Abstract
Transforming growth factor β-activated kinase 1 (TAK1) kinase is an indispensable signaling intermediate in tumor necrosis factor (TNF), interleukin 1, and Toll-like receptor signaling pathways. TAK1-binding protein 2 (TAB2) and its closely related protein, TAB3, are binding partners of TAK1 and have previously been identified as adaptors of TAK1 that recruit TAK1 to a TNF receptor signaling complex. TAB2 and TAB3 redundantly mediate activation of TAK1. In this study, we investigated the role of TAB2 by analyzing fibroblasts having targeted deletion of the tab2 gene. In TAB2-deficient fibroblasts, TAK1 was associated with TAB3 and was activated following TNF stimulation. However, TAB2-deficient fibroblasts displayed a significantly prolonged activation of TAK1 compared with wild type control cells. This suggests that TAB2 mediates deactivation of TAK1. We found that a TAK1-negative regulator, protein phosphatase 6 (PP6), was recruited to the TAK1 complex in wild type but not in TAB2-deficient fibroblasts. Furthermore, we demonstrated that both PP6 and TAB2 interacted with the polyubiquitin chains and this interaction mediated the assembly with TAK1. Our results indicate that TAB2 not only activates TAK1 but also plays an essential role in the deactivation of TAK1 by recruiting PP6 through a polyubiquitin chain-dependent mechanism.
Keywords: JNK, NF-κB, Protein Phosphatase, Signal Transduction, Tumor Necrosis Factor (TNF), Ubiquitin, TAK1
Introduction
Inflammation is an important biological process to prevent invasion of microorganisms and stimulate wound-healing processes. However, prolonged and dysregulated inflammation are associated with human diseases, including inflammatory bowel disease, psoriasis, and cancers. Inflammatory responses ideally are transient and tightly regulated. Transforming growth factor β-activated kinase 1 (TAK1)2 is a member of the mitogen-activated protein kinase (MAPK) kinase kinase family that is activated by inflammatory mediators such as interleukin-1 (IL-1), tumor necrosis factor (TNF), and Toll-like receptor ligands (1, 2). In all of these pathways, TAK1 has shown to be an essential intermediate that transmits the upstream signal from the receptor complex to the downstream MAPKs and to the NF-κB pathway (2–4). Specifically, in the TNF signaling pathway, TAK1 is associated with the TNF receptor complex through TNF receptor-associated factor 2/5 (TRAF2/5) and the kinase receptor-interacting protein 1 (RIP1) to initiate the signaling cascade (5–7). Oligomerization of TAK1 through this complex assembly then induces autophosphorylation of TAK1, thereby activating TAK1. TAK1 in turn activates kinases, including MAPK and extracellular signal-regulated kinase kinases and IκB kinase (IKK), leading to activation of MAPKs, including c-Jun N-terminal kinase (JNK), p38, and NF-κB, respectively. MAPK and NF-κB then cooperatively regulate transcription of a number of genes that are involved in inflammation. TAK1 is typically activated at 5–10 min after IL-1 and TNF stimulation and is quickly inactivated within 10–20 min (Refs. 8, 9 and this report). This reversibility is supposed to be important for preventing prolonged inflammatory responses. The mechanism of TAK1 activation has been studied extensively; however, the mechanism of TAK1 inactivation is still elusive.
Serine/threonine-protein phosphatase 6 (PP6) is a member of type 2A serine/threonine phosphatase family (10). The yeast homologue of PP6, Sit4, is essential for the G1-S transition in the cell cycle as well as transcription, translation, and glycogen metabolism (11). In mammals, PP6 targets several proteins, including IκBϵ (10), DNA-PK (12), and TAK1 (8). PP6 dephosphoryates and inactivates TAK1 following IL-1 stimulation, which is likely to be important for preventing prolonged inflammatory responses. We have shown that TAK1 interacts with PP6; however, the mechanism by which PP6 is recruited to TAK1 complexes has not been determined.
TAK1-binding protein 2 (TAB2) is one of three proteins that are constitutively bound to TAK1. TAK1-binding protein 1 (TAB1) acts as the activation subunit of the TAK1 complex, aiding in the autophosphorylation of TAK1 (13), whereas TAB2 and the closely related protein, TAB3, are adaptors of TAK1 that facilitate the assembly of an active TAK1 complex (6, 7, 14). TAB2 contains three conserved domains: a CUE domain that directly binds to ubiquitin (15), a coiled-coil domain involved in the interaction with TAK1 (14), and a zinc finger domain that is involved in polyubiquitin binding (6). TAB2 and TAB3 bind the polyubiquitin chain and TAK1, thereby bringing TAK1 in close proximity to the polyubiquitinated proteins RIP1 and TRAF and the IKK complex (5, 6, 16). Previous reports have shown that TAB2 is not essential for IL-1 signaling pathways due to the presence of TAB3, which can play a compensatory role (17, 18). However, TAB2 single knock-out mice are embryonic lethal at embryonic day 13.5 (18), indicating that TAB2 has some unique functions that cannot be compensated by TAB3. In this study, we examined the unique role of TAB2 by analyzing fibroblasts having targeted deletion of the tab2 gene.
