Abstract
Mediator is a multisubunit coactivator required for initiation by RNA polymerase II. The Mediator tail subdomain, containing Med15/Gal11, is a target of the activator Gcn4 in vivo, critical for recruitment of native Mediator or the Mediator tail subdomain present in sin4Δ cells. Although several Gal11 segments were previously shown to bind Gcn4 in vitro, the importance of these interactions for recruitment of Mediator and transcriptional activation by Gcn4 in cells was unknown. We show that interaction of Gcn4 with the Mediator tail in vitro and recruitment of this subcomplex and intact Mediator to the ARG1 promoter in vivo involve additive contributions from three different segments in the N terminus of Gal11. These include the KIX domain, which is a critical target of other activators, and a region that shares a conserved motif (B-box) with mammalian coactivator SRC-1, and we establish that B-box is a critical determinant of Mediator recruitment by Gcn4. We further demonstrate that Gcn4 binds to the Gal11 KIX domain directly and, by NMR chemical shift analysis combined with mutational studies, we identify the likely binding site for Gcn4 on the KIX surface. Gcn4 is distinctive in relying on comparable contributions from multiple segments of Gal11 for efficient recruitment of Mediator in vivo.
Keywords: Transcription/Coactivators, Transcription/Yeast, Gal11, Gcn4, KIX, Med15, Mediator, Activation Domain
Introduction
The Mediator complex is an important coactivator for RNA polymerase II (pol II),3 comprised of more than 20 different subunits (1). It is found associated with pol II in a holoenzyme (2–5) but also exists free of pol II (6), and it can be recruited by transcriptional activators to upstream activation sequences (UASs) independently of pol II (7–12). Mediator promotes both basal and activated transcription in vitro (13) and is critical for transcription of most genes in vivo (14). It stimulates preinitiation complex assembly at promoters in vivo by facilitating recruitment of chromatin-modifying complexes (11, 12, 15, 16), TATA-binding protein, and pol II (12, 17–19). Mediator interacts with transcription factors TFIIE and TFIIH and the C-terminal domain (CTD) of pol II, and it stimulates CTD phosphorylation by the Kin28 subunit of TFIIH (3).
Mediator can be divided into three modules based on results obtained from structural, biochemical, and genetic analyses (20–23). The “head” consists of eight subunits and can interact with the pol II CTD, TFIIB, and TATA-binding protein and supports basal, but not activated, transcription in vitro (24), although there is evidence implicating head subunits Med17/Srb4 and Med20/Srb2 in Mediator recruitment by activators Gal4 (25) and Gcn4 (26). The head domain makes multiple contacts with pol II in holoenzyme (20, 27), but the reconstituted head requires TFIIF to form a tight complex with pol II in vitro (13). The middle domain also appears to contact pol II in holoenzyme (27).
The Mediator tail makes limited contacts with pol II (27) and appears to be the principal target of activators. The tail consists of Med15/Gal11, Med16/Sin4, Med2, Med3/Pgd1/Hrs1, and Med5/Nut1, none of which is essential in vivo (21, 28), and is tethered to the middle domain via the Med14/Rgr1-CTD. Deletion of Med16/Sin4 leads to loss of the remaining tail subunits from Mediator (20, 29, 30), and the released subcomplex containing Gal11, Med2, and Pgd1 is stable in cell extracts, capable of binding Gcn4 in vitro, and recruited independently of head subunits by Gcn4 to target genes in vivo (26).
Consistent with these in vivo findings, purified mutant holoenzymes lacking Gal11, Pgd1, or Med2 are impaired for transcriptional activation by Gal4, VP16, and Gcn4 and fail to bind these activators in vitro (4, 31). Interestingly, Gal11 was the only Mediator subunit that photo-cross-linked to Gcn4 or Gal4 in preinitiation complexes reconstituted with these activators in vitro (32). Park et al. (31) showed that several non-overlapping segments of recombinant Gal11 can bind in vitro to Gal4 or Gcn4 and that deletions of the Gal4-binding segments impaired transcriptional activation by Gal4. However, it was not determined whether the Gal11 segments that bind Gcn4 in vitro are important for transcriptional activation or recruitment of Mediator by Gcn4 in vivo. Thus, the physiological relevance of these Gcn4-binding domains in Gal11 was unclear (31).
Interestingly, an N-terminal Gal11 segment (amino acids (aa) 116–255), shown previously to bind Gal4, Gcn4, and the VP16 activation domain in vitro (31), likewise binds a portion of the activation domain of mammalian glucocorticoid receptor (GR) dubbed τ1c. Indeed, Gal11 was the sole Mediator subunit that cross-linked to τ1c in vitro. A sequence motif (B-box) in the aa interval 116–255 of Gal11 is conserved in mammalian coactivator SRC-1 and crucial for the function of Gal11 in transcriptional activation by τ1c in yeast. Several regions in Gal11 are additionally required to support the function of an activation domain from the mammalian androgen receptor in yeast (33).
Surprisingly, a different region of Gal11 is the key target of yeast activators Pdr1 and Oaf1. The solution structures of the extreme N-terminal regions of Gal11 and its mammalian homolog (ARC105) are similar to the activator-binding “KIX” domain of mammalian coactivator CREB-binding protein/p300 (34, 35). The ARC105 and Gal11 KIX domains interact in vitro with the activation domains of mammalian sterol regulatory element-binding protein and yeast Pdr1, although the KIX surfaces differ somewhat for these interactions. Deletion of only the KIX domain of Gal11 nearly abolishes transcriptional activation by Pdr1 and is predicted to impair Mediator recruitment by Pdr1 in vivo. Similar findings were reported for yeast activator Oaf1 (34–36). It is striking that the KIX domain is crucial for activation by Pdr1 and Oaf1 but dispensable for the Gal4 activation domain in a lexA fusion (37) and makes only a small contribution to activation by native Gal4. Moreover, it was reported that neither Gal4, Gcn4, nor VP16 bound stably to the Gal11 KIX domain in vitro (31).
In this study, we sought to define the domains in Gal11 required for recruitment of Mediator by Gcn4 in living cells. Our results indicate that Gcn4 uses a combination of at least three non-contiguous segments in the N-terminal approximately one-third of Gal11, including the KIX domain, to recruit Mediator in vivo. We show that eliminating the KIX domain in combination with one of the other Gcn4-binding regions in Gal11 is required to evoke strong reductions in Gcn4 binding to Mediator tail subcomplex in extracts, in recruitment of the tail subcomplex (or intact Mediator) in vivo, and in transcriptional activation by Gcn4. We also provide evidence that Gcn4 can bind directly to the KIX domain and identify the likely binding surface for Gcn4 by NMR and mutational analysis. Thus, in contrast to certain other activators that primarily target a single region in Gal11, Gcn4 employs a highly redundant set of interactions with different regions of Gal11 for efficient Mediator recruitment in vivo.
EXPERIMENTAL PROCEDURES
Yeast Strain Constructions
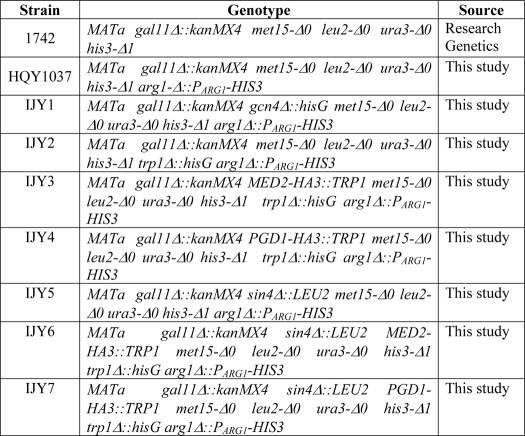
All strains used in this study are listed in Table 1. Strain HQY1037 was generated from gal11Δ::kanMX4 strain 1742 by a two-step gene replacement of ARG1 with PARG1-HIS3, creating the arg1-Δ::PARG1-HIS3 allele in which the ARG1 ORF is replaced by a fusion between the N-terminal 62 codons of ARG1 and the entire HIS3 ORF, using integrative URA3 plasmid pHQ1459 digested at the unique Eco47III site upstream of ARG1. Strain IJY1 was constructed from HQY1037 using the gcn4Δ::hisG plasmid pHQ1240 as described previously (38). IJY2 was constructed from HQY1037 using the trp1Δ::hisG disruption plasmid pNKY1009 (39). sin4Δ:LEU2 strains IJY5, IJY6, and IJY7 were constructed from HQY1037, IJY3, and IJY4, respectively, by transforming with a PCR fragment containing the sin4Δ::LEU2 allele amplified from LEU2 vector YCplac111 (40) using primers 5′-AAGAAGTATAATTTCATTTCAAAAATAAGGTCCAAAGAAAAGAAACTAGCAGACCTGACCTTCTGTTGGTAAATATTAGTTTAACTGTGGGAATACTCAGGT-3′ and 5′-ACTGTCACTCTCATTTCTTTTTATTAAGTCGAGAAGTGAAATGTTTAAAACAATTCTATACAAAACTATGCTATAGTACTAATAATCTACCCTATGAACATATTCCATT-3′.
TABLE 1.
Yeast strains used in this study
All gene replacements were confirmed by PCR analysis of chromosomal DNA using the appropriate primers. Strains IJY3 and IJY4 were generated from IJY2 using the appropriate PCR fragments containing the HA3::TRP1 cassette amplified from pFA6a-3HA-TRP1 (41) with primers 5′-TAAATGATTTCAACGACCTTAATATTGACTGGTCGACCACTGGAGATAATGGCGAATTAGACCTCAGCGGCTTCAATATACGGATCCCCGGGTTAATTAA-3′ and 5′-TCATATATATACCAATTAAATTCACGTAGCGAATGCACAACACGGTTTACAAGTCAATAGTTAACAATAGGAAGACCAAGGAATTCGAGCTCGTTTAAAC-3′ for MED2, and with primers 5′-ACATGAATAACGGGGGGAAAGAACTGGATTCTCTAGACCTGAACAATCTGGAATTAGGTGGTCTGAACATGGATTTCTTGCGGATCCCCGGGTTAATTAA-3′ and 5′-AGCCTTTCGGGTAATAAGGTATAAAGAATAGAAGATTATACAGATAATTACTATCTTGGATACATAGATGCACCAGGAATTCGAGCTCGTTTAAAC-3′ for PGD1. The insertions were confirmed by PCR analysis of chromosomal DNA and Western blot analysis of whole-cell extracts (WCEs) using anti-HA antibodies.
Plasmid Constructions
Plasmid pHQ1459 was constructed in two steps. First, an EcoRI-BamHI PCR fragment containing 734 bp of 5′-non-coding DNA and the first 62 codons of ARG1 was amplified using primers 1094 (5′-CGCGAATTCTTTCCTGTTGCCTCTTTTTC-3′) and 1095 (5′-CGGGGATCCACACAAACGAACTTGCA-3′). A second, BamHI-XbaI PCR fragment containing 737 bp of ARG1 3′-non-coding sequences was amplified using primers 1100 (5′-CGGGGATCCCGCGAGCTCTAAGTCCGCTAGTTCATCGC-3′) and 1097 (5′- CGCTCTAGATGTGAATTCATGGTTAACG-3′). Both fragments were inserted between the EcoRI/XbaI sites of YIplac211 (40) to produce pHQ1457. A BamHI-EcoRV PCR fragment containing the entire HIS3 ORF was amplified using primers 1098 (5′-CGCGGATCCCATGACAGAGCAGAAAGCCCT-3′) and 1099 (5′-CGCGATATCACATAAGAACACCTTTGGTGG-3′) and between the BamHI/Ecl136II sites of pHQ1457 to produce pHQ1459.
