Abstract
Metastasis is a sequential process that allows cells to move from the primary tumor and grow elsewhere. Because of their ability to cleave a variety of extracellular signaling and adhesion molecules, metalloproteases have been long considered key components of the metastatic program. However, the function of certain metalloproteases, such as ADAMTS1, is not clear and seems to depend on the cellular environment and/or the stage of tumor progression. To characterize the function of ADAMTS1, we performed two alternative proteomic approaches, difference gel electrophoresis and stable isotope labeling by amino acids in cell culture, to identify novel substrates of the metalloprotease. Both techniques showed that overexpression of ADAMTS1 leads to the release of semaphorin 3C from the extracellular matrix. Although semaphorins are well known regulators of axon guidance, accumulating evidence shows that they may also participate in tumor progression. Here, we show that the cleavage of semaphorin 3C induced by ADAMTS1 promotes the migration of breast cancer cells, indicating that the co-expression of these molecules in tumors may contribute to the metastatic program.
Keywords: Cancer, Cell/Migration, Extracellular Matrix, Extracellular Matrix/Thrombospondin, Protease/ADAM/ADAMTS, Proteomics
Introduction
Metastasis, the leading cause of death in cancer patients, is a multistep process that allows selected cells to move from the primary tumor and establish secondary tumors in different organs. Metastatic cells are endowed with specific abilities in order to escape from the initial tumor, survive in circulation, arrest in a distant capillary, extravasate, and grow in a remote site. Increased cell migration is required to fulfill several steps of the metastatic program (1).
The initial characterization of cell surface and extracellular zinc endopeptidases showed that they can degrade components of the extracellular matrix (2). This finding along with the fact that, to invade neighboring tissue, migrating malignant cells induce the degradation of the extracellular matrix, led to an intuitive but simplistic hypothesis. Metalloproteases would contribute to tumor progression through the degradation of extracellular structural components. Following up on this view, the therapeutic value of several nonspecific metalloprotease inhibitors was assayed in clinical trials on cancer patients. Unfortunately, these trials failed, showing that the degradation of the extracellular matrix does not explain the role of zinc-dependent metalloproteases in human tumors (3).
Extracellular metalloproteases are classified in large families including the matrix metalloproteases, the ADAMs (proteins containing a disintegrin and a metalloprotease domain), and the ADAMTS (ADAMs with thrombospondin motifs) (4). Contradicting the initial hypothesis, it has become recently clear that individual matrix metalloproteases as well as ADAMs and ADAMTSs fulfill far more sophisticated roles. They show restricted specificity, and their function depends on the substrates they cleave. Whereas some metalloproteases are pro-tumorigenic, others act as tumor suppressors (5). For example, ADAM17 contributes to tumor growth through its ability to cleave and, thus, activate ligands for the epidermal growth factor receptor, a tyrosine kinase causally involved in the progression of many tumors (6). In contrast, by cleaving plasminogen and generating angiostatin (7), matrix metalloprotease-12 exerts the opposite action. It inhibits tumor growth by halting angiogenesis (8). The function of a third category of metalloproteases depends on the cellular environment and/or the stage of tumor progression. This is best exemplified by ADAMTS1; some experimental models clearly show that its expression impairs angiogenesis, tumor growth, and metastasis through different mechanisms (9–12). However, it is acutely up-regulated in highly metastatic breast cancer cells (13) and seems to favor local invasion and lymph node metastasis of pancreatic cancer (14).
ADAMTS1 substrates described to date include the proteoglycans aggrecan (15), versican (16), nidogens-1 and -2 (17), thrombospondin-1 (10), syndecan-4 (18), and TFPI2 (19). It has been recently shown that ADAMTS1 induces a stromal reaction in vivo through the recruitment of fibroblastic cells (20). Clearly the identification of novel substrates may shed light on the role of ADAMTS1 and, hence, on its contribution to tumor spreading.
In search for novel substrates of ADAMTS1, we have analyzed the population of gene products that are secreted (i.e. the secretome (21)) from cells overexpressing the metalloprotease. This proteomic approach identified semaphorin 3C as a putative novel substrate of ADAMTS1. Semaphorins are a large family of secreted and membrane-bound proteins. Although initially semaphorins were identified as regulators of axon guidance during the development of the nervous system, it is nowadays clear that they also contribute positively or negatively to the regulation of different aspects of tumor progression and metastasis, particularly cell migration (22, 23). Semaphorins are classified into eight classes on the basis of their structural elements. Class 3 semaphorins are the only secreted vertebrate semaphorins and are distinguished by a conserved positively charged domain at the carboxyl terminus that serves as an anchor to negatively charged components of the extracellular matrix (24). All class 3 semaphorins analyzed to date (i.e. semaphorins 3A, 3B, 3D, 3E, 3F, and 3G) inhibit cell migration and seem to be endowed with anti-tumor properties (23). For example, semaphorin 3A inhibits the migration of breast cancer cells (25) and blocks the tumor growth in vivo (26). In contrast to other class 3 semaphorins, semaphorin 3C has been poorly characterized.
The results presented here show that, in contrast to other class 3 semaphorins, semaphorin 3C promotes cell migration and that ADAMTS1 regulates its activity through its release from the extracellular matrix. Thus, these results show that both molecules, acting coordinately, increase cell migration and in this way may favor tumor invasion and metastasis.
EXPERIMENTAL PROCEDURES
Cell Lines and Transfections
Cells were grown in Dulbecco's modified Eagle's medium (DMEM)4 /F-12 (1:1) (Invitrogen) supplemented with 10% fetal calf serum and 2 mm l-glutamine. Transient and stable transfections were performed according to the manufacturer's instructions using FuGENE 6 transfection reagent (Roche Diagnostics). For ADAMTS1 overexpression, MCF7TetOff neo cells (Clontech) were transformed with the pUHD10.3 hyg vector, containing ADAMTS1 cDNA. A monoclonal cell line responding optimally to doxycycline was selected by Western blot and grown in larger quantities for use in the proteomic screenings and subsequent Western blot analysis for potential substrates.
Stable shRNA-mediated knockdown clones were obtained using the MISSION® shADAMTS1 lentiviral plasmid vectors (Sigma). Briefly, for lentivirus production, 293T cells were co-transfected with lentivirus envelope elements (pVSV-G and pRSV-Rev), lentivirus gag-pol elements (pMDL RRE, kindly provided by Dr. Juan A. Recio), and the lentiviral plasmid vectors MISSION® shADAMTS1 or MISSION® non-target shRNA control vector. Transfection was performed with FuGENE 6 (Roche Diagnostics) according to the manufacturer's instructions. Lentiviral particles were collected at 24 and 48 h post-transfection. Viruses were used to infect EW-7 cells (kindly provided by Dr. Arjan W. Griffioen) in the presence of 8 μg/ml Polybrene. Different shADAMTS1 clones were selected with 0.5 μg/ml puromycin.
Antibodies
Mouse anti-ADAMTS1 (5D4E11B5) has been described elsewhere (12). Monoclonal anti-SEMA3C was from R&D Systems (Minneapolis, MN). Anti-semaphorin 3A and semaphorin 3B were from Abcam (Cambridge, UK). Additional chemicals were from Sigma.
DIGE
ADAMTS1 MCF7 cells from a single clone were grown in 15-cm dishes in the presence or absence of doxycycline and allowed to attach and reach 90% confluence. Then DMEM/F-12 + 10% fetal bovine serum (FBS) was replaced with serum-free medium, again with or without doxycycline, and the medium was allowed to accumulate shed proteins for 48 h. The media were collected and further processed as described in Bech-Serra et al. (27).