We found that, although TNF induced TAK1 activation in TAB2-deficient fibroblasts, the TAB2 deficiency enhanced and significantly prolonged TAK1 activation following TNF stimulation. We show that TAB2 acts to recruit PP6 to the TAK1 complex. We further show that TAB2-dependent engagement of TAK1·PP6 is mediated through the polyubiquitin chain. Collectively, our results indicate that TAB2 is essential for deactivation of TAK1 by recruiting PP6, which is likely to be important for preventing prolonged inflammatory responses.
EXPERIMENTAL PROCEDURES
Cell Culture
Dermis fibroblasts were isolated from map3k7ip3flox/flox (TAB2-floxed mice) and spontaneously immortalized in culture. TAB2 knock-out fibroblasts were generated by infection of a retroviral vector expressing puromycin-resistant gene alone (control) or puromycin-resistant gene together with Cre recombinase (Cre). Control and TAB2-deleted fibroblast clones were selected by exposure to 10 μg/ml puromycin for a period of 2–3 weeks, and individual clones were isolated. Control puromycin-resistant clones were used as wild type (TAB2+/+), and Cre-expressing clones were used as TAB2-deficient (TAB2−/−) fibroblasts. Fibroblasts and 293 cells were cultured in Dulbecco's modified Eagle's medium with 10% bovine growth serum (Hyclone) and penicillin-streptomycin at 37 °C in 5% CO2.
Antibodies
Anti-JNK1 (FL), anti-p38 (N-20), anti-IκBα (C-21), anti-IKKα (H-744), anti-c-Myc (9E10), and anti-NF-κB p65 (C-20) polyclonal antibodies were purchased from Santa Cruz Biotechnology. Anti-phospho-SAPK/JNK (Thr-183/Tyr-185), anti-phospho-p38 (Thr-180/Tyr-182), anti-phospho-IκBα (Ser-32), and anti-phospho-TAK1 (Thr-187) rabbit polyclonal antibodies (Cell Signaling) were used to detect the phosphorylated forms of JNK, p38, IκBα, and TAK1. Anti-FLAG M5 (Sigma), anti-HA.11 (Covance), and anti-T7 (Novagen) were used for overexpression analysis. The anti-PP6 chicken polyclonal antibody was described previously (19). Anti-TAK1, anti-TAB2, and anti-TAB3 antibodies were described previously (2, 7, 14).
Immunoblots
Whole cell extracts were prepared using an extraction buffer containing 20 mm HEPES (pH 7.4), 150 mm NaCl, 12.5 mm β-glycerophosphate, 1.5 mm MgCl2, 2 mm EGTA, 10 mm NaF, 2 mm dithiothreitol, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 20 μm aprotinin, 0.5% Triton X-100. Cell extracts were resolved on SDS-PAGE and transferred to Hybond-P membranes (GE Healthcare). The membranes were immunoblotted with various antibodies, and the bound antibodies were visualized with horseradish peroxidase-conjugated antibodies against rabbit or mouse IgG using the ECL Western blotting system (GE Healthcare).
Immunoprecipitation and Kinase Assay
Cells were lysed with the extraction buffer described above containing additionally 20 mm iodoacetamide (Sigma) and 1% Triton X-100. 500 μg of protein was incubated with protein G-Sepharose beads (GE Healthcare) as well as 1 μg of the primary antibody overnight at 4 °C. The beads were washed three times with a wash buffer (20 mm HEPES (pH 7.4), 10 mm MgCl2, 0.5 m NaCl) and once with a kinase buffer (10 mm HEPES (pH 7.4), 1 mm dithiothreitol, 5 mm MgCl2). Some samples were subjected to an in vitro IKK kinase assay as described previously (20).
Electrophoresis Mobility Shift Assay
The binding reaction contained a 32P-radiolabeled NF-κB oligonucleotide probe (Promega), 10 μg of cell extracts, 4% glycerol, 1 mm MgCl2, 0.5 mm EDTA, 0.5 mm dithiothreitol, 50 mm NaCl, 10 mm Tris-HCl (pH 7.5), 500 ng of poly(dI·dC) (GE Healthcare), and 10 μg of bovine serum albumin to a final volume of 15 μl. The reaction mixture was incubated at 25 °C for 30 min, separated by 5% (w/v) polyacrylamide gel, and visualized by autoradiography.
Overexpression Analysis in 293 Cells
293 cells were transfected with the standard calcium chloride method. The plasmids for HA-tagged TAB2, T7-tagged TAK1, and Myc-tagged ubiquitin (Myc-Ub) were described previously (14, 15, 21). Wild type, K48R, and K63R mutant versions of ubiquitin were gifts from Dr. Ze'ev Ronai (Burnham Institute), and FLAG-tagged PP6 was described previously (8). The cells were lysed with the extraction buffer described above, and 500 μg of total protein was used for immunoprecipitation analysis.
