Abstract
Secretory proteins are subjected to a stringent endoplasmic reticulum-based quality control system that distinguishes aberrant from correctly folded proteins. The cytoplasmic peptide:N-glycanase cleaves oligosaccharides from misfolded glycoproteins and prepares them for degradation by the 26 S proteasome. In contrast to abundant in vitro data on its enzymatic function, the in vivo relevance of peptide:N-glycanase activity remains unclear. Here we show that the PNG1 ortholog from the filamentous ascomycete Neurospora crassa is an essential protein, and its deletion results in strong polarity defects. PNG1 and its predicted binding partner RAD23 have distinct functions in N. crassa and are involved in cell wall integrity and DNA repair, respectively. Moreover, wild type PNG1 has substitutions in essential catalytic amino acids, and its deglycosylation activity is lost. These substitutions are conserved in many PNG1 orthologs of the fungal kingdom, implying a so far unrecognized enzyme-independent function of PNG1 that may only become apparent in highly polar cells such as fungal hyphae.
Keywords: Carbohydrate/Glycoprotein, Cell/Wall, Glycosylation, Organisms/Neurospora, Protein/Turnover, Cell Polarity
Introduction
Misfolded proteins of the secretory pathway are exported back into the cytosol and degraded by the 26 S proteasome by a mechanism called ER2-associated protein degradation (ERAD (1, 2)). It is thought that glycosylated ERAD substrates are subjected to deglycosylation by the cytoplasmic peptide:N-glycanase (PNGase). This pathway implicates a close functional interaction between the ER export machinery, deglycosylating enzymes, and the proteasome (3). Two interacting proteins, Png1 and Rad23, are suspected to play important roles in maintaining a close interaction between the ER export machinery and the proteasome and in the deglycosylation process itself (4, 5). Png1 is a highly conserved protein whose de-N-glycosylating activity has been determined in yeast (6, 7) (ScPNG1 in this study), animals (8–14), and plants (15). Functional studies indicate that Png1 is the only deglycosylating enzyme acting on N-glycans in the cytosol, and yeast png1Δ cells were found to have no detectable in vitro and in vivo PNGase activity (4, 6). In addition, the genomes of Neurospora crassa and Saccharomyces cerevisiae do not contain obvious orthologs for another cytosolic deglycosylating enzyme called endo-β-N-acetylglucosaminidase (16).
Yeast as well as mouse Png1 binds Rad23, which in turn promotes a direct interaction of this complex with the proteasome. Deletion of yeast Rad23 or down-regulation of its animal/human orthologs results in the accumulation of ubiquitylated proteasome substrates (4). Thus, Rad23 has been proposed to facilitate the substrate transfer to the proteasome. In addition to its assistance in protein deglycosylation, Rad23 forms a complex with Rad4 that recognizes and binds damaged DNA during nucleotide excision repair (17).
Despite the evidence for the involvement of the Png1-Rad23 complex in proteasome-mediated protein degradation, the turnover of most ERAD substrates is not affected by inhibition of PNGase activity (4, 6, 18–20). Furthermore, deletion or down-regulation of Png1 does not result in any observable growth defect, raising questions about the significance of Png1 as an important enzyme in the proteolytic process in vivo. Unfortunately, no mutants in other systems but budding yeast have yet been described, thus precluding a more detailed analysis of Png1 function(s) in vivo.
Filamentous fungi share, along with neurons and pollen tubes, the distinction of being among the most highly polarized cells in nature (21–23). Polarity in these organisms is established during spore germination, which results in the emergence of a highly polarized hypha that continues to grow by apical extension. Here we characterize PNG1 from the filamentous ascomycete N. crassa (designated NcPNG1) as being essential for establishment and maintenance of cellular polarity despite the fact that naturally occurring changes of critical residues within the catalytic center of NcPNG1 preclude any enzymatic activity of the protein. The substitution of these catalytic residues is conserved in many orthologs of the fungal kingdom, implying a so far unrecognized cellular function of PNG1 that is unrelated to protein deglycosylation.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
Strains used in this study are listed in Table 1 (see also Ref. 24). General genetic procedures for N. crassa are available at the Fungal Genetics Stock Center (FGSC). Vogel's minimal medium was supplemented with 1.5% agar, 2% sucrose, and 1% yeast extract. The final inhibitor concentrations for the growth assays were: 5 mg/ml lysing enzymes (Sigma), 0.02 μm caspofungin (MSD Sharp & Dohme GmbH), 5 μm polyoxin D (Kaken Chemical Co.), 0.05 μm latrunculin A (Sigma), 0.1 μg/ml bemonyl (Sigma), 50 μg/ml hygromycin B (Calbiochem), 10 μg/ml nourseothicin (Werner BioAgents), and 10 μm MG-132 (Sigma). Sensitivity to DNA-damaging agents was determined by counting colonies produced on medium supplemented with 2% sorbose to induce the formation of tight colonies and 0.01% methyl methane sulfonate, 10 μm camptothecin, or 10 μg/ml 4-nitrochinoline-N-oxide (all from Sigma). Microscopic documentation of fungal hyphae or colonies was performed with an SZX16 stereomicroscope, equipped with a ColorView III camera and cellD imaging software (Olympus). Images were further processed using PhotoShop CS2 (Adobe).
