Abstract
Centrosome cohesion and segregation are accurately regulated to prevent an aberrant separation of duplicated centrosomes and to ensure the correct formation of bipolar spindles by a tight coupling with cell cycle machinery. CPAP is a centrosome protein with five coiled-coil domains and plays an important role in the control of brain size in autosomal recessive primary microcephaly. Previous studies showed that CPAP interacts with tubulin and controls centriole length. Here, we reported that CPAP forms a homodimer during interphase, and the fifth coiled-coil domain of CPAP is required for its dimerization. Moreover, this self-interaction is required for maintaining centrosome cohesion and preventing the centrosome from splitting before the G2/M phase. Our biochemical studies show that CPAP forms homodimers in vivo. In addition, both monomeric and dimeric CPAP are required for accurate cell division, suggesting that the temporal dynamics of CPAP homodimerization is tightly regulated during the cell cycle. Significantly, our results provide evidence that CPAP is phosphorylated during mitosis, and this phosphorylation releases its intermolecular interaction. Taken together, these results suggest that cell cycle-regulated phosphorylation orchestrates the dynamics of CPAP molecular interaction and centrosome splitting to ensure genomic stability in cell division.
Keywords: Cell/Centrosome, Cell/Mitosis, Cell/Cycle, Cytoskeleton/Microtubules, Protein/Adhesion, Protein/Assembly, CPAP, Dimerization
Introduction
Mammalian centrosomes, the major microtubule-organizing centers, consist of a pair of centrioles normally localized at the periphery of the nucleus and surrounded by a number of proteins collectively referred to as pericentriolar material (1–3). The centrosomes coordinate most microtubule (MT)3 -related processes, including cell shape, polarity, adhesion and motility, cell division and cytokinesis, as well as intracellular transport (4). The centrosome cycle is handled by a “once per cell cycle rule” and “one per centriole rule” to prevent an excess of centrosomes or centrioles (5). The coordination of the centrosome cycle and cell cycle ensures an accurate distribution of genetic material. Daughter cells resulting from cell division possess one centrosome that includes two tightly associated centrioles. These centrioles are barrel-shaped microtubule assemblies but are structurally different from each other, one with a set of appendages at the distal end (mother centriole) and one without appendages (daughter centriole) (3). During late mitosis or early G1 phase, the close link between the two centrioles is lost, a process known as disengagement, and a loose link between the proximal ends of the two centrioles develops (6). The centrosome cohesion is a prerequisite of centriole duplication and keeps the distance between the two centrioles less than 2 μm (4, 7). Centrioles initiate duplication at the onset of S phase, elongate during G2 phase, and complete duplication to form the nascent centrosomes before the onset of mitosis. The G2 phase cell harbors two centrosomes, each of which includes two tightly associated centrioles. This tight association can prevent the reduplication of centrioles termed engagement. In late G2 phase, the pericentriolar material undergoes a phosphorylation-dependent maturation event and recruits several proteins, including γ-tubulin. The daughter centriole from the previous cell cycle then acquires appendages and fully matures. Simultaneously, centrosome cohesion dissipates, and the two centrosomes separate into two functional units to form the bipolar spindle. Perturbation of the spatiotemporal dynamics of centrosome splitting can lead to genomic instability during cell division (5). Malfunctions in centrosome cohesion and separation, in fact, have been seen in many solid tumors (7). How the cohesion and separation of the centrosome are regulated throughout the cell cycle is not yet well understood (8). However, several proteins such as C-Nap1, Rootletin, RanBP1, Dynamin2, Cep68, and Cep215 have been reported to be involved with centrosome cohesion (8–12).
CPAP (centrosomal P4.1-associated protein), also named CENP-J, is a centrosome protein that contains five short coiled-coil segments and a glycine repeated segment termed the G-box. The fifth coiled-coil segment (CC5) and the G-box have 76.6% homology with human TCP10 that plays a role in the transmission ratio distortion phenotype (13). Amino acid residues 311–422 of CPAP (including coiled-coil 2) make up an MT-destabilizing domain, and amino acid residues 423–607 of CPAP (including coiled-coil 3) make up an MT-binding domain. Mutations that disrupt the α-helical structure or the charge property significantly affect the MT-destabilizing ability of CPAP (14, 15). CPAP is a member of the γ-tubulin complex localized within the center of microtubule asters (16). Knockdown of CPAP by CPAP antibody significantly inhibits the formation of microtubule esters (16). Depletion of CPAP by RNA interference destroys centrosome integrity and induces multipolar spindles (17). Furthermore, depletion of CPAP can block the overduplication of centrioles induced by overexpression of PLK4 (18). Overexpression of CPAP causes an elongation of parental and nascent centrioles (19–21). Mutation of CPAP induces a neuro-developmental disorder named MCPH (autosomal recessive primary microcephaly) that causes a great reduction in brain growth in humans (22–26).
The homology proteins of CPAP in Caenorhabditis elegans and Drosophila are SAS4 and DSAS4, respectively. SAS4 and DSAS4 are essential for centrosome duplication and for centrosome size (27–30). Previous studies have illustrated that CPAP in like fashion to SAS4 plays a critical role in centriole biogenesis and centrosome size (19–21). In this study, we found that both partial depletion of CPAP by CPAP siRNA and overexpression of the C terminus of CPAP induced an increase in centrosome splitting, indicating that CPAP functions on centrosome cohesion. Moreover, the CC5 domain of CPAP was identified as being essential for the formation of polymers between CPAP by means of co-immunoprecipitation and pulldown experiments. Furthermore, homodimers of CPAP mediated by the CC5 domain are important for the maintenance of centrosome cohesion. Importantly, CPAP is a mitotic phosphorylation protein, and phosphorylation blocks the self-interaction of CPAP. Collectively, these results demonstrate that negative regulation of CPAP self-interaction by mitotic phosphorylation is required for accurate centrosome cohesion and splitting during the cell cycle.
MATERIALS AND METHODS
Plasmid Construction
Full-length human CPAP gene (GenBankTM accession number NM_018451) was cloned into a T-vector (Takara Biotechnology, Dalian, China). To construct GFP-CPAP and FLAG-CPAP plasmids, the full-length CPAP cDNA was subcloned into pEGFP-C1 vector (Clontech) and p3×FLAG-myc-CMV-24 vector (Sigma) and digested with SalI and BamHI, and GFP-tagged and FLAG-tagged deletion mutants CPAP-ΔCC5, CPAP-CC5, CPAP-CC4, CPAP-N, and CPAP-M and GFP-tagged CPAP-ΔCC5-FKBP were created by standard PCR methods. The FK506-binding protein-coding region was amplified from pC4M-FV2E (ARIAD) by PCR and cloned into the pCS2-FLAG-CPAP vector. All constructs were sequenced in full.