GST-TNF Pulldown Assay
The GST-TNF (a gift from Dr. Zhijian Chen, University of Texas, Southwestern) was isolated from Escherichia coli bacteria cultures. The analysis of the TNF receptor complex was performed as described previously (5).
RESULTS
Activation of MAPK and NF-κB by TNF in TAB2-deficient Fibroblasts
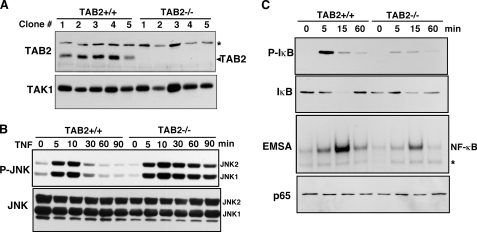
Dermis fibroblasts were isolated from TAB2-floxed mice (18) and immortalized by a standard method. These fibroblasts were exposed to a control or a Cre-expressing retrovirus to delete the tab2 gene. More than 20 independent colonies were isolated and grown in culture. We confirmed tab2 gene deletion in clones used (Fig. 1A) and performed the following experiments using at least three independent clones. We found that MAPK family member JNK was activated in both control and TAB2-deficient fibroblasts following TNF treatment; however, the activation of JNK was prolonged in TAB2−/− cells (Fig. 1B). The IKK-NF-κB pathway was evaluated by IKK kinase assay (supplemental Fig. S1), phosphorylation and degradation of IκB, and electrophoresis mobility shift assay of NF-κB (Fig. 1C). The IKK-NF-κB pathway was activated in response to TNF in control fibroblasts, whereas activation of NF-κB was reduced in TAB2-deficient fibroblasts. We tested activation of JNK and NF-κB in six independent control and TAB2-deficient clones, and we obtained similar results in all clones. We also examined the level of NF-κB target genes, A20 and monocyte chemotactic protein 1 (22) and found that the mRNA levels were increased, but the levels of induction were lower in TAB2-deficient fibroblasts compared with control fibroblasts (data not shown). These results are more or less consistent with the early studies reporting that TAB2 is not essential for TNF-induced TAK1 signaling (17). Our results show that TAB2 is partly involved in activation of NF-κB but is dispensable for activation of JNK. These results suggest that TAK1 can be activated in a TAB2-independent manner.
FIGURE 1.
MAPK and NF-κB pathways are activated in response to TNF independently of TAB2. A, map3k7ip3flox/flox dermis fibroblasts were infected with a control or a Cre-expressing retrovirus. More than 20 independent control or Cre-expressing clones were isolated. The expression levels of TAB2 were confirmed by immunoblot analysis using an anti-TAB2 antibody. Representative immunoblots are shown. Anti-TAK1 immunoblotting was used as a loading control. Asterisk indicates a nonspecific band. B, control (TAB2+/+) and TAB2-deficient (TAB2−/−) fibroblasts were stimulated by TNF (20 ng/ml), and JNK activation was detected by immunoblots with anti-phospho-JNK (upper panel) and anti-JNK (lower panel). C, TAB2+/+ and TAB2−/− fibroblasts were stimulated with TNF (20 ng/ml), and activation of NF-κB was detected by immunoblots with anti-phospho-IκB (top panel) and anti-IκB (second panel) and an NF-κB electrophoresis mobility shift assay (third panel). Total p65 NF-κB levels were determined by anti-p65 immunoblots. Asterisk indicates a nonspecific band.
Activation of TAK1 in TAB2-deficient Fibroblasts
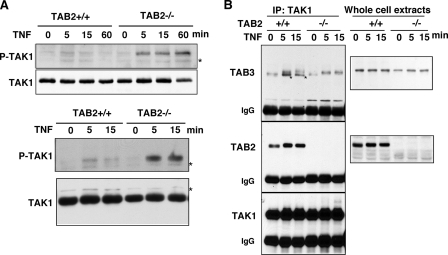
We measured activation of TAK1 by utilizing the phospho-site-specific TAK1 antibody that recognizes only the activated form of TAK1 (8). TAK1 was quickly activated within 5 min following TNF treatment, and the activation was transient as shown in two independent clones (Fig. 2A). In contrast, we found that in TAB2-deficient fibroblasts, TAK1 was not only activated but remained activated for up to 60 min (Fig. 2A). These data indicate that TAB2 is dispensable for activation of TAK1 but may be important for deactivation of TAK1. It is well known that TAB2 and TAB3 redundantly mediate activation of TAK1 (6, 7, 23). Therefore, TAB3 may be sufficient to mediate activation of TAK1 in TAB2-deficient fibroblasts. We examined whether TAK1 binds TAB3 in TAB2-deficient fibroblasts. TAB3 was detected in TAK1 immunoprecipitates in both control and TAB2-deficient fibroblasts (Fig. 2B). The migration of TAB3 in SDS-PAGE was significantly slower after TNF exposure, which is consistent with an earlier observation that TAB3 was phosphorylated concomitantly with TAK1 activation (23). We conclude that TAK1 activation is through TAB3 action in TAB2-deficient fibroblasts.
