Abstract
Nonribosomal peptide synthetases (NRPSs) are large, multidomain proteins that are involved in the biosynthesis of an array of secondary metabolites. We report the structure of the third adenylation domain from the siderophore-synthesizing NRPS, SidN, from the endophytic fungus Neotyphodium lolii. This is the first structure of a eukaryotic NRPS domain, and it reveals a large binding pocket required to accommodate the unusual amino acid substrate, Nδ-cis-anhydromevalonyl-Nδ-hydroxy-l-ornithine (cis-AMHO). The specific activation of cis-AMHO was confirmed biochemically, and an AMHO moiety was unambiguously identified as a component of the fungal siderophore using mass spectroscopy. The protein structure shows that the substrate binding pocket is defined by 17 amino acid residues, in contrast to both prokaryotic adenylation domains and to previous predictions based on modeling. Existing substrate prediction methods for NRPS adenylation domains fail for domains from eukaryotes due to the divergence of their signature sequences from those of prokaryotes. Thus, this new structure will provide a basis for improving prediction methods for eukaryotic NRPS enzymes that play important and diverse roles in the biology of fungi.
Keywords: Enzymes, Enzymes/Multifunctional, Enzymes/Structure, Methods/X-ray Crystallography, Organisms/Fungi, Protein/Domains
Introduction
The nonribosomal peptide synthetases (NRPSs)4 are large, multimodular enzymes, which are ubiquitous in both bacteria and fungi. These enzymes are involved in the synthesis of a wide array of secondary metabolites that have diverse biological roles, including iron sequestration and antimicrobial, insecticidal, and antiviral activity (1, 2). These compounds have had a very significant impact on human health since the discovery and development of penicillin by A. Fleming, H. Florey, and E. B. Chain in the 1920s and 1930s, and they are currently used as antibiotics, antivirals, immunosuppressants, and antitumor agents in humans. NRPSs synthesize peptides by a multiple carrier thiotemplate mechanism similar to that employed by polyketide synthases and fatty-acid synthases (1, 3). In general, NRPSs are modular, with each module catalyzing the incorporation of one amino acid substrate into the growing peptide. NRPS modules are, in turn, made up of independently folding functional domains that catalyze the individual reactions of peptide synthesis. The adenylation domain selects and activates the amino acid substrate before attaching it to a 4′-phosphopantetheine (4′ppant) prosthetic group on the adjacent peptide carrier protein (PCP) domain.
NRPS adenylation domains are members of the acetyl-CoA synthetase-like superfamily (as defined by SCOP (4)). The corresponding Pfam family (PF00501) (5) contains over 17,000 sequences from ∼1550 different species, including bacteria, archaea, and eukarya. Despite low sequence similarity, this superfamily shows a high degree of structural and functional conservation (6). Members of the superfamily activate carboxylic acids by an adenylation reaction, utilizing ATP and forming an enzyme-bound adenylate intermediate. In most cases, these enzymes subsequently catalyze the formation of a thioester bond to either a CoA molecule or a 4′ppant prosthetic group. Members of the superfamily play roles in the biosynthesis and degradation of an array of important primary and secondary metabolites. Different members of the superfamily activate diverse substrates ranging from very small carboxylic acids such as acetate (7) to very large substrates such as 2-amino-9,10-epoxy-8-oxodecanoic acid (8). Thus, the architecture of the binding pocket used to achieve substrate specificity by individual family members is of significant interest.
Following the publication of the first structure of an NRPS adenylation domain, PheA (the phenylalanine-activating adenylation domain from GrsA, the gramicidin S NRPS from Bacillus brevis (9)), a number of methods for predicting the specificity of NRPS adenylation domains have been developed based on the similarity of the binding pocket residues to those of previously characterized domains with known specificity (10–12). These methods work well for predicting the substrates for adenylation domains from prokaryotic NRPSs but are less successful for both eukaryotic NRPSs (13–15) and prokaryotic domains that activate unusual substrates. The three known structures of NRPS adenylation domains (9, 16, 17) are all of prokaryotic origin (as are all current NRPS domain structures). Furthermore, the previously determined structures of members of the acetyl-CoA synthetase-like superfamily bind and activate small and medium sized substrates, and it is expected that the binding pockets for larger substrates are substantially different (13, 18).
Among the largest substrates of the acetyl-CoA synthetase-like superfamily are the Nδ-acyl-Nδ-hydroxyornithines (AHOs) found in the NRPS-synthesized fungal hydroxamate siderophores. These siderophores chelate iron through the bidentate hydroxamate group of the AHOs (19). One class of fungal hydroxamate siderophore are the ferrichromes, which are made up of three AHO residues and, in most cases, three other proteogenic amino acids (19). The particular acyl group of the AHOs differ between various ferrichromes that have been identified and characterized (19).
A novel NRPS gene, sidN, from the Epichloë-Neotyphodium complex (phylum Ascomycota, family Clavicipitaceae) of grass endophytes has recently been cloned and characterized.5 The NRPS encoded by sidN is a siderophore synthetase with a three-module organization that resembles previously characterized NRPSs (13, 20) that synthesize ferrichromes. The mutualistic relationship between the fungal endophytes of the Epichloë-Neotyphodium complex and the agronomic grasses that they colonize plays a vital role in the pastoral agriculture of many parts of the world by improving the tolerance of grass plants to abiotic and biotic stresses (21, 22). SidN is essential for the maintenance of the mutualistic character of this relationship, and the details of the cloning and characterization of sidN will be reported elsewhere.6
Recently, attempts have been made to predict the specificities of the adenylation domains of siderophore synthetases using methods such as homology modeling and inspection of the predicted substrate binding pocket residues (13, 18). These studies have predicted that the third adenylation domain of the three-module siderophore synthetases bind and activate the AHO residues and that these domains have a larger binding pocket than has previously been characterized (13, 18). However, experimental confirmation of these predictions has been hampered by technical difficulties in producing soluble protein of eukaryotic adenylation domains in heterologous expression systems (13).
We have expressed, purified, and determined the three-dimensional structure of the third adenylation domain of SidN (SidNA3) from Neotyphodium lolii Lp19 at 2.0 Å resolution. We have also biochemically characterized this domain, confirming that SidNA3 adenylates the large nonproteogenic amino acid, Nδ-cis-anhydromevalonyl-Nδ-hydroxy-l-ornithine (cis-AMHO). In addition, this amino acid has been confirmed as a component of the fungal siderophore using mass spectroscopy. This is the first reported structure of a eukaryotic NRPS domain and thus provides details of the architecture of the specificity-determining pocket for eukaryotic adenylation domains. The structure and biochemistry of SidNA3 also extend our knowledge of the large acetyl-CoA synthetase-like superfamily for which there are only a handful of structures despite their diversity and ubiquity in nature.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The SidNA3 domain was amplified from N. lolii Lp19 genomic DNA and cloned using the Gateway® system (Invitrogen) into the pDest17 expression vector (Invitrogen). A two-step nested PCR protocol was used to introduce the sequence for a recombinant tobacco etch virus protease cleavage site into the PCR product to enable removal of the polyhistidine fusion tag (23). The plasmid was transformed into Escherichia coli RosettaTM (DE3) cells by electroporation. Expression was performed at 18 °C in ZYP-5052 autoinduction medium (24). The expressed protein (in 25 mm sodium phosphate, pH 6.5, 500 mm NaCl, 10 mm imidazole, 5 mm β-mercaptoethanol) was purified by immobilized metal affinity chromatography. The polyhistidine fusion tag was removed by digestion with recombinant tobacco etch virus protease. The cleaved SidNA3 protein was purified by subtraction immobilized metal affinity chromatography and then further purified by size exclusion chromatography using a HiLoadTM 16/60 SuperdexTM 75 pg column (GE Healthcare) with 25 mm MES, pH 6.5, 500 mm NaCl, 5 mm β-mercaptoethanol as the running buffer. Seleno-methionine-substituted protein was prepared by the same protocol except that PASM-5052 medium (24) was used as the expression medium.
