Abstract
We report the solution NMR structures of the N-domain of the Menkes protein (ATP7A) in the ATP-free and ATP-bound forms. The structures consist of a twisted antiparallel six-stranded β-sheet flanked by two pairs of α-helices. A protein loop of 50 amino acids located between β3 and β4 is disordered and mobile on the subnanosecond time scale. ATP binds with an affinity constant of (1.2 ± 0.1) × 104 m−1 and exchanges with a rate of the order of 1 × 103 s−1. The ATP-binding cavity is considerably affected by the presence of the ligand, resulting in a more compact conformation in the ATP-bound than in the ATP-free form. This structural variation is due to the movement of the α1-α2 and β2-β3 loops, both of which are highly conserved in copper(I)-transporting PIB-type ATPases. The present structure reveals a characteristic binding mode of ATP within the protein scaffold of the copper(I)-transporting PIB-type ATPases with respect to the other P-type ATPases. In particular, the binding cavity contains mainly hydrophobic aliphatic residues, which are involved in van der Waal's interactions with the adenine ring of ATP, and a Glu side chain, which forms a crucial hydrogen bond to the amino group of ATP.
Keywords: Methods/NMR, Nucleotide, Protein/Structure, ATP7A, Menkes Protein, Nucleotide-binding Domain, Copper(I)-transporting ATPases
Introduction
The Menkes protein (ATP7A or MNK) is a human PIB-type ATPase that has a key role in the regulation of copper homeostasis in a number of organs (1, 2). Like all the proteins belonging to the P-type family, MNK uses the energy arising from ATP hydrolysis to transport copper(I) across cell membranes (3). In particular, MNK transfers the metal ion from the cytosol either into the trans-Golgi network or into vesicles (4). In the former case, the metal ion is subsequently incorporated into copper-dependent enzymes within the secretory pathway. For example, tyrosinase, peptidyl-amonooxygenase, and ceruloplasmin co-localize with copper(I)-transporting ATPases in the trans-Golgi network and require the ATPase-mediated copper transport for the formation of the holo-enzymes (5). Instead, the copper(I) ions that are pumped into vesicles are subsequently released into the extracellular environment, through the fusion of the vesicles themselves with the cell membrane. This is the mechanism by which the copper ions ingested with the diet can cross the basolateral membrane of enterocytes (4). Mutations or deletions in the gene encoding the MNK protein are associated with a fatal childhood disorder, Menkes disease (6–8). In Menkes disease, MNK is not functioning, and copper export from the enterocytes is impaired, resulting in copper accumulation in intestinal cells and diminished copper transport to the blood and other tissues (9). Elucidating the consequences of mutations on the structure and function of MNK would help in a better understanding of the molecular mechanism underlying Menkes disease.
All P-type ATPases are multi-domain membrane proteins, sharing a basic core architecture (10). This comprises a hydrophilic region protruding into the cytosol, which contains the Actuator domain (A-domain) and the ATP-binding domain, which in turn can be further separated into two smaller domains named the phosphorylation domain (P-domain) and the nucleotide-binding domain (N-domain). Another region common to all P-ATPases consists of a number of transmembrane helices involved in the formation of an intramembranous channel whose organization leads to the definition of five P-type groups, which are referred to as type I-V (11). Within these groups a number of different subtypes can be distinguished. Copper(I)-transporting ATPases belong to the PIB-(sub)type. In addition to the core structure, their distinguishing feature is the presence of a long N-terminal tail, which contains a variable (between one and six) number of 70-amino acid independently folded domains (12, 13), each of them binding a single copper(I) ion through two cysteines (14).
PIB-ATPases share a common catalytic mechanism with the other P-ATPases (15). A central feature of the catalytic cycle is the formation of a transient acyl-phosphate intermediate upon ATP hydrolysis. This reaction takes place at an invariant aspartate residue in the highly conserved DKTG sequence located in the P-domain. The intermediate is then dephosphorylated by the invariant TGE sequence of the actuator domain (A-domain). The P- and A-domains are overall the most conserved in sequence within all of the P-type ATPases (15). The N-domain, which is the object of the present study, also plays a crucial role with respect to the enzymatic activity, because it is involved in ATP binding. However, the N-domain of PIB-ATPases shares little sequence similarity to the corresponding domains of other P-type ATPases. The structures of the isolated ATP-binding domain of CopA from Archaeoglobus fulgidus in the ATP-free form (16) and complexed with an ATP analogue molecule (AMPPCP)3 (17) and the structure of the N-domain of the human Wilson protein (ATP7B or WLN) (18) in the ATP-bound form are available. In the present work we report the solution structure of both the ATP-free and ATP-bound forms of the N-domain of the human MNK protein. Their comparison provides a detailed analysis of the conformational changes that the N-domain undergoes upon nucleotide binding, which is a crucial requisite for the thorough understanding of the enzymatic cycle of PIB-ATPases.