FIGURE 2.
TNF-induced TAK1 activation is sustained in TAB2-deficient fibroblasts. A, two independent clones of control (TAB2+/+) and TAB2-deficient (TAB2−/−) fibroblasts were stimulated with TNF (20 ng/ml). An activated form of TAK1 was detected by anti-phospho-TAK1 (Thr-187) immunoblotting, and the total amount of TAK1 was determined using an anti-TAK1 antibody. Data are representative of six different clones. Asterisks indicate nonspecific bands. B, TAB2+/+ and TAB2−/− fibroblasts were stimulated with TNF (20 ng/ml), and proteins from cell lysates were immunoprecipitated (IP) with anti-TAK1 antibody. TAB2 and TAB3 co-precipitation was analyzed using anti-TAB2 and anti-TAB3 antibodies. Anti-TAB3 was weakly cross-reacted with TAB2. The bands indicated with asterisks are TAB2 that were reacted with anti-TAB3.
Dephosphorylation of TAK1 Is Impaired in TAB2-deficient Fibroblasts
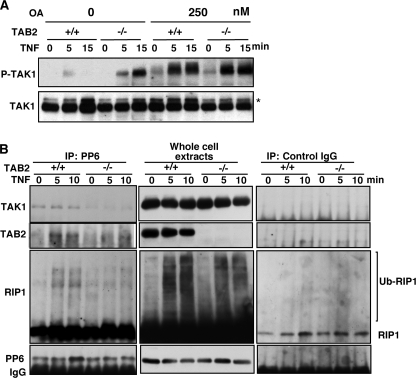
TAK1 is activated by autophosphorylation within its activation loop at Thr-187 and other sites (13, 24). We have previously identified that PP6 dephosphorylates these sites, thereby inactivating TAK1 (8). Kim et al. (25) reported that another type 2A family protein phosphatase PP2A dephosphorylates these sites in some cell types. Therefore, dysregulation of type 2A protein phosphatase-dependent dephosphorylation could be the cause of prolonged activation of TAK1. To determine whether the prolonged activation of TAK1 is due to a defect in dephosphorylation of TAK1, we pretreated control and TAB2-deficient fibroblasts with okadaic acid (OA), a selective inhibitor of type 2 protein phosphatases, including PP6. OA treatment strongly increased phosphorylated TAK1 in control fibroblasts, consistent with our previous report (8). We found that the increased levels of phosphorylated TAK1 by OA treatment was less pronounced in TAB2-deficient fibroblasts, and OA treatment normalized the levels of TAK1 activation in control and TAB2-deficient fibroblasts (Fig. 3A). The results suggest that the prolonged activation of TAK1 in TAB2-deficient fibroblasts might be due to a defect in type 2 protein phosphatase-mediated dephosphorylation of TAK1. Because we observed interaction of PP6 with TAK1 in fibroblasts as shown below, we focused on PP6.
FIGURE 3.
TAB2 is essential for TAK1-PP6 interaction. A, control (TAB2+/+) and TAB2-deficient (TAB2−/−) fibroblasts were pretreated with 250 nm OA for 4 h before TNF treatment (20 ng/ml). Activation of TAK1 was detected by using anti-phospho-TAK1 (Thr-187) antibodies, and the total amounts of TAK1 were determined using an anti-TAK1 antibody. Asterisks indicate nonspecific bands. B, TAB2+/+ and TAB2−/− fibroblasts were stimulated with TNF (20 ng/ml), and proteins from cell lysates were immunoprecipitated (IP) with an anti-PP6 antibody or control IgG. TAK1, TAB2, and RIP1 co-precipitation was determined using anti-TAK1, anti-TAB2, and anti-RIP1 antibodies.
Interaction between TAK1 and PP6 Is TAB2-dependent
To examine the mechanism by which TAK1 is dephosphorylated in control fibroblasts, we first looked at the interaction between endogenous TAK1 and PP6 (Fig. 3B and supplemental Fig. S2). In control fibroblasts, TAK1 was co-precipitated with PP6. However, in TAB2-deficient fibroblasts, PP6 association with TAK1 was diminished (Fig. 3B and supplemental Fig. S2). These results indicate that PP6 is associated with the TAK1 complex in a TAB2-dependent manner. RIP1 is known to be ubiquitinated following TNF stimulation (5). Consistently, we detected ubiquitinated RIP1 only in TNF-treated samples. A small amount of unmodified RIP1 was nonspecifically precipitated with control IgG; however, ubiquitinated RIP1 was specifically co-precipitated with PP6 following TNF stimulation in control but not in TAB2-deficient fibroblasts (Fig. 3B). Interaction of TAB2 with PP6 was also increased following TNF stimulation. TAB2 is a ubiquitin-binding protein and known to bind to the RIP1-anchored polyubiquitin chain (6). Taken together, we postulate that TAB2 recruitment of PP6 involves polyubiquitin chains.
