reported data (17), expression of the HCV genome in human Huh7 hepatoma cells noticeably down-regulated the level of endogenous IFNAR1 (Fig. 5A, top panel). When loading was normalized to yield comparable amounts of IFNAR1 in the immunoprecipitation reaction, we also observed that HCV induced phosphorylation on both Ser535 and Ser532 (Fig. 5A, lower set of panels). This result suggests that cells expressing the HCV genome display an increased priming phosphorylation of IFNAR1 that may lead to down-regulation of the receptor.
FIGURE 5.
Priming phosphorylation of IFNAR1 contributes to regulation of the extent of IFNα/β signaling. A, control (Con) human Huh7 cells and those expressing the HCV replicon were analyzed for IFNAR1 levels by immunoprecipitation-immunoblotting (upper panel). The lower three panels depict the experiments where gel loading was normalized to achieve comparable levels of immunopurified IFNAR1 in each lane. Phosphorylation of IFNAR1 on Ser532 and Ser535 was determined by immunoblotting using the indicated antibodies. B, control human Huh7 cells and those expressing the HCV replicon were transfected with FLAG-tagged STAT1 alone with empty vector (Vec) or FLAG-IFNAR1 (wild type (WT) or S532A mutant) and were untreated or treated with IFNα (60 IU/ml for 30 min) as indicated. Lysates of these cells were immunoprecipitated (IP) using anti-FLAG antibody and analyzed by immunoblotting using antibodies against phospho-STAT1, total STAT1, and IFNAR1. C, mouse embryo fibroblasts from IFNAR1-null animals were reconstituted with murine FLAG-IFNAR1 (wild type or S523A mutant, which is a mouse analogue of human S532A mutant). The cells were treated with indicated doses of murine IFNβ for 1 h, incubated for 8 h in fresh medium, and then infected with VSV (multiplicity of infection of 0.1). Expression of VSV-M protein was analyzed 16 h later by immunoblotting. Levels of β-actin were also determined (lower panel). D, model for ligand-dependent and ligand-independent ubiquitination and degradation of IFNAR1. Both pathways converge at the level of degron phosphorylation (Ser(P)535, pS535). Signaling induced by IFN and dependent on the activity of Tyk2 does not require either CK1α (24) or priming phosphorylation (this study). Ligand-independent pathway initiated by inducers of UPR does not need either ligand or Tyk2 activity but requires CK1α (26) and PERK-dependent priming phosphorylation (this study).
The response of these cells to IFNα was markedly attenuated (17). We further sought to determine whether this inhibition could be rescued by expression of IFNAR1 deficient in Ser532 phosphorylation. Because of limited transfection efficacy in Huh7 cells, we have co-expressed FLAG-tagged STAT1 with FLAG-tagged IFNAR1 proteins and then analyzed STAT1 phosphorylation and levels in FLAG immunoprecipitation reactions. This analysis revealed a decreased phosphorylation of FLAG-STAT1 (Fig. 5B) likely caused by a decreased level of IFNAR1 (as shown in Fig. 5A). Co-expression of a FLAG-IFNAR1S532A mutant that is insensitive to HCV-induced priming phosphorylation restored the efficacy of IFNα-induced FLAG-STAT1 phosphorylation. However, equal amounts of vector for expression of wild type FLAG-IFNAR1 failed to reverse the HCV-mediated inhibition, most likely because of the fact that wild type IFNAR1 is susceptible to ligand-independent ubiquitination and degradation (as seen in Fig. 3, C and D) and, as a result, is expressed at levels markedly lower than that of the priming site phosphorylation-deficient mutant (Fig. 5B, bottom panel).
These data indicate that priming phosphorylation of IFNAR1 may regulate IFNα/β signaling. To further explore this possibility, we reconstituted mouse embryo fibroblasts from IFNAR1 knock-out mice with either wild type murine IFNAR1 or its priming site Ser523 mutant and compared the ability of murine IFNβ to induce an anti-viral state in these cells. Cells that express the priming site mutant exhibited a noticeably higher innate resistance to VSV infection (as judged from lower levels of expression of VSV-M protein in the absence of exogenous IFNβ (Fig. 5C). Furthermore, these cells required an at least five times lower dose of exogenous IFNβ than cells expressing wild type receptor to mount a comparable defense against VSV (compare VSV-M levels at dose 50 IU/ml in wild type cells versus 10 IU/ml in S523A in Fig. 5C). These data together indicate that priming phosphorylation of IFNAR1 contributes to the regulation of the cellular responses to Type I IFN.