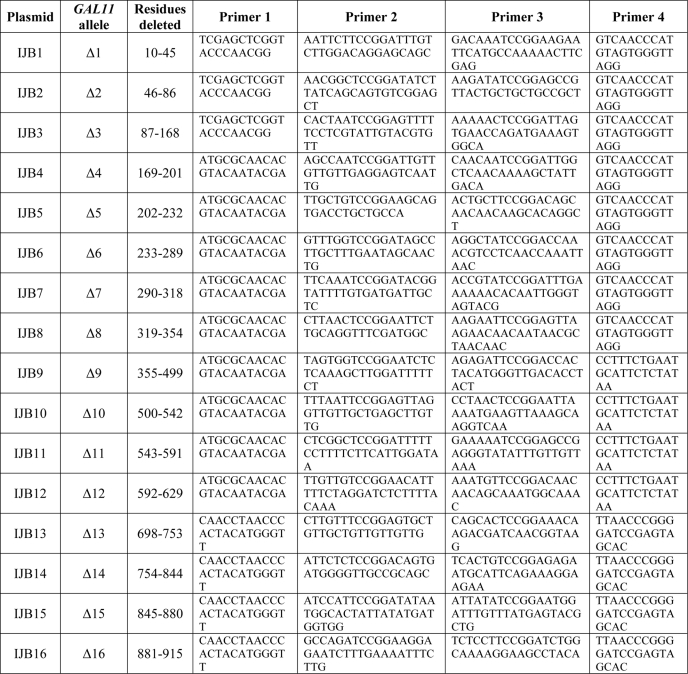
Plasmids containing GAL11 deletion alleles are listed in Table 2 along with the primers used in constructing them by PCR fusion. All such deletions except Δ9 to Δ12 were constructed using as template the single copy URA3 plasmid pMJS15, containing wild-type myc-GAL11 with the native GAL11 promoter and ADH1 transcription terminator. For Δ9 to Δ12, the template was a derivative of pMJS15, pIJB25, with the NsiI site in URA3 eliminated to render the NsiI site in GAL11 unique. In the first stage of PCR fusion, two fragments were amplified with the primer pairs 1 and 2 and 3 and 4, respectively, listed in Table 2. These fragments were then used in 1:1 ratio as templates for a second amplification using primer pairs 1 and 4. For Δ1 to Δ3, the resulting fragments were cloned between the Asp-718 and PflMI sites of pMJS15; for Δ4 to Δ8, they were cloned between the BsaAI and PflMI sites of pMJS15; for Δ9 to Δ12, they were cloned between the BsaAI and NsiI sites of pMJS15; for Δ13 to Δ17, they were cloned between the PflMI and BamHI sites of pMJS15; and for Δ18, they were cloned between the PflMI and SalI sites of pMJS15. For all single deletions, the BspEI restriction site (encoding Ser-Gly) was inserted at the deletion junction. Alleles containing combinations of deletions were constructed as follows, and the primer pairs used are listed in Table 2. pIJB19, containing Δ1Δ2, was generated by PCR fusion using pMJS15 as template, inserting a BspEI site at the Δ1Δ2 deletion junction, and the resulting PCR fragment was cloned between the Asp-718 and PflMI sites of pMJS15. pIJB20 (Δ1Δ2Δ5) was generated by PCR fusion using pIJB19 (Δ1Δ2) as template, inserting an AgeI restriction site (encoding Thr-Gly) at the Δ5 deletion junction; the final PCR fragment was cloned between the Asp-718 and PflMI sites of pMJS15. pIJB21 (Δ1Δ2Δ8) was generated by PCR fusion using pIJB19 as template and inserting an AgeI site at the Δ8 deletion junction. The final PCR fragment was cloned between the Asp-718 and PflMI sites of pMJS15. pIJB22 (Δ1Δ2Δ14) was generated by PCR fusion using pIJB14 as template and inserting an BspEI site at the Δ1Δ2 deletion junction. The final PCR fragment was cloned between the Asp-718 and PflMI sites of pIJB14. pIJB23 (Δ5Δ8) was generated by PCR fusion using pIJB8 as the template and inserting an AgeI site at the Δ5 deletion junction. The final PCR fragment was cloned between Asp-718 and PflMI sites of pMJS15. pIJB24 (Δ8Δ14) was generated using pIJB14 as the template and inserting a BspEI site at the Δ8 deletion junction. The PCR fragment was cloned between BsaAI and PflMI sites of pIJB14. The subcloned fragments of all deletion constructs were confirmed by DNA sequencing.
TABLE 2.
Plasmids containing GAL11 deletion alleles and primers used in their construction by fusion PCR mutagenesis
Plasmids p4913 and p4914, harboring the B-box mutation WQV, were constructed by PCR fusion using pMJS15 (wild type (WT)) or pIJB19 (Δ1Δ2) as templates, respectively, and the following four primers: FZP277, 5′-GAATTCGAGCTCGGTACCCAAC-3′; FZP278, 5′-ACATCCTTTGCAGTAGGTGTCAAC-3′; FZP279, 5′-AACATTCCACCCAATATCAACACCGCTCAGGCTGCTACTGCTTTGGCTCAACAAAAGCTA-3′; FZP280, 5′-TAGCTTTTGTTGAGCCAAAGCAGTAGCAGCCTGAGCGGTGTTGATATTGGGTGGAATGTT-3′. The final PCR fragments were inserted between the KpnI and PflMI sites of pMJS15.
Plasmid pHQ1857 harboring the Ala6 substitution in the KIX domain was constructed as follows. Two PCR fragments were amplified from genomic DNA with the following pairs of primers 1617/1618 and 1619/1620: primer 1617, 5′-GAGCTCGGTACCCAACGGAGCATCAAACATGAC-3′; primer 1618, 5′-CAGCGTCTGCAGCCCCTCCGTTCAGAGTGGCAATGTCCGCGAGCACCTGAAGCAACCCGT-3′; primer 1619, 5′-GAGGGGCTGCAGACGCTGCTGCTAAGATAAGAATTCATGCCAAAA-3′; and primer 1620, 5′-CCTCGTATTGTACGTGTTGCG-3′. The fragment obtained with primers 1617/1618 was digested with Asp-718 and PstI to produce a fragment containing 695 bp of 5′-non-coding region and codons 1–39 of GAL11 with mutations that substitute Ala codons for Met-29, Asn-32, Ser-38, and Ser-39. The fragment obtained with primers 1619/1620 was cut with PstI and BsaAI to produce a fragment containing codons 38–81 with Ala substitutions of Ser-38, Ser-39, Thr-41, and Asp-43, with the S38A and S39A substitutions creating a unique PstI site. The two final fragments were ligated with the Asp-718-BsaAI fragment from pMJS15 to produce pHQ1857. pHQ1858, harboring the Ala4 substitution, was constructed as pHQ1857 except that the PCR fragments do not encode the N32A and T41A substitutions and were made using primer 1621 (5′-CAGTGTCTGCAGCCCCTCCGTTCAGAGTGTTAATGTCCGCGAGCACCTGAAGCAACCCGT-3′) instead of 1618 and primer 1622 (5′-GAGGGGCTGCAGACACTGCTGCTAAGATAAGAATTCATGCCAAAA-3′) instead of 1619.
Plasmids used for in vitro translation of Gal11 polypeptides are listed in Table 3 with the primers used in constructing them. All of the PCR fragments amplified using the indicated primers were cloned between the XhoI and XbaI sites of pTNT (Promega).
TABLE 3.
Plasmids for in vitro translation of Gal11 fragments and primers used in their construction
The primers in Table 4 were used to construct the plasmids for expression in Escherichia coli of the Gal11 KIX domain (residues 2–100), full-length Gcn4, and Gcn4 activation domain (aa 2–151), with each protein containing an N-terminal His6 tag and a TEV protease cleavage site (ENLYFQS). The resulting PCR products were cloned between the NdeI and XhoI sites of pET-28a (Novagen). Plasmid p4915 containing the UASGCRE-CYC1-lacZ reporter in a LEU2 episome was constructed from URA3 plasmid pHYC(14X2) (42) using the marker-swap plasmid pUL9 (43) digested with SmaI.
TABLE 4.
Plasmids expressing Gal11 KIX and Gen4 polypeptides in E. coli and primers used in their construction
Biochemical Methods
For Western blot analysis of Myc-Gal11 expression, yeast strains were grown at 30 °C to an optical density at 600 nm (A600) of 0.6 in synthetic complete medium (SC) lacking uracil (SC-URA) and treated with sulfometuron methyl at 0.6 μg/ml for 30 min to inhibit biosynthesis of isoleucine and valine and induce Gcn4. WCEs were extracted by the trichloroacetic acid method described previously (44) and subjected to Western blot analysis using the following antibodies: anti-Myc (Roche Applied Science), anti-Gcn4 (45), and anti-Gcd6 (46).
GST pulldown assays using yeast WCEs were performed as described previously (26) using bacterial extracts containing GST alone, encoded by p2645 (a modification of pGEX-5x-3, Amersham Biosciences), or GST fusions containing full-length wild-type Gcn4 (GST-GCN4) or the mutant fusion with 10 alanine substitutions in the Gcn4 activation domain (GST-Gcn4-Ala10), encoded by p2865 and p2867, respectively (47). Coimmunoprecipitation analysis with yeast WCEs was conducted as described previously (26) using anti-Myc or anti-HA antibodies conjugated to agarose (Santa Cruz Biotechnology) for the immunoprecipitations. Western blot analysis of immune complexes or fractions bound to immobilized GST proteins was carried out using the following antibodies: anti-Myc (Roche Applied Science), anti-HA (Santa Cruz Biotechnology), anti-Med1 (48), anti-Srb5 (49), anti-Srb7 (50), anti-Srb4 (50), anti-Sin4 (51), anti-Snf6 (provided by Joseph Reese), anti-Spt3 (provided by Fred Winston), and anti-Rpb3 (Neoclone). For GST pulldown assays of recombinant Gal11 fragments, [35S]Gal11 fragments were synthesized in vitro using TnTTM quick-coupled in vitro transcription/translation kits (Promega) and tested for binding to the same GST, GST-GCN4, or GST-Gcn4-Ala10 proteins described above (26). Assays of UASGCRE-CYC1-lacZ reporter expression were conducted as described previously (52) except using LEU2 reporter plasmid p4915.
For chromatin immunoprecipitation (ChIP) experiments, 100-ml cultures were grown in SC-URA to A600 of ∼0.6, and sulfometuron methyl was added to 1 μg/ml for 30 min to induce Gcn4. Cultures were immediately fixed with formaldehyde for 20 min at ambient temperature and quenched with glycine. Cell pellets were sonicated to produce DNA fragments of ∼500 bp, and clarified extracts were used for ChIP experiments as described previously (53), using the anti-Myc and anti-Rpb3 antibodies described above and the primers listed in Table 5. For each primer set employed, we optimized the conditions for PCR analysis to ensure that the amounts of amplified 33P-labeled products being generated are proportional to the amounts of input DNA over the range of concentrations of ARG1 and POL1 sequences present in samples of total or immunoprecipitated chromatin.
TABLE 5.
Primers used for ChIP analysis
Gal11 KIX and Gcn4 Expression and Purification for NMR Studies
Expression in E. coli of isotopically labeled proteins (13C,15N and 2H,15N) was performed as described previously (54). After cell breakage, the His6-tagged fusion proteins were mostly in the soluble cell fraction, but varying amounts of denaturants were included during purification to prevent nonspecific protein interactions with host proteins.