PP6 Interacts with Ubiquitin Chains
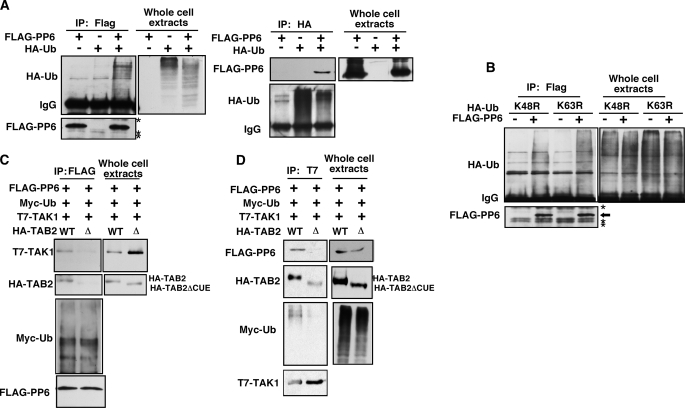
Next, we examined whether PP6 interacts with polyubiquitin chains. Previous studies have shown that TAB2 associates with the RIP1 Lys-63-linked polyubiquitin chain, thereby recruiting TAK1 to the IKK complexes (5, 6). Recently, not only Lys-63-linked but also other types of polyubiquitin chains have been implicated in TNF signaling (26). If PP6 is recruited to the TAK1 complex through TAB2, PP6 might interact with polyubiquitin chains. We exogenously expressed FLAG-tagged PP6 together with wild type, K48R, and K63R mutated versions of HA-tagged ubiquitin (Fig. 4, A and B). Overexpressed HA-Ub spontaneously forms polyubiquitin chains (Fig. 4, A and B, Whole cell extracts). The wild type ubiquitin chain was co-precipitated with PP6 (Fig. 4A). We found that both mutant K48R and K63R ubiquitin chains were co-precipitated with PP6 (Fig. 4B). We note that co-precipitated FLAG-PP6 migrated at a position of at the anticipated protein size (Fig. 4A, top second to right panel). Therefore, FLAG-PP6 was not covalently modified by ubiquitin but interacted with the polyubiquitin chains. We concluded that PP6 interacts with polyubiquitin chains.
FIGURE 4.
Interaction between TAK1 and PP6 is polyubiquitin-dependent. A, 293 cells were transfected with expression vectors for FLAG-tagged PP6 (FLAG-PP6) and HA-tagged ubiquitin (HA-Ub). Cells were lysed, and proteins were immunoprecipitated (IP) with anti-FLAG antibody (left panels) or anti-HA antibody (right panels). Immunoprecipitates and whole cell lysates were analyzed by immunoblotting with an anti-HA antibody and anti-FLAG. Asterisks indicate nonspecific bands. B, 293 cells were transfected with expression vectors for FLAG-tagged PP6 (FLAG-PP6) and HA-tagged K48R or K63R mutant ubiquitin. Cells were lysed, and proteins were immunoprecipitated with anti-FLAG antibody. Immunoprecipitates and whole cell lysates were analyzed by immunoblotting with an anti-HA antibody and anti-FLAG. Asterisks indicate nonspecific bands. C, 293 cells were transfected with expression vectors for T7-tagged TAK1 (T7-TAK1), FLAG-tagged PP6 (FLAG-PP6), Myc-tagged ubiquitin (Myc-Ub), and HA-tagged TAB2 (HA-TAB2) wild-type (WT) or the CUE domain-lacking mutant (Δ). Cells were lysed, and FLAG-PP6 was immunoprecipitated with anti-FLAG antibody. Immunoprecipitates and whole cell lysates were analyzed by immunoblotting with anti-T7, anti-HA, anti-Myc, and anti-FLAG antibodies. D, cell lysates were immunoprecipitated with anti-T7 antibody. The immunoprecipitates and whole cell lysates were analyzed with anti-FLAG, anti-HA, anti-Myc, and anti-T7 antibodies.
