Abstract
Activation of transcription in response to low oxygen tension is mediated by the hypoxia-inducible factor-1 (HIF-1). HIF-1 is a heterodimer of two proteins: aryl hydrocarbon receptor nuclear translocator and the oxygen-regulated HIF-1α. The C-terminal activation domain of HIF-1α has been shown to interact with cysteine/histidine-rich region 1 (CH1) of the coactivator CBP/p300 in a hypoxia-dependent manner. However, HIF forms lacking C-terminal activation domain (naturally occurring or genetically engineered) are still able to activate transcription of target genes in hypoxia. Here, we demonstrate that the N-terminal activation domain (N-TAD) of HIF-1α interacts with endogenous CBP and that this interaction facilitates its transactivation function. Our results show that interaction of HIF-1α N-TAD with CBP/p300 is mediated by the CH3 region of CBP known to interact with, among other factors, p53. Using fluorescence resonance energy transfer experiments, we demonstrate that N-TAD interacts with CH3 in vivo. Coimmunoprecipitation assays using endogenous proteins showed that immunoprecipitation of CBP in hypoxia results in the recovery of a larger fraction of HIF-1α than of p53. Chromatin immunoprecipitation demonstrated that at 1% O2 CBP is recruited to a HIF-1α but not to a p53 target gene. Upon activation of both pathways, lower levels of chromatin-associated CBP were detected at either target gene promoter. These results identify CBP as the coactivator directly interacting with HIF-1α N-TAD and mediating the transactivation function of this domain. Thus, we suggest that in hypoxia HIF-1α is a major CBP-interacting transcription factor that may compete with other CBP-dependent factors, including p53, for limiting amounts of this coactivator, underscoring the complexity in the regulation of gene expression by HIF-1α.
Introduction
Hypoxia-inducible factor-1 (HIF-1)4 regulates the transcription of target genes in response to low oxygen levels (hypoxia). Vascular endothelial growth factor, erythropoietin, and glycolytic enzymes are part of the vast list of genes that are transcriptionally induced under hypoxic conditions by HIF-1 (for review see Refs. 1 and 2). This transcription factor is a heterodimeric complex formed by a constitutively expressed protein, Arnt, and an oxygen-regulated factor, HIF-1α. HIF-1α protein stability is under strict regulation by oxygen levels. At normoxia (21% O2), HIF-1α is hydroxylated at specific proline residues by O2-Fe(II)-dependent prolyl hydroxylases (3, 4). These modified residues mediate the interaction with the von Hippel-Lindau tumor suppressor protein, which is part of a complex with ubiquitin-ligase activity (5–7). Interaction leads to ubiquitylation and proteasome-dependent degradation of HIF-1α (8–12). In response to low levels of molecular oxygen, prolyl hydroxylase activities are inhibited, and the protein is stabilized. After stabilization HIF-1α is retained in the nuclear compartment (13) and forms a transcriptionally active complex with its partner Arnt and coactivators.
CBP/p300 has been shown to have important coactivator functions in HIF-1α-dependent activation of transcription (14–18). CBP/p300 possesses strong histone acetyltransferase activity that regulates remodeling of local chromatin structures and increases DNA accessibility to other regulators (19). CBP/p300 coactivators are involved in multiple physiological processes and are required for correct embryonic development. Homozygous loss of CBP in mice leads to embryonic death caused by numerous defects including deficient blood vessel formation (20, 21), whereas inactivation of p300 leads to embryonic lethality caused by, among other abnormalities, reduced cardiac trabeculation (22). Both CBP and p300 are involved in a number of malignancies. Chromosomal translocations are the cause of hematological disorders, whereas somatic mutations accompanied by the loss of the other allele have been found in tumors. CBP heterozygous germline mutations result in Rubinstein-Taybi syndrome (23).
HIF-1α contains two transactivation domains (see Fig. 1A), the N-terminal and the C-terminal activation domains (N-TAD and C-TAD), which mediate activation of transcription in a hypoxia-dependent manner. Only the C-TAD has been shown to directly interact with coactivator proteins (e.g. CBP through its cysteine/histidine-rich region 1 (CH1); see Fig. 1A) (24, 25). Interaction between the C-TAD and the CH1 domain has been previously demonstrated to regulate the hypoxia-inducible transactivation function of the C-TAD (17, 26). At normoxia, hydroxylation of a specific asparagine residue of the HIF-1α C-TAD by the asparaginyl hydroxylase factor inhibiting HIF-1α inhibits binding to the CH1 region (27–29). In hypoxia asparaginyl hydroxylation is impaired, facilitating the formation of the C-TAD/CH1 interaction interface. In contrast to the C-TAD, the mechanism that regulates the transactivation function of the N-TAD is poorly understood. Most importantly, both naturally occurring (30) and genetically engineered HIF forms (17, 31) lacking the C-TAD are still able to activate transcription of target genes in a hypoxia-dependent manner. The hypoxia-dependent transactivation function of HIF-1α N-TAD seems to be mainly regulated by protein stabilization, because in the context of the minimal transactivation domain, a HIF-1α N-TAD mutant that is not degraded at normoxia (P563A) activates transcription in a constitutive manner (32). In our previous studies we have provided evidence for the involvement of CBP in N-TAD-mediated activation of transcription. We have shown that CBP enhances N-TAD-mediated reporter gene activation and that intranuclear colocalization between CBP and HIF-α could be detected even upon deletion of the C-terminal activation domain (16, 17, 32). Here we characterize the interaction of CBP with HIF-1α N-TAD, and we identify the CH3 domain of CBP as the interaction interface that regulates transcriptional activation mediated by the N-TAD. Furthermore, we show that HIF-1α shares the CH3 domain of CBP with p53, which may affect regulation of target genes in hypoxia.