SILAC Labeling and Sample Preparation
Cells were seeded in SILAC DMEM medium (Invitrogen) supplemented with dialyzed serum (amino acid-free) and the appropriate normal or isotopically labeled amino acids (see also Ref. 28). Cell culture conditions were first optimized for maximal isotopic incorporation and for minimizing arginine to proline conversion by the cell metabolism. We chose [12C,14N]lysine and [12C,14N]arginine “light” (L) amino acids in combination with ADAMTS1 overexpression (no doxycycline) and [13C6,14N]Lys and [13C6,15N4]Arg “heavy” (H) amino acids for the normal condition (doxycycline added). Cells were incubated with heavy or light amino acids in the presence or absence of doxycycline for a total of 7 days, corresponding to ∼6 cell doublings. In the final 48-h serum was left out to reduce serum protein background and to accumulate shed proteins. In addition, to maximize protein yields, the cells were seeded so they reached confluency on day 7. Cells were counted, and the amount of medium was compared with this. Big differences (>5%) in cell counts between treated and untreated situations, possibly due to toxic or proliferative effects of ADAMTS1 overexpression, were not observed. After this, the supernatant of control and overexpressing cells was pooled, and further technical bias, therefore, had equal effects on both heavy and light protein pools, leaving the H/L ratio unaffected. Conditioned media were incubated overnight at 4 °C with wheat germ agglutinin-agarose beads (1 μl of stirred suspension/ml of conditioned medium). Glycoproteins were eluted by rotating for 2 h at 4 °C with 4 ml of 0.5 m n-acetylglucosamine in 10 mm HEPES (pH 7.5), 0.15 m NaCl buffer. The glycoprotein solutions were then concentrated 40-fold by ultrafiltration (Amicon Ultra-4, 5-kDa cutoff; Millipore, Carrigtwohill, Ireland). Urea was then added to the concentrated protein solution to a final 8 m concentration, and pH was adjusted to 8.5. Proteins were reduced by the addition of 50 mm dithiothreitol and incubation for 30 min at room temperature and then carbamidomethylated with 125 mm iodoacetamide for 1 h at room temperature. The protein mixture was then purified by a modified acetone-trichloroacetic acid precipitation (2D-CleanUp kit; GE Healthcare). The resulting protein pellet was resuspended in 40 μl of SDS-PAGE loading buffer and subjected to one-dimensional electrophoresis on a 10% polyacrylamide-SDS gel.
Liquid Chromatography-Mass Spectrometric Analysis
The SDS-PAGE gel was cut into 20 horizontal slices, and individual slices were digested using modified porcine trypsin (Promega). Digests were analyzed on an Esquire HCT IT mass spectrometer (Bruker, Bremen, Germany) coupled to a nano-high performance liquid chromatography system (Ultimate, LC Packings, The Netherlands). Peptide mixtures were initially concentrated on a 300-μm id, 1-mm PepMap nanotrapping column and subsequently loaded onto a 75-μm inner diameter, 15-cm PepMap nanoseparation column (LC Packings). An acetonitrile gradient (0–60% B in 150 min, where B is 80% acetonitrile, 0.1% formic acid in water; flow rate ∼300 nl/min) was used to elute the peptides through a PicoTip emitter nanospray needle (NewObjective, Woburn, MA) onto the nanospray ionization source of the IT mass spectrometer. MS/MS fragmentation (1.9 s, 100–2800 m/z) of two of the most intense ions was carried out, as detected from a 1.2-s MS survey scan (310–1500 m/z) using a dynamic exclusion time of 1.2 min for precursor selection and excluding single-charged ions. An automated optimization of MS/MS fragmentation amplitude beginning at 0.60 V was used.
Protein Identification and Data Analysis
Data processing for protein identification and quantification was performed using WARP-LC 1.1 (Bruker), a software platform integrating liquid chromatography-MS run data processing, protein identification through a data base search of MS/MS spectra and protein quantification based on the integration of the chromatographic peaks of MS-extracted ion chromatograms for each precursor. Proteins were identified using Mascot (Matrix Science, London, UK) to search the International Protein Index Human 3.26 data base (67,665 sequences, 2,846,2007 residues) (29). MS/MS spectra were searched with a precursor mass tolerance of 1.5 Da, fragment tolerance of 0.5 Da, trypsin specificity with a maximum of 1 missed cleavage, cysteine carbamidomethylation set as fixed modification and methionine oxidation, and the corresponding Lys and Arg SILAC labels as variable modifications. Positive identification criterion was set as an individual Mascot score for each peptide MS/MS spectrum higher than the corresponding homology threshold score. For protein quantification, H/L ratios were calculated averaging the measured H/L ratios for the observed peptides after discarding outliers. For selected proteins, quantification data obtained from the automated WARP-LC analysis was manually reviewed.
Generation of Semaphorin Constructs
Wild-type S3C cDNA was a gift from Dr. A Tufro (Albert Einstein College of Medicine, New York); based on this we generated our S3C constructs. Deletion constructs were generated using standard techniques. Semaphorin S3A and S3B constructs were a gift from Dr. G. Neufeld (Rappaport Faculty of Medicine, Haifa, Israel) and Dr. S. Naylor (University of Texas).
Western Blot Analysis
Cells were plated in 60-mm dishes in DMEM/F-12 + 10% FBS. After 24 h cells were incubated with serum-free DMEM (identical to SILAC conditions) and treated in the presence or absence of doxycycline for 48 h. After that the cells were lysed for 30 min in ice-cold 1% Triton X-100 and kept on ice in further steps to avoid unwanted proteolytic activity. Total protein in the lysates was quantified, and equal amounts of protein from cell lysates were concentrated with wheat germ agglutinin-agarose beads (Vector Laboratories, Burlingame, CA) rotating for 2 h at 4 °C. Proteins were eluted directly in SDS-polyacrylamide gel electrophoresis sample buffer (Laemmli buffer). Protein loading of conditioned medium samples for Western blot analysis was adjusted according to the total protein in cell lysates and checked by Ponceau red staining of the blotted nitrocellulose membrane (Bio-Rad). Finally, when analyzing the matrix, cells were detached non-enzymatically using 10 mm EDTA in phosphate-buffered saline, and after washing with phosphate-buffered saline the remaining extracellular matrix was scraped from the plates in sample buffer and loaded onto an SDS-PAGE gel. Detection was performed using the corresponding horseradish peroxidase-conjugated secondary antibody and the SuperSignal chemiluminescence kit from Pierce.
Treatment with Protease Inhibitors
Cells were treated with 5 mm BB94 (a specific metalloprotease inhibitor, British Biotech), for the indicated times, and the processing of S3C was analyzed as described above.
Human Umbilical Vein Endothelial Cell (HUVEC) Repulsion Assay
Chemorepulsion of different semaphorin fragments was analyzed by co-culturing semaphorin expressing HEK293 cells on a HUVEC cell monolayer (gift from Dr. J. Seoane, Vall d'Hebron Institut de Recerca). HEK293 cells were plated and transfected with different semaphorin fragments using FuGENE, and after 12 h they were trypsinized and plated on a HUVEC cell layer (2000 cells/plate). HUVEC cells were used between passages 2 and 8. Endothelial cell medium (ECM-2, Cambrex Bio Science Walkersville, MD) was used to culture HUVEC with the addition of hydrocortisone, human fibroblast growth factor-B, R3-insulin-like growth factor-1, ascorbic acid, FBS (2%), human epidermal growth factor, and GA-1000. Heparin was left out because it causes the release of S3C and ADAMTS1 from the matrix. Some forms of vascular endothelial growth factor are known to compete with semaphorins for their receptors and were, therefore, also omitted from the culture media. The omission of these factors had no observable effect on HUVEC cells during the time of the assay (data not shown). Cell repulsion was assessed after 48 h of co-culture.
