Abstract
PDK4 (pyruvate dehydrogenase kinase 4) regulates pyruvate oxidation through the phosphorylation and inhibition of the pyruvate dehydrogenase complex (PDC). PDC catalyzes the conversion of pyruvate to acetyl-CoA and is an important control point in glucose and pyruvate metabolism. PDK4 gene expression is stimulated by thyroid hormone (T3), glucocorticoids, and long chain fatty acids. The effects of T3 on gene expression in the liver are mediated via the thyroid hormone receptor. Here, we have identified two binding sites for thyroid hormone receptor β in the promoter of the rat PDK4 (rPDK4) gene. In addition, we have investigated the role of transcriptional coactivators and found that the PGC-1α (peroxisome proliferator-activated receptor γ coactivator) enhances the T3 induction of rPDK4. Following T3 administration, there is an increase in the association of PGC-1α with the rPDK4 promoter. Interestingly, this increased association is with the proximal rPDK4 promoter rather than the distal region of the gene that contains the T3 response elements. Administration of T3 to hypothyroid rats elevated the abundance of PGC-1α mRNA and protein in the liver. In addition, we observed greater association of PGC-1α not only with the rPDK4 gene but also with phosphoenolpyruvate carboxykinase and CPT-1a (carnitine palmitoyltransferase 1a) genes. Knockdown of PGC-1α in rat hepatocytes reduced the T3 induction of PDK4, PEPCK, and CPT-1a genes. Our results indicate that T3 regulates PGC-1α abundance and association with hepatic genes, and in turn PGC-1α is an important participant in the T3 induction of selected genes.
Keywords: Gene/Promoters, Gene/Transcription, Hormones/Steroid, Receptors/Steroid/Thyroid, Transcription/Coactivators, PEPCK, Pyruvate Dehydrogenase Kinase, Thyroid Hormone
Introduction
Thyroid hormone (T3)2 plays an important role in various aspects of metabolism, development, and differentiation of cells (1). T3 mediates its effect on gene expression through binding to the thyroid hormone receptors (TR) (2). TRs belong to the superfamily of nuclear hormone receptors, which are a class of ligand-activated transcriptional regulators (3). There are two major TR isoforms encoded on separate genes, designated as TRα and TRβ (2). TRβ is the most abundant isoform in liver and mediates the hepatic actions of T3 (4, 5). The TR binds to specific DNA sequences known as T3-response elements (TRE), which most commonly contain a direct repeat of the AGGTCA sequence separated by four nucleotides (DR4). TR can bind to these elements in the presence or absence of ligand to mediate positive or negative regulation of T3 target genes (2, 6). Generally, TR binds to the TRE as a heterodimer with the retinoid X receptor (RXR) (7).
Lipid and glucose metabolism are among the many physiological processes that are regulated by thyroid hormone (8, 9). In hepatocytes, T3 increases the expression of a number of genes involved in hepatic lipogenesis, including spot 14, fatty acid transporter protein, and fatty-acid synthase (10, 11). Paradoxically, T3 simultaneously induces genes involved in fatty acid oxidation especially CPT-1a (carnitine palmitoyltransferase-1a) (12). With respect to glucose metabolism, T3 stimulates almost all aspects of carbohydrate metabolism, including enhancing gluconeogenesis through elevating the transcription of key gluconeogenic enzymes, such as glucose-6-phosphatase and phosphoenolpyruvate carboxykinase (PEPCK), as well as key enzymes of glycolysis and NADH utilization, including glyceraldehyde-3-phosphate dehydrogenase and mitochondrial α-glycerol phosphate dehydrogenase (13, 14).
The pyruvate dehydrogenase complex (PDC) catalyzes the irreversible oxidative decarboxylation of pyruvate into acetyl-CoA. Regulating PDC is an important step in fuel selection for energy utilization in animals during different nutritional and hormonal states as the modulation of PDC activity impacts fatty acid as well as pyruvate and glucose metabolism (15). Phosphorylation of the PDC on three serine residues of its E1 subunit inhibits PDC activity (15, 16). The regulatory enzymes involved in this covalent modification include the pyruvate dehydrogenase kinases, which inactivate PDC, and the pyruvate dehydrogenase phosphatases, which activate PDC through dephosphorylation (17). Four isoenzymes of pyruvate dehydrogenase kinase have been identified in mammalian tissues (PDK1, PDK2, PDK3, and PDK4) (16). The abundance of the PDK4 isoform, which is highly expressed in heart, skeletal muscle, and liver, is transcriptionally controlled (18). Expression of the PDK4 gene is increased by T3, glucocorticoids, retinoic acid, and long chain fatty acids (18–21). In this study, we focused on characterizing the mechanistic regulation of the rat PDK4 gene by T3.
TR acts in concert with coactivators and other transcription factors to mediate the gene-specific actions of T3. Initially, coactivators, including the cAMP-response element-binding protein-binding protein (CBP/p300), steroid receptor coactivator, and mediator complex, were found to contribute to the regulation of gene expression by T3 (2, 22). These coactivators upon interacting with the liganded TR promote histone modification and transcription activation (23). We have been investigating the role of the PGC-1α (peroxisome proliferator-activated receptor γ coactivator) in hormone responsiveness. PGC-1α is expressed in brown adipose tissue, heart, skeletal muscle, brain, kidney, and liver (24). A unique aspect of PGC-1α is that its abundance is up-regulated by physiological changes, including exercise, fasting, and cold exposure (25, 26). PGC-1α promotes mitochondrial biogenesis in brown adipose tissue and skeletal muscles (27, 28). Moreover, PGC-1α regulates pyruvate oxidation by activating the expression of the PDK4 gene via interactions with the orphan nuclear receptor-estrogen-related receptor α (ERRα) (29, 30). In the liver, PGC-1α promotes increased expression of genes involved in hepatic fatty acid oxidation (31). In addition, PGC-1α drives the expression of genes involved in hepatic gluconeogenesis via interactions with HNF4α (hepatic nuclear factor-4) and FoxO1 (forkhead transcription factor) (32, 33).
Previous studies demonstrated that PGC-1α can coactivate the TRβ (25, 34). Moreover, previous work from our laboratory found that overexpression of PGC-1α enhanced the induction of CPT-1a by T3 (35). These studies suggested a link between the T3 effect on metabolic processes and PGC-1α. Here, we have addressed the question of whether PGC-1α is a coactivator in the T3 induction of the rat PDK4 gene. We identified two TRE sites in the rPDK4 gene promoter. In addition, we found that PGC-1α is associated with rPDK4 and other metabolic hepatic genes in response to T3 administration. Our data demonstrate that PGC-1α enhances the T3 induction of select genes in the liver.
MATERIALS AND METHODS
Transient Transfection of Luciferase Vectors
Rat PDK4-luciferase constructs (rPDK4-luc) were transiently transfected into HepG2 cells by the calcium phosphate method as described previously (35). Transfections included 2 μg of PDK4-luciferase along with SV40-thyroid hormone receptor β (SV40-TRβ), pSV-PGC-1α, and TK-Renilla. Cells were transfected in Dulbecco's modified Eagle's medium containing 5% calf serum, 5% fetal calf serum and incubated overnight at 37 °C. Cells were changed into Dulbecco's modified Eagle's medium containing no serum and treated with 100 nm T3 for 24 h. After T3 treatment, cells were lysed in Passive Lysis Buffer (Promega). Both luciferase and Renilla activity were measured using the Dual-Luciferase reporter kit (catalog no. E1980, Promega). Protein content in each lysate was determined by the BCA protein assay kit (catalog no. 23225, Pierce). Luciferase activity was corrected for both protein content and Renilla activity to account for cell density and transfection efficiency, respectively. Serial deletions of the rat PDK4 promoter were created by PCR amplification as described previously (36).