TABLE 1.
N. crassa strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Wild type | 74-OR23-1A | FGSC 987 |
| png-1(9-1) | png-1(D285N) | Ref. 29 |
| png-1(10-2) | png-1(E239D) | Ref. 29 |
| Δpng-1 (NCU00651) | hph::png-Δ bar::mus-51Δ + png-1+bar::mus-51Δ | FGSC 11474 |
| Δrad-23 (NCU07542) | hph::rad23Δ | FGSC 13431 |
| png-1(9-1);Δrad-23 | png-1(D285N);hph::rad23Δ | This study |
| png-1(10-2);Δrad-23 | png-1(E239D);hph::rad23Δ | This study |
Plasmid Construction
To generate the fungal expression vector, the nourseothricin resistance cassette was amplified using the primers (restriction sites underlined) 5′-GAA TTC GAA AGG CCG TCA GGA GCT G and 5′-GAA TTC TCA GGG GCA GGG CAT G-3′ and pNV1 as template (25) and cloned via EcoRI in pBluescript SKII (Stratagene). The Aspergillus nidulans Pgpd promoter was amplified with primers 5′-GAT CAG ATC TTT TGC CCG GTG TAT GAA ACC GGA AAG G-3′, 5′-GTT AGC GGC CGC GGT GAT GTC TGC TCA AGC GGG G-3′, and pSM1 as template (22, 23) and inserted in BamHI-NotI digested vector. The png-1 genes were amplified using plasmids YEp352png-1 and YEp352png-1-1 that harbored wild type and C191A substituted ScPng1 (23, 24) or genomic DNA from N. crassa as template. The following primers were used: 5′-GCG GCC GCA TGG GAG AGG TAT ACG AAA AAA ATA AC-3′ and 5′-GCG GCC GCC TAT TTA CCA TCC TCC CCA C-3′ or 5′-GCG GCC GCA TGG CAG GAA ACA ATT CAG GG-3′, and 5′-GCG GCC GCT CAA GGT CCA TAC GTC GGC C-3′ for ScPng1 or NcPNG1, respectively. The nourseothricin concentration was adjusted to 30 μg/ml to select for transformants. N. crassa png-1 cDNA was cloned by reverse transcription-PCR with cDNA prepared from vegetative hyphae. The primers used were 5′-ATG GCA GGA AAC AAT TCA G-3′ and 5′-TCA AGG TCC ATA CGT CGG-3′, respectively. Total RNA was obtained by TRIzol extraction (Invitrogen); cDNA was prepared using SuperScript III reverse transcriptase (Invitrogen) and subcloned into TOPO-TA vector (Invitrogen; designated TOPO-TA-png-1). In the process of generating full-length cDNA of png-1, we found that its open reading frame is 90 bp larger than the predicted gene at the BROAD data base. The insertion was amplified with the primers 5′-CGG AAT TCG ATG GCA GGA AAC AAT TCA GGG G-3′ and 5′-CCC AAG CTT AGG TCC ATA CGT CGG CCA G-3′, containing the restriction sites EcoRI and HindIII, respectively, and cloned into pET28b vector (Novagen), resulting in pET-NcPNG1(His)6. Primers used for the mutagenesis of ScPng1 and NcPNG1 are listed in supplemental Table 1.
To construct pRS423GPD-NcPNG1 to express NcPNG1 in S. cerevisiae, N. crassa png-1 was amplified from TOPO-TA-png-1 using the following primers, 5′-CAC Cat ggc agg aaa caa ttc agg-3′ and 5′-cta CTT ATC GTC GTC ATC CTT GTA ATC agg tcc ata cgt cgg cc-3′ (the second primer was designed to add a C-terminal FLAG tag), and was cloned into pENTRTM/D-TOPO (Invitrogen). To construct the destination vector, ccdB gene was amplified from pDEST14 vector (Invitrogen) using the following primers: 5′-cgg ata tct cAC AAG TTT GTA CAA AAA AGC-3′ and 5′-CAG CTT TCT TGT ACA AAG TGG Tga gat atc cg-3′. The amplified fragment was digested with EcoRV and was cloned into the equivalent site of pRS423GPD vector (2μ HIS3; ATCC). The direction of ccdB gene was confirmed by DNA sequencing. The png-1 gene in pENTR vector was then cloned into the destination vector by the LR Clonase II reaction (Invitrogen) to generate pRS423GPD-NcPNG1-FLAG. pRS423GPD-ScPNG1 was constructed in an analogous approach.