Transwell Assays
Chemotaxis toward different semaphorin fragments was analyzed using a 8.0 μm polycarbonate membrane Transwell permeable support (Corning). In the lower compartment, HEK293 cells were plated and transfected using FuGENE with different constructs. In the upper compartment, the membrane was pretreated with DMEM/F-12 + 10% FBS, and subsequently MDA231 or MCF7 cells were plated on the membrane (20,000 cells/well). After 4 h of attachment, these membranes were placed in the wells with mock-transfected or semaphorin-producing HEK293 cells to start the migration assay. After 12 h membranes were removed, the cells were fixed for 30 min in 2.5% glutaraldehyde in phosphate-buffered saline, non-migrated cells on the upper part of the membrane were removed with a cotton swab, and the membranes were subsequently mounted in Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories). Finally the 4′,6-diamidino-2-phenylindole-stained nuclei were counted under the UV microscope (Nikon). A positive (DMEM/F-12, 10% FBS + mock- transfected cells) and a negative (DMEM- F-12, serum-free + mock-transfected cells) control for chemoattraction were included in each experiment.
RESULTS
Generation and Characterization of Cells Conditionally Expressing ADAMTS1
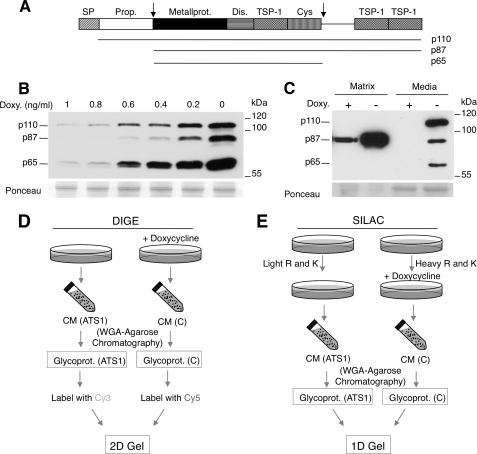
ADAMTS1 is a modular protein composed of signal peptide, propeptide, metalloproteinase, disintegrin, thrombospondin type I-like (TSP-1), and cysteine-rich (Cys-rich) domains (30, 31); see also Fig. 1A. Although ADAMTS1 seems to be down-regulated in samples from primary breast cancers (32), it has been shown that ADAMTS1 is one of the most up-regulated genes in highly metastatic breast cancer cells (13), indicating that it may contribute to tumor spreading.
FIGURE 1.
Generation of a cell line conditionally expressing ADAMTS1 and proteomic approaches to analyze glycosylated secretomes. A, the schematic shows the different domains of ADAMTS1. SP, signal peptide; Prop., propeptide; Metallprot., metalloprotease; TSP-1, thrombospondin-1 type repeat; Dis., disintegrin domain; Cys, cysteine-rich region. Two cleavage sites are marked with arrows; the length of the different ADAMTS1 forms generated through these cleavages, p110, p87, and p65, are indicated. B, MCF7 Tet-Off cells stably transfected with a vector encoding human ADAMTS1 under the control of a promoter repressible by doxycycline (Doxy., MCF7 Tet-Off/ADAMTS1 cells) were treated with different concentrations of the antibiotic for 48 h. The media conditioned by these cells was analyzed by Western blot with anti-ADAMTS1 antibodies. Ponceau S staining was used as loading control. C, MCF7 Tet-Off/ADAMTS1 cells were cultured with or without 1 ng/ml doxycycline for 48 h. Then, cells were detached with phosphate-buffered saline containing 5 mm EDTA. Components of the extracellular matrix were extracted with Laemmli sample buffer. Samples from the conditioned media or from the extracellular matrix were analyzed by Western blot with anti-ADAMTS1 antibodies. Ponceau S staining was used as loading control. D, the schematic shows the DIGE and SILAC protocols. Left, the conditioned media of MCF7 Tet-Off/ADAMTS1 cells treated with or without doxycycline for 48 h (i.e. ADAMTS1-overexpressing or control cells, respectively) were collected. Glycoproteins were purified using wheat germ agglutinin (WGA) chromatography as described under “Experimental Procedures.” Next, purified glycoproteins from cells overexpressing ADAMTS1 or control cells were labeled with the cyanine dyes Cy3 and Cy5, respectively, mixed 1:1, and analyzed by two-dimensional (3D) electrophoresis and mass spectrometry. Right, MCF7 Tet-Off/ADAMTS1 cells were grown with either light or heavy lysine and arginine for 7 days. Labeled cells were treated with or without doxycycline for 48 h, and the media were collected. Isotopically labeled glycoproteins were purified using wheat germ agglutinin chromatography as described under “Experimental Procedures” and mixed 1:1, and the mixture was fractionated through one-dimensional (1D) SDS-PAGE, liquid chromatography (LC), and mass spectrometry.
As a means to identify novel substrates of ADAMTS1, we generated cell lines expressing the metalloprotease in a repressible manner. Analysis of the media conditioned by MCF7 Tet-Off cells permanently transfected with ADAMTS1 and cultured in the absence of doxycycline showed the expected array of ADAMTS1 isoforms (Fig. 1B, right lane). Previous characterization of these isoforms showed that p110 corresponds to the inactive proADAMTS1 precursor, p87 is the product of the processing of p110 by furin-like convertases, and p65 is the product of the subsequent processing of p87 by metalloproteases (Ref. 33 and Fig. 1A).
Doxycycline is a nonspecific metalloprotease inhibitor that could interfere with the identification of ADAMTS1 substrates. For example, collagenases can be inhibited by concentrations of doxycycline of 30 μm (34). To avoid the interference of doxycycline in our assays, we used the minimal concentration that effectively blocks the expression of ADAMTS1, i.e. 2.1 nm (1 ng/ml) (Fig. 1B). This concentration is not expected to affect the activity of metalloproteases known to be inhibited by doxycycline (34).
Secreted ADAMTS1 has been shown to bind to components of the extracellular matrix via its thrombospondin domains (35). Accordingly, the 87-kDa isoform of ADAMTS1 was predominantly found in the protein fraction corresponding to the extracellular matrix (Fig. 1C). These results show that the MCF7 Tet-Off/ADAMTS1 cell line is a useful system to functionally characterize the metalloprotease and that the concentration of doxycycline required to effectively repress the expression of ADAMTS1 is not expected to inhibit its metalloprotease activity.
Identification of ADAMTS1 Substrates Using Different Proteomic Techniques
To identify novel substrates of ADAMTS1, we compared the secretome of MCF7 Tet-Off/ADAMTS1 cells cultured in the presence or absence of doxycycline using two complementary proteomic techniques: DIGE and SILAC.
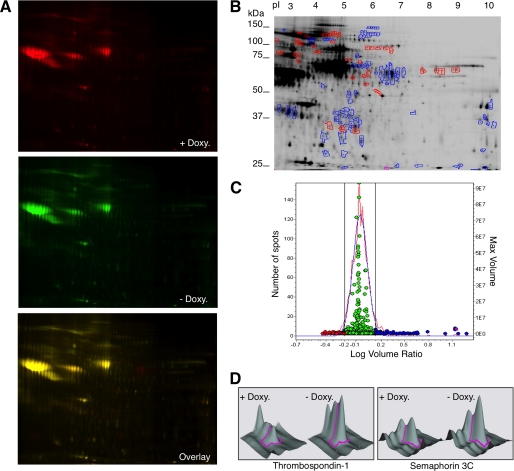
DIGE is a robust quantitative proteomic technique in which two samples are labeled with different fluorochromes and loaded onto a single two-dimensional gel (36), thereby overcoming the lack of reproducibility inherent to different two-dimensional gels. To apply this technology, we purified glycoproteins from the media conditioned by MCF7 Tet-Off/ADAMTS1 cells treated without or with doxycycline and labeled them with the cyanine dyes Cy3 or Cy5, respectively; the labeled glycoproteins were separated in two-dimensional gel electrophoresis (Fig. 1D). To visualize the protein spots, the gel was scanned with fluorophore-specific excitation and emission wavelengths, and two independent images were acquired (Fig. 2A), each corresponding to an individual Cy-labeled probe. Image analysis and determination of significant alterations in spot abundances were performed automatically with the DeCyder software (Fig. 2, B and C).