Mass Spectrometry of the SidN Siderophore
A liquid culture of Epichloë festucae strain Fl1 was grown for ∼2 weeks at 22 °C in modified Mantle media (25), with yeast extract replaced with 0.6 μm thymine. The supernatant was separated by centrifugation and freeze-dried, and 50 g of the dried material was extracted with methanol (500 ml). The extract was evaporated to dryness under reduced pressure, reconstituted in MilliQ water (200 ml), and eluted through a solid phase extraction cartridge (1 g, Isolute ENV+, International Sorbent Technology). The cartridge was washed with water (10 ml), and a siderophore fraction was eluted with methanol (10 ml). The fraction was evaporated to dryness and stored at 0 °C and reconstituted in 1:1 acetonitrile/water with 0.1% formic acid for infusion into a hybrid ion trap Fourier transform ion cyclotron resonance mass spectrometer (LTQ FT, Thermo Fisher Scientific) using static nanospray electrospray ionization in +ve mode. The spray voltage was 5.0 kV, and the capillary temperature was 225 °C. Collision-induced dissociation of isolated parent ions was carried out with a collision energy of 60 units.
Nδ-cis-Anhydromevalonyl-Nδ-hydroxy-l-ornithine Purification
cis-AMHO was obtained by alkaline hydrolysis of fusigen (EMC Microcollections) (26). A solution of fusigen was adjusted to pH 12.0 using 1 n NaOH and incubated at room temperature for 1 h followed by overnight incubation at 4 °C. The solution was neutralized with 1 n HCl. The resulting cis-AMHO was purified by high performance liquid chromatography using a Luna 3-μm C18(2) 100-Å 150 × 3-mm column (Phenomenex) and a water/acetonitrile gradient (no ion-pairing agent) of 0–22% over 20 min using a 0.3 ml min−1 flow rate. Separation was performed on a System Gold instrument (Beckman Coulter), and detection was via absorbance at 220 nm. The cis-AMHO had a retention time of ∼13 min. The cis-AMHO was dried using a Vacufuge Concentrator 5301 (Eppendorf) to remove the acetonitrile and redissolved to the required concentration in water prior to use.
Inorganic Pyrophosphate Release Assay
The release of inorganic pyrophosphate was assayed using an EnzChek® pyrophosphate assay kit (Invitrogen) (27). The assays were carried out at 30 °C in 100-μl volumes in a 96-well microtiter plate. Each reaction contained 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm ATP, 0.25 mm 2-amino-6-mercapto-7-methylpurine ribonucleoside, 1 unit of purine nucleoside phosphorylase, and 0.03 units of inorganic pyrophosphatase, 1 μm SidNA3, 0.2 mm amino acid substrate. Following a 20-min incubation to remove contaminating pyrophosphate and phosphate, the reaction was initiated by addition of the amino acid substrate. The 360 nm absorbance was monitored using an EnVision® 2104 multilabel plate reader (PerkinElmer Life Sciences), reading every 2½ min. The initial rates for steady-state kinetics were measured using the same method except that 0.4 mm 2-amino-6-mercapto-7-methylpurine ribonucleoside and 2 mm ATP were used together with varying amounts of the amino acid substrate, cis-AMHO. For the steady-state kinetics measurements, two replicates were performed for each concentration, and absorbance readings were made every 10 s.
Crystallization
Crystallization was performed using the sitting drop vapor diffusion method at 18 °C. Initial crystallization trials were conducted with drops consisting of 100 nl each of protein (8 mg ml−1 in 25 mm MES, pH 6.5, 200 mm NaCl, 5 mm β-mercaptoethanol, 1 mm MgCl2, 2 mm ATP, and 5 mm CoA) and precipitant solutions. The initial crystals grew in 25.5% polyethylene glycol 4000, 0.17 m ammonium sulfate, and 15% glycerol. Extensive optimization of the crystallization conditions and micro-seeding resulted in diffraction-quality crystals with the best crystals growing in 0.1 m MES, pH 6.5, 13–14% polyethylene glycol 4000, 0.01–0.02 m ammonium sulfate, and 15% glycerol using protein at 5 mg ml−1 in 25 mm MES, pH 6.5, 200 mm NaCl, 5 mm β-mercaptoethanol, 1 mm MgCl2, and 2–5 mm CoA. The crystallization behavior of the seleno-methionine-substituted protein was identical to that of native protein.
Structure Determination
Data collection was performed on flash-cooled crystals at the Stanford Synchrotron Radiation Laboratory, Stanford University, Palo Alto, CA. One high resolution dataset and one multiple wavelength anomalous dispersion dataset from seleno-methionine-substituted protein crystals were collected and used for structure determination. The data were processed with MOSFLM (28) and SCALA (29). AutoSharp (30) was used to determine phase information from multiple wavelength anomalous dispersion. Nineteen seleno-methionine sites were found with a figure of merit for phasing of 0.37. There were two molecules per asymmetric unit, but density was only present for one of the two C-terminal domains.
Automatic model building was carried out using RESOLVE (31) through the PHENIX AutoBuild wizard (32) followed by manual building in Coot (33) and refinement using phenix.refine (34). The high resolution dataset and experimental phases from the multiple wavelength anomalous dispersion dataset were used. Restrained individual coordinates, restrained isotropic B-factors, and TLS parameters were refined with noncrystallographic symmetry restraints applied to the N-terminal domains. For the TLS refinement, three TLS groups consisting of each of the two N-terminal domains and the ordered C-terminal domain were defined.
The final model included a total of 903 residues out of the expected 1124 residues per asymmetric unit (of the missing 221 residues, 136 were from the missing C-terminal domain). Chain A (N-terminal domain only) consisted of residues 13–40, 46–179, and 183–421, and chain B (N- and C-terminal domains) consisted of residues 12–40, 46–180, 183–426, 430–456, 465–482, and 487–535. The single C-terminal domain that was modeled appears to be mobile with weak density and a high average B-factor of 77.9 Å2 compared with 30.9 Å2 for the N-terminal domain in the same molecule (chain B). In the final model, 96.2% of the residues lie in favored regions, and 3.6% of the residues lie within allowed regions of the Ramachandran plot. There is just one Ramachandran outlier in each chain, which is Gly-322. This residue lies at the entrance to the amino acid binding pocket. Data collection and refinement statistics are given in Table 1.
TABLE 1.
X-ray data and refinement statistics
| Data set |
||||
|---|---|---|---|---|
| High resolution | MAD | |||
| Data collection | ||||
| Space group | P1 | P1 | ||
| Cell dimensions | ||||
| a, b, c | 50.6, 75.3, 84.1 Å | 50.7, 75.6, 84.3 Å | ||
| α, β, γ | 114.9, 94.8, 90.2° | 115.1, 94.9, 90.3° | ||
| Peak | Inflection | Remote | ||
|---|---|---|---|---|
| Wavelength | 0.97939 | 0.97939 | 0.97951 | 0.91162 |
| Resolutiona | 43.9 to 2.00 Å (2.1 to 2.00 Å) | 50.4 to 2.55 Å (2.69 to 2.55 Å) | 50.4 to 2.55 Å (2.69 to 2.55 Å) | 50.4 to 2.37 Å (2.50 to 2.37 Å) |
| Completenessa | 96.2% (95.0%) | 97.4% (95.9%) | 97.4% (95.6%) | 97.3% (95.6%) |
| Average I/σIa | 13.2 (3.0) | 14.8 (3.9) | 14.8 (3.7) | 12.0 (2.4) |
| Unique reflectionsa | 72,902 (10,538) | 35,814 (5178) | 35,817 (5179) | 44,562 (6393) |
| Redundancya | 3.7 (3.7) | 3.8 (3.8) | 3.8 (3.8) | 3.8 (3.8) |
| Rsyma,b | 8.4% (55.9%) | 6.8% (28.8%) | 6.9% (30.2%) | 8.5% (46.9%) |
| Refinement | ||||
| Resolution range | 42.6 to 2.0 Å | |||
| No. of reflections | 72,482 | |||
| No. of atoms | ||||
| Protein | 6930 | |||
| Ligand/ions | 12 | |||
| Water | 455 | |||
| R-factorc | 0.194 | |||
| Rfreed | 0.228 | |||
| Wilson B-factor | 24.5 | |||
| Average B-factors (Å2) | ||||
| Protein | 37.0 | |||
| N-terminal A | 30.7 | |||
| N-terminal B | 33.6 | |||
| C-terminal B | 76.4 | |||
| Ligand/ions | 39.8 | |||
| Water | 39.9 | |||
| R.m.s.d. | ||||
| Bond lengths | 0.018 Å | |||
| Bond angles | 1.645° | |||
| Ramachandran analysis (%)e | ||||
| Most favored | 96.2 | |||
| Additionally allowed | 3.6 | |||
| Outliers | 0.2 | |||
a Data in parentheses are for the outermost data shell.
b Rsym = ΣhΣj|I(h;j) − Σ(h)|/ΣhΣjI(h), where I(h;j) is the jth measurement of the intensity of the unique reflection h, and I(h) is the mean over all symmetry-related measurements.
c R-factor = Σ|Fobs − Fcalc|/Fobs, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
d Rfree is the R-factor of the 5% of data (selected randomly) not used in refinement.
e Data are according to the definitions of Lovell et al. (49).