MATERIALS AND METHODS
Preparation of Protein Samples
The DNA segment encoding the N-domain of the MNK protein (hereafter N-MNK), corresponding to amino acids 1051–1231 of ATP7A, was amplified via PCR from human cDNA and cloned in the gateway entry vector pENTR/TEV/d-TOPO (Invitrogen) to include the TEV protease cleavage site at the N-terminal end. This segment was then subcloned into pETG-30A (EMBL-Heidelberg) through the gateway recombination reaction, yielding a plasmid expressing the protein fused with glutathione S-transferase and a His tag at the N terminus. After cleavage of the tag, five amino acids from the restriction site were left at the N terminus of the protein. The plasmids were then transformed in Escherichia coli strain BL21(DE3) Rosetta2 (EMD Bioscience) for protein expression. Protein expression was induced with 0.5 mm isopropyl β-d-thiogalactopyranoside, and the cells were grown at 37 °C for 4 h in minimal medium cultures. For isotope enrichment, (15NH4)2SO4 and [13C]glucose were used. The two proteins were purified by using a HisTrap chelating FF column (GE Healthcare). After this, the His tag was cleaved with AcTEV protease (Invitrogen). The mixture derived from the cleavage step was then applied on a 16/60 Superdex75 size exclusion column. This allowed a complete separation of the cleaved protein from the N-terminal tag. The protein purity was checked by SDS-PAGE and matrix-assisted laser desorption ionization time-of-flight mass spectra. The typical yield was 20 mg of pure protein/liter of culture. Because of the presence of five cysteine residues in the N-domain, the samples were reduced with excess of dithiothreitol in an inert atmosphere chamber (Coy Lab) prior to exchange in the final buffer. NMR samples contained between 0.8 and 1.5 mm protein in 50 mm phosphate buffer at pH 7.0 and 2 mm dithiothreitol. Dynamic Light Scattering analysis confirmed that the protein was monomeric under these conditions. For the preparation of the ATP-bound form of the protein, 3 equivalents of ATP disodium salt (Sigma) were added to the ATP-free N-domain.
NMR Spectroscopy and Structure Determination
NMR spectra were acquired at 298 K on Avance 900, 700, and 500 Bruker spectrometers equipped with triple resonance cryoprobes. The NMR experiments used for the assignment of the backbone and side chain resonances of N-MNK are summarized in supplemental Table S1. The assigned backbone and side chain peaks of both ATP-free and ATP-bound N-MNK are reported in supplemental Tables S2 and S3. Backbone dihedral angle constraints were derived from 15N, 13C', 13Cα, 13Cβ, and Hα chemical shifts, using TALOS (19). Distance constraints for structure determination were obtained from 15N-edited and 13C-edited three-dimensional NOE spectroscopy-HSQC spectra. Additional distance constraints between the ATP molecule and the N-MNK were extracted from a two-dimensional 13C-filtered NOE spectroscopy spectrum (20). One-bond 1H-15N dipolar couplings were measured for both the ATP-free and ATP-bound forms on samples containing 11 mg/ml of filamentous bacteriophage Pf1 strain LP11–92 isolated from wild type Pseudomonas aeruginosa (ASLA, Ltd., Riga, Latvia) at 800 and 900 MHz, respectively, by using the in-phase/anti-phase method (21).
All of the restraints used for structure calculation (upper distance limits, ϕ and ϕ backbone dihedral angles, and residual dipolar couplings) are reported in supplemental Tables S14 and S5. The residual dipolar coupling values were measured for amide signals not overlapped in the 15N HSQC spectra (supplemental Fig. S1). For residual dipolar coupling restraints, the χ tensor parameters were obtained with FANTAORIENT, and they were optimized through iterative cycles of PSEUDOCYANA until convergence (22). For structure calculations, 500 random conformers were annealed in 13,000 steps using the program CYANA-2.1 (23).
The 20 conformers with the lowest residual target function values for both ATP-free and ATP-bound N-MNK domains were subjected to restrained energy minimization with AMBER 10.0 package in explicit water solvent (24). We evaluated the quality of the structures using the PROCHECK NMR (25), WHATIF (26), and PSVS (27) programs for both the ATP-free and ATP-bound forms of N-MNK. The data are summarized in supplemental Tables S4 and S5. The program MOLMOL (28) was used for visualizing the structures.
Acquisition and Analysis of NMR Heteronuclear Relaxation Data
15N longitudinal and traverse relaxation rates (29) and 15N(1H)-NOEs (30) were recorded at 298 K and at 500 MHz at a protein concentration of 1.2 mm. R1 and R2 relaxation rates were obtained by fitting the cross-peak volumes, measured as a function of the relaxation delays, to a single exponential decay as described in the literature (31). Heteronuclear NOE values were calculated as the ratio of peak volumes in spectra recorded with and without 1H saturation. In all experiments the water signal was suppressed with the “water flipback” scheme (30). The relaxation data (R1, R2, and 15N(1H)-NOEs) were analyzed according to the model-free approach of Lipari and Szabo (32, 33), using the program TENSOR2 (34) (supplemental Fig. S2).
Calorimetry
Isothermal Titration Microcalorimetry experiments were performed at 298 K with a VP-ITC microcalorimeter (MicroCal, Inc., Northampton, MA). After an initial injection of 1 μl, aliquots of 10 μl of 5.9 mm ATP were stepwise injected into the sample cell containing a 115 μm solution of ATP-free N-MNK until complete saturation. We performed all of the experiments in 50 mm phosphate buffer at pH 7.0. The heats of dilution were measured by injecting the ligand into buffer and then subtracted from the binding heats. The thermodynamic parameters and affinity constant values were calculated, fitting data to a single binding site model with ORIGIN 7.0 software (Microcal, Inc.).