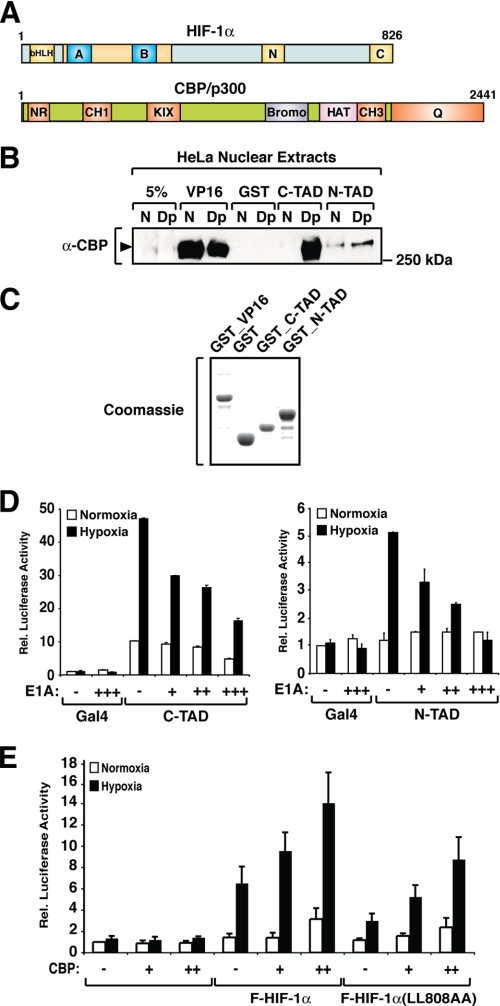
FIGURE 1.
HIF-1α N-TAD interacts with endogenous CBP. A, schematic representation of HIF-1α and CBP/p300 domains. HIF-1α contains an N-terminal DNA-binding domain followed by a helix-loop-helix dimerization interface (bHLH) and the PAS domains (blue boxes labeled A and B). The N- (N) and C-terminal (C) transactivation domains are located in the C-terminal portion of the protein. CBP/p300 contains several domains that mediate interaction with other proteins, such as the NR domain (interaction with nuclear receptors), CH1 and CH3, and the C-terminal glutamine-rich domain (Q). B, HIF-1α N-TAD interacts with endogenous CBP. Nuclear extracts from HeLa cells kept at normoxia (N) or treated with 2,2′-dipyridyl (Dp) were incubated with GST-fused VP16, C-TAD, or N-TAD. Precipitated proteins were separated by SDS-PAGE, and CBP was detected by immunoblot analysis using an anti-CBP antibody (α-CBP). C, Coomassie staining of bacterially expressed GST-fused proteins. D, N-TAD transactivation activity is inhibited by expression of E1A. HEK 293 cells were transfected with 500 ng of GAL4-driven luciferase reporter gene and 10 ng of plasmids encoding FLAG-GAL-C-TAD (left panel) or FLAG-GAL4-N-TAD (right panel) in the absence or presence of increasing concentrations of an E1A expression plasmid (10, 20, or 50 ng). The data are presented as luciferase activity relative to cells transfected with pFLAG-GAL4 and incubated at normoxia. The values represent the means ± S.E. of three independent experiments performed in duplicate. E, full-length HIF-1α mutant with a nonfunctional C-TAD is responsive to CBP. HEK 293 cells were transfected with a HRE-driven luciferase reporter plasmid and plasmids encoding FLAG-HIF-1α or FLAG-HIF-1α(L808A/L809A) (F-HIF-1α(LL808AA)) in the presence or absence of a CBP expressing plasmid. The data are presented as luciferase activity relative to cells transfected with pCMX and incubated at normoxia. The values represent the means ± S.E. of three independent experiments performed in duplicate.
EXPERIMENTAL PROCEDURES
Plasmids and Fusion Proteins
The following plasmids have been previously described: pGEX-pGEX-4T3-CH1, pFLAG-Gal4-C-TAD, pGEMT-mHIF1α, pFLAG-mHIF-1α, pFLAG-mHIF-1α(532Δ584), pFLAG-mHIF-1α(772Δ822) (17), pFLAG-Gal4-N-TAD (32), and pGEX-CBP_Q (33). pGEX-N-TAD and pGEX-C-TAD were generated by inserting a EcoRI/SmaI restriction fragment of pFLAG-Gal4-N-TAD or pFLAG-Gal4-C-TAD into pGEX-4T3 previously digested with the same enzymes. pGEX-CH3 was constructed by inserting a PCR product corresponding to the CH3 region of mouse CBP (amino acids 1757–1854) flanked by EcoRI/SalI sites into pGEX-4T3 previously digested with the same enzymes. For the generation of pFLAG-VP16, an NheI site was created in the multiple cloning site of pCMV2-FLAG, in which a PCR product corresponding to the VP16 transactivation domain and flanked by NheI sites was inserted, generating pFLAG-VP16. pFLAG-CH1-VP16 was constructed by digesting a BamHI/(NotI-Fill-in) fragment of pGEX-CH1 into pFLAG-VP16 previously digested with BglII/(KpnI-Fill-in). pFLAG-CH3-VP16 was created by inserting a EcoRI/(SalI-Fill-in) fragment of pGEX-CH3 in pFLAG-VP16 digested with EcoRI/(KpnI-fill-in). For the generation of pCFP-C-TAD and pCFP-N-TAD, an EcoRI/BamHI fragment of either pFLAG-Gal4-C-TAD or pFLAG-Gal4-N-TAD was cloned into pECFP-C1 (Clontech) digested with the same enzymes. pYFP-CH1 was generated by cloning a pGEX-CH1 BamHI/XhoI fragment into pEYFP-C1 (Clontech) digested with BglII/SalI. pYFP-CH3 was generated by ligation of pEYFP-C1 digested with EcoRI/SalI to a fragment of pGEX-CH3 generated by restriction with the same enzymes. pFLAG-CH3 (used in generation of recombinant baculovirus) was generated by insertion into pFBV-AMG (a modified pVL1393 vector, which contains the FLAG sequence upstream of the multiple cloning site; a kind gift from Armin Gamper, The Rockefeller University) of a pGEX-CH3 BamHI/NotI fragment. All in-frame fusions and constructs were confirmed by sequencing.
Cell Culture and Transient Transfections
Human embryonic kidney (HEK) 293 cells were grown in a 1:1 mixture of Dulbecco's modified Eagle's medium (10% fetal calf serum) and F-12 medium (5% fetal calf serum) containing 50 IU/ml penicillin and 50 μg/ml streptomycin sulfate. All media and growth factors were purchased from Invitrogen. HEK 293 cells were transfected with Lipofectamine (Invitrogen) according to the manufacturer's instructions. After transfection, the cells were allowed to grow for 36 h at normoxia (21% O2) or hypoxia (1% O2). The total protein concentration of whole cell extracts was determined by a colorimetric method (Bio-Rad). Empty pCMV vector was used to keep the DNA amount constant at 1 μg/dish.