Biochemical Assays
Soluble protein prepared from BL21 cells was subjected to PNGase assays as described previously (26). N-Linked glycan binding ability was assayed as described previously (27). Bacterial expression of His-tagged ScPng1 was described (6), and NcPNG1 extracts were prepared by the same extraction method. Enzyme expression was quantified either by Western blotting with anti-His antibody or by Coomassie Brilliant Blue staining, using Multi Gauge version 2.2 software (Fujifilm, Tokyo, Japan). In vivo PNGase activity was assayed as described previously (28). In brief, 108 yeast cells bearing pRS315GPD-RTA and pRS423GPD, pRS423GPD-ScPNG1, or pRS423GPD-NcPNG1-FLAG were harvested and incubated in 0.1 m NaOH at room temperature for 10 min. After centrifugation at 3000 × g, the cell pellet was resuspended in 100 μl of SDS sample buffer (62.5 mm Tris-HCl (pH 6.8), 5% β-mercaptoethanol, 2% SDS, 5% sucrose, 0.04% bromphenol blue) and boiled for 5 min. Cell debris was removed by centrifugation at 10,000 × g. Equal amounts of each sample were analyzed by 15% SDS-PAGE and transferred to BiotraceTM polyvinylidene difluoride membranes (PALL Corp.). Ricin A chain (RTA) was detected using anti-ricin monoclonal antibody (CP75; Abcam) diluted 1:5000 in TBS-T (25 mm Tri-HCl (pH 8.0), 137 mm NaCl, 0.05% Tween 20, 2% skimmed milk) followed by horseradish peroxidase-conjugated anti-mouse secondary antibody diluted 1:5000 in TBS-T. Detection was carried out with the ImmobilonTM Western blot chemiluminescent horseradish peroxidase substrate system in accordance with the manufacturer's instructions (Millipore). To confirm the expression of NcPNG1-FLAG, Western blotting of the extract was carried out using anti-FLAG M2 monoclonal antibody (M2; Sigma).
RESULTS
Neurospora PNG1 Is Required for Maintenance of Cell Wall Integrity
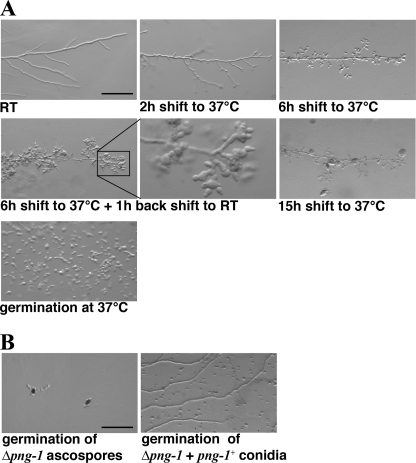
Multiple alleles of png-1 were isolated in a large scale genetic screen for conditional mutants defective in hyphal morphogenesis (29). We focused on png-1(9-1) and png-1(10-2) for a detailed analysis (Fig. 1A). Germination and growth rate as well as hyphal morphology and branching frequency of both conditional mutants were similar to wild type at permissive conditions (25 °C). Within 2 h of transferring them to 37 °C, we observed cessation of tip extension. In contrast to the wild type apex, the tips of png-1(9-1) and png-1(10-2) started to lose polarity, generating balloon-shaped hyphal tips. Concomitant with this growth arrest was the induction of multiple subapical branches in the mutant hyphae that also stopped extension and started to swell apolarly. Transfer back to permissive conditions resulted in growing tips with normal growth rates, diameter, and morphology within 1 h. From one to multiple new tips were generated from each swollen tip, indicating that cell polarity determinants were lost from their apical location and had to be re-established. After prolonged incubation at restrictive temperature, these spherical tips burst, and hyphae lysed. Germination of conidia from both strains under restrictive conditions resulted in apolar and abnormal growth and lysis of large, swollen germlings.
FIGURE 1.
NcPNG1 is essential and required for cell polarization. A, phenotypic characterization of the conditional png-1(9-1) strain grown at the indicated conditions; bar = 200 μm. RT, room temperature. B, asco- and conidiospores of Δpng-1 fail to polarize and grow isotropically or highly abnormal before they lyse; bar = 200 μm.
A deletion strain of png-1 was available through the N. crassa Genome Project (30). Its heterocaryotic nature already suggested an important function of NcPNG1. When we plated asexually derived multinucleate conidiospores on selective medium, we detected a mixture of normal growing germlings and isotropically growing spores (Fig. 1B). Furthermore, we were unable to obtain viable hygromycin-resistant ascospores from wild type × Δpng-1::hph + png-1+ crosses. The majority of the hygR ascospores did germinate in an apolar manner, indicating that NcPNG1 is required for polarity establishment in both types of spores. Rarely, we observed the formation of small hygR colonies, which showed highly abnormal growth and a high rate of cell lysis. In summary, we conclude that NcPNG1 is an essential protein required for establishing and maintaining cellular polarity.