Site-directed Mutagenesis of the rPDK4 Promoter
The QuikChange-XL site-directed mutagenesis kit (Stratagene) was used to generate mutations in the TREs in the −1256/+78 rPDK4-luciferase vector. DR4(−1112) was disrupted by changing the GGT in its second hexamer half-site sequence into AAC, whereas DR4(−1173) was altered by changing the three nucleotides GGT in its second repeat into TTC. Site-directed mutagenesis was used to disrupt the ERRα and FOXO1-binding sites in the −1256/+78 rPDK4-luciferase vector. Primers used to introduce these mutations are listed in supplemental Table S1.
Experimental Animals for in Vivo Electroporation
Eight male hypophysectomized rats weighing 125–150 g were purchased from Harlan Sprague-Dawley. These rats received Tekland iodine-deficient diet and water ad libitum and were housed in a reverse-cycle light-controlled room with a 12-h light period followed by a 12-h dark period. The animals were maintained on the iodine-deficient diet for 21 days prior to commencing experimentation to achieve sufficient turnover of thyroxine. Four of the rats were given an initial subcutaneous injection of 1 mg/kg T3 72 h prior to electroporation and an additional injection of 0.25 mg/kg T3 24 h prior to electroporation. Both the in vivo electroporation and the harvesting of liver punches were performed at the mid-dark part of the reverse-light cycle. The animals were euthanized 24 h after surgery. Serum T3 levels, determined by a solid-phase competitive enzyme-linked immunosorbent assay purchased from Calbiotech (Spring Valley, CA), were 0.9 ± 0.6 pg/ml for the hypophysectomized rats and 3.7 ± 0.6 pg/ml for the T3-treated hypophysectomized rats (means ± S.D.).
In Vivo Electroporation and Luciferase Assays
Electroporation was performed as described previously (37) with some modifications. The liver was surgically exposed as described previously. Fifty μl of sterile saline containing 10 μg of wild type rPDK4 promoter linked to firefly luciferase and 0.01 μg of phRL-CMV Renilla (to correct for transfection efficiency) were introduced by subcapsular injection into different regions of the right lobe in duplicate. A six-needle hexagonal array electrode (0.5 cm in diameter) was placed over the area, and six 150-ms pulses of 150 V/cm with 150-ms rests between pulses were administered with a BTX T830 square-wave electroporator (Harvard Apparatus). Similarly, 10 μg of the rPDK4 promoter linked to firefly luciferase with mutations in the TRE-1 site and 0.01 μg of phRL-CMV were introduced in duplicate in the left lobe. Thus, wild type and mutant promoters were compared in duplicate in the same animal. After electroporation, the liver was placed back in the abdomen, and the wound was closed with surgical staples. The rats were given a single subcutaneous injection of ketoprofen (5 mg/kg) for analgesia. The animals returned to normal activities in a few minutes. They were maintained on warm water blankets following surgery.
Twenty-four h after electroporation, the livers were harvested, and rPDK4 promoter activity was determined from the ratio of luminescence from firefly luciferase to Renilla luciferase (transfection control). Six light needle scars easily defined the circular areas where the cells were transfected. These areas were punched out using a 0.5-cm diameter cork borer. The liver tissue, ∼0.1 g, was placed in 600 μl of passive lysis buffer (Promega, Madison, WI) and homogenized with a Polytron tissue disruptor. The lysate was then centrifuged at 16,000 × g for 5 min. The supernatant was assayed for luciferase activity using the Dual-Luciferase assay kit from Promega and a Turner Designs 20/20 luminometer.
Electrophoretic Mobility Shift
Electrophoretic mobility shift assays were conducted by labeling double-stranded oligonucleotides with Klenow enzyme and [α-32P]dCTP. Oligonucleotides contained sequences representing the TREs (see Fig. 2B for oligomer sequences). His-tagged TRβ and RXR proteins were used to determine the ability of TRβ-RXR to bind to either the wild type or the mutated oligomers (13). The protein-DNA binding mixtures contained labeled probe (30,000 cpm) and the purified protein in 80 mm KCl, 25 mm Tris-HCl, pH 7.4, 0.1 mm EDTA, 1 mm dithiothreitol, 10% glycerol, and poly(dI-dC). The binding reactions were resolved on 5% nondenaturing acrylamide gels (80:1, acrylamide/bisacrylamide) in Tris-glycine running buffer (22 mm Tris and 190 mm glycine). The electrophoresis was conducted at 180 V for 80 min at 4 °C.
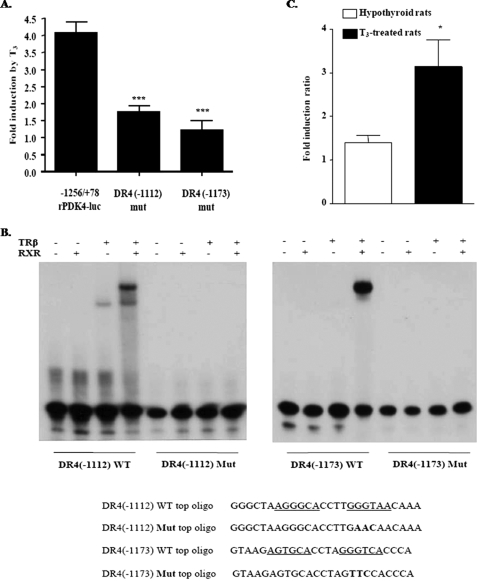
FIGURE 2.
Identification of a thyroid hormone-response element in the rPDK4 promoter. A, HepG2 cells were transiently cotransfected with −1256/+78 rPDK4-luciferase, RSV-TRβ, and TK-Renilla and treated with T3 as described in Fig. 1. Transfections were conducted with −1256/+78 rPDK4-luc in which the DR4 element sites were disrupted. All transfections were performed in duplicate and repeated four times. Results are expressed as the relative induction by T3 ± S.E. by comparing the T3 induction of vectors in the treated cells to the nontreated ones. The significance is calculated relative to the wild type rPDK4 −1256/+78-luciferase (***, p value 0.001 to 0.01). B, ability of TRβ/RXR to bind the two TREs was tested with gel shift mobility assays. Gel shift mobility assay was conducted as described under “Experimental Procedures.” The sequences for the wild type (WT) DR4(−1112) and DR4(−1173) oligomer or the mutated version are shown below the gel shift with the mutated nucleotides shown in boldface, and the sequence for the WT-TRE is underlined. C, hypothyroid rats were transfected in vivo by electroporation of liver as outlined under “Experimental Procedures.” The transfection included the −1256/+78 wild type rPDK4-luciferase or the DR4(−1112) mut rPDK4-luciferase plasmids. CMV-Renilla was included as a control. The values are the average of four hypothyroid untreated rats and four T3-treated rats, and it is represented as fold induction ratio of the wild type rPDK4 luciferase levels to that for the DR4(−1112) mut. *, p value 0.01 to 0.05.
Animals and Treatments for in Vivo ChIP Assays
Adult male Sprague-Dawley rats were housed under controlled conditions (22 °C, constant humidity, 12 h/12 h dark/light cycle) in the animal care facility of the University of Tennessee Health Science Center. Hypothyroidism was induced by propylthiouracil/iodine-free diet (Teklad 95125) for 3 weeks before the experiments. The rats were given a single intraperitoneal injection of T3 (0.33 mg/kg of body weight). Twenty-four h after T3 injection, rats were sacrificed, and livers were isolated for RNA and protein preparation. Thyroid hormone status was verified by measurements of serum T3 levels (hypothyroid, T3 <0.4 μg/liter; hyperthyroid, T3 >6.5 μg/liter).