Ligand Docking
The energy-minimized AHO models were constructed using ChemBio3D Ultra (CambridgeSoft). Using GOLD (35), the ligand was docked into an area of the SidNA3 structure with a radius of 7–8 Å centered on the binding pocket and using the “Detect Cavity” option. The “Standard default settings” genetic algorithm parameters and the default parameters of the ChemScore (36) scoring function were used. Ten docking runs were performed for each ligand model, and the top three solutions were kept.
RESULTS
SidNA3 Expression and Purification
Successful expression and purification of the third adenylation domain of SidN (SidNA3) were achieved by making a range of constructs that were tested for soluble protein expression. Optimization of the N- and C-terminal borders of the domain was of particular importance as differences of just 14 amino acids at either end of the construct lead to either soluble or insoluble protein expression (37). Initially, longer constructs of the SidNA3 domain exhibited a strong tendency to form large soluble aggregates, as determined by size exclusion chromatography. Subsequent optimization of the construct borders and of the purification and storage buffer conditions yielded protein that was suitable for biochemical characterization and crystallization. The final 61-kDa SidNA3 protein domain was comprised of residues 2270–2826 of SidN.
Product of SidN
Evidence that SidN incorporates AMHO residues was provided by mass spectrometric examination of the siderophore produced by SidN from E. festucae Fl1. For these studies, E. festucae Fl1 was used as a model organism for producing the siderophore after mass spectroscopy showed that the siderophores produced by sidN from N. lolii Lp19 and E. festucae Fl1 are identical.7 Furthermore, a comparison of the amino acid sequence of the SidNA3 domain from E. festucae Fl1 shows 99% identity to that from N. lolii Lp19. A siderophore-rich fraction was obtained from an extract of the supernatant of an E. festucae Fl1 culture grown under iron-depleted conditions using solid phase extraction (38). The fraction was examined by high resolution Fourier transform mass spectrometry on a hybrid ion-trap ion cyclotron mass spectrometer. On infusion into the electrospray ionization source, an ion of m/z 1083.53405 ([MH]+, C46H75N12O18, calculated mass 1083.53168) was observed. This ion was not detected in samples from cultures of E. festucae Fl1 in which the sidN gene had been knocked out by gene replacement.5 Collision-induced dissociation of this species in the ion-trap spectrometer gave major product ions from neutral losses of 112.05040 (anhydromevalonyl moiety, C6H8O2, calculated mass 112.05188), 130.07472 (hydroxyornithyl moiety, C5H10N2O2, calculated mass 130.07368), and 242.12498 (AMHO moiety, C11H18N2O4, calculated mass 242.12611), and water. These results show that the acyl group of the three AHO residues in the SidN-produced siderophore is anhydromevalonyl. The anhydromevalonyl group can exist as either the cis or trans isomer, but the mass spectrometry results do not provide any information on which isomer is present in the SidN siderophore. Further work to fully characterize the siderophore, using high resolution NMR, is currently underway.
SidNA3 Activates Nδ-cis-Anhydromevalonyl-Nδ-hydroxy- l-ornithine
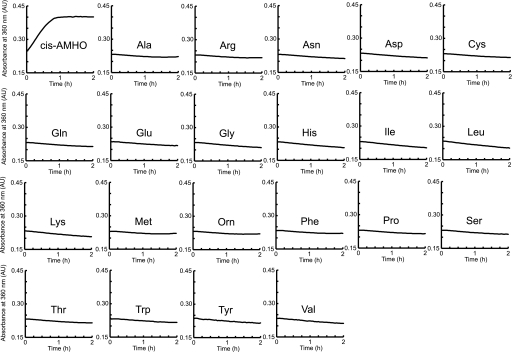
Previous studies have predicted that the third adenylation domain of the three-module siderophore synthetases adenylate the AHO residues (13, 18). To confirm this prediction, a continuous spectroscopic assay (27) that measures the release of inorganic pyrophosphate by the adenylation reaction was employed to investigate the substrate specificity of the SidNA3 domain. cis-AMHO was obtained by hydrolysis of the siderophore fusigen (26), a cyclic trimer of cis-AMHO monomers connected by head-to-tail ester bonds. The activity assays showed that SidNA3 specifically adenylates cis-AMHO but did not adenylate l-ornithine or any of the 20 proteogenic amino acids (Fig. 1). We were unable to source trans-AMHO, and thus we could not investigate whether SidNA3 shows any preference for the cis or trans isomer of the anhydromevalonyl group. By monitoring the steady-state kinetics of pyrophosphate release via this assay, we determined the observed Km and kcat values for adenylation of cis-AMHO to be 40.0 ± 5.0 μm and 0.92 ± 0.04 min−1, respectively. The two auxiliary enzymes used in this enzyme-coupled assay were present in excess and have reported rates that suggest that the adenylation reaction was rate-limiting. Thus, the observed Km and kcat values seen here are likely to reflect the adenylation step catalyzed by SidNA3. This Km value for cis-AMHO and SidNA3 is of the same order of magnitude as the Km values previously determined for other NRPS adenylation domains and their cognate amino acid substrates (39, 40). The low kcat value is the result of the slow catalytic turnover due to slow release of the enzyme-bound adenylate. It is likely that this rate would be greatly enhanced in the presence of the flanking domains of the NRPS, as the 4′ppant prosthetic group on the adjacent PCP domain would immediately react with the adenylated amino acid.
FIGURE 1.
SidNA3 activity assays. Indicative plots of the accumulation of PPi, as monitored by absorbance at 360 nm, for SidNA3 activity using the 20 proteogenic amino acids, cis-AMHO, and ornithine (Orn) as substrates. The assay was conducted three times using different concentrations of enzyme and substrate. In each case, cis-AMHO was the only amino acid that was adenylated. Subsequently, duplicate assays were conducted to calculate observed kcat and Km values (see text for details). AU, absorbance units.
Structure of SidNA3
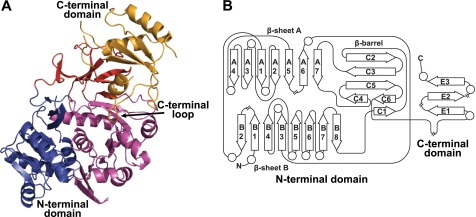
SidNA3 was crystallized by the sitting drop method in the presence of CoA. The structure of SidNA3 was determined by multiple wavelength anomalous dispersion using seleno-methionine-substituted protein. SidNA3 has two domains (Fig. 2), an N-terminal domain (residues 12–421) and a smaller C-terminal domain (residues 422–535) (numbering for SidNA3 used here begins with the first native residue in the expression construct; Fig. 3). The N-terminal domain is made up of a distorted six-stranded antiparallel β-barrel together with an αβαβα structure formed by two predominantly parallel β-sheets (seven- and eight-stranded) flanked by α-helices. The core of the C-terminal domain is made up of a three-stranded antiparallel β-sheet surrounded by three α-helices. A long loop in the C-terminal domain, located between the final β-strand and the final α-helix, loops back toward the N-terminal domain (Fig. 2). The secondary and tertiary structures for each domain are very similar to that of PheA (9), with r.m.s.d. values of 1.6 Å for the N-terminal domain (369 residues aligned) and 1.4 Å for the C-terminal domain (79 residues aligned). Nevertheless, SidNA3 does show some significant differences to the acetyl-CoA synthetase-like superfamily. SidNA3 lacks three α-helices that are found in PheA and, in turn, has an additional two α-helices (Fig. 3). One of the three absent α-helices is at the N terminus, and some of the residues that would be expected to form this α-helix were not included in the SidNA3 construct that was used (Fig. 3). Hence, it is not clear whether this α-helix is present in the native SidNA3 domain. However, the remaining two absent α-helices and one of the two additional α-helices are within the N-terminal domain and are associated with insertions and deletions in the sequence (Fig. 3). The final additional α-helix in SidNA3 is at the C terminus. Apart from PheA, this C-terminal α-helix is present in all of the structures of members of the acetyl-CoA synthetase-like superfamily that contain this region of the protein.