NcPNG1 and NcRAD23 Have Distinct Functions
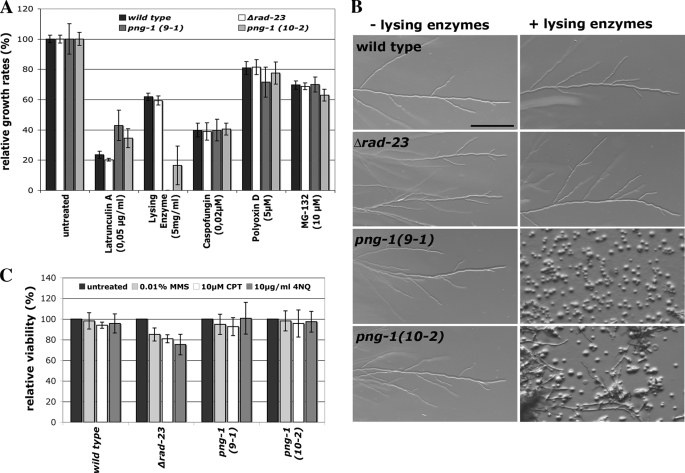
The fungal cell wall and the actin cytoskeleton are key elements to determine cell polarity (31, 32). In line with the observed defects characterized by loss of polarity and cell lysis, we observed hypersensitivity of the conditional strains to lysing enzymes, a general cell wall-degrading enzyme mix containing a mixture of protease, glucanase, and chitinase activities (Fig. 2A). Most notably, polar germination of png-1(9-1) was completely abolished, and only large apolar spheres were generated when conidia were grown under either permissive or semipermissive conditions (Fig. 2B). No altered sensitivity of the png-1 strains to the β1,3-glucan synthase and chitin synthase inhibitors caspofungin and polyoxin D, respectively, was detected. Thus, NcPNG1 is not involved in the organization of the two major structural polysaccharide components of the fungal cell wall. Protein biosynthesis and proteasome function were monitored by growth on hygromycin B, nourseothricin, and MG-132 (data not shown and Fig. 2A). All mutants displayed sensitivities similar to the wild type control, indicating a specific function of NcPNG1 in regulating cell wall integrity and not protein turnover.
FIGURE 2.
NcRAD23 and NcPNG1 have distinct functions. A and B, growth rates of the indicated strains on medium supplemented with various inhibitors at semipermissive conditions (A), and at permissive conditions (B); bar = 200 μm. C, viability of the indicated strains on medium containing DNA-damaging agents at semipermissive conditions. MMS, methyl methane sulfonate; CPT, camptothecin; 4NQ, 4-nitrochinoline-N-oxide.
Png1 has been shown to form a complex with Rad23 in yeast and mouse (5, 33), which may facilitate the transfer of misfolded, glycosylated proteins to the proteasome. N. crassa Δrad-23 displayed wild type characteristics for all parameters tested (germination and growth rate, hyphal morphology, (a)sexual development) and behaved similar to wild type in all growth tests with the above mentioned inhibitors (Fig. 2A). Because of the involvement of Rad23 in DNA repair (17), we compared the sensitivity of wild type, png-1, and Δrad-23 mutants against the DNA-damaging agents methyl methane sulfonate (MMS), camptothecin (CPT), or 4-nitrochinoline-N-oxide (4NQ) (Fig. 2C). We observed a 15, 20, and 25% reduced viability of Δrad-23 against methyl methane sulfonate, camptothecin, and 4-nitrochinoline-N-oxide, respectively, when compared with wild type grown on sublethal concentrations of these drugs. No reduced viability was observed for both conditional png-1 strains. Based on these assays, NcRAD23 and NcPNG1 have distinct functions and are involved in DNA repair and in regulating cell wall integrity, respectively. We were not able to detect any involvement of PNG1 and RAD23 in protein turnover.