FIGURE 2.
Quantitative comparison between the glycosylated secretome of control cells and cells overexpressing ADAMTS1 by DIGE. A, fluorochrome-labeled glycoproteins from MCF7 Tet-Off/ADAMTS1 cells treated with or without doxycycline (Doxy., i.e. control cells or cells overexpressing ADAMTS1, respectively, see Fig. 1, C and D) were analyzed into a two-dimensional gel. The gel was scanned with different wavelengths to visualize proteins labeled with Cy3 or Cy5. B, a total of 1414 spots was analyzed. 132 showed a >1.5-fold change. 83 spots increased in intensity (blue dots/blue-encircled spots), whereas 49 spots decreased (red dots/red-encircled spots) upon ADAMTS1 overexpression. C, experimental (red curve) and normalized model (blue curve) frequency distribution of volume ratios (volume in the Cy5 image divided by volume in the Cy3 image) for the spots detected in the fluorescence images shown in B. The volume of each individual protein spot, represented as a single data point, is plotted in the right axis. The spots in blue and red represent proteins with a higher than 1.5-fold or lower than 0.66 variation, respectively, in the glycoprotein secretome of cells overexpressing ADAMTS1. D, shown is a three-dimensional profile for the spots corresponding to thrombospondin-1 and semaphorin 3C as determined by the DIGE analysis shown in B.
Only those protein spots exhibiting a change in volume ratio exceeding 1.5-fold were selected (Fig. 2, B and C), excised, and subjected to in-gel digestion with trypsin. The resulting peptides were then subjected to peptide mass fingerprinting analysis by matrix-assisted laser desorption ionization mass spectrometry. In a search of the International Protein Index human data base, peptide mass information identified 28 of the 83 protein spots analyzed, assigning them to 5 proteins (Table 1). As expected, several spots corresponded to the ADAMTS1 (Table 1). In addition we identified thrombospondin-1 (Fig. 2D, left), a known substrate of ADAMTS1 (10), peroxidasin, a secreted peroxidase expressed in several tumors, calsyntenin-1, a transmembrane protein whose cytoplasmic domain binds calcium, and semaphorin 3C (Fig. 2D, right), a secreted protein that belongs to a family of regulators of axonal guidance that also contribute to tumor progression and angiogenesis (22).
TABLE 1.
Proteins identified by DIGE and mass spectrometry
| Protein name | UNIPROT accession | Average ratio −/+ doxycycline | S.D. | Number of spots |
|---|---|---|---|---|
| ADAMTS-1 | ATS1_HUMAN | 8.5 | 1.6 | 7 |
| Thrombospondin-1 | TSP1_HUMAN | 3.3 | 0.7 | 4 |
| Peroxidasin homolog (melanoma associated antigen MG50) | PXDN_HUMAN | 2.6 | 0.6 | 6 |
| Calsyntenin-1 | CSTN1_HUMAN | 1,7 | 0.1 | 2 |
| Semaphorin-3C | SEM3C_HUMAN | 1.6 | 1 |
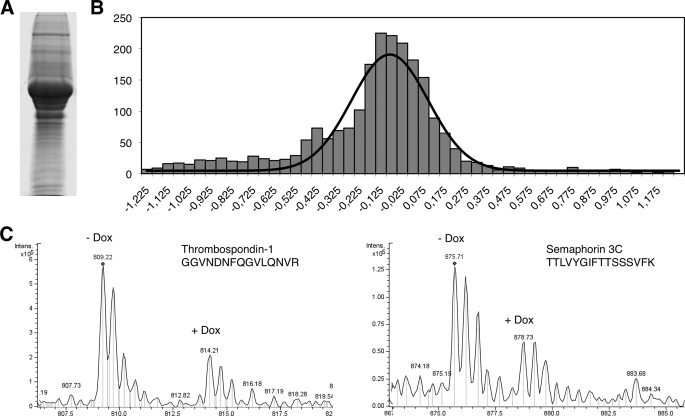
To extend and confirm these results, we repeated the proteomic analysis by SILAC, a technique based on the labeling of proteins with isotopic variants of the same atom (37). To perform the SILAC analysis, we cultured MCF7 Tet-Off/ ADAMTS1 with arginine and lysine labeled with light or heavy isotopes (Fig. 1E and see “Experimental Procedures”). Then the expression of ADAMTS1 was repressed in cells labeled with heavy isotopes by the addition of doxycycline to the culture media. Next, the media conditioned by these cells as well as that corresponding to cells expressing ADAMTS1 (i.e. cells cultured in the presence of light arginine and lysine kept in the absence of doxycycline) were collected, and glycoproteins were purified, mixed 1:1, and subjected to SDS-PAGE. After separation (Fig. 3A), the gel was cut horizontally in 20 slices. These gel segments were separately processed (see “Experimental Procedures”) to identify peptides by mass spectrometry and to compare their relative abundance in lysates from control cells and cells overexpressing ADAMTS1.
FIGURE 3.
Quantitative comparison between the glycosylated secretome of control cells and cells overexpressing ADAMTS1 by SILAC. A, SDS-PAGE separation of the SILAC-labeled mixture of glycoproteins. B, shown is the distribution of the average ratios heavy/light. The average ratios heavy/light of the proteins identified, distributed in the indicated intervals, were plotted against number of proteins in each interval. C, mass spectra of peptide doublets from the selected proteins are shown. The sequences of the peptides are shown in the one-letter code. Dox, doxycycline.
3163 peptides from 749 individual proteins were identified. Peptide coverage analysis from individual transmembrane proteins randomly chosen showed only peptides from the extracellular domain (data not shown), ruling out a significant contamination from the cell fraction. The distribution of the H/L ratios indicated that the levels of the majority of glycoproteins were not affected by the expression of ADAMTS1 (Fig. 3B). Twenty proteins exhibited average ratios H/L below the interval 0.7 (Table 2). To illustrate these results peptide doublets from thrombospondin-1 and semaphorin 3C are shown in Fig. 3C. Given the possible involvement of semaphorin 3C in the regulation of cell migration and metastasis and because it was identified in the two proteomic approaches, we chose this protein for further analysis.
TABLE 2.