RESULTS
The Solution Structures of ATP-free and ATP-bound N-MNK
We overexpressed the soluble nucleotide-binding domain of the human MNK protein (N-MNK) and determined the solution structures of its ATP-free and ATP-bound forms using multidimensional NMR spectroscopy. Their 1H-15N HSQC spectra showed well dispersed amide signals for both forms (supplemental Fig. S3). Only the peaks of the stretch 1125–1176 clustered in the random coil region. 15N relaxation data for these residues featured negative 15N-1H NOE values, indicating that they experienced dynamics on the subnanosecond time scale, as commonly observed for amino acids in random coil conformation (supplemental Fig. S2).
The final bundles of 20 conformers of N-MNK in the two forms have, after energy minimization in explicit solvent, average target functions of 1.17 ± 0.19 and 1.53 ± 0.21 Å2 (CYANA units) (23) and average backbone RMSD values (to the mean structure) of 0.86 ± 0.15 and 0.77 ± 0.08 Å (over residues 1057–1124 and 1177–1230), respectively. The region 1125–1176, which constitutes a long loop between strands β3 and β4, is disordered in both structures consistently with the lack of long range NOEs involving these residues and with its aforementioned dynamics.
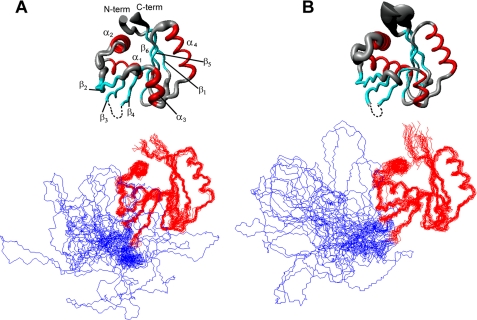
The solution structures of ATP-free and ATP-bound N-MNK (Fig. 1) consist of a twisted six-stranded, antiparallel β-sheet, flanked by two pairs of α-helices, one on the concave side of the sheet (α1 and α2) and the other on the convex one (α3 and α4; Fig. 1). Helices α1 and α4 are tightly packed against the two sides of the β-sheet and, together with it, form the hydrophobic core of the protein. The orientation and position of helix α2 within the protein structure is determined by its hydrophobic contacts with residues of helix α1, specifically involving the aliphatic side chains of Leu1074, Ala1075, and Val1077 (α1) and Ile1092, Tyr1095, and Leu1100 (α2). Helix α3 is constituted by five amino acids; its orientation within the protein structure is defined by the hydrophobic contacts formed by the two most N-terminal residues (Trp1187 and Met1188) and the β-sheet.
FIGURE 1.
Solution structures of ATP-bound N-MNK (A) and ATP-free N-MNK (B). The average backbone RMSD between ATP-free and ATP-bound N-MNK domains is of 1.01 Å. The secondary structure elements comprise residues 1054–1061 (β1), 1070–1081 (α1), 1090–1100 (α2), 1108–1114 (β2), 1118–1124 (β3), 1178–1183 (β4), 1186–1190 (α3), 1197–1209 (α4), 1212–1218 (β5), and 1221–1230 (β6) for ATP-bound N-MNK and residues 1054–1061 (β1), 1070–1080 (α1), 1087–1100 (α2), 1110–1114 (β2), 1118–1123 (β3), 1178–1183 (β4), 1186–1191 (α3), 1197–1209 (α4), 1212–1218 (β5), and 1221–1230 (β6) for ATP-free N-MNK, respectively. Top panels, the radius of the tubes is proportional to the backbone RMSD of each residue. The unstructured loop was omitted for simplicity. Bottom panel, the backbone traces for the twenty lowest energy conformers are superimposed. The unstructured loop is shown in blue.
The Structural Features of the ATP-binding Site of N-MNK
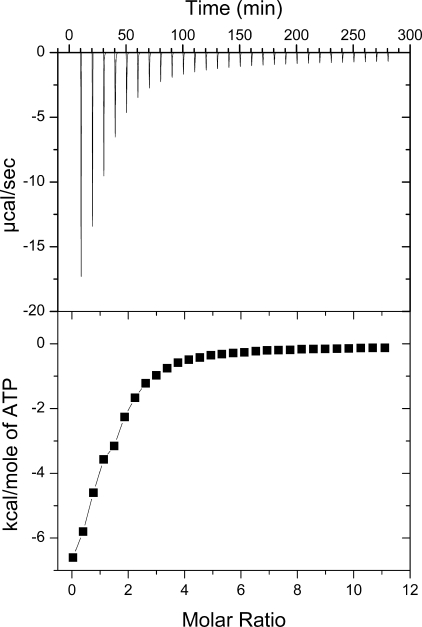
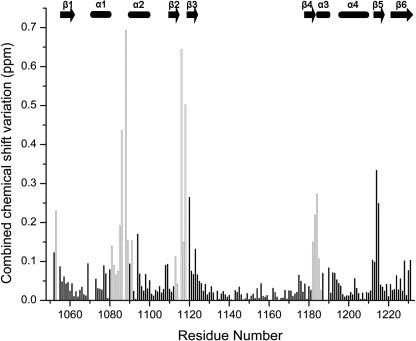
Our ITC data (Fig. 2) could be fitted well to a model assuming a single binding site, with an affinity constant of (1.2 ± 0.1) × 104 m−1. This value is very similar to the value (∼1.4 × 104 m−1) reported for N-WLN (18). The position of the ATP molecule within the N-MNK-binding site was defined through an NMR characterization in solution. The addition of increasing amounts of ATP to N-MNK, either 15N- or 15N-13C-labeled, up to a N-MNK:ATP ratio of 1:3, induced chemical shift changes (Fig. 3) that were monitored for the backbone amide groups through 1H-15N HSQC spectra. The chemical shift changes increased upon increasing ATP concentration, indicating that ATP binds to the protein. The ATP-bound and ATP-free forms of the protein exchanged with one another with a rate fast with respect to the resonance frequency differences in the two species. For a few signals, experiencing the largest changes in chemical shift upon ATP binding, we observed an intermediate exchange regime. The available data allowed us to set the exchange rate between the two forms in the range between 5 × 103 and 3 × 102 s−1. The chemical shift changes are mapped on the protein surface in Fig. 4A. The ATP-binding site comprises residues located in the loops between helices α1 and α2 (hereafter referred to as the α1-α2 loop), between strands β2 and β3 (β2-β3 loop), some residues in strands β4 and β5, and also a few residues in the N- and C-terminal regions (Fig. 3).