FIGURE 2.
Structure and topology of SidNA3. A, ribbon diagram of the SidNA3 structure. The N-terminal domain is made up of a distorted six-stranded antiparallel β-barrel (red) together with an αβαβα structure formed by two predominantly parallel β-sheets flanked by α-helices (blue and purple). The C-terminal domain (orange) is made up of a three-stranded antiparallel β-sheet surrounded by three α-helices. A long loop in the C-terminal domain, located between the final β-strand and the final α-helix, loops back toward the N-terminal domain and is marked. B, topology diagram of SidNA3. Diagram is drawn in the same style as that of the PheA topology diagram in Conti et al. (9) and using the same numbering scheme for the β-strands. The circles represent α-helices and the arrows β-strands. The locations of the chain breaks (because of poor electron density) in the SidNA3 model are marked.
FIGURE 3.
Structure-based sequence alignment. Structure-based sequence alignment of the protein sequences SidNA3 (top) and PheA (bottom) (9). The positions of the β-strands and α-helices for both structures are shown. The numbering scheme for the β-strands is that used in Conti et al. (9). The residues not present in the models are indicated by lowercase letters, and the first five N-terminal residues of SidNA3, which are cloning artifacts, are shown in gray lettering and given negative numbers. The residues involved in binding the amino acid substrate are shaded (dark gray for the standard PheA residues and light gray for the extra SidNA3 residues).
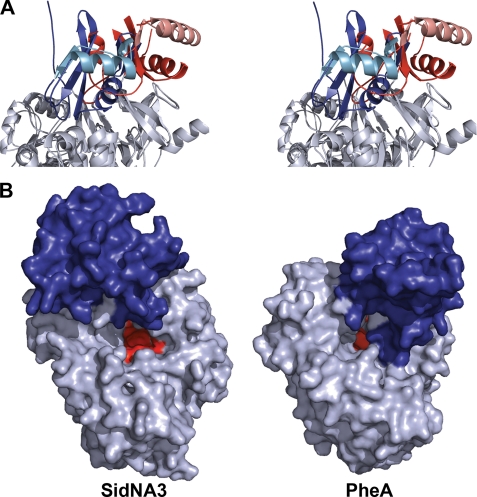
SidNA3 Is in an “Open” Conformation
Most members of the acetyl-CoA synthetase-like superfamily are thought to adopt two distinct conformations to catalyze the two reactions that are performed by these enzymes, the adenylation and thiolation reactions (7, 41). Previous structures show the two conformations differ by an ∼140° rotation of the C-terminal domain relative to the N-terminal domain. SidNA3 was crystallized in a third conformation where the C-terminal domain is rotated ∼45° relative to the orientation seen in the adenylation conformation (Fig. 4A). We describe this as an open conformation as there is a wide separation between the C- and N-terminal domains of SidNA3. There are very few interactions between the two domains or between the C-terminal domain and neighboring molecules in the crystal. The accessibility of the active site, located between the two domains at the surface of the N-terminal domain, is much greater in the open conformation of SidNA3 compared with the adenylation conformation seen in some other structures, such as that of PheA (Fig. 4B).
FIGURE 4.
Orientation of the C-terminal domain for SidNA3. A, stereo ribbon diagram showing the orientation of the C-terminal domain in SidNA3 compared with the adenylation conformation of PheA (9). The C-terminal domain of SidNA3 is red and that of PheA is blue. The N-terminal domains have been aligned and are shown in gray. The second α-helix and subsequent β-strand of the C-terminal domains are colored in lighter shades to aid visualization of the rotation between the conformations. B, molecular surface diagram showing the accessibility of the active sites in the open conformation of SidNA3 compared with the adenylation conformation of PheA (9). The amino acid binding pocket is shown in red, and the C-terminal domain is colored blue.
As would be expected from the lack of molecular interactions between the domains, the C-terminal domain of SidNA3 is not held rigidly in the open conformation and is probably quite mobile, reflected in a high average B-factor (77.9 Å2) compared with the N-terminal domain in the same molecule (30.9 Å2) and has weaker electron density. Electron density is not present for many side chains of residues in this C-terminal domain, and there is no density for several other residues (residues 427–429, 457–464, 483–486, and 536–557). Further support for C-terminal domain mobility is the complete absence of electron density for the C-terminal domain for the second of the two molecules in the asymmetric unit. Significantly, this missing C-terminal domain cannot occupy the same orientation as the ordered C-terminal domain of the other monomer as this would cause clashes with neighboring molecules in the crystal. Similarly, it cannot occupy the adenylation or thiolation conformations previously reported due to the geometry of crystal packing. The missing C-terminal domain is not the result of degradation of the protein as demonstrated by SDS-PAGE and mass spectroscopy (data not shown). The positional disorder of the C-terminal domain makes sense in light of the lack of interactions between the two domains in the open conformation. Similar open conformations with a wide separation between the two domains have been observed in four other members of the superfamily (Protein Data Bank codes 1lci (42), 1ult (43), 2vsq (17), and 3g7s). The orientation of the two domains is different in each of these structures. It is likely that the individual crystal packing arrangements stabilize the exact orientation seen in each of these structures and that the C-terminal domains are all significantly mobile in solution.
Architecture of the SidNA3 Active Site and Substrate-binding Pocket
The ATP and amino acid binding pockets of the active site were identified by comparison with PheA (9). The ATP-binding channel and the amino acid binding pocket are adjacent to each other on the face of the N-terminal domain between it and the C-terminal domain. The ATP binding pocket of SidNA3 shows the archetypal conserved motif 178TSGSTGTPK186, which is similar to the P-loop seen in many ATP-binding proteins (44). This loop is thought to be flexible and has been observed in many conformations in the structures of acetyl-CoA synthetase-like superfamily members. In SidNA3, there is also evidence for flexibility insofar as three amino acids are missing due to poor electron density (Gly-180, Ser-181, and Thr-82).
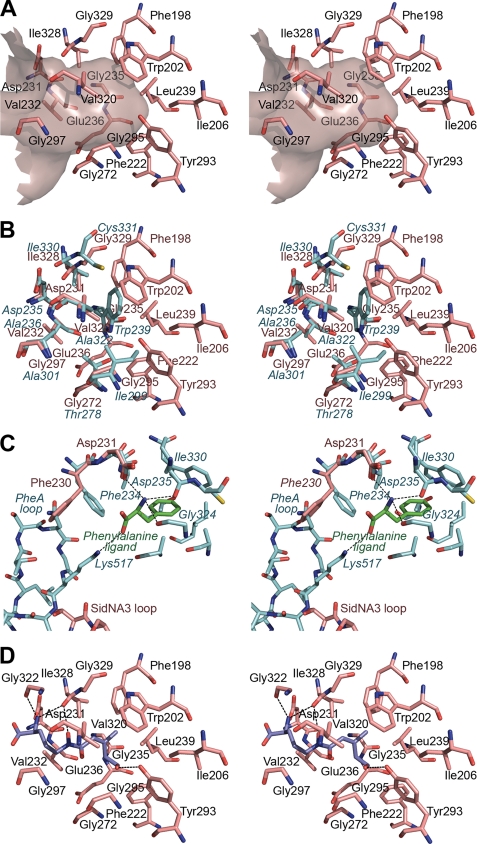
The amino acid binding pocket of SidNA3 is much larger than that of PheA (Fig. 5), consistent with it binding the large amino acid substrate AMHO. In contrast to PheA, the SidNA3 pocket is defined by the main chain and side chains of 17 residues (Phe-198, Trp-202, Ile-206, Phe-222, Asp-231, Val-232, Gly-235, Glu-236, Leu-239, Gly-272, Tyr-293, Gly-295, Val-296, Gly-297, Val-320, Ile-328, and Gly-329). These overlap with the equivalent nine residues that line the PheA substrate binding pocket along with eight additional residues (Phe-198, Trp-202, Ile-206, Phe-222, Glu-236, Leu-239, Tyr-293, and Val-296) that lie deeper in the interior of the N-terminal domain where they bind the hydrophobic anhydromevalonyl moiety. The larger size of the binding pocket in SidNA3 when compared with PheA is the result of the substitution of a tryptophan residue that defines the bottom of the binding pocket (Trp-239 in PheA) with a glycine residue (Gly-235 in SidNA3) (Fig. 5B). There are five glycines in total lining the SidNA3 pocket, giving a large hydrophobic cavity containing only a few potential hydrogen bonding groups.