Proteins identified by SILAC and mass spectrometry
TGFβ, transforming growth factor; PTP, protein-tyrosine phosphatase; ALCAM, activated leukocyte-cell adhesion molecule.
| Protein name | UNIPROT accession | Average ratio H/L | S.D. | No. of peptides |
|---|---|---|---|---|
| Dihydropyridine receptor α2 subunit | Q9UIU0_HUMAN | 0.17 | 0.061 | 3 |
| ADAMTS-1 | ATS1_HUMAN | 0.17 | 0.130 | 62 |
| Fibulin-1 | FBLN1_HUMAN | 0.26 | 0.366 | 6 |
| Cerebellin-4 | CBLN4_HUMAN | 0.27 | 0.141 | 4 |
| SEZ6L2 | SE6L2_HUMAN | 0.40 | 0.095 | 19 |
| Transferrin receptor | TFR1_HUMAN | 0.48 | 0.376 | 3 |
| Thrombospondin-1 | TSP1_HUMAN | 0.50 | 0.134 | 18 |
| Semaphorin-3C | SEM3C_HUMAN | 0.52 | 0.194 | 5 |
| Profilin-1 | PROF1_HUMAN | 0.58 | 0.082 | 5 |
| Latent TGFβ-binding protein 1 | LTBP1_HUMAN | 0.61 | 0.204 | 12 |
| PTP receptor type, K | B7ZMG0_HUMAN | 0.63 | 0.149 | 13 |
| PTP receptor γ | PTPRG_HUMAN | 0.64 | 0.375 | 13 |
| Vitronectin | VTNC_HUMAN | 0.65 | 0.514 | 5 |
| ALCAM | CD166_HUMAN | 0.68 | 0.199 | 14 |
| Galectin-3 | LEG3_HUMAN | 0.69 | 0.063 | 8 |
| Syndecan-1 | SDC1_HUMAN | 0.69 | 0.306 | 3 |
| Attractin | ATRN_HUMAN | 0.70 | 0.240 | 4 |
ADAMTS1 Causes the Release of Extracellular Matrix-bound Semaphorin 3C
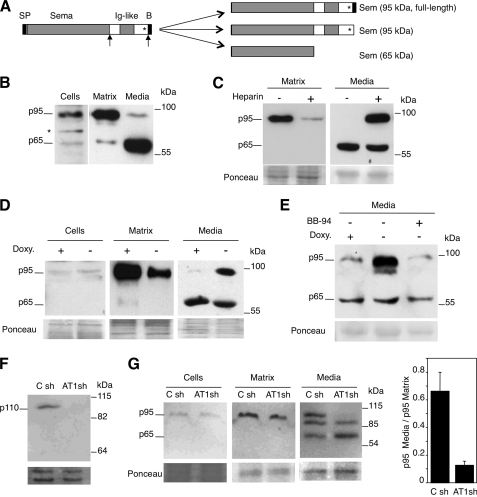
Class 3 semaphorins are modular proteins composed of the Sema domain, essential for signaling, an Ig-like domain, and a carboxyl-terminal conserved basic domain (38). The biosynthesis of class 3 semaphorins is complex; it involves proteolytic processing at at least two sites (Fig. 4A). These cleavages affect only a fraction of semaphorin 3 molecules, occur intra- and extracellularly, and generate at least three species that differ in their activity and their ability to interact with the extracellular matrix. Although the biosynthesis of semaphorin 3A and 3B has been characterized in detail (Adams et al. (39) #2021; De Wit et al. (40) #2020; Varshavsky et al. (41) #2067), much less is known about that of semaphorin 3C. In MCF7 cells semaphorin 3C is expressed as p95 and p65 species (Fig. 4B, Cells). Based on the previous reports on other class 3 semaphorins (39), we identified these isoforms as full-length semaphorin 3C and the product of processing by furin after the Sema domain (see Fig. 4A).
FIGURE 4.
ADAMTS1 releases semaphorin 3C from the extracellular matrix. A, left, the schematic shows the different domains of class 3 semaphorins. SP, signal peptide; Sema, semaphorin domain; Ig-like, immunoglobulin-like domain; B, basic domain. The asterisk denotes the position of a cysteine involved in the establishment of homotypic interactions. The arrows mark the position of the proteolytic cleavage by furin and/or metalloproteases. Right, the schematic shows the different isoforms generated by proteolytic processing of class 3 semaphorins. The approximate molecular weight is indicated. Note that because of its small size, the proteolytic removal of the basic domain does not affect the apparent molecular weight of class 3 semaphorins. B, MCF7 cells were cultured for 48 h, detached with phosphate-buffered saline containing 5 mm EDTA, and lysed. Components of the extracellular matrix were extracted with Laemmli sample buffer. Samples from cell lysates, the conditioned media, or the extracellular matrix were analyzed by Western blot with anti-semaphorin 3C antibodies. C, samples from the extracellular matrix or from the media conditioned by MCF7 cells, cultured in the absence or presence of 0.1 mg/ml heparin, were detached and analyzed as in B. Ponceau S staining was used as the loading control. D, MCF7 Tet-Off/ADAMTS1 cells were cultured with or without 1 ng/ml doxycycline (Doxy.) for 48 h. Then cells were detached with phosphate-buffered saline containing 5 mm EDTA and lysed. Components of the extracellular matrix were extracted with Laemmli sample buffer. Samples from cell lysates, conditioned media, or the extracellular matrix were analyzed by Western blot with anti-semaphorin 3C antibodies. Ponceau S staining was used as the loading control. E, samples from the media conditioned by MCF7 Tet-Off/ADAMTS1 cells cultured with or without 1 ng/ml doxycycline or 10 μm BB94 for 48 h were analyzed by Western blot with anti-semaphorin 3C antibodies. Ponceau S staining was used as the loading control. F, the conditioned media of EW-7 cells expressing control shRNAs (C sh) or shRNAs targeting ADAMTS1 (AT1 sh) were analyzed by Western blot with anti-ADAMTS1 antibodies. Ponceau S staining was used as loading control. G, transiently transfected semaphorin 3C from the same cells as in F was analyzed as in D. The levels of matrix-associated and soluble p95 Semaphorin 3C were quantified. The bars represent the averages (±S.D.) of three independent experiments.
Semaphorin 3A interacts with components of the extracellular matrix via the basic amino acid-rich carboxyl-terminal region (40). Similarly, we found that whereas the p95 form of semaphorin 3C associates with extracellular components (Fig. 4B), the 65-kDa isoform of semaphorin was largely soluble (Fig. 4B). Extracellular matrix-associated semaphorin 3C could be released to the medium by treatment with the highly sulfated glycosaminoglycan heparin (Fig. 4C), supporting that the p95 isoform corresponds to full-length semaphorin 3C and contains the basic residues at the carboxyl terminus of semaphorin 3C. This carboxyl-terminal basic domain is responsible for the electrostatic interaction with the negatively charged proteoglycans of the extracellular matrix. Overexpression of ADAMTS1 did not have an effect on the levels of cell-associated semaphorin 3C (Fig. 4D), but confirming the results of the proteomic analysis induced its release from the extracellular matrix to the extracellular medium (Fig. 4D).
The molecular size of soluble p95 semaphorin 3C is indistinguishable from that of matrix-associated p95 (data not shown), indicating that if ADAMTS1 releases p95 semaphorin 3C from the extracellular matrix through proteolytic cleavage, it occurs near the carboxyl terminus. Supporting this possibility, Adams et al. (39) showed that the carboxyl-terminal fragment proteolytically removed from semaphorin 3D is very small (∼1.3 kDa). To confirm that ADAMTS1 releases p95 from the extracellular matrix through a proteolytic cleavage, we analyzed the effect of BB94, a metalloprotease inhibitor. As shown in Fig. 4E, BB94 prevented the release of p95 semaphorin 3C by ADAMTS1.
The results in Fig. 4, B–E, were obtained with transfected cells. To analyze the role of the endogenous metalloprotease on the release of semaphorin 3C, we used EW-7, a cell line derived from Ewing's sarcoma, because it expresses detectable levels of ADAMTS1 (Fig. 4F). Knockdown of ADAMTS1 from EW-7 cells with specific shRNAs led to a defect in the release of p95 semaphorin 3C (Fig. 4G), further supporting a role for the metalloprotease in the solubilization of this semaphorin isoform. Interestingly, the knockdown of ADAMTS1 did not have an effect on the levels of p65 semaphorin 3C or a ∼87-kDa fragment produced by EW-7 cells, showing that the production of this fragment is independent of ADAMTS1. Collectively, the results shown in Fig. 4 show that ADAMTS1 is endowed with the ability to release semaphorin 3C from the extracellular matrix through a proteolytic cleavage.