DISCUSSION
Ligand-independent phosphorylation of Ser535 within the degron of IFNAR1 is a central event in ubiquitination and ensuing down-regulation of this receptor chain in response to UPR stimuli including viral infection (17). Identification of CK1α, a constitutively active enzyme refractory to further stimulation by UPR, as a kinase responsible for this phosphorylation (24) posed a question of how this phosphorylation is induced by UPR. Here we propose that UPR signaling activates a yet unknown kinase activity that phosphorylates IFNAR1 on Ser532, located proximal to the degron. Phosphorylation of Ser532 in turn functions as a priming event that facilitates CK1α-mediated phosphorylation of the IFNAR1 degron followed by IFNAR1 ubiquitination and degradation. In support of this hypothesis, we show that TG treatment of cells induces a Ser532 kinase that is different from CK1α and that cooperates with CK1α in phosphorylating the IFNAR1 degron in a manner that is dependent on the integrity of Ser532 (Fig. 2D). Our findings further demonstrate that phosphorylation of the priming site indeed occurs in cells where UPR is induced by IFNAR1 overexpression (Fig. 2C) or treatment of cells with TG or infection with VSV (Fig. 3A) or expression of the HCV genome (Fig. 5A). This phosphorylation requires activity of PERK, which by itself is incapable of phosphorylating IFNAR1 (Fig. 4), suggesting the function of another kinase in this process. The data shown in Figs. 1B and 3 (C and D) suggest that basal and TG-induced ubiquitination and degradation of IFNAR1 depends on the priming site, which is also implicated in the regulation of the extent of cellular responses to IFNα/β (Fig. 5, B and C). These findings suggest that priming phosphorylation of IFNAR1 plays an important role in IFNAR1 proteolysis stimulated by UPR.
In addition, the data presented here showed that priming Ser532 phosphorylation is not efficiently induced by IFNα (Fig. 3A) and can occur in cells that lack catalytically active Tyk2 (Fig. 3B). Moreover, an IFNAR1 mutant lacking the priming site remained sensitive to IFNα-inducible phosphorylation of the IFNAR1 degron (Fig. 2A). These data further contribute to the characterization of ligand and JAK-dependent and independent pathways (Fig. 5D). Both pathways converge at the level of phosphorylation of the IFNAR1 degron, which is followed by recruitment of β-Trcp, ubiquitination, endocytosis, and degradation. However, the ligand-inducible pathways require JAK activity but need neither CK1α (24) nor the integrity of the priming site (this study). By contrast, the latter site and CK1α activity are crucial for ligand-independent pathways that can be induced by UPR stimuli in the absence of exogenously added IFNα/β and in cells lacking activity of Tyk2 (15, 17, 24).
The overall mechanism of utilizing inducible priming phosphorylation for subsequent degron phosphorylation in IFNAR1 is reminiscent of a similar regulation that has been described in other substrates of SCFβ-Trcp E3 ubiquitin ligase including β-catenin (38–40), Cdc25a (41–44), and Gli/Ci (45–48). Priming phosphorylation of a substrate at proximal Ser/Thr residues in the n − 3 position has been extensively demonstrated to promote subsequent phosphorylation of this substrate by CK1, sometimes on several consecutive and properly spaced phospho-acceptors (30–37). In the case of IFNAR1, the priming effect seems to be limited to just one specific site (Ser532). Despite the presence of another putative site (Ser529, seen in IFNAR1 of many species but not in chicken; Fig. 1A), mutation of this site did not affect phosphorylation of the IFNAR1 degron on Ser535.3 Another prominent characteristic of priming phosphorylation in IFNAR1 is that this post-translational modification is inducible and seems to underlie the mechanism by which degron phosphorylation, ubiquitination, and degradation can be promoted by UPR signaling in a PERK-dependent manner.
Because PERK itself is not capable of directly phosphorylating IFNAR1, the identity of the priming kinase that acts downstream of PERK to phosphorylate Ser532 in response to UPR (Fig. 5D, Kinase Y) remains to be determined. Our previously reported data that CK1α activity is not induced by UPR (24) and in vitro data presented here (Fig. 2D) rule out the possibility that CK1α itself may function as the priming kinase. In that sense, IFNAR1 is dissimilar from another substrate of β-Trcp, β-catenin, whose degron is phosphorylated by the glycogen synthase kinase 3β upon priming phosphorylation of distal phospho-acceptors by CK1α (40). Future studies aimed at identification and characterization of the priming kinase of IFNAR1 are important for designing the means for inhibition of the ligand-independent pathway and, accordingly, preventing the ability of viruses to decrease the therapeutic effects of Type I IFNs.