Cell Culture and Synchronization
HeLa, U2OS, and 293T cells, from American Type Culture Collection (Manassas, VA), were maintained as subconfluent monolayers in Dulbecco's modified Eagle's growth medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) and 100 units/ml penicillin plus 100 μg/ml streptomycin (Invitrogen) at 37 °C with 10% CO2. Cells were synchronized at G1/S with 5 mm thymidine for 16 h, then washed with phosphate-buffered saline (PBS) three times, and then transfected with various GFP-CPAP constructs. Thirty hours after transfection, HeLa cells were fixed with cold methanol at −20 °C followed by examination under the fluorescence microscope.
Recombinant Protein Expression
The CC5 domain of human CPAP was expressed in bacteria as a glutathione S-transferase fusion protein and purified by using glutathione-agarose as reported previously (31). Briefly, 500 ml of Luria Bertani medium was inoculated with bacteria BL21 (DE3) pLysS transformed with GST-CPAP-CC5. The protein expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 30 °C for 6 h. Bacteria were then collected by centrifugation and resuspended in PBS containing a protease inhibitor mixture (Sigma) followed by sonication. The lysis solution was clarified by centrifugation for 20 min at 10,000 × g.
The soluble fraction was then applied to a column packed with nickel-agarose beads, followed by extensive washes with PBS. His-tagged proteins were then eluted with 200 mm imidazole and dialyzed against appropriate buffers for experimentation.
Pulldown Assay
Glutathione S-transferase pulldown was carried out as described previously (32). The GST-CPAP-CC5 fusion protein-bound Sepharose beads were incubated with purified proteins for 4 h at 4 °C. After the incubation, the beads were extensively washed with PBS and boiled in SDS-PAGE sample buffer, followed by fractionation of bound proteins on a 10% SDS-polyacrylamide gel. Proteins were then transferred onto a nitrocellulose membrane for Western blotting using an appropriate antibody.
Transient Transfection and Immunoprecipitation
293T cells were grown to about 50% confluency in Dulbecco's modified Eagle's medium and co-transfected with FLAG-CPAP and GFP-tagged CPAP full-length or deletion mutants using Lipofectamine 2000 (Invitrogen). Cells were collected 24–36 h after transfection, and proteins were solubilized in lysis buffer (50 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm EGTA, 0.1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml pepstatin A). Lysates were clarified by centrifugation at 16,000 × g for 10 min at 4 °C and incubated with anti-FLAG M2-linked agarose beads (Sigma). Beads were washed five times with lysis buffer and then boiled in protein sample buffer for 2 min. After SDS-PAGE, proteins were transferred onto a nitrocellulose membrane. The membrane was probed with antibodies against the FLAG epitope and GFP. Immunoreactive signals were detected with an ECL kit (Pierce) and visualized by autoradiography on Kodak BioMax film.
CPAP siRNA (target sequence, 5′-AAGGAAGATTGCACCAGTCAA-3′ (18)) was synthesized by Dharmacon Research, Inc. (Boulder, CO). In brief, cells were transfected with a 21-mer siRNA oligonucleotide or a control scramble oligonucleotide, and the efficiency of siRNA-mediated protein suppression was assessed by Western blotting analysis.
Immunofluorescence and Time-lapse Microscopy
For immunofluorescence microscopic analyses, HeLa cells were seeded onto sterile acid-treated 12-mm coverslips in 24-well plates (Corning Glass). Double thymidine-blocked and released HeLa cells were transfected with 1 μl of Lipofectamine 2000 premixed with 1 μg of the various plasmids. In general, 30–48 h after transfection, HeLa cells were rinsed for 1 min with PBS buffer and fixed for 10 min with methanol prechilled at −20 °C followed by rehydration in PBS. Cells on the coverslips were blocked with 0.05% Tween 20 in PBS (TPBS) with 1% bovine serum albumin (Sigma) for 30 min. These cells were incubated with various primary antibodies in a humidified chamber for 1 h and then washed three times in TPBS. Primary antibodies were visualized using fluorescein isothiocyanate-conjugated goat anti-mouse IgG or rhodamine-conjugated goat anti-rabbit IgG. CPAP antibody was raised in a rabbit using a recombinant peptide corresponding to the region of 1318–1331 amino acids of CPAP. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma). Slides were examined with a Zeiss LSM510 confocal scanning fluorescence microscope, and images were collected and analyzed with Image-5 (Carl Zeiss, Germany). For time-lapse microscopy, cells were cultured on coverslips in CO2-independent medium in a sealed chamber heated to 37 °C and observed under a DeltaVision deconvolution microscope (Applied PreScission). Images were acquired at 5- or 10-min intervals and presented in Photoshop.
Molecular Mass Determination
To examine if endogenous CPAP forms dimer in vivo, size exclusion chromatography was carried out using fast protein liquid chromatography with a Superose 6 10/300 GL column (GE Healthcare) previously equilibrated with 50 mm PIPES-K, pH 6.9, 1 mm EGTA, 5 mm MgCl2, and 50 mm NaCl. Elution was performed at a flow rate of 0.5 ml·min−1. The column was calibrated with bovine thyroglobulin (669 kDa; Stokes radius (RS) = 85 Å), ferritin (440 kDa; RS = 61 Å), aldolase (158 kDa; RS = 48 Å), and chicken ovalbumin (44 kDa; RS = 30.5 Å) which were used as standard proteins according to the user's manual. Equal amounts of chromatographic fractions were separated by SDS-PAGE followed by immunoblot analysis of CPAP.
To test if CPAP-FKBP dimerization can be induced in response to addition of AP20187, synchronized HeLa cells transiently transfected with FLAG-CPAP-FKBP were treated with AP20187 36 h after the transfection. Treated HeLa cells were washed twice with buffer containing 5 mm MgCl2, 50 mm NaCl, 0.1 m sucrose, and 50 mm PIPES-K, pH 6.9, and then solubilized in the same buffer containing 0.2% Triton X-100 for 10 min on ice. After centrifugation at 17,000 × g for 10 min at 4 °C, the cleared supernatant (1 ml) was loaded on the top of a 15-ml linear sucrose gradient (5–30%) prepared with buffer containing 50 mm PIPES-K, pH 6.9, 1 mm EGTA, 5 mm MgCl2, and 50 mm NaCl. FLAP-CPAP protein complexes were separated by ultracentrifugation for 24 h at 141,000 × g at 4 °C in an SW 28 rotor (Beckman). The gradients were then fractionated from top to bottom into 21 fractions by a density gradient fraction collector, and equal amounts of fractions were used for further immunoblot analysis of anti-FLAG antibody M2 (Sigma).