Effect of Different Class 3 Semaphorins on HUVECs
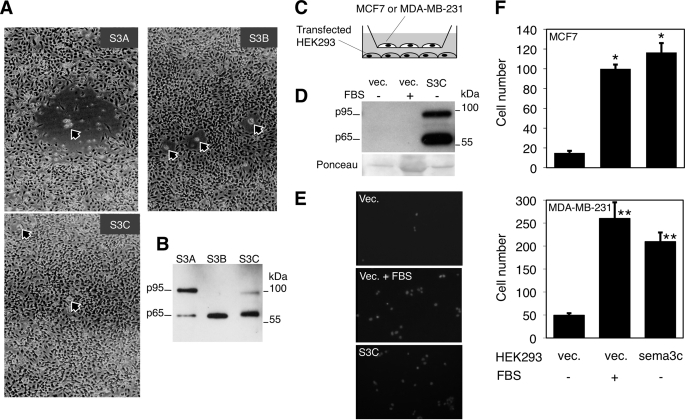
To analyze the effect of semaphorin 3C on cell migration, we used HUVEC, a classical model that readily responds to semaphorin signaling (41). Incubation of HUVECs in the presence of control MCF7 cells or the same cells overexpressing ADAMTS1 did not have an effect on the migration of the former (data not shown). This result was unexpected because MCF7 cells produce semaphorin 3C (Fig. 4), and other class 3 semaphorins have been shown to induce the repulsive response on HUVECs (41).
To assay semaphorin signaling with proper controls, we used the HEK293 cells, which express undetectable levels of class 3 semaphorins (data not shown), transfected with semaphorins 3A, 3B, or 3C. As expected, semaphorins 3A or 3B induced clear repulsive signals as judged by the empty spaces surrounding the transfected cells (Fig. 5A). In contrast and despite the fact that HEK293 cells transfected with semaphorin 3C expressed readily detectable levels of the protein (Fig. 5B), no effect was observed (Fig. 5A). The co-transfection of HEK293 semaphorin 3C with ADAMTS1 resulted also in a lack of effect on the migration of HUVECs (data not shown). These results show that, in contrast to cells expressing semaphorins 3A or 3B, cells expressing semaphorin 3C did not repel HUVECs, arguing that this semaphorin does not affect the migration of these cells or, alternatively, that it exerts an attractive signal.
FIGURE 5.
Effect of different class 3 semaphorins on cell migration. A, HUVECs were seeded in gelatin-coated plates and cultured for 1 day. Then 103 HEK293 cells were transiently transfected with expression vectors encoding semaphorin 3A, 3B, or 3C together with pBABE-GFP and seeded on top of the endothelial cells. Photographs were taken after 48 h. Arrows point to green fluorescent protein-positive HEK293 cells in the different cultures. B, HEK293 cells transiently transfected with expression vectors encoding semaphorin 3A, 3B, or 3C were lysed, and cell lysates were analyzed by Western blot. The nitrocellulose membrane was sequentially incubated with anti-semaphorin 3A, 3B, and 3C antibodies. No cross-reaction of the antibodies was observed. C, the schematic shows the cell types used to analyze the effect of semaphorin 3C on the cell migration in Transwell plates. MCF7 or MDA-MB-231 cells were seeded on the upper chamber of Transwell plates. HEK293 cells transiently transfected with empty vector (vec.) or the vector bearing the sema3C cDNA were seeded in the lower chamber. When indicated, the experiment was performed in the presence of 10% fetal bovine serum. D, the conditioned media of HEK293 cells transfected as in A was analyzed by Western blotting with antibodies against semaphorin 3C. The membrane stained with Ponceau red is shown as loading control. Note that in the second lane the protein excess corresponds to serum components. vec, vector. E, MCF7 and transiently transfected HEK293 cells were seeded and cultured with or without serum on the upper and lower chamber of Transwell plates, respectively, as described in C. After 12 h, cells were fixed and stained with 4′,6-diamidino-2-phenylindole. Representative fields are shown. F, the bars represent the means of the MCF7 (upper graph) or MDA-MB-231 cells (lower graph) counted in three independent experiments performed as in C, each done in triplicate, ±S.D. Analysis of the means using Student's t test showed statistically significant differences (*, p < 0.05; **, p < 0.005).
Semaphorin 3C Promotes Attractive Cell Migration
To further analyze the effect of semaphorin 3C on cell migration, we used the Transwell migration assay, a straightforward assay for attractive cell migration. As targets of semaphorin 3C signaling, we used, in addition to the poorly metastatic MCF7 cells, the highly metastatic cell line MDA-MB-231. Both cell lines express functional receptors for class 3 semaphorins (25).
As shown in Fig. 5, C–F, and compared with HEK293 cells transfected with the empty vector, cells transfected with semaphorin 3C induced a clear increase in the migration of MCF7 or MDA-MB-231 cells, comparable with that induced by serum factors. These results show that semaphorin 3C does regulate cell migration and that, in contrast to semaphorins A and B, it does induce an attractive response.
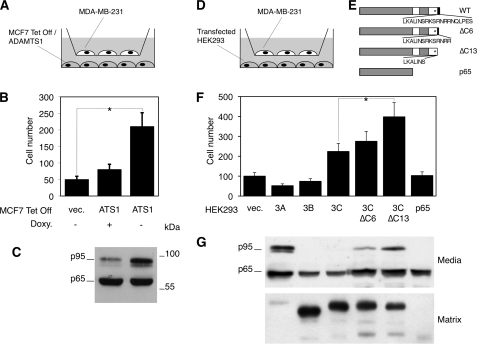
ADAMTS1 Regulates Cell Migration through the Release of Semaphorin 3C from the Extracellular Matrix
According to the results of the Transwell, where there is no direct cell-cell contact, the effect of semaphorin 3C on cell migration seems to be mediated by the soluble forms. Because ADAMTS1 releases semaphorin 3C from the extracellular matrix, we next analyzed the effect of the metalloprotease on cell migration. Concomitant with an increase in the levels of soluble p95 semaphorin 3C, ADAMTS1 induced a clear increase in the number of migrating cells (Fig. 6, A and B).
FIGURE 6.
The p95 form of semaphorin 3C released by ADAMTS1 promotes cell migration. A, the schematic shows the cell types used to analyze the effect of ADAMTS1 on cell migration. B, MDA-MB-231 and MCF7 Tet-Off/ADAMTS1 cells seeded as in A were cultured for 12 h in the presence or absence of doxycycline (Doxy.). Then MDA-MB-231cells were fixed, stained with 4′,6-diamidino-2-phenylindole, and counted. The bars represent the means ±S.D. from three independent experiments, each done in triplicate. Analysis of the means using Student's t test showed statistically significant differences (*, p < 0.05). vec., vector. C, samples from the media conditioned by MCF7 Tet-Off/ADAMTS1 cells were cultured with or without doxycycline were analyzed by Western blot with anti-semaphorin 3C antibodies. D, shown is a schematic showing the cell types used to analyze the effect of different semaphorin 3C deletion mutants on cell migration. E, shown is a schematic representing the proteins encoded by the deletion constructs used to transfect HEK293 cells. WT, wild type. F, bars represent the means ±S.D from three independent experiments, each one done in triplicate. Analysis of the means using Student's t test showed statistically significant differences (*, p < 0.05). G, samples from the media and matrix conditioned by HEK293 cells transiently transfected with expression vectors encoding semaphorin 3A, 3B, or 3C or the deletion constructs shown in C were analyzed by Western blot with anti-semaphorin antibodies as described in Fig. 5D.
ADAMTS1 increases the levels of soluble p95 semaphorin 3C but not those of p65 (Fig. 6C, see also Fig. 4). Thus, if the effect of the metalloprotease on cell migration is mediated by semaphorin 3C, one should conclude that p95, and not p65, is the active form.