Acknowledgments
We thank R. Aldabe, J. Chen, J. Darnell, J. W. Harper, R. Harty, S. Hemmi, D. S. Lyles, S. Pellegrini, and D. Ron for the reagents and the members of the Fuchs and Diehl lab for discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants CA92900 (to S. Y. F.) and CA104838 (to J. A. D.).
J. Liu and S. Y. Fuchs, unpublished data.
- IFN
- interferon
- IFNAR
- IFNα/β receptor chain
- CK1
- casein kinase 1
- L-CK1
- Leishmania casein kinase 1
- TG
- thapsigargin
- VSV
- vesicular stomatitis virus
- PERK
- pancreatic endoplasmic reticulum kinase
- JAK
- Janus kinase
- STAT
- signal transducer and activator of transcription
- HCV
- hepatitis C virus
- UPR
- unfolded protein response
- E3
- ubiquitin-protein isopeptide ligase
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- eIF
- eukaryotic initiation factor.
REFERENCES
- 1.Wallach D. F. (1987) Fundamentals of Receptor Molecular Biology, M. Dekker, New York [Google Scholar]
- 2.Müller U., Steinhoff U., Reis L. F., Hemmi S., Pavlovic J., Zinkernagel R. M., Aguet M. (1994) Science. 264, 1918–1921 [DOI] [PubMed] [Google Scholar]
- 3.Gutterman J. U. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billard C., Sigaux F., Castaigne S., Valensi F., Flandrin G., Degos L., Falcoff E., Aguet M. (1986) Blood. 67, 821–826 [PubMed] [Google Scholar]
- 5.Maxwell B. L., Talpaz M., Gutterman J. U. (1985) Int. J. Cancer 36, 23–28 [DOI] [PubMed] [Google Scholar]
- 6.Aaronson D. S., Horvath C. M. (2002) Science 296, 1653–1655 [DOI] [PubMed] [Google Scholar]
- 7.Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 8.Constantinescu S. N., Croze E., Wang C., Murti A., Basu L., Mullersman J. E., Pfeffer L. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9602–9606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Constantinescu S. N., Croze E., Murti A., Wang C., Basu L., Hollander D., Russell-Harde D., Betts M., Garcia-Martinez V., Mullersman J. E., Pfeffer L. M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10487–10491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar K. G., Tang W., Ravindranath A. K., Clark W. A., Croze E., Fuchs S. Y. (2003) EMBO J. 22, 5480–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar K. G., Krolewski J. J., Fuchs S. Y. (2004) J. Biol. Chem. 279, 46614–46620 [DOI] [PubMed] [Google Scholar]
- 12.Marijanovic Z., Ragimbeau J., Kumar K. G., Fuchs S. Y., Pellegrini S. (2006) Biochem. J. 397, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar K. G., Barriere H., Carbone C. J., Liu J., Swaminathan G., Xu P., Li Y., Baker D. P., Peng J., Lukacs G. L., Fuchs S. Y. (2007) J. Cell Biol. 179, 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar K. G., Varghese B., Banerjee A., Baker D. P., Constantinescu S. N., Pellegrini S., Fuchs S. Y. (2008) J. Biol. Chem. 283, 18566–18572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Plotnikov A., Banerjee A., Suresh Kumar K. G., Ragimbeau J., Marijanovic Z., Baker D. P., Pellegrini S., Fuchs S. Y. (2008) Biochem. Biophys. Res. Commun. 367, 388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.HuangFu W. C., Liu J., Harty R. N., Fuchs S. Y. (2008) FEBS Lett. 582, 3206–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., HuangFu W. C., Kumar K. G., Qian J., Casey J. P., Hamanaka R. B., Grigoriadou C., Aldabe R., Diehl J. A., Fuchs S. Y. (2009) Cell Host Microbe 5, 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waris G., Tardif K. D., Siddiqui A. (2002) Biochem. Pharmacol. 64, 1425–1430 [DOI] [PubMed] [Google Scholar]
- 19.He B. (2006) Cell Death Differ. 13, 393–403 [DOI] [PubMed] [Google Scholar]
- 20.Katze M. G., He Y., Gale M., Jr. (2002) Nat. Rev. Immunol. 2, 675–687 [DOI] [PubMed] [Google Scholar]
- 21.Randall R. E., Goodbourn S. (2008) J. Gen. Virol. 89, 1–47 [DOI] [PubMed] [Google Scholar]