RESULTS
CPAP Participates in Centrosome Cohesion
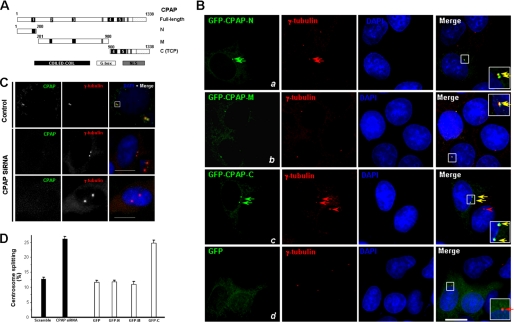
In an attempt to identify novel mitotic spindle proteins essential for chromosome plasticity in cell division, we carried out proteomic analyses of spindle proteins isolated from mitotic HeLa cells. CPAP was recovered in our proteomic analyses as an important protein essential for spindle formation. Recent studies showed that CPAP plays a critical role in regulating centriole duplication and length based on the knockdown and overexpression of CPAP (18–21). To further survey the functional domain of CPAP on the centrosome, the full-length CPAP cDNA was deleted at the N terminus, the middle segment, and the C terminus (Fig. 1A) and inserted into GFP vector to generate GFP-CPAP-N (N terminus), GFP-CPAP-M (middle region), and GFP-CPAP-C (C terminus) plasmids, respectively. These plasmids were then transfected into synchronized HeLa cells, and resulting phenotypes were examined for CPAP and γ-tubulin dual labeling with an immunofluorescence microscope (Fig. 1B). Expression of CPAP deletion mutants exhibited a typical centrosomal location that was superimposed onto that of γ-tubulin (Fig. 1B, panels a–c, green channels). Surprisingly, the expression of GFP-CPAP-C was often displayed as fully separated double dots (Fig. 1B, panel c, arrows). To validate if both dots are centrosome-associated structures, we examined its relationship to the centrosome marker γ-tubulin. Interestingly, γ-tubulin labeling also appeared as separated double dots that co-distribute with GFP-CPAP-C (Fig. 1B, panel c, yellow arrows in merged image), indicating that GFP-CPAP-C labels two fully separated centrosomes.
FIGURE 1.
Human CPAP functions on centrosome cohesion. A, schematic diagram of CPAP and its truncation mutants. N, N terminus; C, C terminus; M, middle region; TCP, t-complex responder gene product. B, overexpression of the C terminus of CPAP induced a centrosome splitting phenotype. Synchronized HeLa cells grown on coverslips were transiently transfected to express GFP-CPAP-N (panel a), GFP-CPAP-M (panel b), GFP-CPAP-C (panel c), and GFP (panel d). Thirty hours after the transfection, HeLa cells were fixed, permeabilized, and stained with GFP antibody (green), γ-tubulin antibody (red), and DNA (blue). Insets show magnified images corresponding to the boxed areas for better visualization of the centrosome splitting phenotype. Bar, 10 μm. C, suppression of CPAP by siRNA resulted in centrosome splitting during interphase. Synchronized HeLa cells grown on coverslips were transfected with CPAP siRNA or scramble control. Thirty hours after transfection, cells were fixed and stained with anti-CPAP antibody (green), anti-γ-tubulin antibody (red), and DAPI (blue). Phenotypes of CPAP-deficient cells show that the distance between two γ-tubulin-labeled centrosome (red spots) was above 2 μm, characteristic of centrosome splitting. Bar, 10 μm. D, percentage of centrosome splitting in CPAP knockdown cells is presented by the filled bars. The percentage of centrosome splitting in CPAP mutant expressing cells is shown in the open bars. Values represent the means ± S.E. of three different experiments.
The separation of centrosomes is a prerequisite for bipolar mitotic spindle assembly in mammalian cells. There are two stages in centrosome separation as follows: the splitting stage, when centrosomes start to move apart, and the elongation stage. The separation of the centrosomes greater than 2 μm in cells of G2 phase was defined as centrosome splitting (18). Careful examination revealed that the centrosomes expressing GFP-CPAP-C were separated about 2.63 μm (judged by γ-tubulin labeling; yellow arrows in Fig. 1B, panel c), suggesting that overexpression of CPAP-C promotes centrosome splitting. To validate this notion, we surveyed about 100 cells positively transfected from three preparations. The percentage of centrosome splitting in GFP-CPAP-C-expressing cells was almost twice that of control GFP-expressing cells (Fig. 1D). Expression of GFP-CPAP-N- or GFP-CPAP-M did not promote centrosome splitting. These results indicate that the overexpression of the C terminus of CPAP perturbs centrosome cohesion and promotes centrosome splitting during interphase.
To further investigate the function of CPAP during centrosome cohesion, we generated a rabbit polyclonal antiserum against CPAP and affinity-purified it. Specific reactivity to endogenous CPAP was confirmed by Western blotting analysis using cell lysates from HeLa cells and cells expression GFP-CPAP. The anti-CPAP antibody specifically recognizes both endogenous and exogenous CPAP in HeLa cells. This specificity was validated by using cell lysates from other cell lines such as Madin-Darby canine kidney cells, U2OS, and 293T (data not shown). We then examined the efficiency of siRNA-mediated CPAP knockdown. Toward this end, cell lysates from HeLa cells transfected with CPAP siRNA and scramble control were fractionated by SDS-PAGE followed by transferring onto nitrocellulose membranes for anti-CPAP Western blotting. Immunoblotting showed that about 70% of CPAP proteins were depleted 72 h after transfection with siRNA, whereas expression of β-tubulin was not altered in these cells. We also examined the localization of endogenous CPAP and exogenous CPAP by immunofluorescence. The anti-CPAP antibody recognizes the localization of centrosomes in HeLa cells throughout the cell cycle. The localization of GFP-CPAP-WT was similar to that of endogenous CPAP.
If CPAP takes part in the centrosome cohesion, suppression of CPAP by siRNA should cause centrosome splitting. As we predicted, knockdown of CPAP by siRNA induced the centrosome splitting in interphase cells (Fig. 1C). As shown in Fig. 1D, in positive transfected interphase cells, the percentage of cells exhibiting the centrosome splitting phenotype in CPAP-suppressed cells was about 30%, which is twice the control level (Fig. 1D), suggesting that CPAP plays an important role in centrosome cohesion. Therefore, we conclude that CPAP participates in centrosome cohesion during interphase, and perturbation of CPAP integrity by overexpression of the C terminus of CPAP induces centrosome splitting.