For Gal11 KIX purification, cells were resuspended in 50 mm HEPES (pH 7.5) containing 4 m urea and protease inhibitor mixture (Roche Applied Science), lysed with a French press (Thermo Scientific), and centrifuged at 25,000 × g for 2 h. The supernatant was loaded on a nickel-Sepharose Fast Flow column (GE Healthcare), washed with 20 mm imidazole, and eluted with 0.5 m imidazole. Pooled fractions were concentrated and applied to a Superdex 200 column (GE Healthcare) equilibrated with 50 mm Tris-HCl (pH 7.5) containing 3 m urea and 3 m guanidine-HCl, and eluted fractions containing the KIX polypeptide were pooled.
For purification of Gcn4 and the Gcn4 activation domain, cells in 20 mm HEPES (pH 7.5) were lysed using a French press and sonication. The lysate was centrifuged for 25,000 × g for 1 h. The pellet was washed with lysis buffer and extracted with 10 mm HEPES (pH 7.5) containing 4 m guanidine-HCl. The solution was clarified by centrifugation at 100,000 × g for 1 h using a Ti45 rotor (Beckman Coulter). The supernatant was applied to a nickel-Sepharose Fast Flow column equilibrated in 10 mm HEPES (pH 7.4), 4 m guanidine-HCl. The column was washed with buffer plus 8 mm imidazole, and then a gradient of 40–400 mm imidazole was applied. Gcn4/Gcn4 activation domain-containing fractions were pooled and applied batch-wise (10 ml) to a SOURCE 15RPC (GE Healthcare) column (1 × 10 cm), and the protein was eluted using a gradient of 10–55% acetonitrile in 0.1% trifluoroacetic acid.
For protein folding and His6 tag removal, proteins (0.5–1.0 mg/ml) were folded by sequential dialysis against 20 mm sodium phosphate (pH 6.8), 30 mm NaCl (dialysis buffer) plus 4 m urea, dialysis buffer plus 2 m urea, and dialysis buffer with no urea. The recovery of folded proteins was greater than 95%. The N-terminal 26-residue sequence containing the His6 tag was removed by TEV protease digestion (AcTEV, Invitrogen). The proteins (0.25–1.0 mg/ml) in 50 mm Tris-HCl (pH 8.0), containing 5 mm dithiothreitol were digested with TEV (1:100 w/w) overnight at 4 °C followed by dialysis against 20 mm sodium phosphate, pH 6.8, 30 mm NaCl. Undigested protein was removed by adding 1 ml of nickel-Sepharose, and the resin was removed by centrifugation at 4,000 × g for 5 min and filtration through a 0.2-μm filter.
The identity and purity (>95%) of proteins were confirmed by SDS-PAGE and mass spectrometry. Concentrations of purified proteins were determined by absorbance at 280 nm using calculated molar absorbance coefficients (55). Gal11 KIX, full-length Gcn4, and Gcn4 activation domain were analyzed by sedimentation equilibrium and determined to be monomers with mass estimates within 2.5% of values predicted from the respective cDNA sequences. Amicon Ultra-15 3,000 molecular weight cut-off centrifugal devices (Millipore) were used to concentrate the proteins for NMR measurements.
NMR Spectroscopy
NMR spectra were recorded at 5 °C on Bruker DMX-600 and DRX-800 MHz spectrometers equipped, respectively, with a room temperature probe with triple-axis gradients and a cryogenic probe with z axis gradients. Spectra were processed using NMRPipe (56) and analyzed in Sparky (57).
The Gal11 KIX domain backbone amide 1H and 15N signals were assigned from three-dimensional HNCA, HN(CO)CA, and HN(CA)CB spectra recorded using pulse schemes based on those reported by Yamazaki et al. (58). The sample used to obtain the resonance assignments consisted of 13C,15N-labeled KIX at a concentration of 1.2 mm in aqueous solution containing 20 mm sodium phosphate, 30 mm NaCl, 7% (v/v) D2O, and 0.02% (w/v) NaN3 at pH 6.8 in a total volume of 300 μl in a Shigemi microcell.
Specific interactions in solution between the Gal11 KIX domain and full-length Gcn4 as well as the Gcn4 activation domain (aa 2–151) were probed by using a series of samples each containing 20 mm sodium phosphate, pH 6.8, 30 mm NaCl, 7% D2O, 225 μm 2H,15N-labeled KIX, and increasing amounts of unlabeled Gcn4 or Gcn4 activation domain. The Gcn4/activation domain concentrations used were 0 (free KIX control), 11.25, 22.5, 45, 90, and 180 μm corresponding to Gcn4/KIX molar ratios of 0, 0.05, 0.1, 0.2, 0.4, and 0.8, respectively. Changes in amide 1H and 15N chemical shifts as a function of increasing Gcn4/activation domain concentration were subsequently monitored by recording two-dimensional 15N-1H heteronuclear single quantum correlation (HSQC) spectra. The overall change in the amide chemical shift for each resolved signal in the two-dimensional HSQC spectra was obtained from the expression
 |
where ΔδHN and ΔδN are the differences in the amide 1H and 15N chemical shifts, respectively, between the free KIX control sample and the sample with the highest Gcn4/KIX ratio.
RESULTS
Three Non-contiguous Regions in Gal11 Mediate Its Efficient Recruitment by Gcn4 in Vivo
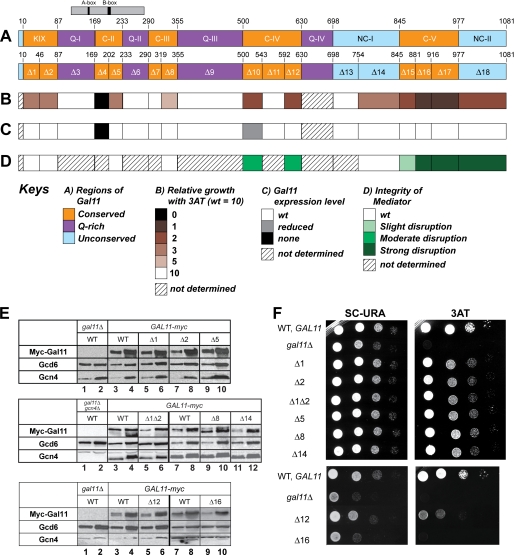
To identify regions in Med15/Gal11 involved in recruitment of Mediator by Gcn4 in vivo, we constructed a set of internal deletions in a Myc epitope-tagged allele (myc-GAL11) on a single copy plasmid and tested the mutant alleles for complementation of the defect in transcriptional activation by Gcn4 in gal11Δ cells. We first constructed a multiple sequence alignment of Gal11 orthologs in four yeast species to identify regions of sequence conservation, which might be candidates for activator-binding regions (supplemental Fig. S1). This alignment revealed five regions of significant sequence conservation (Fig. 1A, regions KIX and C-II to C-V), including the KIX domain at the N terminus. In the N-terminal approximately two-thirds of the protein, the conserved blocks are interspersed with non-conserved blocks variable in length among different species and containing runs of Gln or (less frequently) Asn, with proportions of Gln and Asn ranging from 50 to 66% (Fig. 1A, regions Q-I to Q-IV). In the C-terminal one-third of Gal11, there are two blocks of non-conserved sequences that do not vary substantially in length among different species nor exhibit high levels of Gln/Asn (Fig. 1A, regions NC-I and NC-II).
FIGURE 1.
Identification of regions in GAL11 required to complement the 3ATS/Gcn− phenotype of gal11Δ and a summary of their roles in Gal11 expression and Mediator integrity. A, schematic of Gal11 domain organization (upper bar) and deletions made in a myc-Gal11 allele (lower bar). The region of similarity to the Qr domain of mammalian SRC-1 is indicated above the top bar. The criterion used for designating the KIX and regions C-II, C-III, C-IV, and C-V as conserved blocks was the occurrence of sequence identity or conservative replacements in all four fungal orthologs at >50% of the residues in the block (see “Results” for further details). The color scheme for panels A–D is explained below under Keys. B, summary of the effects of deletions in myc-Gal11 on complementation of the 3ATS phenotype of gal11Δ arg1Δ PARG1 HIS3 strain HQY1037, with relative growth determined as in panel F and scored on a scale from 0 to 10, with 10 and 0 corresponding to growth with wild-type or no myc-Gal11 allele, respectively. C, summary of the effects of deletions on expression of Myc-Gal11 protein determined as in panel E. D, summary of the effects of deletions on Mediator integrity determined as in Fig. 2A. E, transformants of HQY1037 harboring WT myc-Gal11 or the indicated deletion alleles were cultured to mid-logarithmic phase in SC-URA medium and treated with sulfometuron methyl at 0.5 μg/ml for 30 min to induce Gcn4. WCE extracts were prepared by trichloroacetic acid extraction and subjected to Western blot analysis using antibodies against the Myc epitope to detect Myc-Gal11 and antibodies against Gcd6 and Gcn4. Two different amounts of extract, differing by a factor of 3, were loaded in successive lanes for each strain. F, transformants described in E were cultured in SC-URA medium, and serial dilutions were spotted on SC-URA containing 15 mm 3AT and incubated for 3 days at 30 °C.
Based on this analysis, we constructed two or three consecutive deletions of 30–50 amino acids to cover each of the conserved blocks, three deletions that each remove an entire Gln-rich block, and three deletions of 60–100 residues that remove portions of the non-conserved blocks near the C terminus (Fig. 1A, Δ1 to Δ18). Western analysis of WCEs with anti-Myc antibodies revealed that all of the Myc-Gal11 deletion mutants were expressed at WT levels relative to the loading control (Gcd6), except for Δ4 and Δ10 that were undetectable (Δ4) or present at slightly reduced levels (Fig. 1E and data not shown; summarized in Fig. 1C). The Western analysis was conducted under conditions of amino acid starvation, and results in Fig. 1E also indicate that none of the deletions affects induction of Gcn4. Thus, except for Δ4 (which was excluded from the bulk of our analysis), mutant phenotypes conferred by these deletions cannot be attributed to reduced levels of Gal11 or Gcn4.
To determine whether the Gal11 deletions impair activation by Gcn4, we examined sensitivity of the mutants to 3-aminotriazole (3AT), an inhibitor of the histidine biosynthetic enzyme encoded by HIS3. As gcn4Δ mutants are defective for HIS3 induction, they are highly sensitive to this inhibitor (3ATS). The gal11Δ strain we employed lacks the native HIS3 allele and contains instead HIS3 coding sequences replacing those at ARG1, another Gcn4 target gene. Thus, the level of 3AT resistance in this PARG1-HIS3 strain provides a measure of ARG1 promoter activity during activation by Gcn4. We wished to monitor the ARG1 promoter because we found previously that Mediator and other coactivators are recruited by Gcn4 to higher levels above the uninduced levels at ARG1 when compared with HIS3.
Because Gal11 is a coactivator for Gcn4 at ARG1, the parental gal11Δ PARG1-HIS3 strain is more sensitive to 3AT than the isogenic GAL11 strain (Fig. 1F, 3AT). The strongest 3ATS phenotypes were conferred by Δ16 and Δ17 in the conserved C-terminal block C-V followed by Δ18 in the adjacent non-conserved block NC-II and Δ10 and Δ12 in conserved region C-IV (Fig. 1F and summary in Fig. 1B). Intermediate 3ATS phenotypes were observed for both deletions removing different portions of the KIX domain, for Δ5 in region C-II, and for Δ14 in region NC-I, whereas somewhat weaker 3AT sensitivity was conferred by Δ8 in the C-terminal portion of C-III. None of the deletions of Gln-rich blocks Q-I to Q-III had any effect on 3AT resistance (Fig. 1, A, B, and F). These results suggested that the KIX domain is important, but not absolutely required, and that sequences in other conserved and non-conserved blocks are of comparable, or even greater importance, for Gal11 function in transcriptional activation by Gcn4 at ARG1.