FIGURE 5.
Amino acid binding pocket of SidNA3. A, stereo “Connolly” surface (48) diagram of the amino acid binding pocket of SidNA3 and the residues lining it. The entrance to the pocket is to the left of the diagram. B, stereo diagram comparing the residues lining the amino acid binding pockets of SidNA3 and PheA (9). The SidNA3 residues are pink, and the PheA residues are blue. The residues of PheA are labeled in blue italics. C, stereo diagram comparing residues involved in binding the main chain atoms of the amino acid ligand in SidNA3 and PheA (9). The SidNA3 residues are pink; the PheA residues are blue, and the phenylalanine ligand of PheA is green. The C-terminal domain loops of both molecules are also shown. The only binding pocket-lining residues shown are those of PheA. The hydrogen bonds between PheA and the phenylalanine ligand are shown. D, stereo diagram of cis-AMHO docked into the binding pocket of SidNA3. The SidNA3 residues lining the binding pocket are pink. The top-ranked solution for the docking of the cis-AMHO ligand is shown in dark blue. The hydrogen bonds between SidNA3 and the docked ligand are shown (a hydrogen bond between the α-amino group of the ligand and Asp-231 is hidden behind the ligand).
Previous homology modeling had predicted that the AHO-adenylating domains of three-module siderophore synthetases would have larger pockets than PheA (13, 18); however, the observed architecture of the SidNA3 pocket differs from that predicted. Schwecke et al. (13) predicted that the binding pockets of AHO-adenylating domains would be lined by three extra residues, beyond the nine seen in the PheA. One of the three predicted extra residues is seen to line the binding pocket of SidNA3 (Phe-222). Similarly, Bushley et al. (18) predicted that seven residues, in addition to the nine seen in PheA, had the potential to contribute to the pocket of AHO-adenylating domains. For SidNA3, two of these predicted residues (Glu-236 and Leu-239) were observed to be lining the pocket. Thus, the experimental determination of the SidNA3 structure provides a new template to advance our understanding of the determinants of substrate binding in NRPSs that adenylate large amino acids and, in particular, those that synthesize hydroxamate siderophores.
Open Conformation of SidNA3 Has Implications for Substrate Binding
In PheA, the α-amino and α-carboxylate groups of the phenylalanine ligand are held in place by interactions with Asp-235 and Lys-517 (which is also involved in binding ATP) (Fig. 5C) (9). The equivalent residue to Asp-235 in SidNA3, Asp-231, is in an almost identical position (Fig. 5C). However, Lys- 517, and its equivalent in SidNA3, Lys-526, are located on a loop that extends from the C-terminal domain into the active site. Due to the different C-terminal domain orientation in the SidNA3 structure, Lys-526 is shifted ∼11 Å from the equivalent position of Lys-517 in the PheA structure. Another consequence of the difference in the position of the C-terminal domain loop is that in the SidNA3 structure the side chain of the conserved aromatic residue Phe-230 is in a different conformation to that of the equivalent residue in PheA, Phe-234. In PheA (9), the C-terminal domain loop restricts the rotation of the side chain of Phe-234, ensuring that it points toward the amino acid binding pocket where it forms a binding surface for the main chain atoms of the phenylalanine ligand, restricting their movement (Fig. 5C). In contrast, in the SidNA3 structure this surface does not exist as the orientation of the C-terminal domain loop allows the Phe-230 side chain to point away from the amino acid binding pocket, opening up the mouth of the pocket (Fig. 5C). To correctly bind the amino acid ligand, a conformational change from the open conformation to the adenylation conformation would be required to move the C-terminal domain loop into a position such that Phe-230 and Lys-526 can take on their roles in ligand binding.
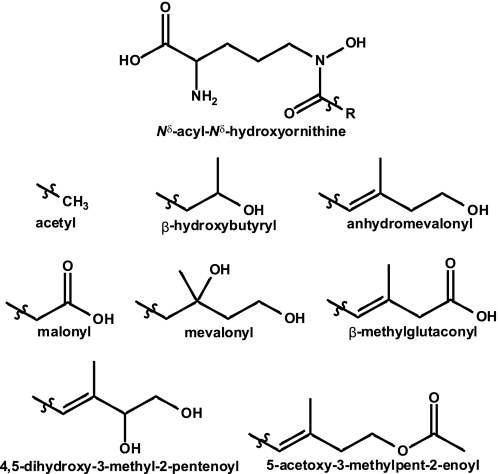
Ligand Docking of Nδ-Acyl-Nδ-hydroxyornithines
To gain insight into the binding specificity of SidNA3, we conducted in silico ligand docking with the SidNA3 structure, using a library of AHOs containing the eight acyl groups that have been observed in siderophores (Fig. 6). Each stereoisomer was modeled separately, resulting in a library of 32 compounds. The ligand docking was carried out with GOLD (35), and binding poses were assessed using the ChemScore (36) scoring function. Consistent with SidNA3 being an AMHO-adenylating domain, the top-ranked ligand from the docking run was cis-AMHO, with Nδ-cis-anhydromevalonyl-Nδ-hydroxy-d-ornithine ranked second (Table 2). Visualization of the binding poses of the top three solutions for the cis-AMHO ligand showed that the ligand is placed in an extended conformation in the binding pocket with all three solutions being well clustered with an r.m.s.d. value of 0.34 Å (Fig. 5D). All of the 10 top-ranked ligands, except Nδ-cis-5-acetoxy-3-methylpent-2-enoyl-Nδ-hydroxy-d-ornithine, showed an identical hydrogen bonding pattern. The terminal hydroxy group of the ligand hydrogen bonds to the side chain hydroxyl of Tyr-293 and the Nδ-hydroxy of the hydroxamate group hydrogen bonds to the main chain carbonyl oxygen of Ile-328 (Fig. 5D). In contrast, the likely hydrogen bond to the carbonyl oxygen of the hydroxamate group remains unsatisfied in all of the docking solutions, which would be energetically unfavorable. The hydroxamate group is able to adopt either the synperiplanar or antiperiplanar conformations (45). This group was allowed to flip during the GOLD docking with the ligand adopting the antiperiplanar conformation in the top-ranked GOLD solutions. In any case, there are no obvious hydrogen bonding partners in the binding pocket for either ligand conformation. However, the pocket is very wide at this point, and it is possible that a water molecule could be involved in satisfying the hydrogen bonding potential of this carbonyl oxygen and bridging between the ligand and main chain atoms. Despite extensive crystallization experiments, we have been unable to determine a structure of SidNA3 with cis-AMHO in the binding pocket.
FIGURE 6.
Nδ-Acyl-Nδ-hydroxy-l-ornithine amino acids found in fungal hydroxamate siderophores. The various R groups are shown below the parent at the top of the figure.
TABLE 2.