Self-interaction of CPAP Is Mediated by Its Fifth Coiled-coil Domain
The aforementioned results show that CPAP is required for centrosome cohesion, and the overexpression of the C-terminal domain of CPAP induces centrosome splitting. We reasoned that the C-terminal domain of CPAP may play a dominant-negative role in this process by preventing the interaction of the full-length CPAP with itself or other accessory proteins. However, there is no available evidence that other proteins interact with CPAP and participate in the regulation of centrosome cohesion. Given that centrosome proteins are often self-associated, we sought to test if the C-terminal domain binds to endogenous CPAP and therefore blocks the self-interaction of CPAP. This might in turn perturb centrosome cohesion by preventing full-length CPAP intermolecular interaction in vivo.
To verify whether CPAP is self-associated, we carried out a co-immunoprecipitation assay using lysates from 293T cells transiently transfected with FLAG-tagged CPAP and GFP-tagged CPAP. As shown in Fig. 2A, Western blotting using anti-FLAG antibody confirmed a successful precipitation of FLAG-CPAP, and GFP antibody blotting demonstrated that GFP-CPAP was co-precipitated with FLAG-CPAP (Fig. 2A, lane 5), although control GFP was not precipitated with FLAG-CPAP (Fig. 2A, lane 2). This indicates that a self-interaction exists between CPAP molecules in vivo. Subsequently, to map the domain that is necessary for the self-interaction, we generated a series of deletion mutants as illustrated in Fig. 1A and used anti-FLAG antibody to immunoprecipitate FLAG-CPAP and GFP-tagged CPAP-N, CPAP-M, and CPAP-C from lysates of 293T cells transiently co-transfected to express FLAG-CPAP and those GFP-tagged CPAP deletion mutants, respectively. Immunoblotting with anti-FLAG antibody confirmed a successful precipitation of FLAG-CPAP, and immunoblotting against GFP confirmed that GFP-CPAP-C was co-precipitated (Fig. 2B, lane 8).
FIGURE 2.
Identification and characterization of a self-interaction domain in CPAP molecule. A, self-association of CPAP in vivo. Extracts from cells transiently transfected to express GFP-CPAP and FLAG-CPAP or GFP and FLAG-CPAP were incubated with antibodies (Ab) against FLAG (lanes 2 and 5), and immunoprecipitates (IP) were resolved by 10% SDS-PAGE. NB, non-binding; HC, heavy chain; LC, light chain; wt, wild type; WB, Western blot. NB, nonbinding fraction. B, self-association of CPAP by C-terminal domain in vivo. Aliquots of 293T cells co-expressing FLAG-tagged CPAP and GFP-tagged deletion mutants of CPAP were lysed and immunoprecipitated with FLAG-M2-agarose (lanes 2, 4, 6, and 8). The beads were washed and boiled in SDS-PAGE buffer prior to being separated on a 10% SDS-PAGE. FLAG antibody and GFP-antibody were used in Western blotting, respectively (upper panel, GFP blot; lower panel, FLAG blot). C, schematic diagram of CPAP-C truncation mutants, which are CC4, CC5, and G-box deletion mutants. NLS, nuclear localization signal. D, self-association of CPAP by CC5 domain in vivo. Lysates from cells transiently transfected to express GFP-CC4, CC5, and G-box deletion mutants and FLAG-CPAP or GFP and FLAG-CPAP were incubated with antibodies against FLAG (lanes 2, 4, 6, and 8). Immunoprecipitates were resolved by 10% SDS-PAGE. E, self-interaction of CC5 domain in vivo. Co-immunoprecipitation of FLAG-tagged CC5 and GFP-tagged CC5 domains. F, self-interaction of CC5 domain in vitro. GST-CC5 affinity matrix absorbs purified His-GFP-CC5 protein expressed in bacteria. Samples were separated by SDS-PAGE. Upper panel, Coomassie Brilliant Blue-stained gel. Lower panel, the same samples were transferred to nitrocellulose membrane and subsequently probed by His antibody.
The C terminus of CPAP consists of two coiled-coil domains (CC4 and CC5) and a G-box domain. Given that coiled-coil domains often mediate protein-protein interactions, GFP-tagged CC4, CC5, as well as G-box truncated colons were constructed to pinpoint the precise region required for self-interaction of CPAP (Fig. 2C). The co-immunoprecipitation assay demonstrated that it is not the CC4 or G-box domain but rather the CC5 domain of the CPAP-C fragment that participates in the CPAP self-interaction (Fig. 2D, lane 6). Indeed, a self-interaction between CC5 domains was confirmed in vivo via a co-immunoprecipitation assay (Fig. 2E, lane 4) and in vitro by pulldown assay (Fig. 2F, lane 5). Based on the in vitro and in vivo analyses, we conclude that CPAP forms intermolecular associations via the CC5 domain.
CC5 Domain of CPAP Is Required for Centrosome Cohesion
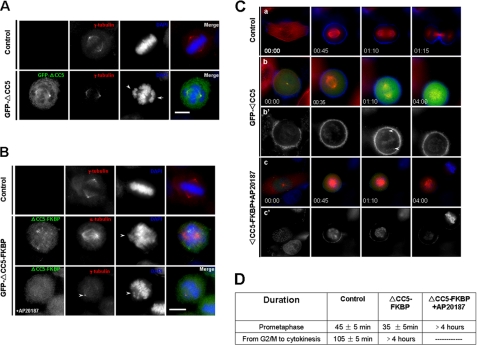
Given the fact that the CC5 domain of CPAP is important for the self-interaction of CPAP and that CPAP participates in the centrosome cohesion, we sought to examine whether the self-interaction of CPAP mediated by the CC5 domain is required for the centrosome cohesion. Toward this aim, we constructed GFP-tagged CPAP that lacks the CC5 domain (GFP-ΔCC5) and transfected it into HeLa cells to examine the effect of overexpression of GFP-ΔCC5 on the centrosome splitting under a fluorescence microscope. Interestingly, overexpression of GFP-ΔCC5 exhibited a centrosome splitting phenotype (Fig. 3A). Quantitative analysis revealed that the percentage of centrosome splitting was increased by 60% in cells expressing GFP-ΔCC5. Given the fact that other coiled-coil domains play minor or major roles in intermolecular association of CPAP, we reasoned that GFP-ΔCC5 mutant interacts with full-length CPAP. Consistent with this notion, our immunoprecipitation experiment confirmed that GFP-ΔCC5 associates with FLAG-CPAP in vivo.
FIGURE 3.