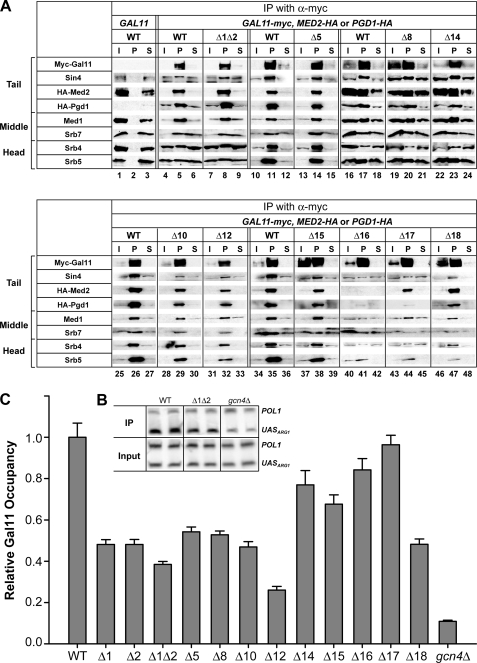
Previous results suggested that residues C-terminal to residue 866 are required for association of Gal11 with Sin4 and most likely the rest of Mediator (26, 59, 60). Hence, the strong 3ATS phenotypes of deletions in the extreme C-terminal regions of Gal11 (Fig. 1B) could result from dissociation of Gal11 or the entire tail domain from the rest of Mediator. To address this possibility, we analyzed coimmunoprecipitation of HA-tagged tail subunits Med2 and Pgd1 and also subunits of the middle (Med1, Srb7) and head (Srb4, Srb5) domains of Mediator, with Myc-tagged WT or mutant Gal11 proteins. These experiments were carried out using independently prepared extracts, with typical results shown in Fig. 2A, and the findings are summarized in Fig. 1D.
FIGURE 2.
Deletion of segments in the N-terminal third of Myc-Gal11 reduce its recruitment to the ARG1 UAS in vivo without affecting Myc-Gal11 expression or Mediator integrity. A, coimmunoprecipitation analysis of Mediator integrity. WCEs from transformants of IJY3 (gal11Δ MED2-HA), IJY4 (gal11Δ PGD1-HA), or HQY1037 (gal11Δ) containing GAL11 or the indicated myc-Gal11 alleles were immunoprecipitated (IP) with Myc antibodies. Immune complexes were subjected to Western analysis with the appropriate antibodies to detect the proteins listed on the left, Myc antibodies to reveal Myc-Gal11, and HA antibodies to reveal HA-Med2 and HA-Pgd1. I, 5% of the input WCE; P, total pellet fraction from immunoprecipitations; S, 10% of supernatant fraction. B and C, ChIP analysis of Myc-Gal11 recruitment to the ARG1 UAS by Gcn4. Transformants of HQY1037 harboring the indicated myc-GAL11 alleles and IJY1 (gcn4Δ) containing WT myc-GAL11 were cultured at 30 °C in SC-URA medium and treated with sulfometuron methyl to induce Gcn4. ChIP analysis was performed using Myc antibodies. ARG1 UAS and POL1 coding sequences were quantified in the immunoprecipitated and input chromatin samples by PCR in the presence of [33P]dATP using primers listed in Table 5. PCR products were resolved by PAGE and either visualized by autoradiography (as illustrated in panel B) or quantified with a PhosphorImager. The ratios of ARG1 to POL1 signals in immunoprecipitated samples were normalized for the corresponding ratios for input samples, and the resulting occupancy values were normalized to the occupancy measured for the WT myc-GAL11 strain to yield the relative Gal11 occupancies. At least three independent cultures and two PCR amplifications for each immunoprecipitation were performed for each strain to yield the means ± S.E. (error bars) (n = 6–16) plotted in C.
As expected, all of the Mediator subunits coimmunoprecipitated with Myc-Gal11 from WCEs of the WT myc-GAL11 strain but not from the strain with untagged GAL11 (Fig. 2A, lanes 1–6). Two of the three deletions affecting C-terminal region C-V, Δ16 and Δ17, greatly reduced coimmunoprecipitation of all subunits tested belonging to the tail, middle, or head domain of Mediator, without reducing the recovery of mutant Myc-Gal11 itself (Fig. 2A, lanes 40–45 versus 34–36). Deletion of extreme C-terminal region NC-II by Δ18 also had a marked effect on association of Gal11 with all subunits except tail subunit Med2 (Fig. 2A, lanes 46–48 versus 34–36). Thus, in agreement with previous findings, amino acids within the entire C-terminal segment, spanning residues 881–1081, are required for the interaction of Gal11 with the rest of Mediator. Two of the three deletions in C-IV (Δ10 and Δ12) consistently reduced the efficiency of coimmunoprecipitation of all subunits tested with Myc-Gal11 (Fig. 2A, lanes 28–33 versus 25–27) but had less dramatic effects when compared with the more C-terminal deletions Δ16 and Δ17. By contrast, Δ15 in C-V moderately reduced the coimmunoprecipitation efficiencies of only the other three tail subunits (Fig. 2A, lanes 37–39 versus 34–36), suggesting that the tail domain is selectively destabilized by this deletion. Importantly, the remaining 3ATS deletions, including Δ1 and Δ2 in the KIX domain, Δ5 and Δ8 in C-II and C-III, respectively, and Δ14 in NC-I, had little or no effect on coimmunoprecipitation of the other Mediator subunits with Myc-Gal11 (Fig. 2A, lanes 7–9 versus 4–6; lanes 13–15 versus 10–12; lanes 19–24 versus 16–18) and thus appear to impair Gal11 function in the context of intact Mediator.
We next employed ChIP analysis to examine whether this last group of 3ATS deletions, which do not affect Gal11 stability or Mediator integrity, reduce the occupancy of Myc-Gal11 at the ARG1 UAS on induction of Gcn4 by starvation for isoleucine and valine. In agreement with previous results (11, 12), GCN4 cells exhibit ∼10-fold greater Myc-Gal11 occupancies when compared with gcn4Δ cells, which exhibit background occupancy values (Fig. 2, B and C). Interestingly, Δ1 and Δ2 in the KIX domain, Δ5 in C-II, and Δ8 in C-III all produce marked reductions in Myc-Gall11 occupancies by 50–60% of the WT values. Δ14 provokes a smaller, but still significant, reduction in Gal11 recruitment (Fig. 2C). These findings suggest that several distinct regions in the N-terminal region of Gal11 contribute to its recruitment by Gcn4 in vivo. As none of the deletions impairs Gal11 recruitment to the degree observed for gcn4Δ, it follows that none of the corresponding regions are essential for Gal11 recruitment by Gcn4.
To identify the complete contribution of the KIX domain to Gal11 recruitment, we combined Δ1, which removes only the N-terminal helix (α1), with Δ2, which removes α2 and α3, to eliminate the entire KIX domain from Myc-Gal11. The Δ1Δ2 double mutation has no effect on Myc-Gal11 expression or its association with other Mediator subunits (data not shown). Although the combined deletion Δ1Δ2 produced a slightly larger decrease in Myc-Gal11 occupancy when compared with the single deletions (Fig. 2C), we did not discern greater 3ATS for Δ1Δ2 when compared with Δ1 and Δ2 (Fig. 1F). Hence, Δ1 and Δ2 each largely disrupts the function of the KIX domain in recruitment of Gal11 by Gcn4.
It is noteworthy that Δ16 and Δ17, which have the most dramatic effects on Gal11 association with other Mediator subunits (Figs. 1D and 2A), have little or no impact on recruitment of Gal11 by Gcn4 (Fig. 2C). This finding is in accordance with our previous determination that deletion of SIN4, which dissociates the tail from the rest of Mediator, had no effect on Gal11 recruitment by Gcn4 (26). The fact that Δ16 and Δ17 also reduce or eliminate Gal11 association with tail subunits Med2 and Pgd1 (Fig. 2A) raises the possibility that Gcn4 can recruit Gal11 as a free subunit to ARG1. If so, then the markedly reduced recruitment of Myc-Gal11 evoked by Δ10 and Δ12 would imply that the regions impaired by these deletions, C-IV, also contribute directly to Gal11 recruitment by Gcn4, a possibility we consider further under the “Discussion.” Δ15 has a smaller effect on Gal11 recruitment when compared with the deletions of similar 3ATS phenotype, such as Δ10 and Δ12 (Fig. 2C), which suggests that the 3ATS phenotype of Δ15 results at least partly from a defect in Mediator function following its recruitment by Gcn4.
The KIX Domain and Regions Impaired by Δ5 and Δ8 Make Independent Contributions to Gal11 Recruitment in Vivo and to Gcn4 Binding in Vitro
To evaluate whether each of the regions removed by deletions Δ1Δ2, Δ5, and Δ8 makes an independent contribution to Gal11 recruitment or, rather, only promotes the function of another region in recruitment, we constructed triple mutants that combine Δ1Δ2 with Δ5 or Δ8. Both triple mutants displayed stronger 3ATS phenotypes when compared with Δ1Δ2 (Fig. 3A). Consistent with this, these triple mutations, and also the Δ5Δ8 double mutation, all provoke greater reductions than the constituent mutations alone in the induction of a Gcn4-dependent reporter, UASGCRE-CYC1-lacZ, with Gcn4-binding sites upstream of a CYC1 promoter (42) (Fig. 3B). These synthetic defects in transcriptional activation do not result from decreased expression of Myc-Gal11 (Fig. 3C) or diminished Mediator integrity in the compound mutant strains (Fig. 3D and data not shown).
FIGURE 3.
Combining deletion of the Gal11 KIX domain (Δ1Δ2) with Δ5 or Δ8 exacerbates Gcn− phenotypes without affecting Myc-Gal11 expression or Mediator integrity. A, complementation of the 3ATS/Gcn− phenotypes of gal11Δ by the indicated plasmid-borne myc-GAL11 alleles was determined as described in the legend for Fig. 1. B, expression of the UASGCRE-CYC1-lacZ reporter in transformants of strain IJY3 harboring the indicated myc-GAL11 alleles was assayed by measuring β-galactosidase-specific activities in WCEs after culturing strains in SC-URA and LEU and treating with sulfometuron methyl at 0.5 μg/ml for 6 h. β-Galactosidase activities were corrected by subtracting the activity measured in transformants harboring empty vector in place of a myc-GAL11 allele and normalizing the resulting values for the corresponding corrected value measured for the WT myc-GAL11 strain to yield the plotted values. The means ± S.E. (error bars) were calculated from two independent assays on three different transformants for each construct (n = 6). C, expression of the indicated myc-GAL11 alleles was determined by Western blot analysis of the transformants in A, as described in the legend for Fig. 1. Two different amounts of extract, differing by a factor of 3, were loaded in successive lanes for each strain. D, integrity of Mediator was analyzed by coimmunoprecipitation analysis of the transformants in A, as described in the legend for Fig. 2.