ChIP Assay
Rat primary hepatocytes were prepared by collagenase perfusion as described previously (38). Rat primary hepatocytes (3 × 106 in 60-mm dishes) were maintained for 12 h in RPMI 1640 media and 10% fetal bovine serum. Following two washes with phosphate-buffered saline, the medium was replaced by RPMI 1640 media without serum. The cells were treated with 100 nm T3 for 24 h. Cross-linking was performed with 1% formaldehyde for 15 min at room temperature and stopped by adding 125 mm glycine to each plate for 5 min. Cross-linked hepatocytes were scraped from the plate and collected by centrifuging for 5 min at 2,000 rpm. ChIP assays were conducted with slight modifications of the protocol provided by the Magna ChIP kit (catalog no. 17-610, Millipore). The cell pellets were resuspended in 200 μl of cell lysis buffer, containing protease inhibitor mixtures (P8340, Sigma). Samples were incubated on ice for 15 min, followed by homogenization using a Dounce homogenizer. The homogenate was pelleted, resuspended in nuclear lysis buffer, and sheared to 400–600 bp with 9 sonications of 30 s, followed by centrifugation at 14,000 rpm for 10 min. Chromatin preparations were immunoprecipitated with the desired antibody or with the control antibody rabbit IgG (sc-2027, Santa Cruz Biotechnology). Antibodies used were anti-TRβ (sc-67122) and anti-PGC-1α (sc-13067). Samples were left rotating overnight at 4 °C with the antibody and the A magnetic beads. The magnetic beads were pelleted and washed with a low salt immune complex wash buffer, high salt immune complex wash buffer, lithium-chloride immune complex wash buffer, and TE buffer. DNA was eluted using the ChIP elution buffer and treated with proteinase K for 4 h at 65 °C followed by 10 min at 95 °C. DNA was purified using the QIAquick PCR purification kit (catalog no. 28104, Qiagen). A total of 3–6 μl of purified sample was used in 35 cycles of PCR. Primers for regions of the target genes are listed in supplemental Table S2. The PCR products were analyzed on 2% NuSieve® 3:1 agarose (catalog no. 50094, Cambrex) and visualized with MultiImage Light Cabinet with Alpha Imager EP software. The relative intensity of the bands was determined using AlphaView version 1.2.0.1 (Alpha Innotech Corp.).
Western Analysis
Western analysis was performed on whole cell extracts from nontreated primary rat hepatocytes or treated with 100 nm T3 for 24 h. Cells were lysed in RIPA buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5% deoxycholate, 5 mm EDTA, pH 8.0, 0.1% SDS, and diluted protease inhibitor mixture). An equal amount of protein was loaded onto a 12% SDS-polyacrylamide gel and transferred to a 0.45-μm pure nitrocellulose membrane (Bio-Rad). Blots were immunoblotted with primary antibodies, anti-PGC-1α (catalog no. sc-13067, Santa Cruz Biotechnology), and anti-PDK4 (catalog no. AP7041b, Abgent), in phosphate-buffered saline containing 5% nonfat dry milk powder and were incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody. Immunoreactive proteins were identified using Super Signal West Femto chemiluminescence substrate (Pierce). The ChemiDocTM XRS gel documentation system (Bio-Rad) was used to quantify the immunoreactive proteins. Actin was used as the loading control for each lane.
Real Time PCR
Real time PCR was used to quantify mRNA abundance. The cDNA was prepared using RNA isolated from primary rat hepatocytes. RNA was isolated with RNA-Stat-60 (Tel-Test) (30). The RNA was then treated with DNase I (2 units) at 37 °C for 1 h followed by addition of DNase inactivation reagent (Ambion). The concentration of each sample containing DNA-free RNA was measured using a NanoDrop machine (Thermo Scientific). Equal amounts of DNA-free RNA were used for first-strand cDNA synthesis. Two μg of DNA-free RNA extracted from different cells was converted to cDNA using Superscript reverse transcriptase III and random hexamers (Invitrogen). The parameters for real time PCR were as follows: 95 °C for 10 min, 40 cycles of 95 °C for 30 s, and 60 °C for 1 min. The final concentration of primers in each well in the PCR plates was 0.1 μm. Four μl of 1:5 of each cDNA was used as a template to assess target genes and normalized with the 18 S gene. The forward and reverse primers used for real time PCR are provided in supplemental Table S3. The primers for PEPCK, fatty-acid synthase, and TRβ were obtained from Qiagen (catalog nos. QT01619975, QT00371210, and QT00193690, respectively).
Adenoviral Infection
Purified adenoviruses encoding shRNA specific for the PGC-1α (Ad-siPGC-1α) and control adenoviruses encoding nontemplate shRNA (Ad-NC) were amplified by ViraQuest Inc. Primary rat hepatocytes were plated at a density of 3 × 106 in a 60-mm dish in RPMI 1640 media (3 plates for each condition). Five hours after plating the cells, media were aspirated, and 2 ml of purified adenoviruses were added to the cells in the presence of 8 μg/ml Polybrene. The media were changed 24 h post-transduction, and the cells were treated with 100 nm T3 for another 24 h in serum-free RPMI 1640 medium. The hepatocytes were harvested, and RNA was isolated using RNA Stat 60.
RESULTS
Identification of Regions within the Rat PDK4 Promoter Required for T3 Responsiveness
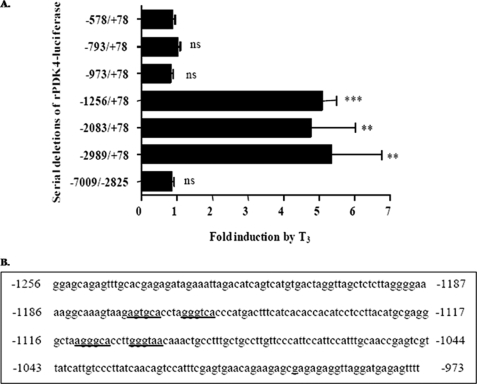
Previous reports indicated that hepatic PDK4 gene expression was stimulated by T3, although the mechanisms by which T3 induced the PDK4 gene were not investigated (20). Our initial experiments were designed to localize the region in the rat PDK4 promoter required for T3 responsiveness. We cotransfected HepG2 hepatoma cells with rPDK4 luciferase reporter genes (rPDK4-luc) containing serial deletions of the PDK4 promoter and an expression vector for TRβ (Fig. 1A). The cells were treated with 100 nm T3 for 24 h. The −1256/+78 luciferase vector was induced 5.1 ± 0.4-fold by T3. Deletion of the nucleotides between −1256 and −973 resulted in complete loss of T3 responsiveness (Fig. 1A). The results are expressed as the relative induction by T3 ± S.E. by comparing the T3 induction of vectors in the T3-treated cells to the nontreated ones. These data suggested that there was a TRE in this region of the rat PDK4 promoter. We identified two potential TREs in the −1256 to −973 region of the rPDK4 promoter. These elements contain direct repeats separated by four nucleotides (DR4) as shown in Fig. 1B.
FIGURE 1.