Self-interaction of CPAP mediated by CC5 domain is required for centrosome cohesion. A, HeLa cells transfected with GFP-ΔCC5 (green) were co-stained with anti-γ-tubulin (red) and DAPI (blue). B, percentage of centrosome splitting in HeLa cells transfected with GFP and GFP-ΔCC5. Values represent the means ± S.E. of three different experiments. C and D, HeLa cells transfected with GFP-tagged ΔCC5-FKBP were synchronized by double thymidine block and released 0 h to G1/S phase (C) and 8 h to G2 phase (D). The percentage of centrosome splitting was calculated before and after adding AP20187. E, analysis of the native molecular sizes of endogenous CPAP by using fast protein liquid and gel filtration chromatography. Cell lysates from synchronized HeLa cells were clarified by centrifugation, and clarified cell lysates were then applied for fast protein liquid and gel filtration chromatography. CPAP was eluted mainly in two fractions as follows: the lower molecular fractions corresponding to the monomer ∼160-kDa PRC1 and the higher molecular fractions corresponding to the dimeric ∼320-kDa CPAP. F, HeLa cells expressing FLAG-CPAP were lysed in the PIPES buffer described under “Materials and Methods,” and cell lysates were subjected to sucrose gradient sedimentation (5 to ≈30%) by ultracentrifugation. The sedimentations were collected and subjected to immunoblotting analysis by using an anti-FLAG antibody.
If CC5 is a major contributor to CPAP intermolecular association and GFP-ΔCC5 induces centrosome splitting due to its association with full-length CPAP, induced dimerization of CPAP-ΔCC5 should override its effect on centrosome splitting. To test this hypothesis, we fused FK506-binding protein to the C terminus of CPAP. Addition of AP20187, a compound that can bind to two FK506-binding protein molecules simultaneously (33), could be expected to induce dimerization of the FLAG-CPAP-FKBP fusion protein. HeLa cells were then transfected with plasmids encoding FLAG-CPAP-FKBP and GFP-ΔCC5-FKBP and treated with AP20187. As shown in Fig. 3F, exogenously expressed FLAG-CPAP forms a dimeric complex based on the sucrose gradient centrifugation assay, which exhibits identical biochemical characteristics seen in endogenous CPAP complex without chemical inducer AP20187. Thus, CPAP-FKBP fusion protein provides a useful means to examine the functional relevance of protein dimerization as demonstrated in other systems (for example see Ref. 33).
To further examine the functional relevance of CPAP dimerization without CC5, HeLa cells were then transfected with plasmid GFP-ΔCC5-FKBP and treated with AP20187. Simultaneously, cells were arrested in G1/S phase by double thymidine blocking. Cells released from G1/S phase for 0 and 8 h were observed by deconvolution microscopy, and the percentage of centrosome splitting is shown in Fig. 3, C and D. The percentage of cells exhibiting centrosome splitting induced by ΔCC5-FKBP was about 23% at G1/S phase and 25% at G2 phase. However, the percentage dropped to 11% at G1/S phase and 14% at G2 phase, respectively, after treating with AP20187. Thus, we conclude that the integrity of CPAP is essential for centrosome cohesion.
CPAP Forms Homodimers in Vivo
To validate if endogenous CPAP forms dimeric complexes, we generated cell lysates from synchronized HeLa cells using PIPES buffer containing 5 mm MgCl2, 50 mm NaCl, and 50 mm PIPES-K, pH 6.9. After centrifugation at 17,000 × g for 10 min (4 °C), the clarified cell lysates were applied to a gel filtration column calibrated with a standard kit. Using immunoblotting analysis of CPAP, the signal was detected in the high molecular mass-containing fractions, which corresponded to the dimeric form of CPAP (≈320 kDa; calculated molecular mass of CPAP is 153 kDa), indicating that CPAP exists as dimers in vivo (Fig. 3E). Using gel filtration chromatography, we have also determined the molecular mass of CC5. Our analysis demonstrated a molecular mass of 45.4 kDa for CC5 (calculated mass is 20.3 kDa). Thus, our results show that CPAP forms homodimers in vitro and that this dimerization is mainly mediated by CC5. We also reason that CC5 domain-mediated self-interaction of CPAP molecules is required for the centrosome cohesion.
Both Monomeric and Dimeric CPAP Are Required for Normal Mitosis
After demonstrating the importance of the CC5 domain in centrosome cohesion in interphase cells, we began to analyze the function of the CC5 domain on mitosis. To this end, we transiently transfected HeLa cells with the GFP-ΔCC5 plasmid and observed the phenotype by immunofluorescence and time-lapse imaging. As shown in Fig. 4A, the absence of the CC5 domain of CPAP results in errors in chromosome alignment at the equator and hinders the process of mitosis. The majority of chromosomes that were not aligned at the equator were located around the centrosomes. The time of prophase and prometaphase was greatly extended for more than 4 h in cells positively transfected with GFP-ΔCC5, although it was only 30 min in control cells. These observations suggest that the monomer state of CPAP promotes centrosome splitting and impairs normal mitosis.
FIGURE 4.
Temporal dynamics of CPAP dimerization is essential for faithful cell division. A, absence of CC5 domain from CPAP resulted in chromosome misalignment to the metaphase plate. Fixed HeLa cells transiently transfected to express GFP-ΔCC5 (green) were stained with γ-tubulin (red) antibodies. DNA was stained with DAPI (blue). Arrows indicate misaligned chromosomes caused by CPAP deletion mutant. The scale bar is 10 μm. B, dimerization of GFP-ΔCC5-FKBP induced by AP20187 led to failure of some chromosomes to align properly at the metaphase plate. HeLa cells transfected to express GFP-ΔCC5-FKBP were treated with AP20187 for 4 h prior to being fixed and stained with γ-tubulin (red) antibodies and DAPI (blue). Arrows indicate misaligned chromosomes. The scale bar is 10 μm. C, time-lapse imaging in HeLa cells expressing GFP-ΔCC5 proteins. mCherry-β-tubulin stably expressing HeLa cells were transiently transfected to express GFP-ΔCC5 and cultured on coverslips in CO2-independent medium, placed into a sealed or open growth chamber heated to 37 °C, and observed under a two-photon excitation microscope using an 1.4 NA plan-Apo×63 oil immersion objective. Chromosomes were stained with Hoechst 2 h before being placed under the microscope. Images were presented using Photoshop software, and different time points were labeled. D, statistical analysis of the time of each phase in the aforementioned cells in C expressing GFP-ΔCC5 proteins. Each value was calculated from at least eight different cells.