Recombinant Protein Expression and Protein-Protein Interaction Assays
GST fusion proteins were expressed in Escherichia coli purified using glutathione-Sepharose. FLAG-tagged CH3 was expressed in SF9 cells and purified by affinity chromatography using M2-agarose (Sigma) followed by specific elution with a synthetic FLAG peptide (Sigma). Full-length HIF-1α and deletion mutants were expressed in the presence of [35S]methionine in a coupled cell-free transcription-translation kit (Promega). Dignam-type nuclear extracts were prepared from HeLa-S3 cells grown to high density in suspension as previously described (34). To mimic hypoxia, the cells were treated with 100 μm 2,2′-dipyridyl for 3 h. For protein-protein interaction assays, the nuclear extracts were dialyzed against buffer containing 180 mm KCl, and the Nonidet P-40 concentration was adjusted to 0.1%. In GST pulldown assays, GST fusion proteins (2.5 μg) were immobilized on 20 μl of glutathione-Sepharose and incubated with nuclear extract (2.5 mg of protein) at 4 °C overnight. Subsequent washing was carried out in buffer containing 200 mm KCl and 0.1% Nonidet P-40. Alternatively, GST fusion proteins (1 μg) were incubated with equal amounts of in vitro translated proteins in a buffer containing 300 mm KCl and 0.1% Nonidet P-40. The washes were carried out with the same buffer. Protein complexes were separated by SDS-PAGE and analyzed by immunoblotting using monoclonal anti-HIF-1α (Abcam) or anti-p53 (Calbiochem) antibodies or by autoradiography.
For immunoprecipitation of CBP-associated complexes, 6 μl of monoclonal anti-CBP antibody (Santa Cruz) was incubated with 20 μl of protein G-Sepharose beads in buffer containing 2 mg/ml bovine serum albumin, followed by incubation with nuclear extract (5 mg of protein) overnight at 4 °C. Incubation and washes were carried out as described for GST pulldown experiments. After separation by SDS-PAGE, the proteins were analyzed by Western blotting using anti-HIF-1α (Abcam), -p53 (Calbiochem), or -CBP (33) antibodies.
FRET Experiments
HEK cells transfected with CFP- and YFP-fused expression plasmids were investigated in a chamber on the microscope stage (Luigs y Neumann) at normoxia at 37 °C, as described previously (35–38). 293 cells were grown on 35-mm dishes (WellcoDish) and transfected using FuGENE 6 according to the manufacturer's instructions. The cells were treated with 100 μm CoCl2 for 4 h. Confocal images were assessed by a dual-laser scanning microscopy system operating with a two-photon 30W Ti:Sapphire laser, 760–920 nm (Coherent Mira 900 F), and a 35 milliwatt helium neon laser line 532 nm. FRET was measured by using the acceptor photobleaching method. In the case of FRET between interacting proteins, selective photobleaching of the acceptor (YFP) leads to an increase in fluorescence emission of the donor (CFP). Bleaching of YFP fluorescence to 4–10% of the original value was achieved by scanning cells with the 532-nm laser line at 100% intensity (20 times iteration). YFP emission was detected using a 590/60-nm band-pass filter. Fluorescence emission of CFP was recorded with the two-photon laser tuned to 800 nm and an emission filter (480/30 nm). Changes in YFP and CFP fluorescence of bleached cells was collected by scanning before and after bleaching. FRET efficiency (E) in percent for a bleached cell was calculated by E = [(1 − (Dbb − bg)/(Dab − bg)] × 100, where D represents the donor fluorescence before (bb) and after (ab) photobleaching. The images were corrected for background fluorescence (bg). FRET efficiency is given as the mean ± S.E. of two independent experiments, each analyzing 10–20 cells.
Chromatin Immunoprecipitation Assays (ChIP)
ChIP assays were performed as described previously (60) with minor changes. Briefly, HCT 166 cells (a kind gift from Dr. Vogelstein, Johns Hopkins, Baltimore, MD) were treated with 2 μm camptothecin and cultured at 21% or 1% O2 for 8 h, before they were fixed with 1% formaldehyde for 20 min. The immunoprecipitation was preformed with antibodies against human HIF-1α (Abcam), p53 (Calbiochem), CBP (Santa Cruz), or IgG (as a negative control), and the DNA were amplified by real time PCR using SYBR Green mix and the 7300 real time PCR system (Applied Biosystems). Primers used for PCR correspond to the putative HRE within the PGK1 promoter (region −317 to −126): 5′-GAT CTT CGC CGC TAC CCT TGT G-3′ and 5′-TAT TGG CCA CAG CCC ATC GC-3′ or the p53 DNA-binding site within the p21cip1 promoter region (−2290 to −2185): 5′-GTG GCT CTG ATT GGC TTT CTG-3′ and 5′-CTG AAA ACA GGC AGC CCA AG-3′. All of the ChIP assays were performed two to three times with representative results presented.
RESULTS
The N-terminal Activation Domain of HIF-1α Interacts with Endogenous CBP
HIF-1α N-TAD is a bifunctional domain that mediates protein degradation at normoxia and activation of transcription at hypoxia. Whereas the function of the N-TAD in regulating HIF-1α degradation has been elucidated in several studies (10–12, 32), not much is known about the mechanism underlying activation of transcription. We have previously proposed that CBP is involved in mediating N-TAD transactivation activity (15–17, 32). In this context we have investigated whether, in analogy to HIF-1α C-TAD, the N-TAD was able to interact with endogenous CBP. To this end, a GST fusion of the minimal HIF-1α N-TAD was expressed in bacteria, purified, and incubated with HeLa nuclear extracts. As positive controls for CBP interaction, we used purified GST fusion proteins spanning the herpes simplex virus VP16 activation domain or the HIF-1α C-TAD. As shown in Fig. 1B, HIF-1α N-TAD interacted with the endogenous CBP present in nuclear extracts prepared from cells kept at normoxia or treated with the iron-chelating, hypoxia-mimicking (prolyl and asparaginyl hydroxylase-inhibiting) agent 2,2′-dipyridyl. The N-TAD (P563A) mutant that mediates transcription in a constitutive manner (32) interacted in a similar manner with endogenous CBP (data not shown). In extracts prepared from 2,2′-dipyridyl-treated cells, a very strong interaction between CBP and C-TAD was observed that was comparable with the constitutive binding of VP16 to CBP. Although interaction of CBP with HIF-1α N-TAD was much weaker than with the C-TAD, the binding observed was specific. Similar amounts of the GST fusion proteins and GST alone were used as indicated (Fig. 1C). Taken together, these results demonstrate that, in analogy to the C-TAD, HIF-1α N-TAD is able to interact with endogenous CBP.