Thyroid hormone stimulates rPDK4 gene expression. A, HepG2 cells were transiently transfected with 2 μg of different rPDK4 luciferase reporters, 1 μg of RSV-TRβ, and 0.1 μg of TK-Renilla. Transfected cells were incubated either in serum-free Dulbecco's modified Eagle's medium or Dulbecco's modified Eagle's medium containing 100 nm T3 for 24 h. All transfections were performed in duplicate and repeated three to six times. Luciferase and Renilla assays were performed in the same tube. Luciferase activity was corrected for both protein content and Renilla activity. Results are expressed as the relative induction by T3 ± S.E. by comparing the T3 induction of vectors in the treated cells to the nontreated ones. The significance is calculated relative to the shortest construct of the rPDK4-luciferase (−578/+78) (**, p value 0.001 to 0.01; ***, p value <0.001). ns, not significant. B, sequence for the region between −1256 and −973 in the rPDK4 promoter is shown with the potential TREs underlined.
To investigate the importance of these elements in the induction of the rPDK4 gene by T3, we disrupted these DR4 sequences by site-directed mutagenesis (Fig. 2A). HepG2 cells were transfected with the DR4(−1112) mutant −1256/+78, DR4(−1173) mutant −1256/+78, or the wild type −1256/+78 PDK4-luc and tested for T3 responsiveness. Disruption of the DR4 element (−1112) caused a significant reduction in the T3 induction (1.7 ± 0.2-fold, 77% less than the wild type 4.0 ± 0.3-fold, p = 0.0019). In addition, mutating the DR4 element (−1173) reduced the T3 response by 93% (1.2 ± 0.3-fold). To test the ability of TRβ to bind this element in vitro, gel shift mobility assays were conducted with the wild type and mutant oligomers representing the DR4 sites. The results indicated that the TRβ can bind as a heterodimer with RXR to the wild type DR4(−1112) site as well as the DR4(−1173) site. However, TRβ did not bind either mutated TRE (Fig. 2B).
To characterize the role of the TRE(−1112) in vivo, we conducted transfections of the wild type −1256/+78 PDK4-luc vector or the TRE mutant (−1112) into the livers of hypothyroid rats. These luciferase reporters were transfected by electroporation into different lobes in duplicate of the same rat liver (37). Four hypothyroid rats were treated with T3, and four additional hypothyroid rats were left untreated prior to introduction of the luciferase vectors. Following T3 treatment, the expression of the wild type −1256/+78 PDK4 luciferase promoter increased significantly, 3.1 ± 0.6-fold (p = 0.01), in comparison with the TRE mutant promoter, and in nontreated rats the 1.4 ± 0.2-fold induction of the wild type promoter was not significantly different from that of the mutant one (Fig. 2C). These data demonstrated that there was a functional TRE localized within the −1112 to −1096 region of the PDK4 gene promoter and that disruption of this element reduced the ability of the T3 to induce the PDK4 gene in vivo.
Association of the TRβ and PGC-1α with the rPDK4 Gene Promoter
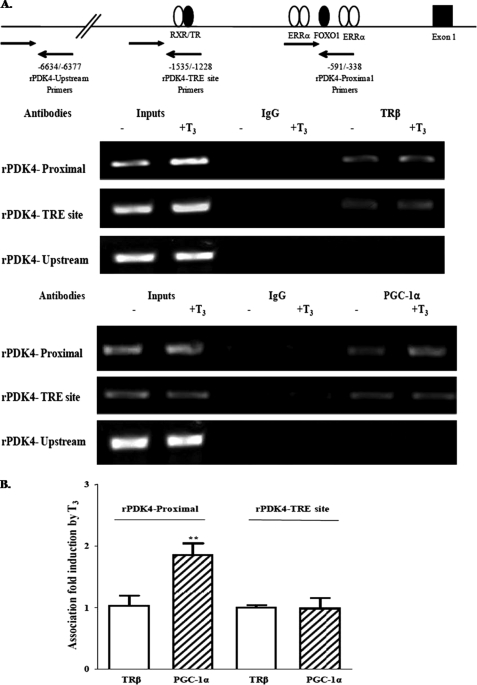
To investigate the role of coactivators in the T3 induction of rat PDK4 expression, we examined the role of PGC-1α that had been shown to stimulate the PDK4 gene (36). To determine whether TRβ and PGC-1α were associated with the rat PDK4 gene in vivo, we conducted ChIP assays (Fig. 3). Rat primary hepatocytes were cultured in serum-free media with or without 100 nm T3 for 24 h. We cross-linked the DNA and proteins in hepatocytes with 1% formaldehyde prior to conducting immunoprecipitations with the TRβ or PGC-1α antibodies. As a control, we immunoprecipitated with rabbit preimmune serum. PCR primers sets were designed to amplify the proximal, the TRE, and the far upstream promoter regions of the rat PDK4 gene as listed in the supplemental material. The upstream primers served as negative controls. The data from the ChIP assays indicated that prior to T3 treatment, the TRβ is associated with both the proximal promoter and the TRE region of the PDK4 gene. However, TRβ was not observed with the upstream region of the PDK4 gene. The addition of T3 to hepatocytes did not increase TRβ binding to any region of the PDK4 gene. In addition, PGC-1α was associated with the proximal promoter and to a lesser extent with the TRE prior to T3 addition. Following treatment of hepatocytes with T3, the association of PGC-1α with the proximal region of the rPDK4 gene increased by 1.85 ± 0.2-fold, but T3 did not increase its association with the TRE region (Fig. 3). These data indicated that only PGC-1α association with the proximal promoter of PDK4 gene is increased following T3 administration.
FIGURE 3.
TRβ and PGC-1α are associated with the rPDK4 gene promoter in vivo. ChIP assays were conducted on primary rat hepatocytes. Hepatocytes were treated with 100 nm T3 for 24 h prior to cross-linking as described under “Experimental Procedures.” A, model of the PDK4 gene and the location of the primers used for the ChIP assay is shown at the top of the figure. Antibodies to TRβ and PGC-1α or immunoglobulin G (IgG) were used for immunoprecipitation. The amplified PCR products using primers for the proximal, TRE, and upstream regions of the rPDK4 gene were resolved on an agarose gel. B, association of TRβ and PGC-1α with PDK4-TRE and -proximal were quantified using Quantity One software. These data are the average ± S.E. of four independent ChIP assays (**, p value 0.001 to 0.01).
Thyroid Hormone Treatment Increases the mRNA of Both the PDK4 and PGC-1α Genes
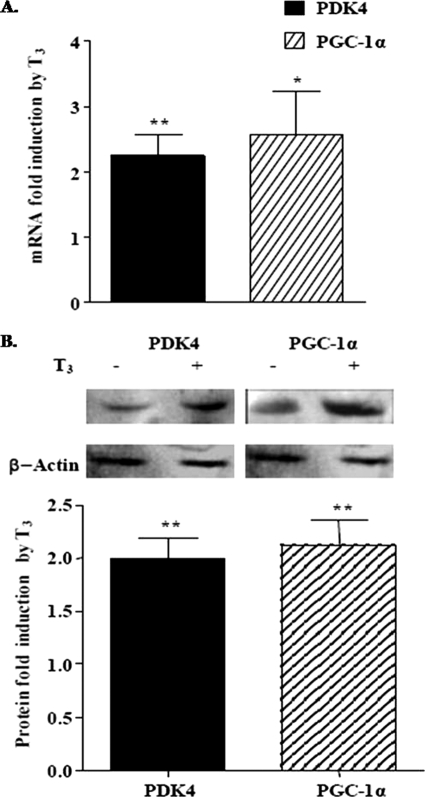
Our next experiments examined the effect of T3 on the abundance of both PDK4 and PGC-1α mRNA and protein in primary rat hepatocytes. Hepatocytes were treated with T3 for 24 h, and then RNA was isolated. Both the PDK4 and the PGC-1α mRNA abundance were increased 2.2 ± 0.3- and 2.6 ± 0.7-fold, respectively, after T3 treatment in rat hepatocytes (Fig. 4A). Using Western analyses, we found that the PDK4 and PGC-1α protein abundance was increased 2 ± 0.2- and 2.2 ± 0.2-fold, respectively, by T3 (Fig. 4B). In addition, we found that PDK2 protein levels were elevated 1.5 ± 0.04-fold by T3 (data not shown). These experiments provide further confirmation that the PGC-1α is a T3-responsive gene and support our hypothesis that PGC-1α is a coactivator in the T3 regulation of the rat PDK4 gene.