Ligands ranked according to in silico docking using GOLD
| Rank | Ligand | Score |
|---|---|---|
| 1 | Nδ-cis-Anhydromevalonyl-Nδ-hydroxy-l-ornithine | 22.35 |
| 2 | Nδ-cis-Anhydromevalonyl-Nδ-hydroxy-d-ornithine | 21.64 |
| 3 | Nδ-trans-Anhydromevalonyl-Nδ-hydroxy-l-ornithine | 20.98 |
| 4 | Nδ-trans-Anhydromevalonyl-Nδ-hydroxy-d-ornithine | 20.31 |
| 5 | Nδ-cis-β-Methylglutaconyl-Nδ-hydroxy-l-ornithine | 19.95 |
| 6 | Nδ-cis-5-Acetoxy-3-methylpent-2-enoyl-Nδ-hydroxy-l-ornithine | 19.05 |
| 7 | Nδ-cis-β-Methylglutaconyl-Nδ-hydroxy-d-ornithine | 19.03 |
| 8 | Nδ-(S)-Mevalonyl-Nδ-hydroxy-l-ornithine | 18.68 |
| 9 | Nδ-(R)-β-Hydroxybutyryl-Nδ-hydroxy-l-ornithine | 18.09 |
| 10 | Nδ-(S)-trans-4,5-Dihydroxy-3-methyl-2-pentenoyl-Nδ-hydroxy-l-ornithine | 18.03 |
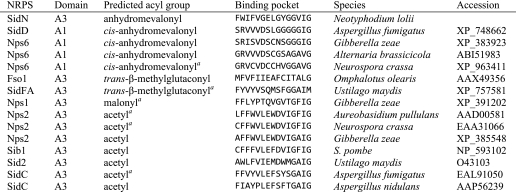
Comparison of the Binding Pockets of Other Siderophore Synthetases
To further investigate the binding of AHOs to the adenylation domains of siderophore synthetases, we compared the pocket-lining residues of a range of domains that adenylate these substrates. AHO-adenylating domains from other fungal siderophore synthetases were identified by searching the relevant literature. A multiple sequence alignment containing SidNA3 and these domains was produced using T-Coffee (46), and 15 residues predicted to line the pocket for these adenylation domains were extracted from the multiple sequence alignment (Table 3). These putative pocket-lining residues were the equivalents of those lining the SidNA3 pocket apart from Asp-231 (invariant in adenylation domains) and Val-296 (only main chain atoms lining the pocket). The domains that adenylate ligands with cis-anhydromevalonyl as the acyl group form a distinct cluster (including, as expected, SidNA3), as do six of the domains that activate ligands with acetyl as the acyl group (Table 3).
TABLE 3.
Amino acid residues lining the binding pockets of Nδ-acyl-Nδ-hydroxy-ornithine-activating adenylation domains
a Data were predicted but experimentally unconfirmed.
Common Binding Pocket Architectures of the Acetyl-CoA Synthetase-like Superfamily
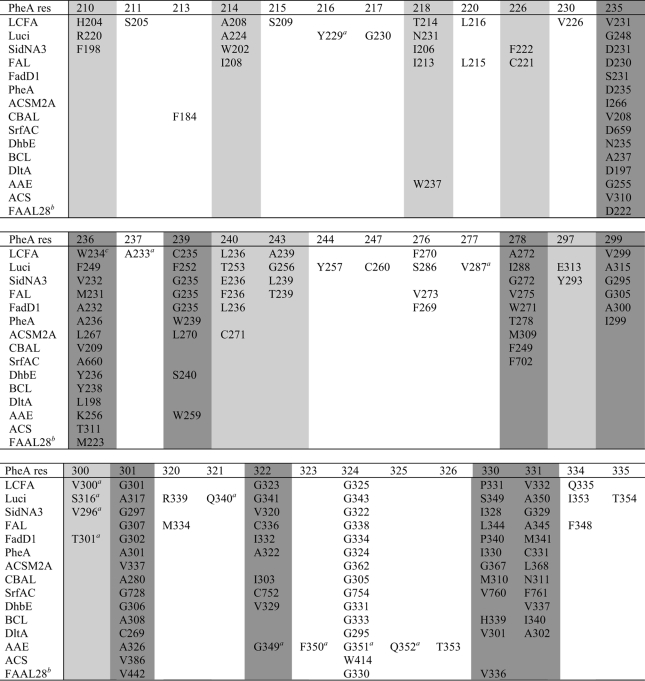
The structures of 18 other members of the acetyl-CoA synthetase-like superfamily are available. To gain insight into the common architecture of the binding pockets of the superfamily, we compared the pockets of these structures with each other and SidNA3 (Table 4). Three of the structures, the long chain fatty acyl-CoA synthetase from Thermus thermophilus (43) and the luciferase enzymes from the Japanese (47) and American (42) fireflies, have larger binding pockets than SidNA3 (Table 4). Strikingly, these larger pockets are lined by the structural equivalents of most of the residues that line the SidNA3 pocket plus some additional residues. The additional size of the long chain fatty acyl-CoA synthetase pocket is unsurprising as it binds and activates myristate and palmitate, substrates that are larger than those of SidNA3. The larger size of the luciferase binding pocket in comparison with that of SidNA3 is somewhat surprising as the substrate, luciferin, is similar in size to cis-AMHO. However, most of the extra binding pocket residues seen, in addition to those in SidNA3, do not appear to be in contact with the luciferin ligand; rather, they play a catalytic role (47). The remaining 14 structures have similar sized or smaller binding pockets than SidNA3 (Table 4), and the pockets of these enzymes are mainly lined by a subset of residues equivalent to those in SidNA3. The binding pocket of SidNA3 defines the upper limits of NRPS adenylation domain binding pockets, as cis-AMHO is one of the largest amino acids to be incorporated into secondary metabolites by NRPSs.
TABLE 4.
The pocket-lining residues in the acetyl-CoA synthetase-like superfamily
The dark gray shading denotes the PheA pocket-lining residues, and the light gray shading denotes the extra SidNA3 pocket-lining residues. The structures are as follows: long chain fatty acyl-CoA synthetase (LCFA) (code 1v26), T. thermophilus long chain fatty acyl-CoA synthetase (43); Luci (code 2d1s), Japanese firefly Luciola cruciata luciferase (47); SidNA3 (this work), N. lolii third adenylation domain from SidN; FAL (code 3gqw), E. coli fatty-acid-AMP ligase; FadD1 (code 3g7s), Archaeoglobus fulgidus long chain fatty-acid-CoA ligase; PheA (code 1amu), B. brevis adenylation domain from gramicidin S synthetase A (9); ACSM2A (code 3eq6), Homo sapiens medium chain acyl-CoA synthetase (50); CBAL (code 1t5d), Alcaligenes sp. AL3007 4-chlorobenzoyl-CoA ligase (51); SrfAC (code 2vsq), B. subtilis adenylation domain from surfactin synthetase C (17); DhbE (code 1md9), B. subtilis DhbE adenylation domain (16); BCL (code 2v7b), Burkholderia xenovorans benzoate-CoA ligase (52); DltA (code 3dhv), Bacillus cereus d-alanyl-carrier protein ligase (53); AAE (code 3etc), Methanosarcina acetivorans acyl-CoA synthetase (54); ACS (code 1pg4), Salmonella enterica acetyl-CoA synthetase (7); and FAAL28 (code 3e53), Mycobacterium tuberculosis fatty acyl-AMP ligase 28 N-terminal domain (55). The American firefly Photinus pyralis luciferase (42, 56), B. subtilis d-alanyl-carrier protein ligase (6), and the Salmonella typhimurium (57) and Saccharomyces cerevisiae (58) acetyl-CoA synthetases are not included in the table due to the high similarity of their binding sites to the homologous structures that are included.
a Only the main chain atoms of these residues line the pocket.
b The binding pocket exposed to solvent in the FAAL28 structure, which was solved in unliganded form, is too small to accommodate its long chain fatty acid substrate. There is an internal cavity in the protein closed off by Met-233. Met-233 is the equivalent of the Trp-234 residue in long chain fatty acid, which closes off the entrance to the binding pocket in the unliganded form and opens up upon ATP binding. Thus, it is likely that Met-233 in FAAL28 is performing a similar function and that the true binding pocket is much larger.
c In long chain fatty acid, sequence alignments place Asn-232 as the equivalent of the PheA residue 236, but structural comparisons show that it is actually Trp-234 that is the structurally equivalent residue due to an alternate main chain route.