E1A Inhibits HIF-1α N-TAD-mediated Activation of Transcription
The viral oncoprotein E1A has been shown to interfere with the function of transactivators that recruit CBP/p300 to form transcriptionally active complexes (39). Inhibition of transcription is mediated by binding of E1A to multiple CBP/p300 domains (40) and interference with the histone acetyltransferase activity of these coactivators (41, 42). E1A has previously been shown to inhibit hypoxia-dependent activation of transcription mediated by HIF-1α and HIF-1α C-TAD (14, 43). In the present study we investigated whether E1A could interfere with the transactivation function of the N-TAD. A GAL4-driven luciferase reporter gene and expression plasmids encoding GAL4-N-TAD or -C-TAD, respectively, were transiently expressed in HEK 293 cells in the absence or presence of increasing concentrations of E1A. As shown in Fig. 1D, E1A inhibited in a concentration-dependent manner both N-TAD- and C-TAD-dependent activation of transcription. The differential effect of E1A on HIF-1α N- and C-TAD activity may reflect the different affinities of these two transactivation domains for CBP/p300, in agreement with the in vitro protein-protein interaction data shown in Fig. 1B. Inhibition of N-TAD-dependent activation of transcription by E1A indicated that CBP/p300 is required for the transactivation function mediated by this domain. These results are in agreement with our prior work showing that CBP enhances the transactivation function of the minimal N-TAD fused to a GAL4 DNA-binding domain (32).
CBP Enhances the Transactivation Function of a HIF-1α Mutant Containing Only the N-TAD as a Functional Transactivation Domain
To investigate whether in the context of full-length HIF-1α, the N-TAD is responsive to CBP, we performed reporter gene assays where we expressed wild-type HIF-1α or a mutant form that corresponds to the inactivation of the C-TAD by point mutation (HIF-1α(L808A/L809A)). We performed transient transfections in HEK 293 cells and evaluated the ability of these proteins to activate HRE-driven luciferase reporter gene in the presence or absence of expressed CBP. As shown in Fig. 1E, the expression of CBP enhances the transactivation function of both wild-type and mutant HIF-1α in a dose-responsive manner. These results show that CBP coactivates the HIF-1α mutant containing only the N-terminal transactivation domain. In conclusion, in the context of the full-length HIF-1α protein, both the N- and the C-TAD are able to respond to the coactivator CBP, indicating that the two domains contribute to the overall transactivation function of the protein.
HIF-1α N-TAD Interacts Directly with the CH3 Domain of CBP
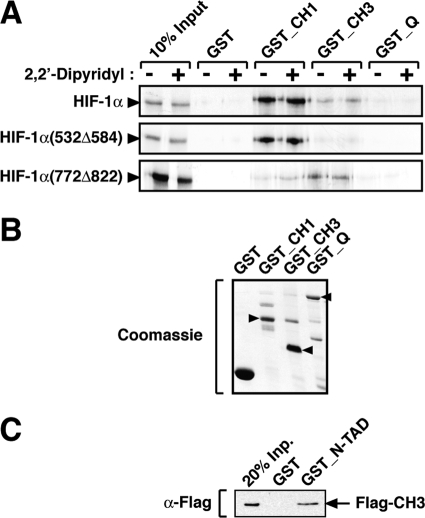
CBP interacts with a large number of functionally diverse proteins through a series of modular protein-binding domains. Two of these domains are the highly homologous CH1 and CH3 regions (residues 349–437 and 1763–1849 of hCBP, respectively), which are major sites for protein-protein interaction (25). Both full-length HIF-1α and the minimal C-TAD have previously been shown to interact with the CH1 domain (14, 17, 24–26). Here we tested whether HIF-1α had the ability to interact with the CH3 region or other CBP domains. HIF-1α was 35S-labeled by in vitro translation in rabbit reticulocyte lysate and incubated with different purified, bacterially expressed GST-fused CBP domains. As observed in Fig. 2A, HIF-1α was precipitated by both the CH1 and CH3 but not by the C-terminal glutamine-rich domain. No interaction was detected between the CBP NR and KIX domains and HIF-1α (data not shown). As shown in Fig. 2A, full-length HIF-1α was able to interact with both the GST-CH1 and -CH3 fusion proteins, although stronger binding to the CH1 domain was displayed. Using the same experimental approach, we next investigated which of the HIF-1α transactivation domains was the major contributor to the interaction with the CH1 and CH3 regions of CBP. In this assay, a mutant form of HIF-1α corresponding to an internal deletion of N-TAD (HIF-1α(532Δ584)) interacted only with the CH1 domain of CBP, and no significant interaction was observed with the CH3 domain. In contrast, deletion of HIF-1α C-TAD (HIF-1α(772Δ822)) resulted in a mutant that displays stronger binding to the CH3 region. These results indicate that the N-TAD is the transactivation domain mainly responsible for the interaction of full-length HIF-1α with the CH3 domain of CBP. In agreement with previous studies (17, 24–26), the C-TAD predominantly determined interaction with the CH1 domain. In vitro translation of HIF-1α and mutant proteins in the presence of 2,2′-dipyridyl (to exclude the possibility of inhibitory enzymatic activities present in rabbit reticulocyte lysate) had no effect on the interaction with CBP domains (Fig. 2A). Similar amounts of the GST fusion proteins and GST alone were used as indicated (Fig. 2B).
FIGURE 2.