FIGURE 4.
Thyroid hormone increases rPDK4 and PGC-1α mRNA and protein abundance. A, primary rat hepatocytes were plated on collagen-coated plates, and T3 was added at a concentration of 100 nm for 24 h. RNA was harvested, and the abundance of rPDK4 and PGC-1α mRNA was determined by real time PCR. The data are presented as the fold induction of mRNA abundance by T3 (average ± S.E.) from four independent hepatocyte preparations. B, expression of the rPDK4 and PGC-1α proteins was monitored by Western blot analysis. The data are presented as the fold induction of protein abundance by T3 (average ± S.E.) from three independent hepatocyte preparations. The control samples were assigned a relative value of 1 (*, p value 0.01 to 0.05; **, p value 0.001 to 0.01).
PGC-1α Enhances the T3 Induction of the rPDK4 Gene
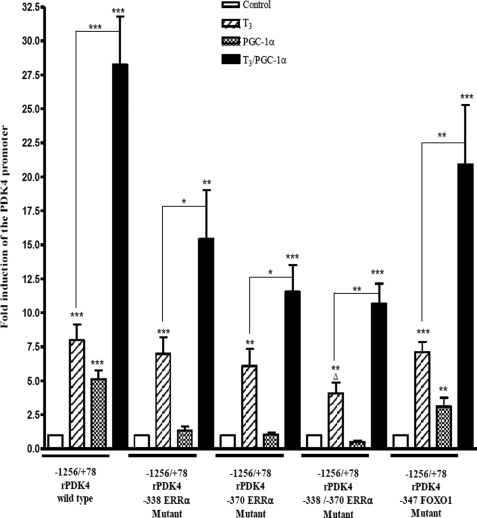
Next, we examined whether PGC-1α could enhance the T3 induction of the PDK4 gene by cotransfecting pSV-PGC-1α with wild type −1256/+78 rPDK4-luc. In addition, we tested rPDK4-luc vectors with mutations in the FoxO1 site at position −347 or the ERRα mutants at positions −338 and −370. The ERR proteins recruit PGC-1α to the PDK4 promoter (30). Addition of 100 nm T3 for 24 h stimulated the wild type version of −1256/+78 rPDK4-luc vector 8.0 ± 1.2-fold, and overexpression of PGC-1α induced it 5.1 ± 0.6-fold. When T3 was added in the presence of PGC-1α, we obtained a synergistic effect with a 28.3 ± 3.5-fold induction. The −338 ERRα mutant, −370 ERRα mutant, and −347 FoxO1 mutant versions of the rPDK4 −1256/+78 all showed similar luciferase induction of 7 ± 1.2-, 6.1 ± 1.3-, and 7.1 ± 0.7-fold, respectively, with T3 treatment. Overexpression of PGC-1α did not induce either of the ERRα mutants. The FoxO1 mutant was only increased 3.1 ± 0.6-fold by T3, which is lower than the wild type (Fig. 5). Surprisingly, when T3 was added in the presence of PGC-1α, we still observed a significant synergistic effect for all the mutant versions of the −1256/+78 PDK4-luc, including a 15.5 ± 3.5-fold for the −338 ERRα mutant, 11.6 ± 2-fold for the −370 ERRα mutant, and 20.1 ± 4.4-fold for the −347 FoxO1 mutant (Fig. 5). Therefore, we created a double mutation of both ERR sites. This double mutant was not induced by PGC-1α but still mediated a reduced synergistic effect with T3. These results indicate that PGC-1α stimulates the PDK4 gene through the −338 to −370 region of the PDK4 promoter. The data also suggest that PGC-1α can act as a coactivator for the T3 induction of the rPDK4 gene in part though association with other factors on the promoter.
FIGURE 5.
PGC-1α enhances the T3 induction of rPDK4 gene. HepG2 hepatoma cells were transiently transfected with 2 μg of different rPDK4-luc constructs, 1 μg of pSV-PGC-1α or pSV, 1.0 μg of RSV-TRβ, and 0.1 μg of TK-Renilla. T3 was added at a concentration of 100 nm for 24 h. All transfections were performed in duplicate and repeated three to six times. Luciferase assays were performed as described in the legend to Fig. 1. Results are expressed as fold induction by PGC-1α or T3 compared with the untreated cells in each data set (*, p value 0.01 to 0.05; **, p value 0.001 to 0.01; ***, p value <0.001; Δ, p value 0.01 to 0.05 between −1256/+78 rPDK4 wild type and −1256/+78 rPDK4 −338/−370 ERRα mutant).
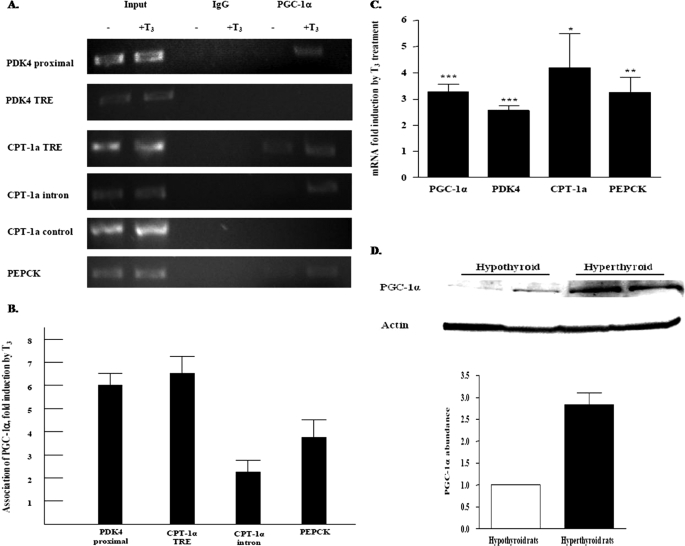
PGC-1α Association with T3-responsive Genes Is Elevated in Hyperthyroid Rats
PGC-1α can stimulate hepatic genes involved in gluconeogenesis and fatty acid oxidation (26). To investigate the role of the PGC-1α in the T3 induction of selected genes, we examined whether there was increased association of PGC-1α with PDK4, CPT-1a, and PEPCK genes in hypothyroid rats treated with T3. Hypothyroid rats were given a single dose of T3, and after 24 h pieces of the liver were analyzed in ChIP assays. We detected PGC-1α in the proximal region of the PDK4 gene promoter, and similarly to the hepatocytes studies in Fig. 3, this association was increased 5.9 ± 0.5-fold by T3 treatment. In contrast with our ChIP experiments in primary hepatocytes, we did not observe any association of PGC-1α with the TRE region of the PDK4 gene promoter. We speculate that this may reflect differences of factor association in the whole liver and primary hepatocytes at the 24-h time point. In addition, we found that there was increased association of PGC-1α with both the first intron and TRE region of the CPT-1a gene promoter following T3 treatment as well as increased association of the PGC-1α with the PEPCK promoter (Fig. 6, A and B). T3 not only promoted the association of PGC-1α with these genes but also significantly elevated the abundance of PGC-1α mRNA (Fig. 6C). In addition, PGC-1α protein levels were induced in the hyperthyroid rats by 2.8 ± 0.3-fold (Fig. 6D). The increased abundance and association of the PGC-1α with different metabolic genes after T3 treatment suggest that PGC-1α participates in the T3 activation of several genes.