To test whether the polymer forms of CPAP also have effects on mitosis, we transiently transfected HeLa cells to express GFP-ΔCC5-FKBP and treated them with AP20187. Treatment also resulted in errors of chromosome alignment at the equator and delayed the process of mitosis (Fig. 4B), suggesting that the self-interaction of CPAP stops the normal process of mitosis. Time-lapse images show that it takes 35 min for cells expressing GFP-ΔCC5 to form a bipolar spindle. In contrast, it takes 45 min for control cells and more than 4 h for cells that expressed GFP-ΔCC5-FKBP and were treated with AP20187 to form a bipolar spindle (Fig. 4C). Cells expressing GFP-ΔCC5 required more than 3 h to align their chromosomes at the equator (Fig. 4C, panels b and b′). Statistical analysis of the time of each phase is shown in Fig. 4D.
Thus, we conclude that both the absence of the CC5 domain of CPAP during interphase and the dimerization of CPAP in mitosis influence the chromosome alignment in mitosis and prolong the process of mitosis. These findings suggest that both monomeric and dimeric CPAP are required for normal mitosis, and the status of monomerization and dimerization of CPAP may be regulated in the cell cycle.
Mitotic Phosphorylation Negatively Regulates the Self-interaction of CPAP
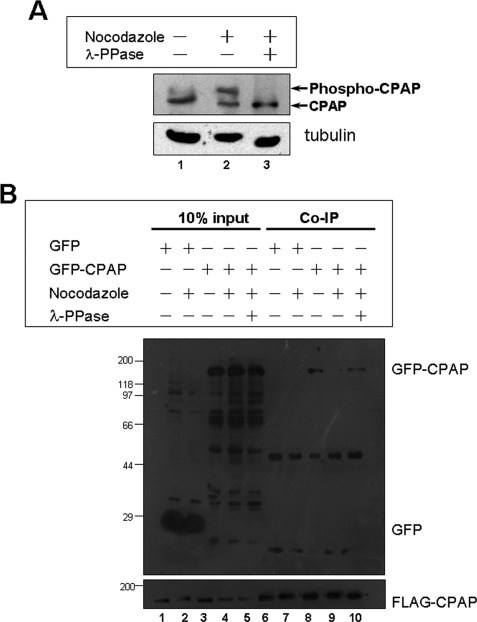
To investigate the possible modification change of CPAP from interphase to mitosis, we treated 293T cells subcultured in 24-well plates with or without the microtubule depolymerizing drug, nocodazole, followed by an immunoblotting assay with CPAP antiserum. Interestingly, CPAP was detected as a doublet in asynchronized cells and prometaphase cells that had been synchronized by treating with nocodazole. However, the slower migrating CPAP form was predominant in prometaphase cells compared with asynchronized cells (Fig. 5A, lanes 1 and 2). To test whether the migration change was induced by phosphorylation, 293T cells were treated with nocodazole and λ-phosphatase (λ-PPase). As shown in Fig. 5A, lane 3, Western blotting with CPAP antibody revealed that CPAP shifted to the faster migrating form. These findings indicate that the mitosis-specific form is phosphorylated, and CPAP is regulated by phosphorylation during the cell cycle.
FIGURE 5.
Self-interaction of CPAP was negatively regulated by mitotic phosphorylation. A, lysates from asynchronous cells (lane 1), cells treated with nocodazole (lane 2), and cells treated with both nocodazole and λ-PPase (lane 3) were resolved by 6% SDS-PAGE and immunoblotted with anti-CPAP antibody. B, extracts of asynchronous cells, nocodazole-synchronized mitotic cells, and mitotic cells treated with λ-PPase co-expressing FLAG-CPAP and GFP-CPAP or FLAG-CPAP and GFP were immunoprecipitated using FLAG-M2 antibody (lanes 6–10). Immunoprecipitates were then fractionated by 10% SDS-PAGE and transferred onto a nitrocellulose membrane and immunoblotted with GFP (upper panel) and FLAG (lower panel) antibody respectively. co-IP, co-immunoprecipitation.
A common function of phosphorylation is to regulate protein-protein interactions. Thus, we reasoned that the self-interaction of CPAP might be regulated by phosphorylation. To confirm our speculation, we performed a co-immunoprecipitation assay using lysates of 293T cells transiently transfected to express FLAG-tagged CPAP and GFP-tagged CPAP, following nocodazole or nocodazole plus λ-PPase treatment. As shown in Fig. 5B, Western blotting using anti-FLAG antibody confirmed a successful precipitation of FLAG-CPAP, and GFP antibody blotting demonstrated that GFP-CPAP was co-precipitated with FLAG-CPAP (Fig. 5B, lanes 8–10), whereas control GFP was not precipitated with FLAG-tagged CPAP (Fig. 5B, lanes 6 and 7). The level of GFP-CPAP co-precipitated with FLAG-CPAP decreased significantly in cells treated with nocodazole in Fig. 5B, lane 9, and recovered to almost two-thirds of the level of asynchronized cells in cells treated with nocodazole and λ-PPase in lane 10. Thus, we conclude that the self-interaction of CPAP was negatively regulated by mitotic phosphorylation.
DISCUSSION
CPAP was first identified as a centrosomal resident protein associated with P4.1. It also associates with the γ-tubulin complex and inhibits microtubule nucleation from the centrosome (16). Further research revealed that CPAP plays an important role in centrosome function, including centrosome duplication, centriole assembly, and control of centriole length (27–30). In this study, we demonstrated that CPAP plays a role in centrosome cohesion.
Here, we showed that suppression of CPAP by siRNA induces a distinct centrosome splitting during interphase. This phenotype is also seen when the C-terminal CPAP is overexpressed, suggesting that CPAP integrity is essential for centrosome cohesion. Our experimental results also show that CPAP molecules exist as dimers, and this dimerization is mainly mediated by the fifth coiled-coil domain. Importantly, this intermolecular association is critical for the function of CPAP in centrosome cohesion. It was interesting to see that overexpression of GFP-ΔCC5 resulted in centrosome splitting phenotype (Fig. 3), and this phenotype is rescued upon GFP-ΔCC5 homodimerization induced by AP20187. We reason that CPAP dimerization is mainly achieved via the CC5 domain, whereas CPAP-ΔCC5 mutant also forms a weaker intermolecular association with itself or endogenous CPAP. In this case, the CPAP-ΔCC5 mutant forms a heterodimer with endogenous CPAP to prevent formation of functional homodimer of wild type CPAP and becomes a homodimer upon addition of AP20187 to allow endogenous CPAP to form a functional homodimer. Importantly, this self-association of CPAP is negatively regulated by mitotic phosphorylation as the dimerization is abolished when CPAP is phosphorylated during mitosis, suggesting that mitotic phosphorylation regulates the conversion of CPAP from dimer to monomer and reconciles centrosome cohesion and separation during the cell cycle.