NcPNG1 and Other Fungal Orthologs Lack Residues Critical for Peptide:N-Glycanase Activity
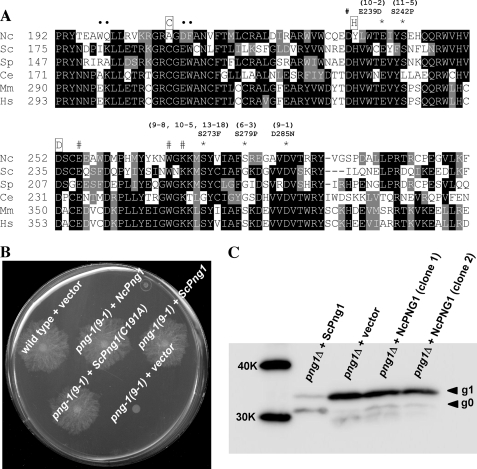
To determine residues that are important for NcPNG1 function, we sequenced several conditional png-1 mutants. All mutations identified resulted in changes in well conserved residues within the core domain of PNG1 (Fig. 3A). Among the mutations observed, Glu-239 (corresponding to Glu-222 in ScPng1) was previously reported to be important for the enzymatic activity of ScPng1 (34), suggesting that the mutant defects may be a result of impaired enzyme activity. However, two out of three residues that constitute the catalytic triad of the enzyme and are essential for PNGase activity in yeast and animals (8, 13, 18, 34) were not conserved in NcPNG1. Specifically, amino acids 208 and 235 (corresponding to ScPng1 Cys-191 and His-218) were substituted for Ala and Tyr, making it unlikely that PNG1 is an active enzyme in vivo. To confirm that the essential function of PNG1 is unrelated to protein deglycosylation, we complemented png-1(9-1) with wild type ScPng1 and ScPng1(C191A). Both yeast proteins, and NcPNG1 as control, complemented the growth defects of png-1(9-1) (Fig. 3B), indicating that enzyme activity of PNG1 is not required for its morphogenetic function. In a second approach to support this surprising finding that NcPNG1 is not a functional enzyme, we utilized yeast png1Δ cells expressing RTA as a known in vivo substrate of PNGase (4). NcPNG1 was expressed well in yeast, as judged by Western blotting analysis (data not shown). As shown in Fig. 3C, expression of NcPNG1 did not result in conversion of the glycosylated g1 form of RTA to the deglycosylated g0 form, providing additional support that NcPNG1 lacks deglycosylating activity in vivo.
FIGURE 3.
PNGase activity of NcPNG1 is not required for its cellular function. A, alignment of NcPNG1 with functionally characterized PNGase orthologs. Amino acids corresponding to catalytic triad (Cys, His, and Asp) are marked with boxed letters. Substitutions determined in conditional mutants of Ncpng-1 are marked with *. Amino acids Ile-193, Trp-194, Ile-181, and Lys-182 (shown to disrupt the enzymatic activity of ScPng1) are marked with ●. The residues marked with # were reported to be important for the binding to chitobiose structure within N-linked glycans (35). Sp = Schizosaccharomyces pombe; Ce = C. elegans; Mm = Mus musculus; and Hs = Homo sapiens. B, growth of png-1(9-1) transformed with the indicated plasmids at restrictive conditions. Catalytically inactive ScPng1(C191A) is able to complement the growth defects of N. crassa png-1(9-1). C, assessment of in vivo PNGase activity of NcPNG1 co-expressed with the PNGase substrate RTA (non-toxic mutant (4)) in yeast. To maximize the detection of RTA, a proteasome mutant (TSY216; cim5-1 png1Δ (28)) was used. The arrows indicate unmodified RTA (g0) or RTA modified with glycan (g1). The g0 form can be either deglycosylated or non-glycosylated RTA. The reduced stability of RTA as well as the dominant occurrence of g0 form indicates in vivo PNGase activity (lane 1), which is absent in png1Δ (lane 2) and png1Δ-expressing NcPNG1 (lanes 3 and 4). Expression of NcPNG1 was verified by anti-FLAG Western blot experiments (not shown). K, kilodalton.
Thus, we tested whether substitutions of the catalytic residues were enough to render NcPNG1 an active enzyme. However, PNGase activity was still absent in NcPNG1 carrying a functional catalytic triad (NcPNG1(A208C,Y235H); Table 2). Thus, we performed an extensive mutagenesis analysis in which PNGase-active ScPng1 was mutagenized to residue(s) found at corresponding position(s) in NcPNG1 to determine additional residues required for PNGase activity. We identified amino acid Glu-193 of ScPng1 (equivalent to Asp-210 in NcPNG1); the N. crassa substitution abolished the activity of ScPng1(E210D) (Table 2). In addition, double substitution of ScPng1 Ile-181 and Lys-182 to Trp and Gln (the corresponding N. crassa residues) also caused loss of enzyme activity (Table 2), clearly indicating that other residues in addition to substitutions in catalytic triad are the cause for enzyme-inactive NcPNG1. These data confirm that the cellular importance of NcPNG1 is not dependent on deglycosylation activity.
TABLE 2.
PNGase activity in point mutated fungal PNG1 proteins
| Relative activitya | |
|---|---|
| ScPng1 | |
| Wild type | +++ |
| N178T | ++ |
| D179E,P180A | ++ |
| I181W,K182Q | ND |
| I181W | +++ |
| K182Q | +++ |
| E185R | ++ |
| E185R,T186V | ++ |
| R187K,K188R | +++ |
| E193D,W194F | ND |
| E193D | ND |
| W194F,C195A | ++ |
| L198V | +++ |
| L200M | +++ |
| L200M,I201L | +++ |
| G206D,L207I | ++ |
| D208R,V209A | ++ |
| Y211W | +++ |
| N214C,R215Q | ++ |
| V219L | + |
| C221T | +++ |
| Y223I | ++ |
| F224Y | +++ |
| N266E,F227H | +++ |
| NcPNG1 | |
| Wild type | ND |
| A208C | ND |
| Y235H | ND |
| A208C,Y235H | ND |
a Relative activity was calculated by quantifying the specific activity (PNGase activity per enzyme amount). The value of wild type ScPng1 was set to 100%; +++, >50%; ++, 10–50%; +, 1–10%. ND, not detected (<1%).