FIGURE 6.
PGC-1α association with T3-responsive genes is elevated in T3-treated rats. A, ChIP assays were conducted in liver samples from hypothyroid rats that were untreated or exposed to T3 for 24 h. The ChIP assays were conducted as outlined under “Experimental Procedures.” Various portions of the rPDK4, CPT-1a, and PEPCK genes were analyzed for the binding of PGC-1α. Representative PCRs are shown. B, binding of PGC-1α to the various genes was quantified using Quantity One software. The results are the summary of four independent hypothyroid and T3-treated rats. C, RNA was extracted from the livers and the mRNA abundance of the indicated genes was quantified by real time PCR. D, abundance of PGC-1α protein was quantified using Western blot analysis. The PGC-1α abundance was assessed using protein extracts from the same rats that were used for the ChIP assays and real time PCR.*, p value 0.01 to 0.05; **, p value 0.001 to 0.01; ***, p value <0.001.
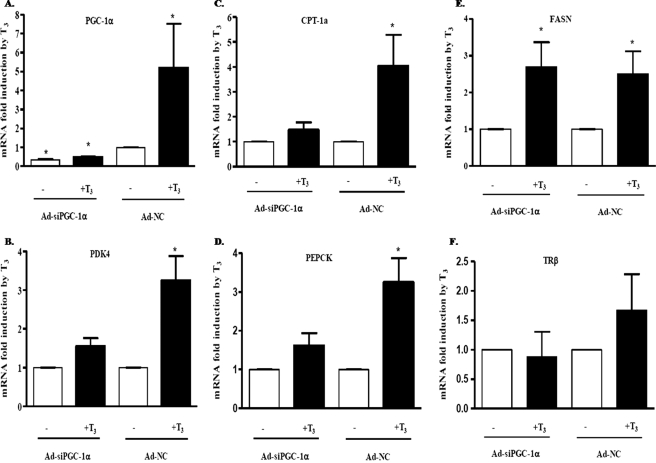
Knockdown of PGC-1α Affects the T3 Induction of Specific Metabolic Genes
To test the role of the PGC-1α in the T3 induction of hepatic metabolic gene expression, we infected rat primary hepatocytes with adenovirus encoding shRNA to silence PGC-1α (Ad-siPGC-1α) prior to treatment of cells with T3. Adenovirus encoding control shRNA, which did not alter other rat genes, was used as a control (Ad-NC). Ad-siPGC-1α significantly decreased the expression of PGC-1α by 70% compared with the Ad-NC in hepatocytes not treated with T3. In addition, the induction of PGC-1α by T3 was reduced in hepatocytes infected with Ad-siPGC-1α. The PGC-1α mRNA in hepatocytes infected with Ad-NC was increased 5.2 ± 2.3-fold by T3 (Fig. 7A). The knockdown of the PGC-1α in primary hepatocytes decreased the T3 induction of PDK4, CPT-1a, and PEPCK, whereas T3 stimulated these genes in hepatocytes infected with Ad-NC by 3.3 ± 0.6-, 4 ± 1.3-, 3.3 ± 0.6-fold, respectively (Fig. 7, B–D). This impairment of the T3 induction for these metabolic genes after PGC-1α silencing indicates that PGC-1α is an important coactivator in the T3 induction process of these genes in primary hepatocytes. To determine whether PGC-1α would increase expression of genes not regulated by PGC-1α, we measured the induction of fatty-acid synthase and the thyroid receptor β (TRβ) mRNA levels by T3. There was no significant change in their induction levels by T3 after knocking down PGC-1α (Fig. 7, E and F). Our data demonstrated that PGC-1α is an important coactivator for the T3 induction of specific genes in the liver.
FIGURE 7.
Effect of PGC-1α knockdown on various T3-responsive genes. Rat primary hepatocytes were infected with adenoviral vectors expressing shRNA for PGC-1α (Ad-siPGC-1α) or expressing control-shRNA (Ad-NC). Hepatocytes were treated with T3 at a concentration of 100 nm for 24 h. The mRNA abundance of the indicated genes was measured by real time PCR. The infections were repeated three to four times on independent plates of cells. A, fold induction of the PGC-1α gene expression in hepatocytes in comparison with hepatocytes infected with Ad-NC and nontreated with T3. B, rPDK4; C, CPT-1a; D, PEPCK; E, fatty-acid synthase (FASN); and F, TRβ mRNA abundance was also measured. The data are expressed as the mean of the fold induction ± S.E. of mRNA abundance relative to cells nontreated with T3 (*, p value 0.01 to 0.05; **, p value 0.001 to 0.01).
DISCUSSION
Pyruvate dehydrogenase kinases decrease PDC activity by phosphorylation, thereby inhibiting the conversion of pyruvate to acetyl-CoA (15). The importance of PDK4 in glucose homeostasis has been demonstrated in PDK4 knock-out (PDK4−/−) mice (39). With fasting, the blood concentrations of glucose, lactate, pyruvate, and alanine were lower in PDK4−/− mice (39). These mice exhibited elevated PDC activity and increased glucose utilization. Moreover, a recent study with PDK4−/− mice found that the knock-out mice, when placed on a high fat diet, had improved glucose tolerance and greater insulin sensitivity (40). Changes in PDK4 abundance in part mediate the long term changes in PDC activity. PDK4 levels are transcriptionally regulated by a variety of hormones and dietary lipids (18, 41). In these studies, we have investigated the mechanisms by which T3 induces PDK4 gene expression in the liver. Our data demonstrate that T3 stimulates PDK4 gene expression in vitro in hepatoma cells and in vivo in hyperthyroid rats. This stimulation is mediated through two TREs in the PDK4 promoter. Previously, we had found that the coactivator PGC-1α induces PDK4 (30). Here, we report that PGC-1α is recruited to the rat PDK4 gene following T3 administration through interactions with ERRα and that PGC-1α is an important participant in the T3 stimulation of PDK4 expression. However, PGC-1α likely contributes to the T3 induction of only a limited subset of T3-responsive genes.
Previous studies found that overall pyruvate dehydrogenase kinase activity was elevated in the liver of hyperthyroid rats (42). Priestman et al. (43) reported that PDK2 protein was increased 1.5-fold in the liver of hyperthyroid rats. Subsequently, Sugden et al. (44) reported that hyperthyroidism induced the expression of the PDK4 but not PDK2 in hearts of rats. In the liver, hyperthyroidism increased both hepatic PDK2 and PDK4 protein levels (20). In addition to elevated pyruvate dehydrogenase kinase activity, there was a suppression of the pyruvate dehydrogenase activity in hyperthyroid rats (20). These results are consistent with our observations regarding the induction of PDK4 protein abundance by T3 and indicate that T3 enhances PDC activity.