FIGURE 2.
ATP binding by N-MNK. Top panel, raw ITC data demonstrating saturable exothermic evolution of heat upon sequential additions of ATP to N-MNK. Bottom panel, normalized ITC data for ATP titration plotted versus the molar ratio of nucleotide/N-MNK.
FIGURE 3.
Combined chemical shift variations of signals between ATP-free N-MNK and ATP-free N-MNK in the presence of 3.0 equivalents of ATP. Empty columns correspond to residues whose backbone experienced significant structural changes upon ATP binding; these residues are shown in red/pink in Fig. 5B. Combined chemical shift variations are calculated from the experimental 1H and 15N chemical shift changes (Δδ(1H) and Δδ(15N), respectively) between corresponding peaks in the two forms, through the following equation (43), . The missing assignment for residue Ile1119 of the ATP-free N-MNK prevented the determination of its chemical shifts perturbations. The position of secondary structure elements is shown.
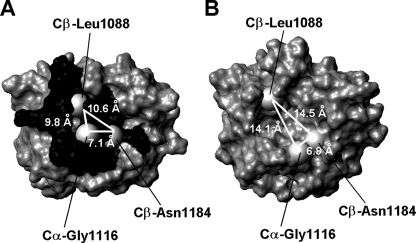
FIGURE 4.
Chemical shift mapping of the surface of N-MNK. A, mapping of chemical shift changes on the molecular surface of the ATP-bound N-MNK domain. B, molecular surface of ATP-free N-MNK domain. The Cβ atoms of Leu1088 and Asn1184 and the Cα atom of Gly1116 are shown as gray spheres. Distances between such atoms are also indicated. The unstructured loop was omitted for simplicity.
We obtained 32 ATP/N-MNK NOEs (supplemental Table S6) from the analysis of the spectra acquired on protein samples where the N-MNK:ATP molar ratio was 1:3. An extensive network of NOEs between the H2 and H8 adenine protons of ATP and the protein residues indicated a single conformation for the base ring of bound ATP. The NOEs between the H2 proton and residues Ile1119 and Ile1182 and between the H8 proton and residues His1086, Pro1087, Leu1088, and Gly1089 defined the orientation of the ring. We observed a lower number of restraints between the ATP ribose protons and the protein resonances. In particular, NOEs were detected between the ribose and the side chains of Leu1088 and Asn1184 and with the amide proton of Gly1116.
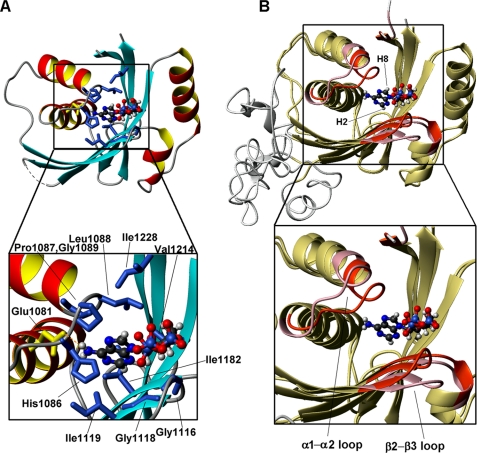
The adenine ring of ATP is located in a hydrophobic cavity formed by His1086, Pro1087, Leu1088, Gly1089, (α1-α2 loop), Ile1119 (β3), Ile1182 (β4), Val1214 (β5), and Ile1228 (β6) (Fig. 5A). At the bottom of this cavity, the charged side chain of Glu1081 forms a hydrogen bond (in the majority of the conformers of the bundle) with the NH2 group of the ATP adenine ring. All of the aforementioned amino acids are conserved in copper-transporting PIB-type ATPases (Fig. 6). The ATP ribose ring is partially exposed with the H1′ proton being more buried within the protein structure, whereas the H5′ and H5″ protons are the most exposed. Finally, the γ-phosphate group protrudes into the solvent (Fig. 5). This is consistent with its subsequent transfer to the invariant Asp residue of the DKTG motif of the P-domain during the catalytic cycle, from the E1 to the E1P state of the enzyme (35). The present structural features are in agreement with previously reported titrations of the N-domain of the homologous human copper(I)-transporting Wilson ATPase (N-WLN) with ATP, ADP, or AMP (18, 36), which suggested that there is very little or no interaction of the γ-phosphate group with the protein.
FIGURE 5.
ATP-binding mode of N-MNK. A, the structure of ATP-bound N-MNK showing the side chains of hydrophobic amino acids in blue and of Glu1081 in yellow, which are in contact with the ATP molecule. The unstructured loop was omitted for simplicity. In the zoom on the ATP-binding cavity, the amino acids are labeled. B, overlay of the structures of ATP-free (pink) and ATP-bound (red) N-MNK, highlighting the regions of structural variation.