Interaction between TAK1 and PP6 Depends on Polyubiquitin
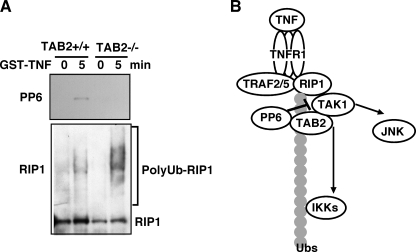
To examine whether TAB2 mediates interaction of TAK1 with PP6 through a ubiquitin-dependent mechanism, we utilized a truncated version of TAB2, TAB2ΔCUE, which is lacking the CUE domain, one of the ubiquitin-binding domains of TAB2 (15). We anticipated that full-length TAB2, but not TAB2ΔCUE, facilitates interaction of TAK1 with PP6. We co-expressed vectors for T7-TAK1, HA-TAB2 or HA- TAB2ΔCUE, FLAG-PP6, and Myc-tagged wild type ubiquitin in 293 cells. TAK1, TAB2, and polyubiquitin were co-precipitated with PP6 (Fig. 4C). However, the interaction between TAK1 and PP6 was reduced in TAB2ΔCUE-expressing cells. In a reciprocal experiment, PP6 and polyubiquitin were co-precipitated with TAK1 in a TAB2 CUE domain-dependent manner (Fig. 4D). These indicate that TAB2 recruits PP6 to the TAK1 complex through polyubiquitin chains. Finally, we examined whether endogenous PP6 is recruited to polyubiquitin chains. Upon TNF stimulation, TNF receptor binds to polyubiquitinated RIP1 and TRAF2/5, which recruit the TAB2·TAK1 complex. Therefore, if PP6 is recruited to the TAK1 complex through polyubiquitin chains, PP6 should be detected in the TNF receptor complex. We utilized a GST-TNF fusion protein to isolate the TNF receptor complex (5). Control and TAB2-deficient fibroblasts were stimulated with GST-TNF, and the proteins from the cell lysates were subjected to a GST pulldown assay (Fig. 5A). PP6 and ubiquitinated RIP1 were precipitated with GST-TNF in control fibroblasts. However, PP6 was not detected in the TNF receptor complex in TAB2-deficient fibroblasts. This suggests that PP6 is associated with the TNF receptor complex through the TAB2-polyubiquitin chains.
FIGURE 5.
PP6 is recruited to TNF receptor complex. A, control (TAB2+/+) and TAB2-deficient (TAB2−/−) fibroblasts were left untreated or stimulated with GST-TNF (1 μg/ml) for 5 min. GST pulldown was performed, and the precipitates were analyzed with anti-PP6 and anti-RIP1 antibodies. B, model.
DISCUSSION
TAB2 and its closely related protein, TAB3, have been identified as essential molecules in TNF and IL-1 signaling pathways. They recruit the TAK1 kinase complex to the TNF and IL-1 signaling complex that activates IKK-NF-κB and MAPK pathways through a ubiquitin chain-dependent mechanism (5–7, 27). TAB2 and TAB3 can bind to ubiquitin through the CUE domain (15) and bind to polyubiquitin chains through the zinc finger domain (6), both of which are important for TAB2/TAB3-mediated NF-κB activation. Multiple types of ubiquitin chains participate in TNF signaling pathways. Lys-48-linked ubiquitination of IκB targets it for degradation and induces activation of NF-κB. Lys-63-linked ubiquitination of RIP1 is important for TNF signaling, which tethers several signaling molecules leading to activation of IKK-NF-κB (5). RIP1 is also modified with a Lys-48-linked ubiquitin chain and degraded at later time point after TNF stimulation, which is one of the negative feed regulatory mechanisms of TNF signaling (28). Recently, a novel type of ubiquitin chain, a linear ubiquitin chain, has been identified as an essential mediator of TNF signaling (29–31). The head-to-tail-linked ubiquitin chain binds to and/or is formed on NEMO, which appears to play an important role in IKK activation. TAB2 and TAB3 bind to the polyubiquitin chains of RIP1 (5) and can interact a linear ubiquitin chain of NEMO (31). Therefore, ubiquitin binding of TAB2 and TAB3 might mediate recruitment of TAK1 to both RIP1 and NEMO. In this article, PP6 is identified as a ubiquitin-binding protein, which is recruited to the TAK1 complex through TAB2. Therefore, ubiquitin binding of TAB2 participates not only in an assembly for activation complex but also in assembly of deactivation signaling complex in TNF signaling pathway (Fig. 5B).
We have found that activation of IKK-NF-κB was reduced in TAB2-defieictent fibroblasts, whereas activation of JNK was prolonged (Fig. 1). JNK activation was very much correlated with the activity of TAK1, whereas IKK activation was diminished even though TAK1 was still activated. As described above, TAB2 is likely to participate in recruitment of not only RIP1 but also of the NEMO·IKK complex. Therefore, we speculate that TAB2 deficiency partly interrupts the close proximity between TAK1 and the NEMO·IKK complex, thereby diminishing IKK activation. On the other hand, TAK1 is likely to engage in an interaction with MAP2K-JNK pathway through a polyubiquitin-independent mechanism as summarized in Fig. 5B.
PP6 binds to polyubiquitin chains in overexpression experiments (Fig. 4, A and B). However, the interaction of polyubiquitinated RIP1 with PP6 is TAB2-dependent (Fig. 3B). Why does endogenous PP6 not bind to the polyubiquitin chain of RIP1 in TAB2-deficient fibroblasts? We speculate that TAB2 may be important to facilitate binding of PP6 to a polyubiquitin chain under physiological conditions (Fig. 5B).