Centrosome cohesion is orchestrated by several proteins that can be categorized into two classes. One type of protein is able to bind the proximal ends of the centrioles, and the other type is able to bind the walls of the centrioles in a tubulin-polyglutamylation-dependent manner. Glutamylated tubulin is a framework for anchoring the tubulin minus ends (34). Previous studies have identified several proteins involved in centrosome cohesion, including C-Nap1, Rootletin, pericentrin, Dynamin2, Cep68, and Cep215 (8–12). Immunoelectron microscopy has shown that both Rootletin and Cep68 localize to striking fibers linked to centrioles (8, 10, 12), although Cep215 is associated with the centriolar cylinders and C-Nap1 is confined to the proximal end domains of centrioles (9, 12). We have known that CPAP interacts with tubulin and residues 423–607 within CPAP that constitutes the MT-binding domain (14, 15). Moreover, immunoelectron microscopy using antibody against the C terminus of CPAP has revealed that CPAP is concentrated within the proximal lumen of both parental centrioles and procentrioles (18). Thus, CPAP recruits the centriole MTs through the MT-binding domain within the middle segment, although its C terminus may act as an inner linker anchoring itself in the lumen of centriolar cylinders by polymerization. Therefore, overexpression of CPAP leads to abnormally elongated centrioles (19–21), whereas depletion of CPAP impairs centriole duplication (27). It is reasonable to postulate that knockdown of CPAP will shorten the centriolar cylinder, resulting indirectly in centrosome splitting. Consistent with this view, we have shown that the cells expressing GFP-ΔCC5 show decreased accumulation of glutamylated tubulin on centrosomes, suggesting that dimeric CPAP may maintain the centrosome cohesion through recruiting glutamylated tubulin.
There is another possibility that the N terminus of CPAP may penetrate through the centriolar cylinder and cross-link with other centrosomal proteins as part of a linker. This rationale is consistent with a previous hypothesis that additional proteins could collaborate with C-Nap1 to form linker structures of centrosome cohesion (9). However, to date no one has identified a centrosomal cohesion protein that interacts with CPAP. It will be of interest to identify the target protein(s) of CPAP that directly interact(s) with CPAP and function(s) on centrosome cohesion. Because a close spatial relationship of CPAP with Cep215 and C-Nap1 exists, it would be worthwhile to investigate whether CPAP binds to Cep215 and/or C-Nap1 and cooperates in maintaining centrosome cohesion. Interestingly, functional perturbation of both CPAP and Cep215 are implicated in MCPH disease (23).
Previous research has shown that CPAP is a part of the γ-tubulin complex, and most of the total protein is soluble (16, 34), suggesting that CPAP may be a component of pericentriolar material. In fact, a fraction of CPAP is found outside of the centriolar cylinder (18). Recent work demonstrates that partial CPAP depletion results in asymmetric distribution of the pericentriolar material (20). All these findings point to the fact that CPAP may participate directly in centrosome cohesion.
Structural and functional transitions during centrosome duplication, maturation, and separation are orchestrated in synchrony with cell cycle by both kinase and phosphatase (3, 35). To date, many mitotic kinases have been found to play at least some roles in centrosome cohesion and separation, including cAMP-dependent protein kinase, Nek2, Plk1, Aurora-A, and Cdk2/cyclin A and E (4, 36). Nek2, a member of the NIMA family, was identified as centrosome-associated kinase and found to be involved in centrosome separation (37, 38). Further research showed that Nek2 interacts with and phosphorylates directly or indirectly C-Nap1, β-catenin, and Rootletin to orchestrate centrosome cohesion and separation (8, 9, 39). However, centrosome cohesion is regulated precisely by a balance of kinase and phosphatase activities, phosphorylation and proteolysis. As a physiological antagonist of Nek2, PP1α (protein phosphatase 1 α-isoform) is associated with and down-regulates Nek2 activity in vitro and in vivo (4, 40, 41). Our data reveal that CPAP is phosphorylated during mitosis, which is critical to negatively regulate the intermolecular association of CPAP. Interestingly, predictions based on bioinformatics suggest that there are many potential phosphorylation sites of Nek2, PLK1, and Aurora-A within CPAP (data not shown). It will be significant to identify the kinase responsible for the phosphorylation of CPAP and the site that is phosphorylated. Future biochemical characterization and visualization of the spatiotemporal dynamics of CPAP intermolecular association will further our understanding of the molecular mechanisms underlying centrosome cohesion regulation.
Taken together, we conclude that CPAP exerts its function in centrosome cohesion via an intermolecular association of CC5, and this dimerization is released by mitotic phosphorylation. The mitotic phosphorylation of CPAP regulates centrosome cohesion and separation during the cell cycle. Our future work will focus on identifying the kinase that phosphorylates CPAP and that governs the interaction between CPAP molecules.
Acknowledgments
We thank the members of our research groups for their insightful discussion and technical assistance during the course of this study.
This work was supported, in whole or in part, by National Institutes of Health Grants DK-56292, CA89019, CA92080, and CA118948 and Grant G12RR03034 from the NCRR for work performed in the facilities used. This work was also supported by Chinese Academy of Science Grants KSCX1-YW-R65 and KSCX2-YW-H10, Chinese 973 Project Grants 2002CB713700, 2006CB943600, 2007CB914503, and 2010CB912103, Chinese 863 Project Grant 2006AA02A247, Chinese Natural Science Foundation Grants 30270654, 30070349, 90508002, 30121001, and 30871236 (to C. J.), 90813008 (to Z. W.), and 30870990 (to X. D.), China National Key Projects for Infectious Disease Grant 2008ZX10002-021, American Cancer Society Grant RPG-99-173-01, a research grant from Georgia Cancer Coalition Breast Cancer, and Atlanta Clinical and Translational Science Award Chemical Biology Grant P20RR011104.
- MT
- microtubule
- GFP
- green fluorescent protein
- PBS
- phosphate-buffered saline
- siRNA
- small interfering RNA
- DAPI
- 4′,6-diamidino-2-phenylindole
- PIPES
- 1,4-piperazinediethanesulfonic acid
- λ-PPase
- λ-phosphatase.