To confirm that the soluble p95 species generated by ADAMTS1 is the active semaphorin 3C isoform that increases cell motility, we transfected HEK293 cells with serial carboxyl-terminal deletion constructs (Fig. 6, D and E) and analyzed their effect on the migration of MDA-MB-231 cells (Fig. 6F). These deletions truncate the basic amino acid-rich region (Fig. 6E), mimicking the fragment generated by ADAMTS1. As expected, the partial truncation of the basic amino acid region (ΔC6 construct) led to an increase in the levels of soluble semaphorin 3C, a concomitant decrease in the levels of the matrix associated isoform (Fig. 6G), and promoted cell migration (Fig. 6F). A longer truncation (ΔC13) led to similar but more profound effects (Fig. 6, F and G). As previously shown (25), semaphorin 3A or 3B inhibited cell migration (Fig. 6F). Collectively, these results demonstrate that the release of semaphorin 3C induced by ADAMTS1 leads to increased cell migration.
DISCUSSION
Metastasis is the leading cause of mortality in patients with cancer. It is a multistep process that allows cells to move from a primary tumor and grow elsewhere. Although extracellular metalloproteases have been long considered key components of the metastatic program, their specific roles are still far from clear. The initial hypothesis of metalloproteases as passive degraders of the extracellular matrix has been progressively replaced by a far more complex view (42). Currently, in addition to remodelers of the extracellular matrix, metalloproteases are considered components of signaling pathways that regulate cell proliferation, adhesion, and migration. In fact, whereas some metalloproteases are still considered potent pro-metastatic factors, others seem to be endowed with tumor suppressor abilities (5).
The establishment of the role of a particular metalloprotease requires the detailed knowledge of its array of substrates as well as the characterization of the functional consequences of their cleavages (43). This task is particularly necessary in the case of metalloproteases, such as ADAMTS1, that seem to have pro- or anti-tumor properties depending on the cellular environment and the substrates co-expressed. Although substrates of some metalloproteases have been identified through hypothesis-driven research, the recently developed proteomic techniques represent a unique opportunity to identify substrates of proteases in an unbiased way (44). Using these techniques we have identified several new candidate substrates of ADAMTS1. We further characterized one of them, semaphorin 3C, because it may help to explain the role of ADAMTS1 during metastasis.
The simplest explanation for our results is that ADAMTS1 cleaves semaphorin 3C near the carboxyl terminus. However, alternative mechanisms may explain our results. ADAMTS1 could cleave the components of the extracellular matrix to which semaphorin 3C binds or it may activate another protease endowed with the ability to cleave semaphorin 3C. However, even in this situation, the results presented here clearly show the ability of ADAMTS1 directly or indirectly to solubilize the active form of semaphorin 3C so it can diffuse and promote cell migration.
Initially semaphorins along with their receptors, the neuropilins and plexins, were identified as axon-guidance cues during the establishment of neural circuitries (24, 45). However, it is currently acknowledged that semaphorins, which are expressed in many cancer cells, also play roles in the regulation of the tumor microenvironment and contribute positively or negatively to many aspects of cancer progression, including the induction of cell migration, one of the hallmarks of metastasis (22, 23). The semaphorins are not unique in displaying dual activities as axon guidance factors and as modulators of angiogenesis and tumor progression. Particular axon guidance factors from the Slit, Netrin, and Ephrin families also display this pattern (46, 47). This is not surprising because there are significant similarities between an extending axon and the leading edge of a migrating cell (38).
Although ADAMTS1 is not significantly expressed in primary breast tumors (32), it is one of the most up-regulated genes in highly metastatic cells derived from the MDA-MB-231 cell line (13), indicating that it may contribute to the metastatic program. Thus, whereas the levels of ADAMTS1 are low in the primary tumor, its overexpression in a subset of cancer cells may contribute to the spreading of the tumor. The results shown here indicate that semaphorin 3C can mediate at least in part the effect of ADAMTS1 on cell migration and, thus, favor the spreading of the tumor when co-expressed with semaphorin 3C, which, importantly, is also expressed in breast cancer cells (49). Furthermore, semaphorin 3C is a transcriptional target of HER2, a tyrosine kinase receptor causally involved in the progression of a subset of aggressive breast cancers (50). Thus, the fact that semaphorin 3C is frequently expressed in breast cancer samples along with the fact that ADAMTS1 can be expressed in highly metastatic breast cancer cells indicates that both molecules can cooperate to promote breast cancer metastasis.
Up-regulation of semaphorin 3C has also been detected in recurrent squamous cell carcinomas (51), in highly metastatic lung cancer cells (52), and in ovarian cancer samples (49). Interestingly, lung cancer cells also express ADAMTS1 (53), indicating that, also in this type of tumors, both molecules may functionally interact to promote cell migration.
Acknowledgments
We kindly thank Dr. A Tufro (Albert Einstein College of Medicine) for the semaphorin 3C construct. We thank Dr. G. Neufeld (Rappaport Faculty of Medicine) and Dr. S. Naylor (University of Texas) for the Semaphorin 3A and 3B constructs. The Proteomics Laboratory is a member of the National Spanish Institute for Proteomics (PROTEORED).
This work was supported by grants from the Instituto de Salud Carlos III (Intrasalud PI081154 and the network of cooperative cancer research (RTICC-RD06/0020/0022)) (to J. A.) and CP06/00304 and P1071058 (to F. C.) and the Breast Cancer Research Foundation and La Marató de TV3 (to J. A.) and Grants CP06/00304 and PI071058 (to F. C.).
- DMEM
- Dulbecco's modified Eagle's medium
- shRNA
- short hairpin RNA
- MS
- mass spectroscopy
- H
- heavy
- L
- light
- DIGE
- difference gel electrophoresis
- SILAC
- stable isotopes labeling by amino acids in cell culture
- FBS
- fetal bovine serum
- HUVEC
- human umbilical vein endothelial cell(s).