Interestingly, by ChIP analysis, we observed a marked decrease in Myc-Gal11 occupancy at the ARG1 UAS for the Δ1Δ2Δ5 triple mutant versus the Δ1Δ2 and Δ5 mutants (Fig. 4A). A much smaller decrease in occupancy was observed for the Δ1Δ2Δ8 triple mutant versus the Δ1Δ2 strain (Fig. 4B). However, combining Δ5 and Δ8 also produced a strong additive reduction in Myc-Gal11 occupancy in the Δ5Δ8 double mutant (Fig. 4C). Consistent with these results, we observed a strong additive reduction in the occupancy of pol II subunit Rpb3 in the promoter and coding sequences at ARG1 in the Δ1Δ2Δ5 triple mutant when compared with the Δ1Δ2 and Δ5 strains (Fig. 4D) and in the Δ5Δ8 double mutant versus the Δ5 and Δ8 single mutants (Fig. 4F). Again, less pronounced additivity in the reduction of Rpb3 occupancy was observed on combining Δ1Δ2 and Δ8 in the Δ1Δ2Δ8 triple mutant (Fig. 4E). These results strongly suggest that the region deleted by Δ5 acts independently of the KIX domain to support Gal11 recruitment by Gcn4 and the attendant recruitment of pol II to the promoter at ARG1. Evidence suggesting that the region deleted by Δ8 also acts independently of the KIX and Δ5 regions in recruitment by Gcn4 is presented below.
FIGURE 4.
The KIX domain and regions encompassing Δ5 and Δ8 contribute independently to Myc-Gal11 and Rpb3 (pol II) recruitment in vivo. A–C, recruitment of Myc-Gal11 to the ARG1 UAS by Gcn4 was measured by ChIP analysis in transformants of gal11Δ strain HQY1037 harboring the indicated myc-GAL11 alleles, as described in the legend for Fig. 2. D–G, pol II recruitment to PARG1-HIS3 was measured by ChIP analysis of transformants of HQY1037 as described in the legend for Fig. 2 except using antibodies against Rpb3p and primers to amplify the ARG1 TATA element or 5′ end of the HIS3 coding region. Rpb3 occupancies calculated as in described in the legend for Fig. 2 were corrected by subtracting the Rpb3 occupancy measured in transformants harboring empty vector in place of a myc-GAL11 allele and normalizing the resulting values for the corresponding corrected value measured for the WT myc-GAL11 strain. At least three independent cultures and two PCR amplifications for each immunoprecipitation were performed for each strain to yield the means ± S.E. (error bars) (n = 6–16) plotted in the figure.
As indicated above, Δ16 and Δ17 disrupt the association of Gal11 with Mediator (Fig. 2A) but do not impair recruitment of Gal11 by Gcn4 (Fig. 2C). On the other hand, we found that Δ16 and Δ17 produce strong reductions in Rpb3 occupancy in the ARG1 promoter and coding region (Fig. 4G). Δ18, which has a relatively smaller effect on Mediator integrity, produces a correspondingly smaller decrease in Rpb3 occupancy (Fig. 4G). The marked reductions in pol II (Rpb3) occupancy produced by Δ16 and Δ17, along with their strong 3ATS phenotypes (Fig. 1, B and F), indicate that the high level Gal11 recruitment observed for these mutant proteins (Fig. 2C) does not provide significant coactivator function at ARG1, presumably because the mutant Gal11 proteins are not tightly associated with other Mediator subunits (Fig. 2A).
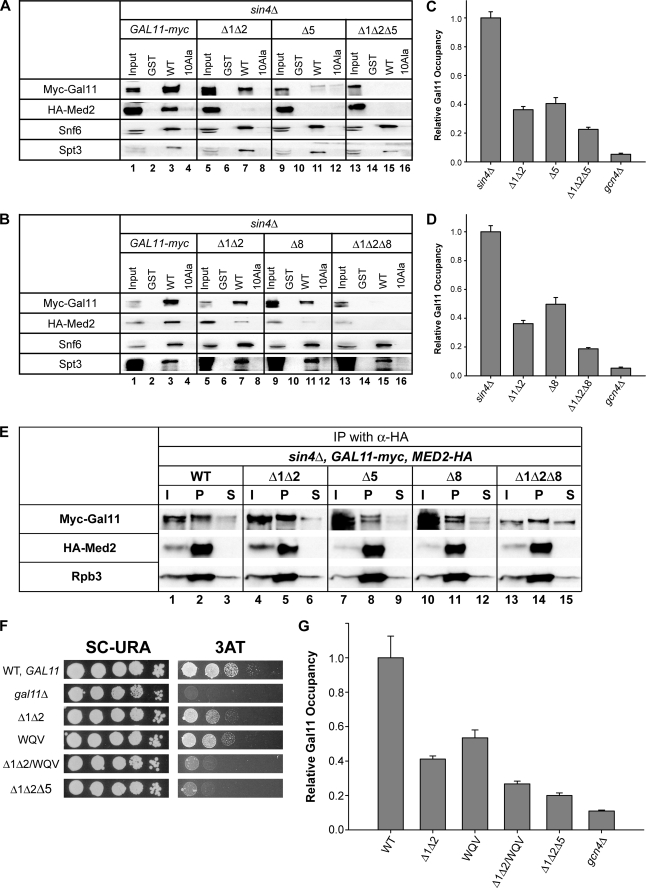
We next addressed whether the regions removed by Δ1Δ2, Δ5, and Δ8 contribute to a direct interaction of the Mediator tail domain with Gcn4 in vitro. We showed previously that the tail subcomplex containing Med2/Gal11/Pgd1 is dissociated from the rest of Mediator in WCEs of a strain lacking Sin4, and this stable subcomplex binds specifically to recombinant GST-Gcn4 in glutathione-Sepharose pulldown assays dependent on the critical hydrophobic residues in the Gcn4 activation domain (26). Accordingly, we investigated the effects of deleting KIX and the regions encompassing Δ5 and Δ8 on binding of Myc-Gal11 and HA-Med2 in sin4Δ WCEs to GST-Gcn4. Multiple pulldown assays were conducted for each mutant strain, and typical results are shown in Fig. 5, A and B.
FIGURE 5.
Three segments in the Gal11 N-terminal region function additively to promote binding to Gcn4 in vitro and recruitment to ARG1 in vivo of the Mediator tail in sin4Δ cells, and B-box mutation WQV mimics the Gcn− phenotype of Δ5 in SIN4 cells. A and B, WCEs from transformants of sin4Δ strain IJY6 harboring the indicated myc-GAL11 alleles were incubated with equal amounts of GST, GST-Gcn4 (WT), and mutant GST-Gcn4-Ala10 (10Ala) proteins immobilized on glutathione-Sepharose resin. Bound fractions and 10% of the input yeast WCEs were subjected to Western analysis with antibodies against Snf6 or Spt3, Myc antibodies to detect Myc-Gal11, and HA antibodies to detect HA-Med2. C and D, ChIP analysis of Myc-Gal11 occupancy of the ARG1 UAS in transformants of sin4Δ strain IJY6 harboring the indicated myc-GAL11 alleles, conducted as described in the legend for Fig. 2. At least three independent cultures and two PCR amplifications for each immunoprecipitation were performed for each strain to yield the means ± S.E. (error bars) (n = 6–16) plotted here. E, coimmunoprecipitation analysis of the Mediator tail subdomain in transformants of gal11Δ sin4Δ MED2-HA strain IJY6 containing the indicated myc-GAL11 alleles. WCEs were immunoprecipitated with HA antibodies, and immune complexes were subjected to Western analysis with c-Myc antibodies to detect Myc-Gal11, HA antibodies to detect HA-Med2, or Rpb3 antibodies. I, 5% of input WCE; P, total immunoprecipitate; S, 10% of supernatant. F, complementation of the 3ATS/Gcn− phenotype of gal11Δ by the indicated myc-GAL11 alleles in transformants of strain IJY3, as described in the legend for Fig. 1. G, ChIP analysis of Myc-Gal11 occupancy of the ARG1 UAS in transformants of SIN4 strain IJY3 harboring the indicated myc-GAL11 alleles, as described in the legend for Fig. 2. At least three independent cultures and two PCR amplifications for each immunoprecipitation were performed for each strain to yield the means ± S.E. (error bars) (n = 6–16) plotted here.
In agreement with previous results, we found that Myc-Gal11 and HA-Med2 bound to WT GST-Gcn4 but not GST alone or GST-Gcn4-Ala10, which harbors multiple Ala substitutions in the critical hydrophobic residues of the activation domain (Fig. 5A, lanes 1–4). Equivalent amounts of all three GST fusions were immobilized on the beads (data not shown). Importantly, deletion of the KIX domain and Δ5 both conferred marked reductions in binding of Myc-Gal11 and HA-Med2 to GST-Gcn4 without affecting the binding of SWI/SNF (Snf6) or SAGA (Spt3) subunits in the WCEs to GST-Gcn4 (Fig. 5A, cf. lanes 3, 7, and 11). Similar results were obtained for Δ8 (Fig. 5B, cf. lanes 3 and 11). Furthermore, the Δ1Δ2Δ5 and Δ1Δ2Δ8 triple mutants showed no binding whatsoever to GST-Gcn4 (Fig. 5, A and B), indicating a strong additive effect of combining either Δ5 or Δ8 with elimination of the KIX domain in this binding assay. These findings support the idea that the KIX domain and the regions removed by Δ5 and Δ8 all contribute to direct and independent interactions of the Mediator tail subcomplex with the Gcn4 activation domain.
To provide an in vivo counterpart to these in vitro binding data, we conducted ChIP assays in the sin4Δ background. We showed previously that the Mediator tail subcomplex, but not Mediator subunits from the middle or head domains, is recruited efficiently to ARG1 in sin4Δ cells (26). Thus, we observed the expected Gcn4-dependent recruitment of Myc-Gal11 to the ARG1 UAS in the sin4Δ background on starvation for Ile/Val (Fig. 5C). Importantly, Δ1Δ2, Δ5, and Δ8 all produced marked reductions in Myc-Gal11 recruitment, and combining Δ1Δ2 with either Δ5 or Δ8 led to substantial further reductions in Myc-Gal11 recruitment in the two triple mutants (Fig. 5, C and D). Coimmunoprecipitation analysis revealed that Δ1Δ2, Δ5, Δ8, and the Δ1Δ2Δ8 triple mutant do not reduce the amount of HA-Med2 that coimmunoprecipitates with Myc-Gal11 from sin4Δ extracts, indicating that the deletions do not disrupt the free tail subcomplex present in sin4Δ cells (Fig. 5E).
The results in Fig. 5, A–E, provide compelling evidence that the KIX domain and the distinct regions impaired by Δ5 and Δ8 mediate independent interactions with the Gcn4 activation domain and make additive contributions to recruitment of the Mediator tail subcomplex by Gcn4 in vivo. We currently do not understand why involvement of the Δ8 region in KIX-independent Mediator recruitment appears to be more pronounced in sin4Δ cells, where Δ8 produces a large decrease in recruitment when combined with Δ1Δ2 (Fig. 5D), when compared with the situation in SIN4 cells where Δ8 produces only a slight additional reduction in Gal11 recruitment in the Δ1Δ2 background (Fig. 4B).