FIGURE 6.
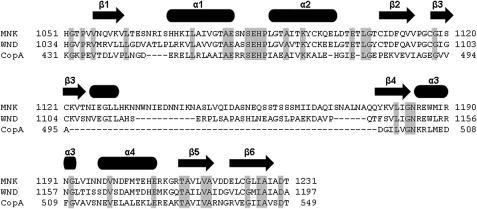
Alignment of the sequences of the N-domains of the MNK, WLN, and A. fulgidus CopA ATPases. The position of secondary structure elements is shown. Conserved residues are shaded.
Fig. 5B shows an overlay of the ATP-free and ATP-bound N-MNK structures where regions experiencing meaningful differences, i.e. outside the uncertainty of the two bundles, are colored in pink and red, respectively. The effect of ATP binding was already evident from chemical shift variation data (Fig. 3). All of the residues experiencing structural rearrangement of their backbone indeed featured appreciable combined chemical shift changes. The backbone conformation of residues 1214–1215, whose amide signals had variations larger than 0.2 ppm (Fig. 3), was not affected by ATP binding, whereas the side chains of these residues were rearranged. The α1-α2 loop and the N-terminal region of helix α2 undergo the largest conformational changes upon ATP binding. The establishment of tight contacts between the side chain of Leu1088 and the ATP adenine ring induces conformational changes in the backbone of the α1-α2 loop that are instrumental to allow the ATP molecule to bind inside the cavity. The stretch SEHP (residues 1084–1087) of this loop, fully conserved in all organisms, has conformation disorder in ATP-free N-MNK, whereas it is more ordered in the ATP-bound form. In ATP-free N-MNK, His1086, whose mutation to Gln in the ATP7B protein causes Wilson disease, has RMSD values of 1.4 and 2.2 Å, for backbone and side chain atoms, respectively, whereas these values drop to 0.5 and 0.8 Å in the ATP-bound form. Indeed, the imidazole ring of His1086 interacts with the adenine moiety of ATP. Also the δNH2 group of Asn1083, which experiences a large chemical shift variation upon ATP binding, is more ordered in the ATP-bound form (side chain RMSD of 0.71 Å for the ATP-bound form and 2.16 Å for the ATP-free form) because of contacts with the side chains of Cys1108 and Ile1109 both located in strand β2. Such interactions are absent in ATP-free N-MNK structure.
On the side of the adenine ring H2 proton (Fig. 5B), the ATP-protein interaction further involves residues located in the β2-β3 loop, the N-terminal region of strand β3, and the C terminus of strand β4. The backbone chemical shifts of the residues located in these regions, especially the invariant Gly1116 and Gly1118, are indeed affected by the interaction with the ATP molecule (Fig. 3). Furthermore, the NH2 group of Asn1184, located at the C terminus of strand β4, also shows meaningful chemical shift variation upon ATP binding and a number of contacts, which are absent in the spectra of ATP-free N-MNK. In the ATP-bound structure its side chain is oriented toward the ribose group of the ATP molecule, whereas it is solvent-exposed in the free form.
The size of the ATP-binding cavity is considerably affected by the presence of the nucleotide molecule, resulting in a more open conformation in the ATP-free form than in the ATP-bound state. In the ATP-free structure the Cβ of Leu1088 is 14.1 ± 2.0 and 14.5 ± 1.6 Å away from the Cα of Gly1116 and the Cβ of Asn1184, respectively (Fig. 4B), whereas these distances are significantly smaller in ATP-bound N-MNK (9.8 ± 1.6 and 10.6 ± 1.3 Å) (Fig. 4A). The decrease in size of the adenine ring binding cavity, caused by the movement of both the α1-α2 and β2-β3 loops, allows the structure of ATP-bound N-MNK to tightly pack around the ATP molecule.
DISCUSSION
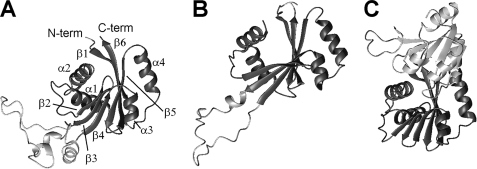
The fold of N-MNK structure is close to that of the corresponding N-WLN domain, which was solved by NMR (Protein Data Bank code 2ARF (18)), and of CopA from A. fulgidus, which was solved by x-ray (Protein Data Bank codes 2B8E (16), 3A1C, and 3A1D (17)). The structures of CopA contain also the P-domain. The backbone RMSD values with respect to the above structures are, respectively, 3.5 and 2.0 Å (values calculated with respect to ATP-free N-MNK). The sequence identity of N-MNK to the same domain of the two other proteins is 51 and 22%, respectively (Fig. 6). Notably, the CopA N-domain lacks the long unstructured loop involving residues 1125–1176 of N-MNK. Indeed this loop is present only in higher eukaryotes, suggesting that it could play a functional role distinct from the catalytic cycle. This solvent-exposed loop is rich in hydrophobic residues and thus might be involved in interdomain or intermolecular interactions, or both.