PDK4 is highly regulated at the transcriptional level. In addition to T3, the abundance of PDK4 is transcriptionally up-regulated by retinoic acid, glucocorticoids, and long chain fatty acids (18, 19). Multiple nuclear receptors, including the retinoic acid receptor, the glucocorticoid receptor, ERRα, and HNF4 bind the PDK4 gene (18, 30, 36). retinoic acid receptor-α binds within the first 200 bp of the human PDK4 promoter (46). Huang and co-workers (19, 47) reported that in 7800C1 hepatoma cells, glucocorticoids profoundly stimulate PDK4 expression, and a glucocorticoid-response element was identified in the human PDK4 promoter. In addition, high fat diets induce PDK4 expression, but no induction of the PDK4 gene was observed in PPARα knock-out mice (48, 49). However, to our knowledge, a PPARα-binding site has not been identified on the PDK4 promoter. By demonstrating the binding of TR, we have expanded the repertoire of nuclear receptors that interact with the PDK4 gene.
In addition to the transcriptional activation via the binding of liganded nuclear receptors, the actions of these receptors are modulated in part through their interactions with other transcription factors bound to the promoter. These accessory factors and nuclear receptors include hormone-response units. For example, T3 induces expression of the PEPCK gene through a TRE in the proximal promoter, but the CCAAT enhancer-binding protein is required for the activation of PEPCK by T3 (50). With respect to the PDK4 gene, both Sp1 and CBF are needed for the retinoic acid induction of PDK4 as mutation of the binding sites for these factors eliminated the retinoic acid stimulation (46). Along with the liganded glucocorticoid receptor, FoxO1 participates in the glucocorticoid induction of the PDK4 and PEPCK genes (47, 51). Having previously identified ERRα and FoxO1 as regulators of PDK4, we tested whether ERRα and FoxO1 contributed to the T3 induction of PDK4. Mutation of both ERR sites reduced the T3 stimulation of PDK4 suggesting that ERRα participates in T3 action. These observations suggest that different combinations of transcription factors mediate the various hormone responses of the PDK4 gene. We found that HNF4 stimulated the PDK4 gene although HNF4 did not appear to recruit PGC-1α to the PDK4 gene (36). Interestingly, an HNF4 site is located adjacent to the TRE in the PDK4 gene raising the possibility that HNF4 contributes to the T3 induction.
Previous studies had identified links between T3 and PGC-1α. First, PGC-1α can physically interact with and coactivate T3 receptors (25, 34). These interactions occurred via the AF-2 domain of the TRβ and the LXXLL motif (L2) in PGC-1α (34). Additional links between T3 and PGC-1α were reported by Weitzel et al. (52) who showed that the hepatic PGC-1α mRNA expression was induced by T3 treatment of hypothyroid rats. In these studies, it was found that both the PGC-1α and PGC-1β isoforms were rapidly induced within 6 h by T3. Many additional hepatic genes were stimulated 24 h later leading to the suggestion that PGC-1α might contribute to their induction. We also observed that PGC-1α was increased by T3 in primary rat hepatocytes. However, we did not observe any increase in PGC-1α in ventricular myocytes following T3 administration suggesting that the induction of PGC-1α might be limited to the liver (data not shown). It was found by Irrcher et al. (53) that PGC-1α was increased by T3 in the liver and soleus muscle but not the heart. In contrast, it was reported by others that PGC-1α was elevated in the heart of rats treated with thyroxine for 15 days but not at earlier time points (54). Finally, Crunkhorn and Patti (8) examined the potential linkage between PGC-1α and T3 action on mitochondrial genes. It was noted that small interfering RNA to PGC-1α decreased the abundance of TRα in C2C12 cells (8). In our studies, we did not observe any effect of PGC-1α silencing on TRβ levels in hepatocytes suggesting that this may be a tissue-specific action.
We found that overexpression of PGC-1α showed a significant synergistic effect with T3 in inducing PDK4 gene expression. PGC-1α stimulates PDK4 expression via interactions with ERRα (29). In hepatocytes and rat liver, our ChIP assays showed an association of PGC-1α with the PDK4 promoter. The key aspect of these assays was that the association of PGC-1α with the proximal PDK4 promoter was increased in both systems following T3 administration. We observed association of PGC-1α with the TRE region in hepatocytes but not the rat liver possibly reflecting differences in the timing of PGC-1α association with the promoter in primary hepatocytes as opposed to the liver. In luciferase assays, the T3 and PGC-1α synergism was reduced by mutating the ERRα-binding sites, suggesting that PGC-1α participates in T3 action not only via interactions with ERRα but also by interacting with liganded TRβ or additional factors. Our ChIP assays indicated that T3 increased PGC-1α association with other T3 responsiveness genes further implicating this coactivator in T3 action.
Adenovirus-mediated silencing of PGC-1α was utilized to investigate its role in the T3 induction of PDK4 and other metabolic genes. Silencing PGC-1α decreased the T3 stimulation of PDK4, CPT-1a, and PEPCK gene expression, but it did not affect the induction of fatty-acid synthase and TRβ mRNA. These results suggest that whereas PGC-1α is an important coactivator for the T3 induction of selected genes in hepatocytes, genes and pathways that are not normally affected by PGC-1α will not utilize this coactivator in the T3 response. We reported that PGC-1α participated in the T3 induction of the CPT-1a gene (35). These previous observations, which were based primarily on overexpression approaches, were confirmed by the knockdown of PGC-1α. It was reported by other investigators that the T3 induction of gene expression was independent of PGC-1α (45). However, these studies were conducted in GC-1 cells, and we speculate that the interrelationship we have observed with T3 and PGC-1α may occur in the liver and with a limited group of genes. Interestingly, CPT-1a is induced in the liver by T3 but not in the heart (12). On the other hand, PDK4 is increased in both the heart and the liver by T3 suggesting that PDK4 and CPT-1a use different repertoires of transcription factors to respond to T3.
Based on our studies, we conclude that T3 increases the abundance of the PGC-1α and that more PGC-1α will be available to stimulate expression of PDK4 and other target genes. PGC-1α can induce the PDK4 gene expression in part through the interaction with the ERRα at the proximal part of the PDK4 promoter (29, 30). Finally, our data suggest that PGC-1α is an important coactivator for the T3 activation of some hepatic genes.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK0059368 (to E. A. P.), DK075504 (to M. B. E.), and DK075414 (to G. C. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3.
- T3
- thyroid hormone
- ChIP
- chromatin immunoprecipitation assay
- DR4
- direct repeat separated by 4 nucleotides
- ERR
- estrogen-related receptor
- HNF4
- hepatic nuclear factor 4
- PDC
- pyruvate dehydrogenase complex
- PDK
- pyruvate dehydrogenase kinase
- PEPCK
- phosphoenolpyruvate carboxykinase
- RXR
- retinoid X receptor
- TR
- thyroid hormone receptor
- TRE
- T3-response element
- shRNA
- short hairpin RNA.