Conserved B-box Motif in the SRC-1-related Region of Gal11 Is Required for Efficient Gal11 Recruitment by Gcn4
While this work was in progress, it was reported that Gal11 residues Gln-198 and Val-199 are required for its physical association in vitro with a portion of the AF-1 activation domain of mammalian glucocorticoid receptor (GR) known as τ1c and that Gln-198/Val-199 further support the activation function of τ1c in yeast cells. Interestingly, these residues occur in an ∼160-aa stretch of Gal11 that exhibits some sequence similarity to the Qr domain of the mammalian coactivator SRC-1, and the SRC-1 Qr domain can substitute for the corresponding region in Gal11 to support transcriptional activation by τ1c in yeast cells. Of two conserved motifs in this region, dubbed A- and B-boxes, only the latter, containing Gln-198 and Val-199, is required for τ1c activation in yeast (33). The Qr-related region of Gal11 extends from within Gln-rich segment Q-1, across conserved region C-II (eliminated by Δ4 and Δ5), and into Gln-rich segment Q-2 (Fig. 1A). The B-box is located at the extreme C-terminal end of the segment removed by Δ4 (Fig. 1A), whose product is unstable in yeast (Fig. 1C). To assess whether the B-box might be important for activation by Gcn4, we tested a 3-residue Ala substitution of B-box residues Trp-196, Gln-198, and Val-199 (dubbed WQV) for their effects on activation and Gal11 recruitment by Gcn4.
The WQV substitution confers a 3ATS phenotype similar to that of Δ1Δ2 and, interestingly, combining the WQV and Δ1Δ2 mutations produces a greater 3ATS phenotype than given by the individual mutations alone (Fig. 5F). Moreover, WQV and Δ1Δ2 provoked comparable and additive reductions in Gcn4-dependent activation of the UASGCRE-CYC1-lacZ reporter (Fig. 3B) and in Myc-Gal11 recruitment (Fig. 5G), similar to the effect of combining Δ5 with Δ1Δ2 (Figs. 3B and 5G). Western analysis of WCEs revealed that WQV alone or in combination with Δ1Δ2 did not significantly affect the steady-state level of Myc-Gal11 (data not shown). These findings indicate that the B-box motif is required for high level recruitment of Myc-Gal11 by Gcn4 and that the B-box and KIX domains act independently in Myc-Gal11 recruitment and attendant transcriptional activation by Gcn4. We speculate that the B-box and residues removed by Δ5 comprise a single, Gcn4-binding domain.
Evidence That Gcn4 Interacts Directly with the KIX Domain
Park et al. (31) showed previously that Gcn4 interacts in vitro with three separate fragments of recombinant Gal11 (Fig. 6A, boxes labeled Gcn4). Consistent with our results, two of these fragments encompass the regions deleted by Δ5 and Δ8 (Fig. 6A). However, the Gal11 fragment containing the entire KIX domain was not observed to bind Gcn4, Gal4, or VP16 in the previous study. Hence, we sought evidence that Gcn4 can interact specifically with a recombinant KIX domain and to confirm the ability of other recombinant regions encompassing Δ5 and Δ8 to bind Gcn4 in vitro.
FIGURE 6.
Multiple recombinant Gal11 regions interact with Gcn4p in vitro. A, schematic representation of Gal11 indicating regions (labeled Gcn4) shown previously to interact with Gcn4 in vitro (31) (upper bar), locations of GAL11 deletions analyzed here (middle bar), and the recombinant Gal11 segments tested here for binding to Gcn4 (three lower bars labeled with amino acid coordinates). B, 35S-labeled Gal11 fragments synthesized in rabbit reticulocyte lysate were incubated with equal amounts of GST, GST-Gcn4p (WT), or GST-Gcn4-Ala10 (10Ala) expressed in E. coli and immobilized on glutathione-Sepharose resin. The bound fractions and 20% of the input 35S-labeled Gal11 fragments were separated by SDS-PAGE, stained with GelCode Blue stain reagent (Pierce), and subjected to autoradiography or phosphorimaging analysis. Arrows indicate the full-length Gal11 polypeptides, which exhibit the predicted electrophoretic mobilities.
To this end, we tested 35S-labeled fragments of Gal11 translated in vitro for binding to GST-Gcn4 in glutathione-Sepharose pulldown assays. The fragment Gal111–168, containing the KIX domain but lacking the region encompassing B-box/Δ5, showed stronger binding to GST-Gcn4 than to equal amounts of GST alone or GST-Gcn4-Ala10 (Fig. 6B, lanes 1–4), consistent with a specific interaction between the Gcn4 activation domain and the Gal11 KIX domain. Similar results were obtained for fragment Gal1187–289, encompassing B-box/Δ5 (Fig. 6B, lanes 5–8), and for the full-length and various truncated forms of Gal11233–499 (generated by in vitro translation), encompassing Δ8 (Fig. 6B, lanes 9–12). These results suggest that Gcn4 can interact directly with the KIX domain, as well as two other regions encompassing B-box/Δ5 and Δ8, dependent on the critical residues in the Gcn4 activation domain. As the yield of complex formation in all of these assays was quite low, it appears that Gcn4 makes low affinity interactions with multiple segments in Gal11. The biological implications of this aspect of Gcn4-Gal11 association are discussed below.
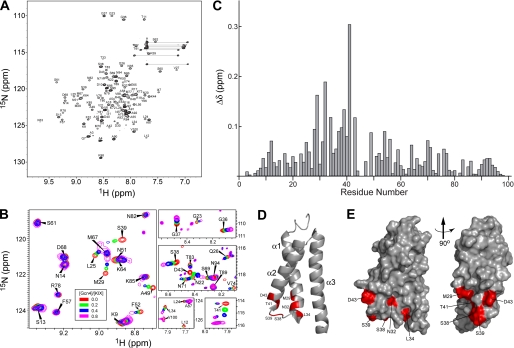
Solution NMR spectroscopy has shown that the Gal11 KIX domain (residues 6–90) forms a tight three-helix bundle packing around a hydrophobic core (35) (Fig. 7D). To provide direct evidence that the KIX domain interacts specifically with Gcn4 and to identify potential interaction sites, we used NMR chemical shift mapping for a similar construct (residues 2–100). All of the backbone assignments were made for our KIX domain construct, confirming the presence of a three-helix bundle and enabling the chemical shifts observed on binding to Gcn4 to be mapped to the structure. Spectra of the 15N-labeled KIX polypeptide were determined in the absence and presence of increasing amounts of unlabeled full-length Gcn4 or the Gcn4 activation domain (aa 2–151), up to an ∼1:1 molar ratio of the two polypeptides, and the residues experiencing significant changes in amide chemical shifts (Δδ) were identified as described under “Experimental Procedures.”
FIGURE 7.
NMR analysis of the interactions between the Gal11 KIX domain (residues 2–100) and the Gcn4 activation domain (residues 2–151). A, assigned 800-MHz two-dimensional 15N-1H HSQC spectrum of 15N,2H-labeled Gal11 KIX domain. B, representative regions of overlaid HSQC spectra of free 15N,2H-Gal11 KIX (red contours) and 15N,2H-Gal11 KIX in the presence of increasing amounts of unlabeled Gcn4 activation domain, corresponding to Gcn4/KIX molar ratios of 0.2 (green), 0.4 (blue), and 0.8 (magenta). C, the plot of amide chemical shift changes (see Equation 1 under “Experimental Procedures”) of Gal11 KIX observed upon the addition of the Gcn4 activation domain. D, ribbon representation of Gal11 KIX (Protein Data Bank (PDB) entry 2K0N) with residues displaying amide chemical shift changes. Δδ ≥ 0.10 ppm is highlighted in red. E, left, surface representation of the ribbon diagram in panel D, and right, the same surface representation rotated by 90° about the axis indicated.
For the Gcn4 activation domain, we observed significant dose-dependent chemical shift changes at 7 residues, displaying Δδ values of 0.1 ppm or greater: Met-29, Asn-32, Leu-34, Ser-38, Ser-39, Thr-41, and Asp-43 (Fig. 7, A–C). These Δδ values are ∼8-fold higher than the average Δδ value (0.012 ppm), calculated for a set residues whose spectral shifts are not appreciably affected by Gcn4 binding, e.g. Phe-57, Ser-61, Arg-78, Asn-82, Lys-85, and Thr-89 (Fig. 7B), which we regarded as experimental noise. The 7 residues displaying Δδ values of 0.1 ppm or greater map near the end of helix α1, the beginning of α2, and the loop connecting α1 and α2 (Fig. 7, D and E). We also considered residues that display shift changes between 0.05 and 0.1 ppm and found an additional 15 residues that satisfy this criterion. About half of these additional residues are located in the same regions containing the 7 residues with Δδ values greater than 0.1 ppm (supplemental Fig. S5). Thus, overall, 14 of 22 residues displaying amide chemical shift changes of 0.05 ppm or greater map to the region between aa 25 and 49. Using full-length Gcn4, the overall magnitude of the chemical shift changes upon Gcn4 binding was slightly reduced relative to that seen with the Gcn4 activation domain; however, essentially the same set of KIX domain residues in the region aa 25–49 displayed the largest chemical shift changes (supplemental Fig. S6). Together, these results indicate that specific regions of the KIX domain interact with the Gcn4 activation domain, with a prominent contribution of surface-exposed residues near the end of α1, beginning of α2, and the loop connecting these two helices (Fig. 7, D and E). This region of the KIX domain partially overlaps, but is also distinct from, the binding sites identified by this same technique for segments of the activation domains of yeast Pdr1 and mammalian sterol regulatory element-binding protein (35).
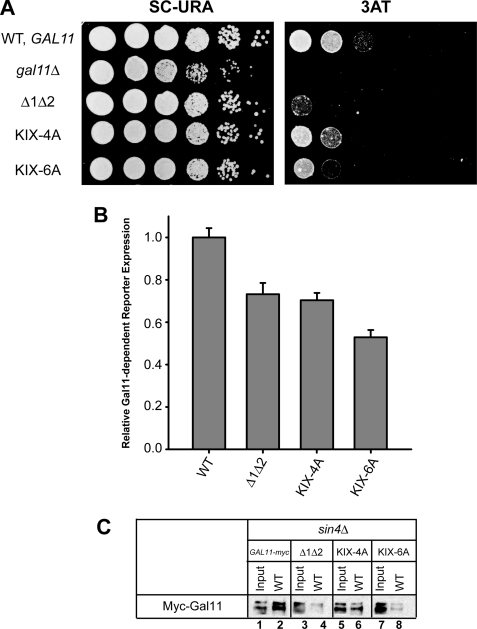
To provide evidence that residues undergoing chemical shift changes in the NMR analysis are involved in Gcn4-KIX interaction, we constructed multiple Ala substitutions in 4 or 6 of the residues that show the largest shift changes, that are surface-exposed, and that do not contribute significantly to the hydrophobic core of the structure (35). The Ala6 substitution of Met-29, Asn-32, Ser-38, Ser-39 Thr-41, and Asp-43 in the myc-Gal11 allele conferred 3AT sensitivity (Fig. 8A), reduced activation of the UASGCRE-CYC1-lacZ reporter in vivo (Fig. 8B), and reduced binding of the Mediator tail subdomain in WCEs to recombinant GST-Gcn4 in pulldown assays (Fig. 8C) to an extent comparable with the effects of the KIX deletion (Δ1Δ2) in these assays. The Ala4 substitution of Met-29, Asn-32, Ser-38, and Ser-39 similarly reduced UASGCRE-CYC1-lacZ expression and produced smaller, but still significant, reductions in 3AT resistance and binding of the tail subdomain to GST-Gcn4 when compared with the Ala6 substitution (Fig. 8). Neither substitution affected Myc-Gal11 expression in vivo (data not shown). Hence, the cluster of surface-exposed KIX residues displaying the largest chemical shift changes on interaction with Gcn4 are important for Gcn4 binding to the Mediator tail subdomain, consistent with a role in direct KIX-Gcn4 interaction.
FIGURE 8.