DISCUSSION
We have determined the structure of SidNA3, the first structure of a eukaryotic NRPS domain. Activity assays clearly show that SidNA3 adenylates cis-AMHO but not any of the 20 proteogenic amino acids or l-ornithine. This is the first experimental evidence in support of the predictions that the third module of three-module siderophore synthetases adenylate AHO residues (13, 18). Consistent with it binding cis-AMHO, one of the largest substrates of the acetyl-CoA synthetase-like superfamily, the amino acid binding pocket of SidNA3 is much larger than that of PheA (9). The SidNA3 binding pocket is lined by 17 rather than 9 residues (as seen for PheA), and it defines the upper limits of NRPS adenylation domain binding pockets. Previous homology modeling predicted that the AHO-activating domains would have larger pockets than PheA (13, 18) but do not correctly identify all of the residues involved in substrate binding. This highlights the value of an experimentally determined structure in understanding substrate specificity for the large family of eukaryotic NRPS enzymes.
The N- and C-terminal domains in the SidNA3 structure are arranged in an open conformation previously unseen in NRPS adenylation domains. This conformation is distinct from the two catalytically active conformations thought to be adopted by members of the acetyl-CoA synthetase-like superfamily (7). However, similar open conformations have previously been observed in four other members of the superfamily (Protein Data Bank codes 1lci (42), 1ult (43), 2vsq (17), and 3g7s). The exact orientation of the C-terminal domain relative to the N-terminal domain is different in each of these structures. Nevertheless, the structures are similar in that there is a wide separation between the two domains and a highly accessible active site. Given the lack of interactions between the two domains in the open conformation, it is likely that the exact orientation of the C-terminal domain seen in these structures is stabilized by crystal packing and that the C-terminal domain is mobile in solution. This is supported by the high mobility of the C-terminal domains observed in the SidNA3 structure. Furthermore, in the structure of the open conformation of the long chain fatty acyl-CoA synthetase (43), the C-terminal domain of each monomer of the enzyme, which forms a domain-swapped dimer, is in a different open orientation (43).
The recent structure of the surfactin synthetase termination module (SrfAC) from Bacillus subtilis (17) has provided further insights into the relevance of the open conformations for NRPS adenylation domains. This structure shows that the condensation domain and the N-terminal part of the adenylation domain associate closely to form a catalytic platform. The PCP domain and the C-terminal part of the adenylation domain rearrange on this platform to move the tethered peptide product between the reaction centers (17). It seems probable that the conformational changes of the adenylation domain would be coordinated with rearrangements in the positioning and conformation of the PCP domain. This is further reinforced by the fact that the 4′ppant arm in SrfAC is not long enough to reach each of the catalytic centers, implying considerable mobility among the domains of NRPSs. Thus, in addition to the adenylation and thiolation conformations, the adenylation domain may also adopt at least two other stable conformations that allow the PCP domain and 4′ppant to be positioned in the reaction centers of the preceding and successive domains in the peptide synthesis reaction pathway. In the SrfAC structure, the PCP domain is positioned such that the 4′ppant arm would be able to reach into the active site of the preceding condensation domain, suggesting that the module is in a conformation suitable for catalyzing the condensation reaction (17). The adenylation domain is in an open conformation with the C-terminal domain stabilized by interactions with the neighboring condensation domain (17). SidNA3 was crystallized in isolation from its neighboring domains and, hence, cannot form similar interactions, which would explain the high mobility of the C-terminal domain.
The residues lining the substrate binding pocket of PheA (9) have previously been used to develop methods to predict the specificities of newly identified adenylation domains (10–12). The specificity of a novel adenylation domain is predicted based on its similarity to the signature sequences of adenylation domains with known substrates. These methods work well for prokaryotic NRPSs but are generally unsuccessful for eukaryotic NRPSs (13). Of significant interest is whether the SidNA3 binding pocket is able to discriminate between acyl-hydroxyornithines with the various acyl groups seen in siderophores (Fig. 6). In silico, ligand docking of AHOs into the SidNA3 structure was undertaken and showed cis-AMHO as the top-ranked substrate, consistent with the fact that AMHO is the in vivo substrate. Visualization of the in silico-bound cis-AMHO shows that the terminal alcohol of the ligand is hydrogen-bonded to the side chain hydroxyl of Tyr-293. Tyr-293 is one of the few hydrogen bonding partners in the SidNA3 binding pocket and is located at the very base of the pocket, which is otherwise a largely hydrophobic site. Furthermore, comparisons of the pocket-lining residues of other AHO-adenylating domains showed that the SidNA3 domain clustered with those known to activate cis-AMHO (Table 3). The discrimination in the AHO that are used to synthesize the siderophore in vivo may also be due to limitations in the pool of AHOs available in the cell. Further experimental investigation is necessary to resolve the questions of the specificity of the SidNA3 domain for AHO residues. Nevertheless, our results demonstrate the specificity of SidNA3 for AHOs over ornithine or any of the other 20 common amino acids.
The structure of SidNA3 shows that although the substrate binding pockets of eukaryotic NRPS adenylation domains are constructed along fundamentally similar principles to their prokaryotic counterparts, their elaboration to accommodate more complex substrates is not readily predictable from sequence comparison alone. The low success rate of specificity-prediction methods for eukaryotic domains is due to the divergence in their signature sequences from those of prokaryotes. The structure of SidNA3 therefore provides a springboard for the development of more accurate specificity prediction methods for the abundant eukaryotic NRPS adenylation domains.
It is possible that eukaryotes utilize a more diverse set of signature sequences than the prokaryotes. Furthermore, it is perhaps rather simplistic to attempt prediction of NRPS substrate specificities using just the nine residues that line the PheA pocket. It is obvious from the comparisons of the substrate binding pockets of superfamily members that the size of the pocket varies considerably. By limiting the comparisons to the nine PheA residues, some of the important specificity-determining residues may be missed, especially for larger substrates. Expanding the number of residues considered to match those found in the SidNA3 pocket will serve to improve the coverage of the possible specificity-determining residues. This would be at the expense of increasing the noise for smaller pockets, however, and the pocket size itself is probably a key specificity determinant for many of these enzymes. Thus, it is likely that the greater use of structurally based tools such as homology modeling based on appropriate templates, employed to inform comparisons of the unknown domains with those of known specificity, will prove more fruitful than simple sequence comparisons. Development of an accurate substrate prediction method for domains of unknown specificity is now the subject of current work.
Acknowledgments
We thank Jeremy Owen and David Ackerley for assistance in implementing the pyrophosphate release assays and Karl Fraser and Scott Harrison for separation of the siderophore fraction and measurement of its mass spectrum. We are grateful to Christine Voisey and Damien Fleetwood for providing N. lolii genomic DNA and Victoria Money for assistance with the x-ray data collection. Data collection was performed at beamline BL9-2 of the Stanford Synchrotron Radiation Laboratory, Stanford University, Palo Alto, CA.
This work was supported in part by the New Zealand Foundation for Research Science and Technology Contract CX10X0203.
The atomic coordinates and structure factors (code 3ITE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
L. J. Johnson, unpublished data.
L. J. Johnson, M. Christensen, A. Koulman, G. A. Lane, K. Fraser, R. D. Johnson, G. T. Bryan, and S. Rasmussen, manuscript in preparation.