NcPNG1 Is Unable to bind Man8GlcNAc2
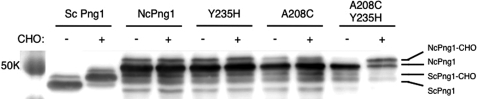
ScPng1 exhibits high affinity toward glycans that contain an N,N′-diacetylchitobiose (chitobiose) structure (26, 34–36). Binding of ScPng1 to glycans such as iodoacetamide-Man8GlcNAc2 can be monitored by its increase in molecular weight and can be detected by a shift in migration in SDS-PAGE (27). The amino acids important for binding to chitobiose were conserved in NcPng1 (Fig. 3A) except for the catalytic His-235 and the probe-reacting Cys-208 (35). Thus, we tested whether NcPNG1 was able to bind to carbohydrate probes after substituting the two critical residues (Fig. 4). Wild type NcPNG1 as well as NcPNG1(Y235H) or NcPNG1(A208C) did not bind to the probe. When we introduced both mutations generating NcPNG1(A208C,Y235H), its carbohydrate binding property was partially regained, indicating that NcPNG1 has lost not only its PNGase activity but also the chitobiose binding property.
FIGURE 4.
NcPNG1 does not bind to the carbohydrate probe Man8GlcNAc2-IAc. E. coli cell extracts of overnight cultures expressing ScPng1, NcPNG1, or their mutants forms were mixed with Man8GlcNAc2-IAc and incubated for 10 min at 37 °C. The reaction was then subjected to SDS-PAGE. Binding of protein to the carbohydrate probe results in increased molecular weight and can be detected by a shift in migration in SDS-PAGE. Proteins were visualized by Western blotting using anti-His-probe antibody. CHO, carbohydrate probe. K, kilodalton.
Interestingly, the catalytic Cys and His residues are not conserved in most filamentous fungal species (supplemental Fig. 1). For example, png-1 genes of most sequenced mold genomes have Cys to Ala/Val and His to Tyr substitutions in their catalytic centers. Moreover, the second CXXC motif that is required for zinc chelating and for PNGase activity (18, 34–36) is also substituted in many mold PNGase orthologs.
DISCUSSION
Here, we describe the functional characterization of PNG1 in N. crassa, a highly conserved protein that was previously thought to be primarily involved in the degradation of misfolded glycosylated proteins. However, NcPNG1 is an essential protein required for establishing and maintaining cell polarity despite its lack of enzymatic activity and inability to bind N-linked glycans such as iodoacetamide-Man8GlcNAc2. This may be achieved by regulating the fungal cell (which may be considered as having analogous functions as the animal extracellular matrix) by a mechanism that does not affect the major structural polysaccharides β-glucan and chitin. It is interesting that most PNG1 relatives of the fungal kingdom lack residues required for enzymatic function, implying a so far unrecognized cellular function of PNG1 that is unrelated to protein deglycosylation. We propose that this is a conserved aspect of PNG1 in fungi and potentially also in higher eukaryotes. However, its non-enzymatic function may only become apparent in highly polar cells such as fungal hyphae or neuronal cells in animals.
The surprising finding of this study is that NcPNG1 is most likely not acting as PNGase in N. crassa and related fungi. This conclusion is based on the following experiments and observations. (a) The catalytic residues cysteine and histidine are substituted to alanine and tyrosine, respectively, in NcPNG1. These residues are essential for transglutaminase/PNGase activity in all so far tested enzymes from various organisms (budding yeast, Caenorhabditis elegans, and mammalian cells) using a range of different substrates (heat-denatured ovalbumin, S-alkylated RNase B, fetuin-derived glycopeptides as in vitro substrates; non-toxic ricin A-chain and T cell receptor α subunit as in vivo substrates) (8, 13, 34, 37, 38). Moreover, the catalytic cysteine was found to be modified by the PNGase inhibitors Z-Vad-fmk and haloacetamidyl-oligosaccharides in yeast and mammalian PNGases (19, 27, 39). These compounds were widely used as in vivo inhibitors for cytoplasmic PNGase (40–42). Also, structural analyses have identified this cysteine as the sole catalytic nucleophile required for the deglycosylation reaction (27, 35, 36). (b) The lack of in vitro deglycosylation activity and carbohydrate binding property of NcPNG1 was determined by two independent assays. Using these assays, we have identified additional NcPNG1 residues that abolish in vitro PNGase activity when introduced into ScPNG1. Furthermore, NcPNG1 is inactive when expressed in the yeast context. (c) Most importantly, the N. crassa mutant can be complemented with an enzymatically inactive version of the S. cerevisiae protein in vivo, making the requirement of species-specific cofactors or interacting proteins unlikely. In summary, we believe that these data are convincing arguments that the in vivo importance of the PNGase ortholog in N. crassa is not related to its enzymatic activity.