Ala substitutions of surface-exposed KIX residues displaying the largest chemical shift changes on interaction with Gcn4 impair activation by Gcn4 in vivo and Gcn4 binding to the Mediator tail subdomain in vitro. A, complementation of the 3ATS/Gcn− phenotypes of gal11Δ by the indicated plasmid-borne myc-GAL11 alleles was determined as described in the legend for Fig. 1. B, expression of the UASGCRE-CYC1-lacZ reporter in transformants of strain IJY3 harboring the indicated myc-GAL11 alleles was assayed by measuring β-galactosidase-specific activities in WCEs as described in the legend for Fig. 3. The means ± S.E. (error bars) were calculated from two independent assays on 4–6 different transformants for each construct (n = 8–12). C, WCEs from transformants of sin4Δ strain IJY7 harboring the indicated myc-GAL11 alleles were incubated with GST-Gcn4 (WT) immobilized on glutathione-Sepharose resin, as described in the legend for Fig. 5.
DISCUSSION
In this study, we have shown that Gal11 employs multiple distinct regions to interact efficiently with the Gcn4 activation domain, including the N-terminal KIX domain (aa 10–86), a second region encompassed by the Qr-related segment (aa 116–277) containing the B-box motif (aa 196–203) and residues removed by Δ5 (aa 202–232), and a third region encompassing the residues removed by Δ8 (aa 319–354). Deleting each of these segments individually impairs binding of the Med2/Gal11/Pgd1 tail subcomplex present in sin4Δ extracts to recombinant Gcn4 and, importantly, also reduces the recruitment of both the tail subcomplex and the native Mediator by Gcn4 to the ARG1 UAS in living cells. Furthermore, combining mutations in these three regions produced additive reductions in binding of the tail subcomplex to Gcn4 in vitro, in recruitment of the tail subcomplex or intact Mediator to ARG1 in vivo, and in the ability of Gcn4 to activate transcription. Regarding this last point, we observed additive reductions in resistance to 3AT, in activation of a Gcn4-dependent lacZ reporter, and in pol II occupancy of the ARG1 promoter and coding region when a deletion of the KIX domain was combined with Δ8 or Δ5. Similar results were obtained when the WQV substitution in the B-box motif was combined with deletion of the KIX domain. None of these mutations that impaired Mediator recruitment had any effect on Gal11 stability or integrity of the Mediator tail subdomain, either singly or in combination. Together, our findings suggest that each of these Gal11 segments interacts directly with Gcn4 and contributes to the efficiency of Mediator recruitment and attendant preinitiation complex assembly in vivo.
It could be argued that the deletion mutations defining the three distinct Gcn4-binding domains in Gal11, Δ1, Δ2, Δ5, and Δ8, alter the interaction with Gcn4 by producing gross alterations in Gal11 structure rather than excising discrete interaction modules. There are several persuasive arguments against this possibility. First, these are limited deletions of 30–50-amino acid segments that have no effect on Gal11 stability or its incorporation into the Mediator tail subdomain, even when they are combined in double or triple mutants. Second, the fact that removing the intervening Gln/Asn-rich segments had no effect on Gal11 function argues strongly in favor of a modular structure of Gal11. Third, Δ1Δ2 precisely excises the KIX domain, which folds autonomously in solution, and there is no reason to suppose that its removal would alter the structures of other domains in Gal11. Fourth, the point mutations we made in the B-box motif and the KIX domain closely approximate the phenotypes of deletions of the cognate domains, indicating that the effects of the deletions are confined to the specific segments of Gal11 they eliminate. Fifth, the deletions in the C terminus that actually do disrupt Mediator integrity had little or no effect on Gal11 recruitment, reflecting our previous findings that the Mediator tail can be recruited by Gcn4 independently of the rest of the complex (26).
Supporting the conclusion that Gal11 contains independently functioning interaction modules, we found that distinct recombinant Gal11 polypeptides containing the KIX domain, a region altered by Δ5 and the WQV substitution, and a region encompassing Δ8 all bind to recombinant Gcn4 in vitro in a manner enhanced by the hydrophobic residues in Gcn4 necessary for Mediator binding to Gcn4 in vitro and transcriptional activation by Gcn4 in vivo (47, 61, 62). Furthermore, we used NMR chemical shift mapping to provide evidence that the recombinant Gcn4 activation domain interacts specifically with the KIX domain in solution, contacting a surface that overlaps with, but is distinct from, surfaces contacted by segments of other yeast or mammalian activation domains.
Previous studies by Park et al. (31) showed that three non-overlapping segments of Gal11 can bind individually to recombinant GST-Gcn4 in vitro (Fig. 6A), but it was unknown whether these interactions are dependent on the key hydrophobic residues in the Gcn4 activation domain. Moreover, it was not determined whether mutating the relevant Gal11 segments impairs activation by Gcn4, the binding of Mediator to Gcn4 in vitro, or Mediator recruitment by Gcn4 in vivo. Two of these Gal11 segments, aa 116–255 and aa 256–410, include the B-box/Δ5 and Δ8 regions, respectively (Fig. 6A), which we implicated in Gal11 recruitment by Gcn4 with both in vitro and in vivo experiments (Fig. 5). The segment encompassing Δ8 is of additional interest because it includes a residue (Thr-322) shown previously to be required for binding and transcriptional activation by a hydrophobic 8-aa peptide dubbed P201 (37). In addition, the GAL11-P mutation, which creates a novel target in Gal11 for the Gal4 dimerization domain, also maps in the Δ8 interval at Asn-342 (59). These findings support the idea that this region of Gal11 is surface-exposed and available for interactions with activation domains.
The third Gal11 segment found by Park et al. (31) to bind Gcn4 in vitro, aa 701–864, encompasses Δ13, Δ14, and a portion of Δ15. We found that both Δ14 and Δ15 confer statistically significant decreases in Gal11 recruitment by Gcn4 to ARG1; however, the effects were smaller than observed for removal of the KIX domain, Δ5, or Δ8 (Fig. 2C). In addition, as documented in the supplemental material, we found that Δ14 has little or no effect on binding of recombinant Gcn4 to the Mediator tail subcomplex in sin4Δ extracts or in recruiting the tail subcomplex by Gcn4 in vivo, even when combined with deletion of the KIX domain (supplemental Fig. S2, C and D). On the other hand, Δ14 does confer a 3ATS phenotype (Fig. 1F and supplemental Fig. S2A) and impairs activation of the UASGCRE-lacZ reporter, and Δ14 exacerbates these same phenotypes conferred by elimination of the KIX domain (supplemental Fig. S2B). Thus, the region defined by Δ14 contributes to the coactivator function of Gal11, but it appears to function primarily after recruitment of Mediator by Gcn4.
It is noteworthy that Δ10 and Δ12 produced substantial reductions in Gal11 recruitment by Gcn4 in vivo (Fig. 2C). Because both mutations appeared to partially disrupt the interaction of Gal11 with the rest of Mediator (Fig. 2A), we did not explore intensively the possibility that the conserved region C-IV interacts directly with Gcn4. However, our finding that Δ16 and Δ17 in the extreme C terminus provoke stronger disruptions of Gal11 association with Mediator than do Δ10 and Δ12 (Fig. 2A), but confer no reduction in Gal11 recruitment by Gcn4 (Fig. 2C), suggests that the recruitment defects produced by Δ10 and Δ12 go beyond their moderate effects on Mediator integrity. Accordingly, region C-IV might contain an additional interaction module for Gcn4. Supporting this possibility, additional results shown in the supplemental material indicate that GST-Gcn4 can bind to recombinant Gal11 segment aa 355–697, containing C-IV and its flanking Gln-rich regions (supplemental Fig. S3). Hence, it is likely that all four conserved blocks flanked by Gln/Asn-rich segments in Gal11, i.e. KIX, C-II, C-III, and C-IV, provide independent binding surfaces for the Gcn4 activation domain in vivo.
Because the Gal11 KIX domain did not bind to GST-Gcn4 in the study of Park et al. (31), involvement of this highly conserved domain in Mediator recruitment by Gcn4 was unexpected. Indeed, only a small fraction of the KIX-containing polypeptide in our study was recovered in a complex with GST-Gcn4, although the interaction was specifically dependent on the key hydrophobic residues of the Gcn4 activation domain. Our NMR chemical shift data indicated a specific interaction between KIX and Gcn4 involving primarily a cluster of ∼10–15 residues at the end of α1, beginning of α2, and the loop connecting these two helices in the three-helix bundle of the Gal11 KIX domain. Although residues that exhibit NMR chemical shifts on ligand binding are not necessarily sites of direct contact, we showed that Ala substitutions of the surface-exposed KIX residues associated with the largest chemical shift changes impaired interaction of the Mediator tail subdomain with Gcn4 in vitro and the ability of Gal11 to support activation by Gcn4 in vivo. Hence, it seems likely that these residues contribute to a binding site for Gcn4 on the KIX surface.
Interactions of the KIX domains of Gal11 and mammalian MED15/ARC105 and CREB-binding protein have been mapped by NMR for segments of different mammalian and yeast activators. The results indicate overlap, but also significant differences, in the surfaces of the KIX domains that appear to interact with different activators (34–36). Our results extend this conclusion to include the Gcn4-KIX interaction. The putative Gcn4-interacting residues in the KIX domain are located in a relatively hydrophobic portion of the KIX surface (supplemental Fig. S4), consistent with the possibility of interactions with critical hydrophobic residues in the Gcn4 activation domain.
In our NMR studies, we observed fast exchange between unbound KIX and KIX in complex with Gcn4, which is consistent with a high off-rate for the Gcn4-KIX interaction. Indeed, NMR is one of the few techniques capable of mapping such transitory interactions. A high off-rate probably also characterizes the interactions between Gcn4 and the other Gal11 segments we examined, accounting for the low yields of complex formation detected in all of the pulldown assays (Fig. 6 and supplemental Fig. S3). Hence, we propose that the Gcn4 activation domain makes multiple, low affinity interactions with several regions throughout Gal11, which are cumulative and probably cooperative in nature, resulting in the specific and stable association of full-length Gcn4 with Mediator tail that we observe in GST pulldown assays (Fig. 5, A and B).
Recent studies have led to quite different conclusions about the domains in Gal11 that are critical for interacting with particular activation domains in yeast cells. Yeast activators Pdr1 and Oaf1 bind tightly to the Gal11 KIX domain in vitro and are highly dependent on this interaction for the activation of target genes in vivo (35, 36). By contrast, the activation domains in Gal4, VP16, or GR-τ1c interact weakly or not at all with the recombinant Gal11 KIX domain, and eliminating KIX has little or no effect on activation by Gal4 (31, 37) or τ1c (33) in cells. By contrast, τ1c binds tightly to a recombinant Gal11 fragment containing the B-box, and substituting the B-box completely impairs its coactivator function for τ1c (33).
Our results indicate that Gcn4 differs from both Pdr1/Oaf1 and GR-τ1c in requiring both the KIX and the B-box for wild-type levels of Mediator recruitment and transcriptional activation but is not strongly dependent on either one of these Gal11 domains for substantial Mediator recruitment and coactivator function at Gcn4 target genes in vivo. Moreover, Gcn4 binds to Gal11 segments encompassing the regions affected by Δ8 and Δ10/Δ12, which also contribute to the efficiency of Mediator recruitment by Gcn4. All of these interactions appear to be of lower affinity than those involving the KIX domain and Pdr1 or Oaf1 (35, 36). Thus, although Gcn4 appears to make multiple, low affinity interactions with several Gal11 regions, Pdr1 and Oaf1 interact more tightly, and perhaps exclusively, with the KIX domain. This view is consistent with our previous finding that the Gcn4 activation domain has a random coil structure in solution (63) and contains seven different clusters of hydrophobic residues scattered throughout the acidic activation domain, whose functions in Gcn4-Mediator interactions and transcriptional activation are highly redundant (47, 61, 62).