As shown in Fig. 7, the β-strands of the three structures could be superimposed well, whereas the relative orientation of the α-helices is variable. In particular, in the N-WLN domain the relative orientation of helices α1 and α2 and of helices α3 and α4 differ from that observed for the same pairs of helices in N-MNK and in the N-domain of CopA. The packing of the helices against the β-sheet is also different in N-WLN with respect to that of the other two proteins. These differences are not related to the fact that the N-WLN structure is in the ATP-bound form, because the present data on N-MNK and those reported for the AMPPCP-free and -bound forms of CopA (16, 17) show that the relative orientation of the helices is not affected by substrate binding. The helices in N-WLN have a different orientation with respect to those in the present N-MNK structure despite the high sequence similarity of the two proteins. The packing quality of the ordered residues (as automatically defined by the PSVS server (27)) in the two structures shows a significantly better packing for the present one; in particular, the clash score for the N-WLN structure is very poor (Molprobity (37) Z-score around −9.0 versus +1.0 for the present structure), indicating a number of bad van der Waal's contacts. All of the various quality parameters computed by the software validation tools are somewhat better for the present structure and in line with the average values obtained for high quality x-ray structures. When comparing N-MNK and CopA (Fig. 7), the largest variation is in the orientation of the α1-α2 pair with respect to the β-sheet, which is partly justified by the fact that the human protein contains a four-amino acid insertion in the loop between strand β1 and helix α1.
FIGURE 7.
Comparison of the structures of various copper(I)-transporting ATPases. ATP-bound N-MNK (A) is compared with the structures of ATP-bound N-WLN (18) (B) and of the ATP-bound PN-domains of A. fulgidus CopA (17) (C). The common core of the N-domain is in dark gray in all panels. The unstructured loop of N-MNK and N-WLN, which is absent in CopA, and the P-domain in the structure of A. fulgidus CopA are in light gray. The structures in the three panels were superimposed by best fitting the coordinates of the backbone atoms of the corresponding residues in the alignment of Fig. 6. Only residues involved in secondary structure elements were taken into account. For clarity, the structures have been translated horizontally after superimposition. C-term, C-terminal; N-term, N-terminal.
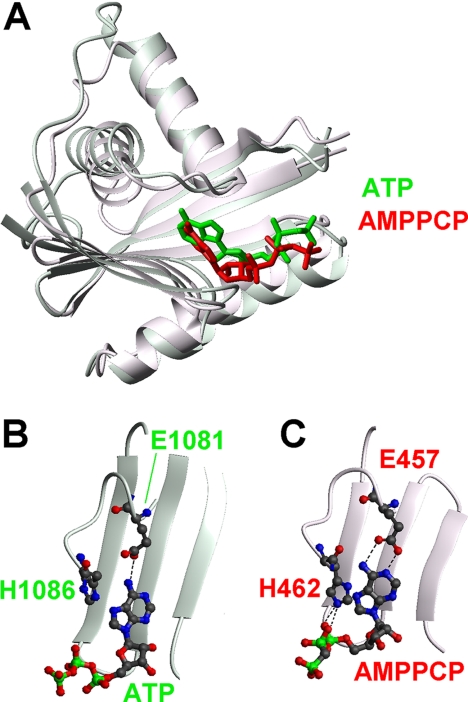
In the structure of CopA in complex with the AMPPCP molecule (17), the protein-substrate interactions involve the same regions as found in N-MNK (α1-α2 and β2-β3 loops) (Fig. 8). The orientation of the AMPPCP within the protein scaffold is slightly different with respect to N-MNK. In particular, the adenine ring of the AMPPCP molecule lies against the β-sheet, and both the N1 and the N6 atom of the amino group are in a position allowing the formation of hydrogen bonds with the side chain of Glu457 (CopA numbering). Instead, only one hydrogen bond involving the amino group is present in N-MNK. In the bacterial protein, the Nϵ2 atom of the imidazole ring of His462 (His1086 in N-MNK numbering) forms hydrogen bonds with the α and β phosphates of the AMPPCP molecule (Fig. 8C). The resonance of the Hϵ2 proton of the His1086 ring was not observable in the NMR spectra of ATP-bound N-MNK, thus preventing us from observing the aforementioned hydrogen bond. The position of His462 is defined also by van der Waal's interactions between its side chain and the adjacent proline residue (Pro463 in CopA numbering corresponding to Pro1087 in N-MNK). In turn, the latter amino acid is in van der Waal's contact with the side chain of Thr426 of the P-domain. The absence of this domain in the present structure can be responsible for the different orientation, within the binding cavity, of the fully conserved His1086 (Fig. 8B). Unfortunately, it is not possible to perform a comparison of the binding mode of ATP also with ATP-bound N-WLN, as the deposited 2ARF structure does not contain the coordinates for the ATP molecule.
FIGURE 8.
Comparison of the ATP-bound structure of N-MNK and of the N-domain of AMPPCP-bound A. fulgidus CopA showing in detail the protein-substrate interactions. A, superimposition of the overall structures of the N-domain of CopA (molecule A in 3A1C, red) and N-MNK (green). The substrates are shown as sticks. The unstructured loop of N-MNK was omitted for simplicity. Details of the nucleotide-binding regions are shown in B for N-MNK and in C for CopA.