A. Koulman and G. A. Lane, unpublished data.
- NRPS
- nonribosomal peptide synthetase
- 4′ppant
- 4′-phosphopantetheine
- PCP domain
- peptide carrier protein domain
- AHO
- Nδ-acyl-Nδ-hydroxyornithine
- SidNA3
- the third adenylation domain of SidN
- cis-AMHO
- Nδ-cis-anhydromevalonyl-Nδ-hydroxy-l-ornithine
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Finking R., Marahiel M. A. (2004) Annu. Rev. Microbiol. 58, 453–488 [DOI] [PubMed] [Google Scholar]
- 2.Fleetwood D. J., Scott B., Lane G. A., Tanaka A., Johnson R. D. (2007) Appl. Environ. Microbiol. 73, 2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sieber S. A., Marahiel M. A. (2005) Chem. Rev. 105, 715–738 [DOI] [PubMed] [Google Scholar]
- 4.Murzin A. G., Brenner S. E., Hubbard T., Chothia C. (1995) J. Mol. Biol. 247, 536–540 [DOI] [PubMed] [Google Scholar]
- 5.Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) Nucleic Acids Res. 36, D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonus H., Neumann P., Zimmermann S., May J. J., Marahiel M. A., Stubbs M. T. (2008) J. Biol. Chem. 283, 32484–32491 [DOI] [PubMed] [Google Scholar]
- 7.Gulick A. M., Starai V. J., Horswill A. R., Homick K. M., Escalante-Semerena J. C. (2003) Biochemistry 42, 2866–2873 [DOI] [PubMed] [Google Scholar]
- 8.Scott-Craig J. S., Panaccione D. G., Pocard J. A., Walton J. D. (1992) J. Biol. Chem. 267, 26044–26049 [PubMed] [Google Scholar]
- 9.Conti E., Stachelhaus T., Marahiel M. A., Brick P. (1997) EMBO J. 16, 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challis G. L., Ravel J., Townsend C. A. (2000) Chem. Biol. 7, 211–224 [DOI] [PubMed] [Google Scholar]
- 11.Rausch C., Weber T., Kohlbacher O., Wohlleben W., Huson D. H. (2005) Nucleic Acids Res. 33, 5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stachelhaus T., Mootz H. D., Marahiel M. A. (1999) Chem. Biol. 6, 493–505 [DOI] [PubMed] [Google Scholar]
- 13.Schwecke T., Göttling K., Durek P., Dueñas I., Käufer N. F., Zock-Emmenthal S., Staub E., Neuhof T., Dieckmann R., von Döhren H. (2006) ChemBioChem 7, 612–622 [DOI] [PubMed] [Google Scholar]
- 14.Stack D., Neville C., Doyle S. (2007) Microbiology 153, 1297–1306 [DOI] [PubMed] [Google Scholar]
- 15.von Döhren H. (2009) Fungal Genet. Biol. 46, S45–S52 [DOI] [PubMed] [Google Scholar]
- 16.May J. J., Kessler N., Marahiel M. A., Stubbs M. T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12120–12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanovic A., Samel S. A., Essen L. O., Marahiel M. A. (2008) Science 321, 659–663 [DOI] [PubMed] [Google Scholar]
- 18.Bushley K. E., Ripoll D. R., Turgeon B. G. (2008) BMC Evol. Biol. 8, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson L. (2008) Mycol. Res. 112, 170–183 [DOI] [PubMed] [Google Scholar]
- 20.Eisendle M., Oberegger H., Zadra I., Haas H. (2003) Mol. Microbiol. 49, 359–375 [DOI] [PubMed] [Google Scholar]
- 21.Gimenez C., Cabrera R., Reina M., Gonzalez-Coloma A. (2007) Curr. Org. Chem. 11, 707–720 [Google Scholar]
- 22.Rodriguez R., Redman R. (2008) J. Exp. Bot. 59, 1109–1114 [DOI] [PubMed] [Google Scholar]
- 23.Moreland N., Ashton R., Baker H. M., Ivanovic I., Patterson S., Arcus V. L., Baker E. N., Lott J. S. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 1378–1385 [DOI] [PubMed] [Google Scholar]
- 24.Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 25.Mantle P. G., Nisbet L. J. (1976) J. Gen. Microbiol. 93, 321–334 [DOI] [PubMed] [Google Scholar]
- 26.Diekmann H., Zähner H. (1967) Eur. J. Biochem. 3, 213–218 [DOI] [PubMed] [Google Scholar]
- 27.Upson R. H., Haugland R. P., Malekzadeh M. N., Haugland R. P. (1996) Anal. Biochem. 243, 41–45 [DOI] [PubMed] [Google Scholar]
- 28.Leslie A. G. (1992) Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 29.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 30.Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) in Macromolecular Crystallography Protocols (Doublié S., ed) Vol. 2., pp. 215–230, Humana Press Inc., Totowa, NJ [Google Scholar]
- 31.Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1174–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34.Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) CCP4 Newsletter 42, Contribution 8 [Google Scholar]
- 35.Cole J. C., Nissink J. W., Taylor R. (2005) in Virtual Screening in Drug Discovery (Alvarez J., Shoichet B. eds) pp. 379–415, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 36.Eldridge M. D., Murray C. W., Auton T. R., Paolini G. V., Mee R. P. (1997) J. Comput.-Aided Mol. Des. 11, 425–445 [DOI] [PubMed] [Google Scholar]
- 37.Meekhof A. E., Hamill S. J., Arcus V. L., Clarke J., Freund S. M. (1998) J. Mol. Biol. 282, 181–194 [DOI] [PubMed] [Google Scholar]
- 38.McCormack P., Worsfold P. J., Gledhill M. (2003) Anal. Chem. 75, 2647–2652 [DOI] [PubMed] [Google Scholar]
- 39.Luo L., Burkart M. D., Stachelhaus T., Walsh C. T. (2001) J. Am. Chem. Soc. 123, 11208–11218 [DOI] [PubMed] [Google Scholar]
- 40.Villiers B. R., Hollfelder F. (2009) Chem. Bio. Chem. 10, 671–682 [DOI] [PubMed] [Google Scholar]
- 41.Reger A. S., Wu R., Dunaway-Mariano D., Gulick A. M. (2008) Biochemistry 47, 8016–8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conti E., Franks N. P., Brick P. (1996) Structure 4, 287–298 [DOI] [PubMed] [Google Scholar]
- 43.Hisanaga Y., Ago H., Nakagawa N., Hamada K., Ida K., Yamamoto M., Hori T., Arii Y., Sugahara M., Kuramitsu S., Yokoyama S., Miyano M. (2004) J. Biol. Chem. 279, 31717–31726 [DOI] [PubMed] [Google Scholar]
- 44.Saraste M., Sibbald P. R., Wittinghofer A. (1990) Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 45.Larsen I. (1988) Acta Crystallogr. B Struct. Sci. 44, 527–533 [Google Scholar]
- 46.Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 47.Nakatsu T., Ichiyama S., Hiratake J., Saldanha A., Kobashi N., Sakata K., Kato H. (2006) Nature 440, 372–376 [DOI] [PubMed] [Google Scholar]
- 48.Connolly M. (1983) J. Appl. Crystallogr. 16, 548–558 [Google Scholar]
- 49.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins Struct. Funct. Genet. 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 50.Kochan G., Pilka E. S., von Delft F., Oppermann U., Yue W. W. (2009) J. Mol. Biol. 388, 997–1008 [DOI] [PubMed] [Google Scholar]
- 51.Gulick A. M., Lu X., Dunaway-Mariano D. (2004) Biochemistry 43, 8670–8679 [DOI] [PubMed] [Google Scholar]
- 52.Bains J., Boulanger M. J. (2007) J. Mol. Biol. 373, 965–977 [DOI] [PubMed] [Google Scholar]
- 53.Du L., He Y., Luo Y. (2008) Biochemistry 47, 11473–11480 [DOI] [PubMed] [Google Scholar]
- 54.Shah M. B., Ingram-Smith C., Cooper L. L., Qu J., Meng Y., Smith K. S., Gulick A. M. (2009) Proteins Struct. Funct. Bioinformat. 77, 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arora P., Goyal A., Natarajan V. T., Rajakumara E., Verma P., Gupta R., Yousuf M., Trivedi O. A., Mohanty D., Tyagi A., Sankaranarayanan R., Gokhale R. S. (2009) Nat. Chem. Biol. 5, 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franks N. P., Jenkins A., Conti E., Lieb W. R., Brick P. (1998) Biophys. J. 75, 2205–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reger A. S., Carney J. M., Gulick A. M. (2007) Biochemistry 46, 6536–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jogl G., Tong L. (2004) Biochemistry 43, 1425–1431 [DOI] [PubMed] [Google Scholar]