We observed that PP6 was constitutively associated with the TAK1·TAB2 complex (Fig. 3B and supplemental Fig. S2) and that polyubiquitinated RIP1 was further recruited to TAK1·TAB2·PP6 complex following TNF stimulation (Figs. 3B and 5A). We also showed that overexpressed PP6 bound to polyubiquitin chains, and TAB2 facilitated interaction between PP6-polyubiquitin chains and TAK1 (Fig. 4). Taken together, we speculate that the TAK1·TAB2 complex is associated with PP6 through polyubiquitin chains even in unstimulated cells and that the polyubiquitin chains may be different in unstimulated and TNF-stimulated cells. In TNF-stimulated cells, polyubiquitinated RIP1 and TRAF2/5 are presumably the polyubiquitin chains for TAK1·TAB2·PP6 assembly. An unidentified polyubiquitinated protein may function for the basal assembly of TAK1·TAB2·PP6 in unstimulated cells. We assume that this basal TAK1·TAB2·PP6 complex blocks TAK1 activation and that TAK1 is activated by switching polyubiquitin chains to the TNF-induced polyubiquitin chains.
We have demonstrated previously that TAK1 deletion results in accumulation of reactive oxygen species and greatly increases sensitivity to TNF-induced apoptosis (32, 33). Furthermore, it has been shown that TNF-induced JNK and NF-κB pathways are critically involved in cell death and survival (34–37). Therefore, we have tested the TNF-induced cell death in the TAB2-deficient fibroblasts. We have observed that all clones of TAB2-deficient fibroblasts are hypersensitive to TNF-induced apoptosis compared with the wild type fibroblasts.3 Because TAB2 deletion does not impair but rather activates TAK1 as shown in this article, the mechanisms by which TAK1 or TAB2 deletion causes hypersensitivity to TNF-induced apoptosis are completely different. We show here that TAB2-deficient fibroblasts induce less NF-κB but prolong activation of JNK following TNF treatment. NF-κB is known to activate antiapoptotic genes, whereas prolonged activation of JNK is known to be proapoptotic (34–37). Taken together, we speculate that the cause of TNF hypersensitivity in TAB2-deficient fibroblasts is a combination of decreased NF-κB and prolonged activation of JNK. Germ line deletion of TAB2 causes liver degeneration involving cell death during embryogenesis. In fetal liver, TAB2 deficiency may cause hyperactivation of TAK1·JNK and less effective activation of NF-κB resulting in cell death and liver degeneration. Our results demonstrate that TAB2-dependent deactivation of TAK1 is important for TNF signaling, which may be critically involved in cell survival.
Supplementary Material
Acknowledgments
We thank Drs. Ronai and Chen for materials.
This work was supported, in whole or in part, by National Institutes of Health Grant GM068812 (to J. N.-T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
P. Broglie, unpublished observation.
- TAK1
- transforming growth factor β-activated kinase
- MAPK
- mitogen-activated protein kinase
- IL-1
- interleukin 1
- TNF
- tumor necrosis factor
- TRAF
- TNF receptor-associated factor
- RIP1
- receptor-interacting protein 1
- IKK
- IκB kinase
- JNK
- c-Jun N-terminal kinase
- PP6
- protein phosphatase 6
- TAB
- TAK1-binding protein
- HA
- hemagglutinin
- Ub
- ubiquitin
- GST
- glutathione S-transferase
- OA
- okadaic acid.