REFERENCES
- 1.Kedrin D., van Rheenen J., Hernandez L., Condeelis J., Segall J. E. (2007) J. Mammary Gland Biol. Neoplasia 12, 143–152 [DOI] [PubMed] [Google Scholar]
- 2.Basbaum C. B., Werb Z. (1996) Curr. Opin. Cell Biol. 8, 731–738 [DOI] [PubMed] [Google Scholar]
- 3.Coussens L. M., Fingleton B., Matrisian L. M. (2002) Science 295, 2387–2392 [DOI] [PubMed] [Google Scholar]
- 4.Stöcker W., Bode W. (1995) Curr. Opin. Struct. Biol. 5, 383–390 [DOI] [PubMed] [Google Scholar]
- 5.López-Otín C., Matrisian L. M. (2007) Nat. Rev. Cancer 7, 800–808 [DOI] [PubMed] [Google Scholar]
- 6.Blobel C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 7.Dong Z., Kumar R., Yang X., Fidler I. J. (1997) Cell 88, 801–810 [DOI] [PubMed] [Google Scholar]
- 8.Gorrin-Rivas M. J., Arii S., Furutani M., Mizumoto M., Mori A., Hanaki K., Maeda M., Furuyama H., Kondo Y., Imamura M. (2000) Clin. Cancer Res. 6, 1647–1654 [PubMed] [Google Scholar]
- 9.Kuno K., Bannai K., Hakozaki M., Matsushima K., Hirose K. (2004) Biochem. Biophys. Res. Commun. 319, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 10.Lee N. V., Sato M., Annis D. S., Loo J. A., Wu L., Mosher D. F., Iruela-Arispe M. L. (2006) EMBO J. 25, 5270–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y. J., Xu Y., Yu Q. (2006) Oncogene 25, 2452–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luque A., Carpizo D. R., Iruela-Arispe M. L. (2003) J. Biol. Chem. 278, 23656–23665 [DOI] [PubMed] [Google Scholar]
- 13.Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordón-Cardo C., Guise T. A., Massagué J. (2003) Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 14.Masui T., Hosotani R., Tsuji S., Miyamoto Y., Yasuda S., Ida J., Nakajima S., Kawaguchi M., Kobayashi H., Koizumi M., Toyoda E., Tulachan S., Arii S., Doi R., Imamura M. (2001) Clin. Cancer Res. 7, 3437–3443 [PubMed] [Google Scholar]
- 15.Kuno K., Okada Y., Kawashima H., Nakamura H., Miyasaka M., Ohno H., Matsushima K. (2000) FEBS Lett. 478, 241–245 [DOI] [PubMed] [Google Scholar]
- 16.Sandy J. D., Westling J., Kenagy R. D., Iruela-Arispe M. L., Verscharen C., Rodriguez-Mazaneque J. C., Zimmermann D. R., Lemire J. M., Fischer J. W., Wight T. N., Clowes A. W. (2001) J. Biol. Chem. 276, 13372–13378 [DOI] [PubMed] [Google Scholar]
- 17.Canals F., Colomé N., Ferrer C., Plaza-Calonge Mdel C., Rodríguez-Manzaneque J. C. (2006) Proteomics 6, S28–S35 [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Manzaneque J. C., Carpizo D., Plaza-Calonge Mdel C., Torres-Collado A. X., Thai S. N., Simons M., Horowitz A., Iruela-Arispe M. L. (2009) Int. J. Biochem. Cell Biol. 41, 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres-Collado A. X., Kisiel W., Iruela-Arispe M. L., Rodríguez-Manzaneque J. C. (2006) J. Biol. Chem. 281, 17827–17837 [DOI] [PubMed] [Google Scholar]
- 20.Rocks N., Paulissen G., Quesada-Calvo F., Munaut C., Gonzalez M. L., Gueders M., Hacha J., Gilles C., Foidart J. M., Noel A., Cataldo D. D. (2008) Cancer Res. 68, 9541–9550 [DOI] [PubMed] [Google Scholar]
- 21.Antelmann H., Tjalsma H., Voigt B., Ohlmeier S., Bron S., van Dijl J. M., Hecker M. (2001) Genome Res. 11, 1484–1502 [DOI] [PubMed] [Google Scholar]
- 22.Neufeld G., Kessler O. (2008) Nat. Rev. Cancer 8, 632–645 [DOI] [PubMed] [Google Scholar]
- 23.Capparuccia L., Tamagnone L. (2009) J. Cell Sci. 122, 1723–1736 [DOI] [PubMed] [Google Scholar]
- 24.Luo Y., Raible D., Raper J. A. (1993) Cell 75, 217–227 [DOI] [PubMed] [Google Scholar]
- 25.Bachelder R. E., Lipscomb E. A., Lin X., Wendt M. A., Chadborn N. H., Eickholt B. J., Mercurio A. M. (2003) Cancer Res. 63, 5230–5233 [PubMed] [Google Scholar]
- 26.Kigel B., Varshavsky A., Kessler O., Neufeld G. (2008) PLoS ONE 3, e3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bech-Serra J. J., Santiago-Josefat B., Esselens C., Saftig P., Baselga J., Arribas J., Canals F. (2006) Mol. Cell. Biol. 26, 5086–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esselens C. W., Malapeira J., Colome N., Moss M., Canals F., Arribas J. (2008) Biol. Chem. 389, 1075–1084 [DOI] [PubMed] [Google Scholar]
- 29.Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 30.Kuno K., Kanada N., Nakashima E., Fujiki F., Ichimura F., Matsushima K. (1997) J. Biol. Chem. 272, 556–562 [DOI] [PubMed] [Google Scholar]
- 31.Vázquez F., Hastings G., Ortega M. A., Lane T. F., Oikemus S., Lombardo M., Iruela-Arispe M. L. (1999) J. Biol. Chem. 274, 23349–23357 [DOI] [PubMed] [Google Scholar]
- 32.Porter S., Scott S. D., Sassoon E. M., Williams M. R., Jones J. L., Girling A. C., Ball R. Y., Edwards D. R. (2004) Clin. Cancer Res. 10, 2429–2440 [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Manzaneque J. C., Milchanowski A. B., Dufour E. K., Leduc R., Iruela-Arispe M. L. (2000) J. Biol. Chem. 275, 33471–33479 [DOI] [PubMed] [Google Scholar]
- 34.Smith G. N., Jr., Mickler E. A., Hasty K. A., Brandt K. D. (1999) Arthritis Rheum. 42, 1140–1146 [DOI] [PubMed] [Google Scholar]
- 35.Kuno K., Matsushima K. (1998) J. Biol. Chem. 273, 13912–13917 [DOI] [PubMed] [Google Scholar]
- 36.Van den Bergh G., Arckens L. (2004) Curr. Opin. Biotechnol. 15, 38–43 [DOI] [PubMed] [Google Scholar]
- 37.Everley P. A., Zetter B. R. (2005) Ann. N. Y. Acad. Sci. 1059, 1–10 [DOI] [PubMed] [Google Scholar]
- 38.Tamagnone L., Comoglio P. M. (2004) EMBO Rep. 5, 356–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams R. H., Lohrum M., Klostermann A., Betz H., Püschel A. W. (1997) EMBO J. 16, 6077–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Wit J., De Winter F., Klooster J., Verhaagen J. (2005) Mol. Cell. Neurosci. 29, 40–55 [DOI] [PubMed] [Google Scholar]
- 41.Varshavsky A., Kessler O., Abramovitch S., Kigel B., Zaffryar S., Akiri G., Neufeld G. (2008) Cancer Res. 68, 6922–6931 [DOI] [PubMed] [Google Scholar]
- 42.Brinckerhoff C. E., Matrisian L. M. (2002) Nat. Rev. Mol. Cell Biol. 3, 207–214 [DOI] [PubMed] [Google Scholar]
- 43.Overall C. M., Blobel C. P. (2007) Nat. Rev. Mol. Cell Biol. 8, 245–257 [DOI] [PubMed] [Google Scholar]
- 44.López-Otín C., Overall C. M. (2002) Nat. Rev. Mol. Cell Biol. 3, 509–519 [DOI] [PubMed] [Google Scholar]
- 45.Kolodkin A. L., Matthes D. J., Goodman C. S. (1993) Cell 75, 1389–1399 [DOI] [PubMed] [Google Scholar]
- 46.Klagsbrun M., Eichmann A. (2005) Cytokine Growth Factor Rev. 16, 535–548 [DOI] [PubMed] [Google Scholar]
- 47.Hinck L. (2004) Dev. Cell 7, 783–793 [DOI] [PubMed] [Google Scholar]
- 48.Deleted in proof
- 49.Galani E., Sgouros J., Petropoulou C., Janinis J., Aravantinos G., Dionysiou-Asteriou D., Skarlos D., Gonos E. (2002) Anticancer Res. 22, 2275–2280 [PubMed] [Google Scholar]
- 50.Beckers J., Herrmann F., Rieger S., Drobyshev A. L., Horsch M., Hrabé de Angelis M., Seliger B. (2005) Int. J. Cancer 114, 590–597 [DOI] [PubMed] [Google Scholar]
- 51.Yamada T., Endo R., Gotoh M., Hirohashi S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14713–14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martín-Satué M., Blanco J. (1999) J. Surg. Oncol. 72, 18–23 [DOI] [PubMed] [Google Scholar]
- 53.Rocks N., Paulissen G., Quesada Calvo F., Polette M., Gueders M., Munaut C., Foidart J. M., Noel A., Birembaut P., Cataldo D. (2006) Br. J. Cancer 94, 724–730 [DOI] [PMC free article] [PubMed] [Google Scholar]