Acknowledgments
We thank Mark Swanson for pMJS15, Rick Young, David Stillman, Fred Winston, Stefan Bjorklund, and Joe Reese for antibodies, and Karin Musier-Forsyth and Thomas Magliery for discussion and access to equipment.
This work was supported in part by the Intramural Research Program of the National Institutes of Health.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- pol II
- RNA polymerase II
- UAS
- upstream activation sequence
- CTD
- C-terminal domain
- aa
- amino acids
- WCE
- whole-cell extract
- HA
- hemagglutinin
- SC
- synthetic complete medium
- SC-URA
- SC lacking uracil
- GST
- glutathione S-transferase
- ChIP
- chromatin immunoprecipitation
- TEV
- tobacco etch virus
- WT
- wild type
- ORF
- open reading frame
- GR
- glucocorticoid receptor
- 3AT
- 3-aminotriazole
- HSQC
- heteronuclear single quantum correlation
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1.Bourbon H. M., Aguilera A., Ansari A. Z., Asturias F. J., Berk A. J., Bjorklund S., Blackwell T. K., Borggrefe T., Carey M., Carlson M., Conaway J. W., Conaway R. C., Emmons S. W., Fondell J. D., Freedman L. P., Fukasawa T., Gustafsson C. M., Han M., He X., Herman P. K., Hinnebusch A. G., Holmberg S., Holstege F. C., Jaehning J. A., Kim Y. J., Kuras L., Leutz A., Lis J. T., Meisterernest M., Naar A. M., Nasmyth K., Parvin J. D., Ptashne M., Reinberg D., Ronne H., Sadowski I., Sakurai H., Sipiczki M., Sternberg P. W., Stillman D. J., Strich R., Struhl K., Svejstrup J. Q., Tuck S., Winston F., Roeder R. G., Kornberg R. D. (2004) Mol. Cell 14, 553–557 [DOI] [PubMed] [Google Scholar]
- 2.Gustafsson C. M., Myers L. C., Beve J., Spåhr H., Lui M., Erdjument-Bromage H., Tempst P., Kornberg R. D. (1998) J. Biol. Chem. 273, 30851–30854 [DOI] [PubMed] [Google Scholar]
- 3.Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. (1994) Cell 77, 599–608 [DOI] [PubMed] [Google Scholar]
- 4.Lee Y. C., Park J. M., Min S., Han S. J., Kim Y. J. (1999) Mol. Cell. Biol. 19, 2967–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Ranish J. A., Aebersold R., Hahn S. (2001) J. Biol. Chem. 276, 7169–7175 [PubMed] [Google Scholar]
- 6.Takagi Y., Kornberg R. D. (2006) J. Biol. Chem. 281, 80–89 [DOI] [PubMed] [Google Scholar]
- 7.Bhoite L. T., Yu Y., Stillman D. J. (2001) Genes Dev. 15, 2457–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosma M. P., Panizza S., Nasmyth K. (2001) Mol. Cell 7, 1213–1220 [DOI] [PubMed] [Google Scholar]
- 9.Park J. M., Werner J., Kim J. M., Lis J. T., Kim Y. J. (2001) Mol. Cell 8, 9–19 [DOI] [PubMed] [Google Scholar]
- 10.Kuras L., Borggrefe T., Kornberg R. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13887–13891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu H., Hu C., Zhang F., Hwang G. J., Swanson M. J., Boonchird C., Hinnebusch A. G. (2005) Mol. Cell. Biol. 25, 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govind C. K., Yoon S., Qiu H., Govind S., Hinnebusch A. G. (2005) Mol. Cell. Biol. 25, 5626–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagi Y., Calero G., Komori H., Brown J. A., Ehrensberger A. H., Hudmon A., Asturias F., Kornberg R. D. (2006) Mol. Cell 23, 355–364 [DOI] [PubMed] [Google Scholar]
- 14.Thompson C. M., Young R. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4587–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaudreau L., Schmid A., Blaschke D., Ptashne M., Hörz W. (1997) Cell 89, 55–62 [DOI] [PubMed] [Google Scholar]
- 16.Lemieux K., Gaudreau L. (2004) EMBO J. 23, 4040–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuras L., Struhl K. (1999) Nature 399, 609–613 [DOI] [PubMed] [Google Scholar]
- 18.Li X. Y., Virbasius A., Zhu X., Green M. R. (1999) Nature 399, 605–609 [DOI] [PubMed] [Google Scholar]
- 19.Qiu H., Hu C., Yoon S., Natarajan K., Swanson M. J., Hinnebusch A. G. (2004) Mol. Cell. Biol. 24, 4104–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dotson M. R., Yuan C. X., Roeder R. G., Myers L. C., Gustafsson C. M., Jiang Y. W., Li Y., Kornberg R. D., Asturias F. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14307–14310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers L. C., Kornberg R. D. (2000) Annu. Rev. Biochem. 69, 729–749 [DOI] [PubMed] [Google Scholar]
- 22.Guglielmi B., van Berkum N. L., Klapholz B., Bijma T., Boube M., Boschiero C., Bourbon H. M., Holstege F. C., Werner M. (2004) Nucleic Acids Res. 32, 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadick J. Z., Asturias F. J. (2005) Trends Biochem. Sci. 30, 264–271 [DOI] [PubMed] [Google Scholar]
- 24.Kang J. S., Kim S. H., Hwang M. S., Han S. J., Lee Y. C., Kim Y. J. (2001) J. Biol. Chem. 276, 42003–42010 [DOI] [PubMed] [Google Scholar]
- 25.Koh S. S., Ansari A. Z., Ptashne M., Young R. A. (1998) Mol. Cell 1, 895–904 [DOI] [PubMed] [Google Scholar]
- 26.Zhang F., Sumibcay L., Hinnebusch A. G., Swanson M. J. (2004) Mol. Cell. Biol. 24, 6871–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis J. A., Takagi Y., Kornberg R. D., Asturias F. A. (2002) Mol. Cell 10, 409–415 [DOI] [PubMed] [Google Scholar]
- 28.Béve J., Hu G. Z., Myers L. C., Balciunas D., Werngren O., Hultenby K., Wibom R., Ronne H., Gustafsson C. M. (2005) J. Biol. Chem. 280, 41366–41372 [DOI] [PubMed] [Google Scholar]
- 29.Myers L. C., Gustafsson C. M., Hayashibara K. C., Brown P. O., Kornberg R. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Bjorklund S., Jiang Y. W., Kim Y. J., Lane W. S., Stillman D. J., Kornberg R. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10864–10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J. M., Kim H. S., Han S. J., Hwang M. S., Lee Y. C., Kim Y. J. (2000) Mol. Cell. Biol. 20, 8709–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fishburn J., Mohibullah N., Hahn S. (2005) Mol. Cell 18, 369–378 [DOI] [PubMed] [Google Scholar]
- 33.Kim D. H., Kim G. S., Yun C. H., Lee Y. C. (2008) Mol. Cell. Biol. 28, 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F., Vought B. W., Satterlee J. S., Walker A. K., Jim Sun Z. Y., Watts J. L., DeBeaumont R., Saito R. M., Hyberts S. G., Yang S., Macol C., Iyer L., Tjian R., van den Heuvel S., Hart A. C., Wagner G., Näär A. M. (2006) Nature 442, 700–704 [DOI] [PubMed] [Google Scholar]
- 35.Thakur J. K., Arthanari H., Yang F., Pan S. J., Fan X., Breger J., Frueh D. P., Gulshan K., Li D. K., Mylonakis E., Struhl K., Moye-Rowley W. S., Cormack B. P., Wagner G., Näär A. M. (2008) Nature 452, 604–609 [DOI] [PubMed] [Google Scholar]
- 36.Thakur J. K., Arthanari H., Yang F., Chau K. H., Wagner G., Näär A. M. (2009) J. Biol. Chem. 284, 4422–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Z., Ansari A. Z., Lu X., Ogirala A., Ptashne M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8591–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon S., Qiu H., Swanson M. J., Hinnebusch A. G. (2003) Mol. Cell. Biol. 23, 8829–8845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alani E., Cao L., Kleckner N. (1987) Genetics 116, 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gietz R. D., Sugino A. (1988) Gene 74, 527–534 [DOI] [PubMed] [Google Scholar]
- 41.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 42.Hinnebusch A. G., Lucchini G., Fink G. R. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cross F. R. (1997) Yeast 13, 647–653 [DOI] [PubMed] [Google Scholar]
- 44.Reid G. A., Schatz G. (1982) J. Biol. Chem. 257, 13056–13061 [PubMed] [Google Scholar]
- 45.Zhang F., Gaur N. A., Hasek J., Kim S. J., Qiu H., Swanson M. J., Hinnebusch A. G. (2008) Mol. Cell. Biol. 28, 6796–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cigan A. M., Foiani M., Hannig E. M., Hinnebusch A. G. (1991) Mol. Cell. Biol. 11, 3217–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drysdale C. M., Jackson B. M., McVeigh R., Klebanow E. R., Bai Y., Kokubo T., Swanson M., Nakatani Y., Weil P. A., Hinnebusch A. G. (1998) Mol. Cell. Biol. 18, 1711–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balciunas D., Gälman C., Ronne H., Björklund S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson C. M., Koleske A. J., Chao D. M., Young R. A. (1993) Cell 73, 1361–1375 [DOI] [PubMed] [Google Scholar]
- 50.Hengartner C. J., Thompson C. M., Zhang J., Chao D. M., Liao S. M., Koleske A. J., Okamura S., Young R. A. (1995) Genes Dev. 9, 897–910 [DOI] [PubMed] [Google Scholar]
- 51.Jiang Y. W., Dohrmann P. R., Stillman D. J. (1995) Genetics 140, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moehle C. M., Hinnebusch A. G. (1991) Mol. Cell. Biol. 11, 2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Govind C. K., Zhang F., Qiu H., Hofmeyer K., Hinnebusch A. G. (2007) Mol. Cell 25, 31–42 [DOI] [PubMed] [Google Scholar]
- 54.Yamazaki T., Hinck A. P., Wang Y. X., Nicholson L. K., Torchia D. A., Wingfield P., Stahl S. J., Kaufman J. D., Chang C. H., Domaille P. J., Lam P. Y. (1996) Protein Sci. 5, 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wetlaufer D. B. (1963) Adv. Protein Chem. 17, 303–390 [DOI] [PubMed] [Google Scholar]
- 56.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 57.Goddard T., Kneller D. (1993) SPARKY 3, University of California, San Francisco, CA [Google Scholar]
- 58.Yamazaki T., Lee W., Arrowsmith C., Muhandiram D., Kay L. (1994) J. Am. Chem. Soc. 116, 11655–11666 [Google Scholar]
- 59.Barberis A., Pearlberg J., Simkovich N., Farrell S., Reinagel P., Bamdad C., Sigal G., Ptashne M. (1995) Cell 81, 359–368 [DOI] [PubMed] [Google Scholar]
- 60.Nishizawa M. (2001) Yeast 18, 1099–1110 [DOI] [PubMed] [Google Scholar]
- 61.Drysdale C. M., Dueñas E., Jackson B. M., Reusser U., Braus G. H., Hinnebusch A. G. (1995) Mol. Cell. Biol. 15, 1220–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson B. M., Drysdale C. M., Natarajan K., Hinnebusch A. G. (1996) Mol. Cell. Biol. 16, 5557–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huth J. R., Bewley C. A., Jackson B. M., Hinnebusch A. G., Clore G. M., Gronenborn A. M. (1997) Protein Sci. 6, 2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]