Although the cytoplasmic deglycosylation activity performed by PNG1 is apparently missing in molds, a different PNGase activity has been observed in Aspergillus tubingensis (Ref. 43 corresponding to NCU04643 in N. crassa). Although the cellular function of this enzyme is still unstudied, its enzymatic properties (e.g. requirement of acidic pH for enzyme activity) as well as its character as N-glycosylated protein make it unlikely that this acidic PNGase can substitute the deglycosylation function of the cytoplasmic PNGase in the ERAD process.
The identified point mutations in our conditional mutants are substitutions in conserved residues that seem primarily involved in the stabilization of protein structure and thus do not allow functional conclusions without structural information for NcPNG1. Because residues involved in the Rad23-Png1 interaction in yeast and mammals are distinct, the identified mutations may affect species-specific interactions of this complex in N. crassa, but our current data argue against a connection between PNG1 and RAD23 in N. crassa. However, we found that Δrad-23;png-1(9-1) and Δrad-23;png-1(10-2) double mutants displayed weak synthetic growth defects (23 and 27% reduced growth, respectively), making some degree of overlapping functions of the two proteins possible.
In summary, the enzymatic properties of PNGase have been studied for many years in vitro, but most ERAD substrates susceptible for deglycosylation are efficiently degraded even in the absence of Png1 (6, 19, 20). The essential nature and the presence of conditional mutants make N. crassa PNG1 a prime model for deciphering the non-enzymatic function of PNG1. We speculate that this PNG1 function may also exist in other highly polar cells such as neurons.
Supplementary Material
Acknowledgments
We thank Drs. Yukishige Ito (RIKEN ASI) and Ichiro Matsuo (Gunma University) for providing iodoacetamide-Man8GlcNAc2-IAc and Dr. Hiroto Hirayama (RIKEN ASI) for critically reading this manuscript. Caspofungin was kindly provided by MSD Sharp & Dohme GmbH.
This work was supported by grants from the CREST, JST, and Global COE Program (Osaka University) (to T. S.) and by the Deutsche Forschungsgemeinschaft (DFG) through the DFG Research Center of Molecular Physiology of the Brain and the DFG Priority Program Cell Polarity (Grant SP1111) (to S. S.).
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) AB509452.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated protein degradation
- PNGase
- peptide:N-glycanase
- RTA
- Ricin A chain
- chitobiose
- N,N′-diacetylchitobiose
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- Nc
- N. crassa
- Sc
- S. cerevisiae.
REFERENCES
- 1.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahner A., Brodsky J. L. (2004) Trends Cell Biol. 14, 474–478 [DOI] [PubMed] [Google Scholar]
- 3.Suzuki T. (2007) Semin. Cell Dev. Biol. 18, 762–769 [DOI] [PubMed] [Google Scholar]
- 4.Kim I., Ahn J., Liu C., Tanabe K., Apodaca J., Suzuki T., Rao H. (2006) J. Cell Biol. 172, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki T., Park H., Kwofie M. A., Lennarz W. J. (2001) J. Biol. Chem. 276, 21601–21607 [DOI] [PubMed] [Google Scholar]
- 6.Suzuki T., Park H., Hollingsworth N. M., Sternglanz R., Lennarz W. J. (2000) J. Cell Biol. 149, 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki T., Park H., Kitajima K., Lennarz W. J. (1998) J. Biol. Chem. 273, 21526–21530 [DOI] [PubMed] [Google Scholar]
- 8.Hirsch C., Blom D., Ploegh H. L. (2003) EMBO J. 22, 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato T., Kawahara A., Ashida H., Yamamoto K. (2007) J. Biochem. 142, 175–181 [DOI] [PubMed] [Google Scholar]
- 10.Römisch K., Ali B. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6730–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T., Seko A., Kitajima K., Inoue Y., Inoue S. (1993) Biochem. Biophys. Res. Commun. 194, 1124–1130 [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T., Seko A., Kitajima K., Inoue Y., Inoue S. (1994) J. Biol. Chem. 269, 17611–17618 [PubMed] [Google Scholar]
- 13.Suzuki T., Tanabe K., Hara I., Taniguchi N., Colavita A. (2007) Biochem. Biophys. Res. Commun. 358, 837–841 [DOI] [PubMed] [Google Scholar]
- 14.Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. (1996) Cell 84, 769–779 [DOI] [PubMed] [Google Scholar]