REFERENCES
- 1.Blagden S. P., Glover D. M. (2003) Nat. Cell Biol. 5, 505–511 [DOI] [PubMed] [Google Scholar]
- 2.Doxsey S., McCollum D., Theurkauf W. (2005) Annu. Rev. Cell Dev. Biol. 21, 411–434 [DOI] [PubMed] [Google Scholar]
- 3.Meraldi P., Nigg E. A. (2002) FEBS Lett. 521, 9–13 [DOI] [PubMed] [Google Scholar]
- 4.Meraldi P., Nigg E. A. (2001) J. Cell Sci. 114, 3749–3757 [DOI] [PubMed] [Google Scholar]
- 5.Nigg E. A. (2007) Trends Cell Biol. 17, 215–221 [DOI] [PubMed] [Google Scholar]
- 6.Barr A. R., Gergely F. (2007) J. Cell Sci. 120, 2987–2996 [DOI] [PubMed] [Google Scholar]
- 7.Faragher A. J., Fry A. M. (2003) Mol. Biol. Cell 14, 2876–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahe S., Stierhof Y. D., Wilkinson C. J., Leiss F., Nigg E. A. (2005) J. Cell Biol. 171, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayor T., Stierhof Y. D., Tanaka K., Fry A. M., Nigg E. A. (2000) J. Cell Biol. 151, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J., Adamian M., Li T. (2006) Mol. Biol. Cell 17, 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Fiore B., Ciciarello M., Mangiacasale R., Palena A., Tassin A. M., Cundari E., Lavia P. (2003) J. Cell Sci. 116, 3399–3411 [DOI] [PubMed] [Google Scholar]
- 12.Graser S., Stierhof Y. D., Nigg E. A. (2007) J. Cell Sci. 120, 4321–4331 [DOI] [PubMed] [Google Scholar]
- 13.Peng B., Sutherland K. D., Sum E. Y., Olayioye M., Wittlin S., Tang T. K., Lindeman G. J., Visvader J. E. (2002) Mol. Endocrinol. 16, 2019–2033 [DOI] [PubMed] [Google Scholar]
- 14.Hung L. Y., Chen H. L., Chang C. W., Li B. R., Tang T. K. (2004) Mol. Biol. Cell 15, 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu W. B., Hung L. Y., Tang C. J., Su C. L., Chang Y., Tang T. K. (2008) Exp. Cell Res. 314, 2591–2602 [DOI] [PubMed] [Google Scholar]
- 16.Hung L. Y., Tang C. J., Tang T. K. (2000) Mol. Cell. Biol. 20, 7813–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho J. H., Chang C. J., Chen C. Y., Tang T. K. (2006) Biochem. Biophys. Res. Commun. 339, 742–747 [DOI] [PubMed] [Google Scholar]
- 18.Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y. D., Nigg E. A. (2007) Dev. Cell 13, 190–202 [DOI] [PubMed] [Google Scholar]
- 19.Kohlmaier G., Loncarek J., Meng X., McEwen B. F., Mogensen M. M., Spektor A., Dynlacht B. D., Khodjakov A., Gonczy P. (2009) Curr. Biol. 19, 1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt T. I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S. B., Stierhof Y. D., Nigg E. A. (2009) Curr. Biol. 19, 1005–1011 [DOI] [PubMed] [Google Scholar]
- 21.Tang C. J., Fu R. H., Wu K. S., Hsu W. B., Tang T. K. (2009) Nat. Cell Biol. 11, 825–831 [DOI] [PubMed] [Google Scholar]
- 22.Bond J., Roberts E., Springell K., Lizarraga S. B., Lizarraga S., Scott S., Higgins J., Hampshire D. J., Morrison E. E., Leal G. F., Silva E. O., Costa S. M., Baralle D., Raponi M., Karbani G., Rashid Y., Jafri H., Bennett C., Corry P., Walsh C. A., Woods C. G. (2005) Nat. Genet. 37, 353–355 [DOI] [PubMed] [Google Scholar]
- 23.Woods C. G., Bond J., Enard W. (2005) Am. J. Hum. Genet. 76, 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J., Jackson A. P., Bond J., Woods C. G. (2006) Trends Mol. Med. 12, 358–366 [DOI] [PubMed] [Google Scholar]
- 25.Evans P. D., Vallender E. J., Lahn B. T. (2006) Gene 375, 75–79 [DOI] [PubMed] [Google Scholar]
- 26.Gul A., Hassan M. J., Hussain S., Raza S. I., Chishti M. S., Ahmad W. (2006) J. Hum. Genet. 51, 760–764 [DOI] [PubMed] [Google Scholar]
- 27.Leidel S., Gönczy P. (2003) Dev. Cell 4, 431–439 [DOI] [PubMed] [Google Scholar]
- 28.Kirkham M., Müller-Reichert T., Oegema K., Grill S., Hyman A. A. (2003) Cell 112, 575–587 [DOI] [PubMed] [Google Scholar]
- 29.Salisbury J. L. (2003) Trends Cell Biol. 13, 340–343 [DOI] [PubMed] [Google Scholar]
- 30.Basto R., Lau J., Vinogradova T., Gardiol A., Woods C. G., Khodjakov A., Raff J. W. (2006) Cell 125, 1375–1386 [DOI] [PubMed] [Google Scholar]
- 31.Yuan K., Hu H., Guo Z., Fu G., Shaw A. P., Hu R., Yao X. (2007) J. Biol. Chem. 282, 27414–27423 [DOI] [PubMed] [Google Scholar]
- 32.Zhu M., Wang F., Yan F., Yao P. Y., Du J., Gao X., Wang X., Wu Q., Ward T., Li J., Kioko S., Hu R., Xie W., Ding X., Yao X. (2008) J. Biol. Chem. 283, 18916–18925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang J., Chen Y., Zhao Y., Yu H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20232–20237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bornens M. (2002) Curr. Opin. Cell Biol. 14, 25–34 [DOI] [PubMed] [Google Scholar]
- 35.Mayor T., Meraldi P., Stierhof Y. D., Nigg E. A., Fry A. M. (1999) FEBS Lett. 452, 92–95 [DOI] [PubMed] [Google Scholar]
- 36.Lutz W., Lingle W. L., McCormick D., Greenwood T. M., Salisbury J. L. (2001) J. Biol. Chem. 276, 20774–20780 [DOI] [PubMed] [Google Scholar]
- 37.Fry A. M., Schultz S. J., Bartek J., Nigg E. A. (1995) J. Biol. Chem. 270, 12899–12905 [DOI] [PubMed] [Google Scholar]
- 38.Fry A. M., Meraldi P., Nigg E. A. (1998) EMBO J. 17, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bahmanyar S., Kaplan D. D., Deluca J. G., Giddings T. H., Jr., O'Toole E. T., Winey M., Salmon E. D., Casey P. J., Nelson W. J., Barth A. I. (2008) Genes Dev. 22, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helps N. R., Luo X., Barker H. M., Cohen P. T. (2000) Biochem. J. 349, 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mi J., Guo C., Brautigan D. L., Larner J. M. (2007) Cancer Res. 67, 1082–1089 [DOI] [PubMed] [Google Scholar]