- 22.Lang K., Weiner D. B. (2008) Expert Rev. Vaccines 7, 915–923 [DOI] [PubMed] [Google Scholar]
- 23.Hartwell D., Shepherd J. (2009) Int. J. Technol. Assess Health Care 25, 56–62 [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Carvalho L. P., Bhattacharya S., Carbone C. J., Kumar K. G., Leu N. A., Yau P. M., Donald R. G., Weiss M. J., Baker D. P., McLaughlin K. J., Scott P., Fuchs S. Y. (2009) Mol. Cell. Biol. 29, 6401–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirogane T., Jin J., Ang X. L., Harper J. W. (2005) J. Biol. Chem. 280, 26863–26872 [DOI] [PubMed] [Google Scholar]
- 26.Chen L., Li C., Pan Y., Chen J. (2005) Mol. Cell. Biol. 25, 6509–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cullinan S. B., Zhang D., Hannink M., Arvisais E., Kaufman R. J., Diehl J. A. (2003) Mol. Cell. Biol. 23, 7198–7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luquin E., Larrea E., Civeira M. P., Prieto J., Aldabe R. (2007) Antiviral Res. 76, 194–197 [DOI] [PubMed] [Google Scholar]
- 29.Goldman L. A., Zafari M., Cutrone E. C., Dang A., Brickelmeier M., Runkel L., Benjamin C. D., Ling L. E., Langer J. A. (1999) J. Interferon Cytokine Res. 19, 15–26 [DOI] [PubMed] [Google Scholar]
- 30.Knippschild U., Gocht A., Wolff S., Huber N., Löhler J., Stöter M. (2005) Cell Signal. 17, 675–689 [DOI] [PubMed] [Google Scholar]
- 31.Umphress J. L., Tuazon P. T., Chen C. J., Traugh J. A. (1992) Eur. J. Biochem. 203, 239–243 [DOI] [PubMed] [Google Scholar]
- 32.Roach P. J. (1991) J. Biol. Chem. 266, 14139–14142 [PubMed] [Google Scholar]
- 33.Meggio F., Donella-Deana A., Pinna L. A. (1979) FEBS Lett. 106, 76–80 [DOI] [PubMed] [Google Scholar]
- 34.Flotow H., Graves P. R., Wang A. Q., Fiol C. J., Roeske R. W., Roach P. J. (1990) J. Biol. Chem. 265, 14264–14269 [PubMed] [Google Scholar]
- 35.Donella-Deana A., Grankowski N., Kudlicki W., Szyszka R., Gasior E., Pinna L. A. (1985) Biochim. Biophys. Acta 829, 180–187 [DOI] [PubMed] [Google Scholar]
- 36.Bustos V. H., Marin O., Meggio F., Cesaro L., Allende C. C., Allende J. E., Pinna L. A. (2005) Biochem. J. 391, 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bustos V. H., Ferrarese A., Venerando A., Marin O., Allende J. E., Pinna L. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19725–19730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagen T., Vidal-Puig A. (2002) Biochem. Biophys. Res. Commun. 294, 324–328 [DOI] [PubMed] [Google Scholar]
- 39.Amit S., Hatzubai A., Birman Y., Andersen J. S., Ben-Shushan E., Mann M., Ben-Neriah Y., Alkalay I. (2002) Genes Dev. 16, 1066–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002) Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 41.Jin J., Ang X. L., Ye X., Livingstone M., Harper J. W. (2008) J. Biol. Chem. 283, 19322–19328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin J., Shirogane T., Xu L., Nalepa G., Qin J., Elledge S. J., Harper J. W. (2003) Genes Dev. 17, 3062–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donzelli M., Busino L., Chiesa M., Ganoth D., Hershko A., Draetta G. F. (2004) Cell Cycle 3, 469–471 [PubMed] [Google Scholar]
- 44.Kang T., Wei Y., Honaker Y., Yamaguchi H., Appella E., Hung M. C., Piwnica-Worms H. (2008) Cancer Cell 13, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang J. (2006) Cell Cycle 5, 2457–2463 [DOI] [PubMed] [Google Scholar]
- 46.Bhatia N., Thiyagarajan S., Elcheva I., Saleem M., Dlugosz A., Mukhtar H., Spiegelman V. S. (2006) J. Biol. Chem. 281, 19320–19326 [DOI] [PubMed] [Google Scholar]
- 47.Pan Y., Bai C. B., Joyner A. L., Wang B. (2006) Mol. Cell. Biol. 26, 3365–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia J., Zhang L., Zhang Q., Tong C., Wang B., Hou F., Amanai K., Jiang J. (2005) Dev. Cell 9, 819–830 [DOI] [PubMed] [Google Scholar]