- 15.Diepold A., Li G., Lennarz W. J., Nürnberger T., Brunner F. (2007) Plant J. 52, 94–104 [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T., Yano K., Sugimoto S., Kitajima K., Lennarz W. J., Inoue S., Inoue Y., Emori Y. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prakash S., Prakash L. (2000) Mutat. Res. 451, 13–24 [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T., Park H., Lennarz W. J. (2002) FASEB J. 16, 635–641 [DOI] [PubMed] [Google Scholar]
- 19.Misaghi S., Pacold M. E., Blom D., Ploegh H. L., Korbel G. A. (2004) Chem. Biol. 11, 1677–1687 [DOI] [PubMed] [Google Scholar]
- 20.Blom D., Hirsch C., Stern P., Tortorella D., Ploegh H. L. (2004) EMBO J. 23, 650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palanivelu R., Preuss D. (2000) Trends Cell Biol. 10, 517–524 [DOI] [PubMed] [Google Scholar]
- 22.Harris S. D. (2006) Int. Rev. Cytol. 251, 41–77 [DOI] [PubMed] [Google Scholar]
- 23.Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., Read N. D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Plamann M., Goodrich-Tanrikulu M., Schulte U., Mannhaupt G., Nargang F. E., Radford A., Selitrennikoff C., Galagan J. E., Dunlap J. C., Loros J. J., Catcheside D., Inoue H., Aramayo R., Polymenis M., Selker E. U., Sachs M. S., Marzluf G. A., Paulsen I., Davis R., Ebbole D. J., Zelter A., Kalkman E. R., O'Rourke R., Bowring F., Yeadon J., Ishii C., Suzuki K., Sakai W., Pratt R. (2004) Microbiol. Mol. Biol. Rev. 68, 1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCluskey K. (2003) Adv. Appl. Microbiol. 52, 245–262 [DOI] [PubMed] [Google Scholar]
- 25.Seiler S., Vogt N., Ziv C., Gorovits R., Yarden O. (2006) Mol. Biol. Cell 17, 4080–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki T. (2005) Methods 35, 360–365 [DOI] [PubMed] [Google Scholar]
- 27.Suzuki T., Hara I., Nakano M., Zhao G., Lennarz W. J., Schindelin H., Taniguchi N., Totani K., Matsuo I., Ito Y. (2006) J. Biol. Chem. 281, 22152–22160 [DOI] [PubMed] [Google Scholar]
- 28.Tanabe K., Lennarz W. J., Suzuki T. (2006) Methods Enzymol. 415, 46–55 [DOI] [PubMed] [Google Scholar]
- 29.Seiler S., Plamann M. (2003) Mol. Biol. Cell 14, 4352–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunlap J. C., Borkovich K. A., Henn M. R., Turner G. E., Sachs M. S., Glass N. L., McCluskey K., Plamann M., Galagan J. E., Birren B. W., Weiss R. L., Townsend J. P., Loros J. J., Nelson M. A., Lambreghts R., Colot H. V., Park G., Collopy P., Ringelberg C., Crew C., Litvinkova L., DeCaprio D., Hood H. M., Curilla S., Shi M., Crawford M., Koerhsen M., Montgomery P., Larson L., Pearson M., Kasuga T., Tian C., Ba°türkmen M., Altamirano L., Xu J. (2007) Adv. Genet. 57, 49–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park H. O., Bi E. (2007) Microbiol. Mol. Biol. Rev. 71, 48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moseley J. B., Goode B. L. (2006) Microbiol. Mol. Biol. Rev. 70, 605–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park H., Suzuki T., Lennarz W. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11163–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katiyar S., Suzuki T., Balgobin B. J., Lennarz W. J. (2002) J. Biol. Chem. 277, 12953–12959 [DOI] [PubMed] [Google Scholar]
- 35.Zhao G., Li G., Zhou X., Matsuo I., Ito Y., Suzuki T., Lennarz W. J., Schindelin H. (2009) Glycobiology 19, 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J. H., Choi J. M., Lee C., Yi K. J., Cho Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9144–9149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yee V. C., Pedersen L. C., Le Trong I., Bishop P. D., Stenkamp R. E., Teller D. C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7296–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi S., Katiyar S., Lennarz W. J. (2005) FEBS Lett. 579, 823–826 [DOI] [PubMed] [Google Scholar]
- 39.Miyazaki A., Matsuo I., Hagihara S., Kakegawa A., Suzuki T., Ito Y. (2009) Glycoconj. J. 26, 133–140 [DOI] [PubMed] [Google Scholar]
- 40.Ostankovitch M., Altrich-Vanlith M., Robila V., Engelhard V. H. (2009) J. Immunol. 182, 4830–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagihara S., Miyazaki A., Matsuo I., Tatami A., Suzuki T., Ito Y. (2007) Glycobiology 17, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 42.Altrich-VanLith M. L., Ostankovitch M., Polefrone J. M., Mosse C. A., Shabanowitz J., Hunt D. F., Engelhard V. H. (2006) J. Immunol. 177, 5440–5450 [DOI] [PubMed] [Google Scholar]
- 43.Ftouhi-Paquin N., Hauer C. R., Stack R. F., Tarentino A. L., Plummer T. H., Jr. (1997) J. Biol. Chem. 272, 22960–22965 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.