The global fold of the N-domains of copper-transporting ATPases is shared by the corresponding domains of other P-type ATPases, such as the mammalian SERCA calcium-transporting ATPase (Protein Data Bank codes 1IWO, 1SU4, and 1VFP (38, 39)), the mammalian Na+,K+-ATPase (Protein Data Bank code 1MO8 (40)) and the bacterial Kdp-ATPase (Protein Data Bank code 2A00 (41)), despite their low sequence similarity. However, there are notable variations in the way each of these domains binds ATP. In the bacterial Kdp-ATPase, the crucial protein-ATP interactions involve hydrogen bonding of the adenine NH2 group to the side chain of Asp344, a π-π stacking interaction between the adenine ring and Phe377 and a π-cation interaction between the adenine ring and the side chain of conserved Lys395 (41). In the N-domain of rat Na+,K+-ATPase, the amino protons of the base form a hydrogen bond to the Oϵ1 atom of Gln482, whereas the adenine ring is again involved in a π-π stacking interaction with Phe475 (40). Similarly, in the SERCA complex with an ATP analogue, the substrate molecule forms a hydrogen bond with Glu442 and a π-π stacking interaction with Phe487 (38). At the rim of the ATP-binding cavity, there are positively charged amino acids whose side chains interact with the phosphate groups. However, these interactions with the N-domain appear to be relatively loose. Indeed the N-domain of Kdp-ATPase, similarly to the N-WLN protein as already mentioned (36), does not discriminate between AMP, ADP, and ATP, indicating that the energetics of the phosphate-protein interactions is modest (41). In the 1VFP structure of the full-length SERCA protein, the γ-phosphate group of the bound ATP analogue interacts with the P-domain (38). The substrate therefore acts as a bridge between the N- and P-domains. In the present structure, the amino group of ATP forms a hydrogen bond with Glu1081, which is three amino acids before the Asp344 of Kdp-ATPase, and it is located at the C terminus of helix α1, whereas Asp344 is located in the α1-α2 loop. Moreover, in the binding pocket there are neither an aromatic residue to form π-π stacking interactions nor a positively charged residue to give rise to π-cation interactions. These are replaced by an extensive ensemble of hydrophobic interactions (Fig. 5). We can speculate that the formation of the hydrogen bond is somehow “mandatory” as the amino group of the adenine ring becomes fully buried upon binding. At the same time, this very specific interaction may be instrumental to orient the ring within the cavity, because the other interactions appear in the present structure to be less specific. Finally, at the level of the entire protein backbone, the rearrangements that we observed in N-MNK upon ATP binding are similar to those reported for the N-domain Kdp-ATPase (41), involving the same loop regions (Fig. 5), and instead differ from those reported for the N-domain of Na+,K+-ATPase (40), where a rearrangement of the β-sheet structure occurs.
In summary, the comparison of the solution structures of ATP-free and ATP-bound N-MNK provides interesting insights in the ATP-binding mode that is presumably general to all eukaryotic copper(I)-transporting ATPases. In particular, the ATP-protein interactions comprise a hydrogen bond between a carboxylate group (that of Glu1081 in the case of N-MNK) and the amino group of the adenine ring, an interaction that is common to the other structurally characterized N-domains but features a network of hydrophobic interactions instead of the commonly observed π-π stacking interaction with an aromatic side chain, present in the other P-type ATPases (40–42). This is in agreement with very recent data on a bacterial homologue of MNK, that is A. fulgidus CopA (17). At a global level, the backbone rearrangements observed are small and mainly limited to two loops in the vicinity of the ATP molecule. In line with this, the N-MNK protein dynamics is also affected only at a local level, i.e. for a few amino acids and amino acidic side chains within the binding pocket. It therefore appears that the atomic level mechanism of ATP binding by the various members of the broad family of P-type ATPases features a mixture of common and subfamily specific behavior.
Supplementary Material
This work was supported by Ministero dell'Istruzione e dell' Universita' e della Ricerca Fondo per gli Investimenti della Ricerca di Base-Proteomica Grant RBRN07BMCT, Ente Cassa di Risparmio di Firenze Project “Relazione varianti proteiche strutturali-malattie genetiche,” and European Commission Project SPINE2-COMPLEXES 031220.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S6.
The atomic coordinates and structure factors (codes 2kmx and 2kmv) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AMPPCP
- adenosine 5′-(β,γ-methylene)triphosphate
- NOE
- nuclear Overhauser effect
- HSQC
- heteronuclear single quantum coherence
- RMSD
- root mean square deviation.