REFERENCES
- 1.Yen P. M. (2001) Physiol. Rev. 81, 1097–1142 [DOI] [PubMed] [Google Scholar]
- 2.Zhang J., Lazar M. A. (2000) Annu. Rev. Physiol. 62, 439–466 [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazar M. A. (1993) Endocr. Rev. 14, 184–193 [DOI] [PubMed] [Google Scholar]
- 5.Yen P. M., Ando S., Feng X., Liu Y., Maruvada P., Xia X. (2006) Mol. Cell. Endocrinol. 246, 121–127 [DOI] [PubMed] [Google Scholar]
- 6.Rastinejad F., Perlmann T., Evans R. M., Sigler P. B. (1995) Nature 375, 203–211 [DOI] [PubMed] [Google Scholar]
- 7.Glass C. K. (1994) Endocr. Rev. 15, 391–407 [DOI] [PubMed] [Google Scholar]
- 8.Crunkhorn S., Patti M. E. (2008) Thyroid 18, 227–237 [DOI] [PubMed] [Google Scholar]
- 9.Heimberg M., Olubadewo J. O., Wilcox H. G. (1985) Endocr. Rev. 6, 590–607 [DOI] [PubMed] [Google Scholar]
- 10.Ness G. C., Zhao Z. (1994) Arch. Biochem. Biophys. 315, 199–202 [DOI] [PubMed] [Google Scholar]
- 11.Lopez D., Ness G. C. (2006) Biochim. Biophys. Acta 1761, 492–500 [DOI] [PubMed] [Google Scholar]
- 12.Mynatt R. L., Park E. A., Thorngate F. E., Das H. K., Cook G. A. (1994) Biochem. Biophys. Res. Commun. 201, 932–937 [DOI] [PubMed] [Google Scholar]
- 13.Park E. A., Song S., Olive M., Roesler W. J. (1997) Biochem. J. 322, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno M., de Lange P., Lombardi A., Silvestri E., Lanni A., Goglia F. (2008) Thyroid 18, 239–253 [DOI] [PubMed] [Google Scholar]
- 15.Harris R. A., Bowker-Kinley M. M., Huang B., Wu P. (2002) Adv. Enzyme Regul. 42, 249–259 [DOI] [PubMed] [Google Scholar]
- 16.Patel M. S., Korotchkina L. G. (2001) Exp. Mol. Med. 33, 191–197 [DOI] [PubMed] [Google Scholar]
- 17.Holness M. J., Sugden M. C. (2003) Biochem. Soc. Trans. 31, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 18.Kwon H. S., Harris R. A. (2004) Adv. Enzyme Regul. 44, 109–121 [DOI] [PubMed] [Google Scholar]
- 19.Huang B., Wu P., Bowker-Kinley M. M., Harris R. A. (2002) Diabetes 51, 276–283 [DOI] [PubMed] [Google Scholar]
- 20.Holness M. J., Bulmer K., Smith N. D., Sugden M. C. (2003) Biochem. J. 369, 687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris R. A., Huang B., Wu P. (2001) Adv. Enzyme Regul. 41, 269–288 [DOI] [PubMed] [Google Scholar]
- 22.Lonard D. M., O'Malley B. W. (2006) Cell 125, 411–414 [DOI] [PubMed] [Google Scholar]
- 23.Rosenfeld M. G., Lunyak V. V., Glass C. K. (2006) Genes Dev. 20, 1405–1428 [DOI] [PubMed] [Google Scholar]
- 24.Finck B. N., Kelly D. P. (2006) J. Clin. Invest. 116, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. (1998) Cell 92, 829–839 [DOI] [PubMed] [Google Scholar]
- 26.Lin J., Handschin C., Spiegelman B. M. (2005) Cell Metab. 1, 361–370 [DOI] [PubMed] [Google Scholar]
- 27.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 28.Patti M. E., Butte A. J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., Landaker E. J., Goldfine A. B., Mun E., DeFronzo R., Finlayson J., Kahn C. R., Mandarino L. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wende A. R., Huss J. M., Schaeffer P. J., Giguère V., Kelly D. P. (2005) Mol. Cell. Biol. 25, 10684–10694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Ma K., Sadana P., Chowdhury F., Gaillard S., Wang F., McDonnell D. P., Unterman T. G., Elam M. B., Park E. A. (2006) J. Biol. Chem. 281, 39897–39906 [DOI] [PubMed] [Google Scholar]
- 31.Leone T. C., Lehman J. J., Finck B. N., Schaeffer P. J., Wende A. R., Boudina S., Courtois M., Wozniak D. F., Sambandam N., Bernal-Mizrachi C., Chen Z., Holloszy J. O., Medeiros D. M., Schmidt R. E., Saffitz J. E., Abel E. D., Semenkovich C. F., Kelly D. P. (2005) PLoS Biol. 3, e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 33.Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., Newgard C. B., Spiegelman B. M. (2001) Nature 413, 131–138 [DOI] [PubMed] [Google Scholar]
- 34.Wu Y., Delerive P., Chin W. W., Burris T. P. (2002) J. Biol. Chem. 277, 8898–8905 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Ma K., Song S., Elam M. B., Cook G. A., Park E. A. (2004) J. Biol. Chem. 279, 53963–53971 [DOI] [PubMed] [Google Scholar]
- 36.Ma K., Zhang Y., Elam M. B., Cook G. A., Park E. A. (2005) J. Biol. Chem. 280, 29525–29532 [DOI] [PubMed] [Google Scholar]
- 37.Lagor W. R., Heller R., de Groh E. D., Ness G. C. (2007) Exp. Biol. Med. 232, 353–361 [PubMed] [Google Scholar]
- 38.Deng X., Cagen L. M., Wilcox H. G., Park E. A., Raghow R., Elam M. B. (2002) Biochem. Biophys. Res. Commun. 290, 256–262 [DOI] [PubMed] [Google Scholar]
- 39.Jeoung N. H., Wu P., Joshi M. A., Jaskiewicz J., Bock C. B., Depaoli-Roach A. A., Harris R. A. (2006) Biochem. J. 397, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeoung N. H., Harris R. A. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E46–E54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowker-Kinley M. M., Davis W. I., Wu P., Harris R. A., Popov K. M. (1998) Biochem. J. 329, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugden M. C., Fryer L. G., Priestman D. A., Orfali K. A., Holness M. J. (1996) Mol. Cell. Endocrinol. 119, 219–224 [DOI] [PubMed] [Google Scholar]
- 43.Priestman D. A., Donald E., Holness M. J., Sugden M. C. (1997) FEBS Lett. 419, 55–57 [DOI] [PubMed] [Google Scholar]
- 44.Sugden M. C., Langdown M. L., Harris R. A., Holness M. J. (2000) Biochem. J. 352, 731–738 [PMC free article] [PubMed] [Google Scholar]
- 45.Wulf A., Harneit A., Weitzel J. M. (2007) Mol. Cell. Endocrinol. 270, 57–63 [DOI] [PubMed] [Google Scholar]
- 46.Kwon H. S., Huang B., Ho Jeoung N., Wu P., Steussy C. N., Harris R. A. (2006) Biochim. Biophys. Acta 1759, 141–151 [DOI] [PubMed] [Google Scholar]
- 47.Kwon H. S., Huang B., Unterman T. G., Harris R. A. (2004) Diabetes 53, 899–910 [DOI] [PubMed] [Google Scholar]
- 48.Sugden M. C., Bulmer K., Gibbons G. F., Holness M. J. (2001) Arch. Biochem. Biophys. 395, 246–252 [DOI] [PubMed] [Google Scholar]
- 49.Sugden M. C., Bulmer K., Gibbons G. F., Knight B. L., Holness M. J. (2002) Biochem. J. 364, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park E. A., Song S., Vinson C., Roesler W. J. (1999) J. Biol. Chem. 274, 211–217 [DOI] [PubMed] [Google Scholar]
- 51.Wang J. C., Strömstedt P. E., Sugiyama T., Granner D. K. (1999) Mol. Endocrinol. 13, 604–618 [DOI] [PubMed] [Google Scholar]
- 52.Weitzel J. M., Radtke C., Seitz H. J. (2001) Nucleic Acids Res. 29, 5148–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irrcher I., Adhihetty P. J., Sheehan T., Joseph A. M., Hood D. A. (2003) Am. J. Physiol. Cell Physiol. 284, C1669–C1677 [DOI] [PubMed] [Google Scholar]
- 54.Goldenthal M. J., Weiss H. R., Marín-García J. (2004) Mol. Cell. Biochem. 265, 97–106 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.