REFERENCES
- 1.Kawai T., Akira S. (2007) Trends Mol. Med. 13, 460–469 [DOI] [PubMed] [Google Scholar]
- 2.Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. (1999) Nature 398, 252–256 [DOI] [PubMed] [Google Scholar]
- 3.Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 1087–1095 [DOI] [PubMed] [Google Scholar]
- 4.Takaesu G., Surabhi R. M., Park K. J., Ninomiya-Tsuji J., Matsumoto K., Gaynor R. B. (2003) J. Mol. Biol. 326, 105–115 [DOI] [PubMed] [Google Scholar]
- 5.Ea C. K., Deng L., Xia Z. P., Pineda G., Chen Z. J. (2006) Mol. Cell 22, 245–257 [DOI] [PubMed] [Google Scholar]
- 6.Kanayama A., Seth R. B., Sun L., Ea C. K., Hong M., Shaito A., Chiu Y. H., Deng L., Chen Z. J. (2004) Mol. Cell 15, 535–548 [DOI] [PubMed] [Google Scholar]
- 7.Ishitani T., Takaesu G., Ninomiya-Tsuji J., Shibuya H., Gaynor R. B., Matsumoto K. (2003) EMBO J. 22, 6277–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kajino T., Ren H., Iemura S., Natsume T., Stefansson B., Brautigan D. L., Matsumoto K., Ninomiya-Tsuji J. (2006) J. Biol. Chem. 281, 39891–39896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takaesu G., Ninomiya-Tsuji J., Kishida S., Li X., Stark G. R., Matsumoto K. (2001) Mol. Cell Biol. 21, 2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefansson B., Brautigan D. L. (2006) J. Biol. Chem. 281, 22624–22634 [DOI] [PubMed] [Google Scholar]
- 11.Zabrocki P., Van Hoof C., Goris J., Thevelein J. M., Winderickx J., Wera S. (2002) Mol. Microbiol. 43, 835–842 [DOI] [PubMed] [Google Scholar]
- 12.Mi J., Dziegielewski J., Bolesta E., Brautigan D. L., Larner J. M. (2009) PLoS ONE 4, e4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kishimoto K., Matsumoto K., Ninomiya-Tsuji J. (2000) J. Biol. Chem. 275, 7359–7364 [DOI] [PubMed] [Google Scholar]
- 14.Takaesu G., Kishida S., Hiyama A., Yamaguchi K., Shibuya H., Irie K., Ninomiya-Tsuji J., Matsumoto K. (2000) Mol. Cell 5, 649–658 [DOI] [PubMed] [Google Scholar]
- 15.Kishida S., Sanjo H., Akira S., Matsumoto K., Ninomiya-Tsuji J. (2005) Genes Cells 10, 447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. (2001) Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 17.Shim J. H., Xiao C., Paschal A. E., Bailey S. T., Rao P., Hayden M. S., Lee K. Y., Bussey C., Steckel M., Tanaka N., Yamada G., Akira S., Matsumoto K., Ghosh S. (2005) Genes Dev. 19, 2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanjo H., Takeda K., Tsujimura T., Ninomiya-Tsuji J., Matsumoto K., Akira S. (2003) Mol. Cell. Biol. 23, 1231–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefansson B., Brautigan D. L. (2007) Cell Cycle 6, 1386–1392 [DOI] [PubMed] [Google Scholar]
- 20.Kim J.-Y., Omori E., Matsumoto K., Núñez G., Ninomiya-Tsuji J. (2008) J. Biol. Chem. 283, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uemura N., Kajino T., Sanjo H., Sato S., Akira S., Matsumoto K., Ninomiya-Tsuji J. (2006) J. Biol. Chem. 281, 7863–7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixit V. M., Green S., Sarma V., Holzman L. B., Wolf F. W., O'Rourke K., Ward P. A., Prochownik E. V., Marks R. M. (1990) J. Biol. Chem. 265, 2973–2978 [PubMed] [Google Scholar]
- 23.Mendoza H., Campbell D. G., Burness K., Hastie J., Ronkina N., Shim J. H., Arthur J. S., Davis R. J., Gaestel M., Johnson G. L., Ghosh S., Cohen P. (2008) Biochem. J. 409, 711–722 [DOI] [PubMed] [Google Scholar]
- 24.Singhirunnusorn P., Suzuki S., Kawasaki N., Saiki I., Sakurai H. (2005) J. Biol. Chem. 280, 7359–7368 [DOI] [PubMed] [Google Scholar]
- 25.Kim S. I., Kwak J. H., Wang L., Choi M. E. (2008) J. Biol. Chem. 283, 10753–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu M., Skaug B., Zeng W., Chen Z. J. (2009) Mol. Cell 36, 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z. J., Sun L. J. (2009) Mol. Cell 33, 275–286 [DOI] [PubMed] [Google Scholar]
- 28.Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., Dixit V. M. (2004) Nature 430, 694–699 [DOI] [PubMed] [Google Scholar]
- 29.Komander D., Reyes-Turcu F., Licchesi J. D., Odenwaelder P., Wilkinson K. D., Barford D. (2009) EMBO Rep. 10, 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 31.Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., Iwai K. (2009) Nat. Cell Biol. 11, 123–132 [DOI] [PubMed] [Google Scholar]
- 32.Omori E., Morioka S., Matsumoto K., Ninomiya-Tsuji J. (2008) J. Biol. Chem. 283, 26161–26168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omori E., Matsumoto K., Sanjo H., Sato S., Akira S., Smart R. C., Ninomiya-Tsuji J. (2006) J. Biol. Chem. 281, 19610–19617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 35.Sakon S., Xue X., Takekawa M., Sasazuki T., Okazaki T., Kojima Y., Piao J. H., Yagita H., Okumura K., Doi T., Nakano H. (2003) EMBO J. 22, 3898–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pham C. G., Bubici C., Zazzeroni F., Papa S., Jones J., Alvarez K., Jayawardena S., De Smaele E., Cong R., Beaumont C., Torti F. M., Torti S. V., Franzoso G. (2004) Cell 119, 529–542 [DOI] [PubMed] [Google Scholar]
- 37.Ventura J. J., Cogswell P., Flavell R. A., Baldwin A. S., Jr., Davis R. J. (2004) Genes Dev. 18, 2905–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.