The N-terminal transactivation domain of HIF-1α interacts with the CH3 region of CBP. A, HIF-1α N-TAD is the major contributor for the interaction of full-length HIF-1α with the CH3 domain of CBP. In vitro translated [35S]methionine-labeled wild-type or mutant HIF-1α proteins were incubated with GST-fused CBP domains, as indicated. Precipitated proteins were separated by SDS-PAGE and detected by autoradiography. B, Coomassie staining of bacterially expressed GST-fused proteins. Arrowheads indicate the bands with the correct molecular sizes. C, HIF-1α N-TAD interacts directly with the CH3 domain of CBP. Purified baculovirus-expressed FLAG-CH3 domain was precipitated by bacterially expressed GST-N-TAD. After separation by SDS-PAGE, the proteins were analyzed by Western blotting using an anti-FLAG (α-Flag) antibody.
We next analyzed the nature of the interaction between HIF-1α N-TAD and the CH3 region of CBP. To establish whether HIF-1α N-TAD and CBP CH3 can interact directly, FLAG-tagged CH3 (FLAG-CH3) was expressed in SF9 cells using a baculovirus system and purified by immunoaffinity chromatography. Bacterially expressed GST-N-TAD was then immobilized using glutathione-Sepharose beads and incubated with purified FLAG-CH3. FLAG-CH3 was precipitated by GST-N-TAD but not by GST alone, indicating a direct interaction between the two domains (Fig. 2C). In conclusion, these results show that CBP interacts directly with HIF-1α N-TAD and that this interaction is mediated by the CH3 domain of the coactivator.
HIF-1α Transactivation Domains Interact in Vivo with the Cysteine/Histidine-rich Regions of CBP
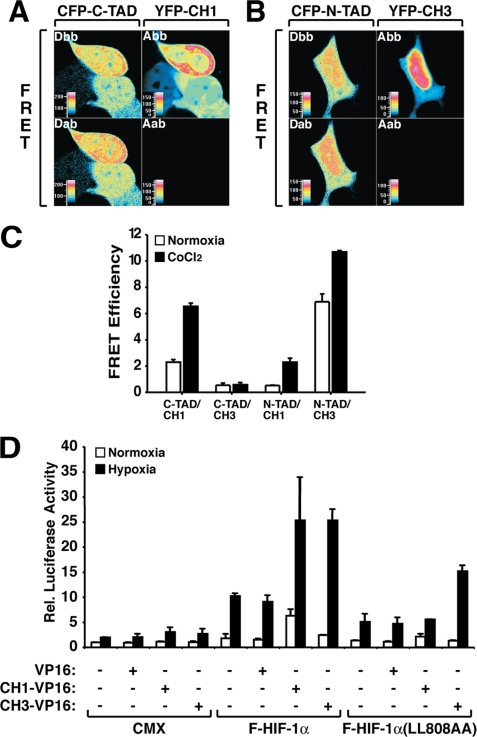
Based on the in vitro protein-protein interaction assays, we next investigated whether direct C-TAD/CH1 or N-TAD/CH3 interactions also occur in vivo in cultured cells. HEK 293 cells were transfected with plasmids encoding CFP-fused C-TAD or N-TAD, together with pYFP-CH1 or -CH3 constructs. The cells were then kept at normoxia or treated with the hypoxia-mimicking (prolyl and asparaginyl hydroxylase-inhibiting) agent CoCl2 and analyzed by FRET using laser-scanning confocal microscopy. In agreement with previously published in vitro studies (17, 24–26), an interaction between HIF-1α C-TAD and CBP CH1 was observed in living cells. This binding was inducible by CoCl2 treatment and presented a FRET signal of 2.3% at normoxia and 6.55% in the presence of CoCl2 (Fig. 3, A and C). In contrast, expression of CFP-C-TAD together with YFP-CH3 showed a very low FRET signal, indicating that these two protein domains do not interact in vivo. As a negative control for the C-TAD/CH1 in vivo interaction, we investigated the FRET efficiency using a C-TAD mutant (C-TAD(L808A/L809A)) previously shown to be deficient in transactivation and CH1 binding (17). The FRET signal observed was of 1.3 and 1.65% efficiency at normoxia or after CoCl2 treatment, respectively (data not shown). We next investigated the interaction between HIF-1α N-TAD and the CH3 region of CBP by FRET analysis. Cells expressing CFP-N-TAD and YFP-CH3 generated a positive FRET signal of 6.9 and 10.7% efficiency in normoxic and CoCl2-treated cells, demonstrating that these protein domains are able to interact in vivo. The FRET signal observed with CFP-N-TAD and YFP-CH1 showed a much more reduced efficiency (0.53% at normoxia and 2.3% in the presence of CoCl2), indicating that the affinity of the N-TAD to the CH1 region is much lower when compared with the affinity to the CH3 domain. Fig. 3C summarizes the results from the FRET experiments. A N-TAD mutant (N-TAD(P563A)) functioning as a constitutive transactivator (32) was also able to generate a positive FRET signal when expressed together with CH3 (normoxia, 5.7%; CoCl2, 6.25%) (data not shown), supporting the proposed model that interaction of the N-TAD with CH3 is relevant for the activity of the transactivation domain. Taken together these results demonstrate specific in vivo interactions between the N-TAD and CH3 and between the C-TAD and CH1 of CBP.
FIGURE 3.
In vivo interaction between HIF-1α transactivation domains and cysteine/histidine-rich regions of CBP. A and B, FRET analysis shows in vivo interaction between CFP-C-TAD/YFP-CH1 and CFP-N-TAD/YFP-CH3. HEK 293 cells were transfected with 600 ng of CFP and 400 ng of YFP-fused expression plasmids. The images show representative cells of donor (D) and acceptor (A) before (bb) and after (ab) selective photobleaching. The cells are shown in false color representing different fluorescence intensities as indicated by the color table shading from black (lowest intensity), through blue, green, yellow, orange, red, and purple to white (highest intensity). C, analysis of FRET efficiency (%) of the corresponding CFP/YFP fusion proteins. HEK 293 cells expressing the different fusion proteins were kept at normoxia or treated with 100 μm CoCl2 for 4 h. The values represent the means ± S.E. of two independent experiments each of 10–20 cells. D, expression of CH3-VP16 protein increases luciferase activity mediated by a HIF-1α mutant bearing a nonfunctional C-TAD. HEK 293 cells were transfected with 300 ng of HRE-driven luciferase reporter gene and 50 ng of HIF-1α or the corresponding mutant expression plasmids in the absence or presence of 400 ng of pFLAG-CH1-VP16 or pFLAG-CH3-VP16. The data are presented as luciferase activity relative to cells transfected with pFLAG and incubated at normoxia. The values represent the means ± S.E. of three independent experiments performed in duplicate.