REFERENCES
- 1.Lutsenko S., Barnes N. L., Bartee M. Y., Dmitriev O. Y. (2007) Physiol. Rev. 87, 1011–1046 [DOI] [PubMed] [Google Scholar]
- 2.Cox D. W., Moore S. D. P. (2002) J. Bioenerg. Biomembr. 34, 333–338 [DOI] [PubMed] [Google Scholar]
- 3.Solioz M., Vulpe C. D. (1996) Trends Biochem. Sci. 21, 237–241 [PubMed] [Google Scholar]
- 4.Monty J. F., Llanos R. M., Mercer J. F., Kramer D. R. (2005) J. Nutr. 135, 2762–2766 [DOI] [PubMed] [Google Scholar]
- 5.Petris M. J., Strausak D., Mercer J. F. (2000) Hum. Mol. Genet. 9, 2845–2851 [DOI] [PubMed] [Google Scholar]
- 6.Vulpe C., Levinson B., Whitney S., Packman S., Gitschier J. (1993) Nat. Genet. 3, 7–13 [DOI] [PubMed] [Google Scholar]
- 7.Chelly J., Tümer Z., Tønnesen T., Petterson A., Ishikawa-Brush Y., Tommerup N., Horn N., Monaco A. P. (1993) Nat. Genet. 3, 14–19 [DOI] [PubMed] [Google Scholar]
- 8.Mercer J. F., Livingston J., Hall B., Paynter J. A., Begy C., Chandrasekharappa S., Lockhart P., Grimes A., Bhave M., Siemieniak D. (1993) Nat. Genet. 3, 20–25 [DOI] [PubMed] [Google Scholar]
- 9.Kodama H., Murata Y. (1999) Pediatr. Int. 41, 430–435 [DOI] [PubMed] [Google Scholar]
- 10.Møller J. V., Juul B., le Maire M. (1996) Biochim. Biophys. Acta 1286, 1–51 [DOI] [PubMed] [Google Scholar]
- 11.Lutsenko S., Kaplan J. H. (1995) Biochemistry 34, 15607–15613 [DOI] [PubMed] [Google Scholar]
- 12.Arnesano F., Banci L., Bertini I., Ciofi-Baffoni S., Molteni E., Huffman D. L., O'Halloran T. V. (2002) Genome Res. 12, 255–271 [DOI] [PubMed] [Google Scholar]
- 13.Sharma S., Rosato A. (2009) J. Chem. Inf. Model. 49, 76–83 [DOI] [PubMed] [Google Scholar]
- 14.Lutsenko S., Petrukhin K., Cooper M. J., Gilliam C. T., Kaplan J. H. (1997) J. Biol. Chem. 272, 18939–18944 [DOI] [PubMed] [Google Scholar]
- 15.Kühlbrandt W. (2004) Nat. Rev. Mol. Cell. Biol. 5, 282–295 [DOI] [PubMed] [Google Scholar]
- 16.Sazinsky M. H., Mandal A. K., Argüello J. M., Rosenzweig A. C. (2006) J. Biol. Chem. 281, 11161–11166 [DOI] [PubMed] [Google Scholar]
- 17.Tsuda T., Toyoshima C. (2009) EMBO J. 28, 1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dmitriev O., Tsivkovskii R., Abildgaard F., Morgan C. T., Markley J. L., Lutsenko S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5302–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 20.Zwahlen C., Legault P., Vincent S. J., Greenblatt J., Konrat R., Kay L. E. (1997) J. Am. Chem. Soc. 119, 6711–6721 [Google Scholar]
- 21.Ottiger M., Delaglio F., Bax A. (1998) J. Magn. Reson. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 22.Banci L., Bertini I., Huber J. G., Luchinat C., Rosato A. (1998) J. Am. Chem. Soc. 120, 12903–12909 [Google Scholar]
- 23.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 24.Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Wang B., Pearlman D. A., Crowley M., Brozell S., Tsui V., Gohlke H., Mongan J., Hornak V., Cui G., Beroza P., Schafmeister C. E., Caldwell J. W., Ross W. S., Kollman P. A. (2008) AMBER 10, version 8.0, University of California, San Francisco, CA [Google Scholar]
- 25.Laskowski R. A., Rullmann J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 26.Vriend G. (1990) J. Mol. Graph. 8, 52–56, 29 [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya A., Tejero R., Montelione G. T. (2007) Proteins 66, 778–795 [DOI] [PubMed] [Google Scholar]
- 28.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 29.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 30.Grzesiek S., Bax A. (1993) J. Am. Chem. Soc. 115, 12593–12594 [Google Scholar]
- 31.Mandel M. A., Akke M., Palmer A. G., III (1995) J. Mol. Biol. 246, 144–163 [DOI] [PubMed] [Google Scholar]
- 32.Lipari G., Szabo A. (1982) J. Am. Chem. Soc. 104, 4546–4559 [Google Scholar]
- 33.Lipari G., Szabo A. (1982) J. Am. Chem. Soc. 104, 4559–4570 [Google Scholar]
- 34.Dosset P., Hus J. C., Marion D., Blackledge M. (2001) J. Biomol. NMR 20, 223–231 [DOI] [PubMed] [Google Scholar]
- 35.Voskoboinik I., Mar J., Strausak D., Camakaris J. (2001) J. Biol. Chem. 276, 28620–28627 [DOI] [PubMed] [Google Scholar]
- 36.Morgan C. T., Tsivkovskii R., Kosinsky Y. A., Efremov R. G., Lutsenko S. (2004) J. Biol. Chem. 279, 36363–36371 [DOI] [PubMed] [Google Scholar]
- 37.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyoshima C., Mizutani T. (2004) Nature 430, 529–535 [DOI] [PubMed] [Google Scholar]
- 39.Toyoshima C., Nomura H. (2002) Nature 418, 605–611 [DOI] [PubMed] [Google Scholar]
- 40.Hilge M., Siegal G., Vuister G. W., Güntert P., Gloor S. M., Abrahams J. P. (2003) Nat. Struct. Biol. 10, 468–474 [DOI] [PubMed] [Google Scholar]
- 41.Haupt M., Bramkamp M., Heller M., Coles M., Deckers-Hebestreit G., Herkenhoff-Hesselmann B., Altendorf K., Kessler H. (2006) J. Biol. Chem. 281, 9641–9649 [DOI] [PubMed] [Google Scholar]
- 42.Pedersen B. P., Buch-Pedersen M. J., Morth J. P., Palmgren M. G., Nissen P. (2007) Nature 450, 1111–1114 [DOI] [PubMed] [Google Scholar]
- 43.Garrett D. S., Seok Y. J., Peterkofsky A., Clore G. M., Gronenborn A. M. (1997) Biochemistry 36, 4393–4398 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.