Expression of a CH3-VP16 Fusion Protein Enhances the Transactivation Function of a HIF-1α Mutant Containing an Inactive C-TAD
To investigate the functional interaction between CBP cysteine/histidine-rich regions and the N-TAD of HIF-1α in the context of the full-length protein, we performed reporter gene assays using wild-type HIF-1α or the mutant HIF-1α(L808A/L809A), where the C-TAD is not functional. These proteins were tested for their ability to activate transcription of an HRE-driven luciferase reporter gene in transiently transfected cells, in the absence or presence of VP16 transactivation domain fusion proteins spanning either the CH1 or CH3 domains. As demonstrated in Fig. 3D, the relative luciferase activity resulting from expression of wild-type HIF-1α was further increased by expression of either the CH1- and CH3-VP16 fusion proteins but not the VP16 transactivation domain alone, indicating a functional interaction between these two CBP domains and HIF-1α. In contrast, hypoxia-induced luciferase activity mediated by the HIF-1α(L808A/L809A) mutant was increased only in the presence of CH3-VP16, consistent with an interaction between this HIF-1α mutant with the CH3 but not with the CH1 domain. The C-TAD deletion mutant of HIF-1α (HIF-1α(772Δ822)) used in Fig. 2A was also investigated in functional interaction assays together with the isolated cysteine/histidine-rich regions of CBP fused to the VP16 transactivation domain. HIF-1α(772Δ822) showed the same pattern of functional interaction with the expressed VP16 fusion proteins (data not shown). Thus, these data strongly support the conclusion that the interaction between full-length HIF-1α and the CH3 domain of CBP is mediated by the N-TAD.
Endogenous HIF-1α Interacts Strongly with the CH3 Domain of CBP
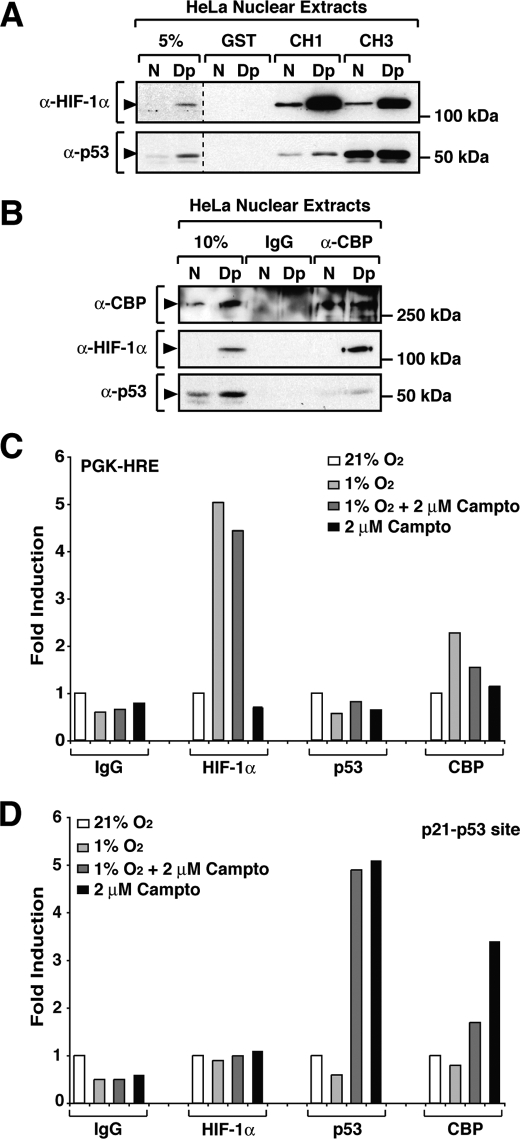
To characterize the interaction of endogenous HIF-1α with the cysteine/histidine-rich domains of CBP, bacterially expressed GST-CH3 and -CH1 were incubated with HeLa nuclear extracts prepared from cells grown under normoxic or 2,2′-dipyridyl-treated conditions. After separation by SDS-PAGE, CH1- and CH3-associated protein complexes were analyzed by immunoblotting using specific anti-HIF-1α and anti-p53 antibodies. Because the interaction between p53 and CBP has been extensively characterized (33, 44–46) and given the obvious relevance for cancer biology, we extended our studies to include p53. At normoxia, low p53 protein levels were observed in HeLa cells. However, as expected (47, 48), treatment of cells with the hypoxia-mimicking agent led to an increase in p53 protein levels (Fig. 4, A and B). As observed in Fig. 4A, both GST-CH1 and -CH3 fusion proteins were able to efficiently precipitate endogenous HIF-1α in a 2,2′-dipyridil-inducible manner. Furthermore HIF-1α showed binding to both the CH1 and CH3 domains of CBP, further illustrating the importance of both CBP domains for the interaction with HIF-1α. In contrast to HIF-1α, p53 interacted preferentially with the CH3 domain of CBP and displayed weak binding to the CH1 region.
FIGURE 4.
HIF-1α binding to CBP displays a high affinity interaction. A, endogenous HIF-1α interacts with both the CH1 and the CH3 regions of CBP. HeLa nuclear extracts kept at normoxia (N) or treated with 2,2′-dipyridyl (Dp) were incubated with bacterially expressed GST-fused CH1 and CH3 proteins. Precipitated proteins were separated by SDS-PAGE and detected by Western blot analysis using anti-HIF-1α (α-HIF-1α) and anti-p53 (α-p53) antibodies. B, recruitment of CBP to HIF-1α and p53. Endogenous CBP was immunoprecipitated with an anti-CBP (α-CBP) antibody, and interacting proteins were separated by SDS-PAGE and detected by Western blot analysis using specific antibodies. C and D, recruitment of CBP to target genes promoters is impaired by simultaneously activation of HIF-1α- and p53-regulated pathways. HCT 166 cells were fixed with formaldeyde after 8 h of treatment as described. ChIP was performed using antibodies against IgG, HIF-1α, p53, and CBP. Binding sites of HIF-1α and p53 on the target gene promoters PGK-1 and p21, respectively, were amplified using quantitative reverse transcription-PCR as described under “Experimental Procedures.” The ChIP experiments were repeated three times, and the figure shows the results of a representative experiment.
Interaction of HIF-1α and p53 with CBP in the Presence of 2,2′-Dipyridyl
Because we have observed that both p53 and HIF-1α target common CBP domains (i.e. the CH1 and CH3 regions), we decided to investigate the interaction of these transcription factors with endogenous CBP. HeLa nuclear extracts prepared from cells kept under normoxia or treated with 2,2′-dipyridyl were used to immunoprecipitate CBP-associated protein complexes using anti-CBP antibodies. Precipitated proteins were analyzed by SDS-PAGE followed by Western blot analysis. As shown in Fig. 4B, both HIF-1α and p53 are able to interact with endogenous CBP. Quantitative analysis demonstrated that in extracts from 2,2′-dipyridyl-treated cells, 20% of HIF-1α versus 2% of p53 protein was recovered in CBP immunoprecipitation assays from HeLa nuclear extracts, indicating a stronger binding of CBP to HIF-1α when compared with p53.
Recruitment of CBP to Target Gene Promoters upon Activation of HIF-1α and p53 Pathways
The existence of negative cross-talk between the HIF-1α and p53 pathways has been described in several studies. However, the proposed mechanisms and functional outcomes of this negative mode of cross-regulation are quite contradictory (48–54). We therefore investigated whether the negative cross-talk between HIF-1α and p53 could be the result of competition for the common coactivator CBP/p300. To this end we have performed ChIP analyses and investigated the recruitment of CBP to HIF-1α and p53 target gene promoters when either or both pathways are activated. As presented in Fig. 4C, in HCT166 cells HIF-1α was recruited to the PGK-1 promoter when the cells were treated with 1% hypoxia, and this binding was unaffected by activation of the p53 pathway by camptothecin. In contrast, recruitment of p53 to the p53 target gene promoter p21 was not observed in response to 1% hypoxia (Fig. 4D). Activation of the p53 pathway by camptothecin led to the binding of p53 protein to the p21 promoter, and this recruitment was not changed by concomitant treatment of the cells with hypoxia (Fig. 4D). In these experimental conditions the maximum binding of CBP (Fig. 4, C and D) to the PGK1 promoter was observed in response to hypoxia treatment, whereas maximum binding to the p21 promoter was detected in cells treated with camptothecin, indicating that activation of each pathway is correlated with CBP recruitment to the target gene promoter. When both pathways are simultaneously activated by concomitant treatment with hypoxia and camptothecin, the levels of CBP on target gene promoters are reduced when compared with the maximum binding levels. Our results suggest that both HIF-1α and p53 may compete for recruitment of CBP to their target gene promoters upon activation by their specific signaling pathways. These observations demonstrate that CBP/p300 is a limiting factor for both HIF-1- and p53-mediated transcriptional responses (49, 50).
DISCUSSION
In this study we have characterized the interaction of CBP/p300 with the N-TAD of HIF-1α, and we show that this interaction is relevant to HIF-1α function. In strong support of this conclusion, we have previously observed in reporter gene assays that CBP significantly enhances N-TAD-dependent activation of transcription (15, 16, 32). Furthermore, the loss of interaction between HIF-1α and CBP in subcellular colocalization studies and immunoprecipitation assays is only achieved following inactivation of both transactivation domains by mutation (17, 18). Here, we have identified a new HIF-1α/CBP interaction interface that is formed by the N-TAD of HIF-1α and the CH3 region of CBP.
In the present study binding of HIF-1α to CBP proved to be more complex than the currently accepted model. To date, HIF-1α has only been shown to interact with the CH1 domain of CBP/p300. Here we demonstrate that distinct HIF-1α transactivation domains are able to bind distinct cysteine/histidine-rich regions of CBP, possibly contributing to a more stable and functionally active complex. Some of the results obtained when HIF-1α fragments corresponding to the minimal N- or C-TAD were used in our assays suggest that interaction of the N-TAD with CBP is weaker when compared with the C-TAD. However, our analysis of CBP interaction with full-length HIF-1α suggests that both N-TAD and C-TAD contribute in a similar way to the recruitment of CBP. More importantly, the current results may explain why genetically engineered mice carrying deletions of the CH1 domains of CBP and p300 show only a slight reduction of hypoxia-induced transcription of HIF target genes (31). Based on our results, we speculate that in these animals HIF-1α N-TAD is recruiting CBP by binding to the CH3 domain and that this interaction is sufficient to ensure the formation of an active transcription complex in vivo.
Several proteins, including viral oncoproteins and transcription factors, have been shown to interact with distinct CBP domains (25). The tumor suppressor protein p53 belongs to the group of proteins interacting with the CH1, CH3, and C-terminal glutamine-rich domains of CBP. Interaction with CH1 has been associated with Mdm2-dependent degradation of p53 (45, 55), whereas binding to CH3 has been correlated with the activation of transcription by recruitment of CBP to target gene promoters and histone acetylation (44, 56, 57). Mdm2-mediated degradation of p53 plays a major role in cells not infected with the high risk human papillomavirus. In contrast, in cells infected with human papillomavirus such as the HeLa cells used in the present study, the key regulator of p53 protein levels is the E6 oncoprotein. The weak interaction of p53 with the CH1 region of CBP in HeLa cells may therefore reflect low levels of p53 protein associated with Mdm2-dependent degradation. On the other hand, the strong interaction with the CH3 domain reflects a high affinity of p53 for this region that is associated with the transactivation function of the protein (56, 57). In contrast to p53, the two interaction interfaces of HIF-1α with CBP/p300 are involved in the transactivation function of HIF-1α. Our results show that in the context of the full-length protein, HIF-1α N-TAD may play a much more important role in HIF-1α transactivation function than previously predicted. This observation provides an explanation for the strong transactivation activity of HIF proteins in von Hippel-Lindau tumor suppressor protein-deficient cells (58) at normoxia in conditions where the HIF-1α C-TAD function is inhibited by factor inhibiting HIF-1α. von Hippel-Lindau tumor suppressor protein deficiency leading to stabilization of HIF proteins is enough to render HIF transcriptionally active, indicating that a functional N-TAD is enough to mediate up-regulation of HIF target genes at normoxia.
The cross-talk between HIF-1α and p53 pathways has been extensively studied, but the functional consequences of this potentially regulatory mechanism remain unclear. It has been shown that overexpression of p53 is able to inhibit HIF-dependent activation of transcription (49, 50), and p53-deficient cells show higher levels of HIF transactivation activity and higher inducibility of HIF target genes (51, 52). These studies are consistent with a model where HIF-1α and p53 compete for a common factor important for activation of transcription. Other studies have indicated that at low oxygen levels p53 binds to target gene promoters but is not able to activate transcription because of inefficient recruitment of CBP (48, 53, 54). It is important to note that stabilization and eventual activation of p53 only occurs in response to hypoxia-mimicking agents or at anoxia (near 0% O2) (47, 48, 53, 54) and not at the degree of hypoxia used here for activation of HIF signaling. Our studies show that in the presence of 1% oxygen, a concentration known to stabilize and activate HIF-1α, there is no recruitment of either p53 or CBP to a p53 target promoter, indicating that at this concentration of oxygen there is no activation of the p53 pathway. Furthermore it has been shown that CSB (Cockayne syndrome B), a protein belonging to the SWI/SNF2 family of chromatin remodelers, is a HIF-1α target gene and impairs recruitment of CBP to p53 target promoters in response to the hypoxia-mimicking agent desferrioxamine (59). In the CSB study it was shown that only in cells where CSB is not functional, the p53 pathway is activated by desferroxamine and competes with HIF-1α for CBP recruitment (59). In our study CBP recruitment to target promoters was observed when both pathways were independently activated. Altogether, the presented observations are consistent with a model in which CBP/p300 is a limiting factor for both HIF-1- and p53-mediated transcriptional responses. However, in what conditions are low oxygen levels sufficient to activate the p53 pathway is a question that requires further investigation.
In conclusion, interaction with the CH3 region of CBP is required for N-TAD-mediated transactivation and thereby contributes significantly to the modulation of HIF-1α activity by the CBP coactivator. Thus, this interaction is an important mechanism for gene regulation in hypoxic cells. Moreover, in therapeutic efforts to inhibit HIF-1α function, e.g. in anti-angiogenic tumor therapy, it may not suffice to impair the interaction between the HIF-1α C-TAD and the CBP CH1 region but may also require the development of strategies disrupting HIF-1α N-TAD-CBP CH3 complex formation.
This work was supported, in whole or in part, by National Institutes of Health Grants DK060764 (to S. M.) and DK071900 and CA129325 (to R. G. R.). This work was also supported by grants from the Swedish Medical Research Council (to T. P. and L. P.) and from the Swedish Cancer Foundation, the Swedish Heart-Lung Foundation, and the European Union (to L. P.).
- HIF-1
- hypoxia-inducible transcription factor-1
- C-TAD
- C-terminal activation domain
- N-TAD
- N-terminal activation domain
- CREB
- cAMP-responsive element-binding protein
- CBP
- CREB-binding protein
- CH
- cysteine/histidine-rich domain
- FRET
- fluorescence resonance energy transfer
- HEK
- human embryonic kidney
- GST
- glutathione S-transferase
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Ruas J. L., Poellinger L. (2005) Semin. Cell Dev. Biol. 16, 514–522 [DOI] [PubMed] [Google Scholar]
- 2.Lendahl U., Lee K. L., Yang H., Poellinger L. (2009) Nat. Rev. Genet. 10, 821–832 [DOI] [PubMed] [Google Scholar]
- 3.Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 4.Bruick R. K., McKnight S. L. (2001) Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 5.Kamura T., Sato S., Iwai K., Czyzyk-Krzeska M., Conaway R. C., Conaway J. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10430–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 7.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 8.Huang L. E., Gu J., Schau M., Bunn H. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kallio P. J., Wilson W. J., O'Brien S., Makino Y., Poellinger L. (1999) J. Biol. Chem. 274, 6519–6525 [DOI] [PubMed] [Google Scholar]
- 10.Ohh M., Park C. W., Ivan M., Hoffman M. A., Kim T. Y., Huang L. E., Pavletich N., Chau V., Kaelin W. G. (2000) Nat. Cell Biol. 2, 423–427 [DOI] [PubMed] [Google Scholar]
- 11.Tanimoto K., Makino Y., Pereira T., Poellinger L. (2000) EMBO J. 19, 4298–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cockman M. E., Masson N., Mole D. R., Jaakkola P., Chang G. W., Clifford S. C., Maher E. R., Pugh C. W., Ratcliffe P. J., Maxwell P. H. (2000) J. Biol. Chem. 275, 25733–25741 [DOI] [PubMed] [Google Scholar]
- 13.Kallio P. J., Okamoto K., O'Brien S., Carrero P., Makino Y., Tanaka H., Poellinger L. (1998) EMBO J. 17, 6573–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arany Z., Huang L. E., Eckner R., Bhattacharya S., Jiang C., Goldberg M. A., Bunn H. F., Livingston D. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12969–12973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ema M., Hirota K., Mimura J., Abe H., Yodoi J., Sogawa K., Poellinger L., Fujii-Kuriyama Y. (1999) EMBO J. 18, 1905–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrero P., Okamoto K., Coumailleau P., O'Brien S., Tanaka H., Poellinger L. (2000) Mol. Cell. Biol. 20, 402–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruas J. L., Poellinger L., Pereira T. (2002) J. Biol. Chem. 277, 38723–38730 [DOI] [PubMed] [Google Scholar]
- 18.Ruas J. L., Poellinger L., Pereira T. (2005) J. Cell Sci. 118, 301–311 [DOI] [PubMed] [Google Scholar]
- 19.Bannister A. J., Kouzarides T. (1996) Nature 384, 641–643 [DOI] [PubMed] [Google Scholar]
- 20.Tanaka Y., Naruse I., Hongo T., Xu M., Nakahata T., Maekawa T., Ishii S. (2000) Mech. Dev. 95, 133–145 [DOI] [PubMed] [Google Scholar]
- 21.Oike Y., Hata A., Mamiya T., Kaname T., Noda Y., Suzuki M., Yasue H., Nabeshima T., Araki K., Yamamura K. (1999) Hum. Mol. Genet. 8, 387–396 [DOI] [PubMed] [Google Scholar]
- 22.Yao T. P., Oh S. P., Fuchs M., Zhou N. D., Ch'ng L. E., Newsome D., Bronson R. T., Li E., Livingston D. M., Eckner R. (1998) Cell 93, 361–372 [DOI] [PubMed] [Google Scholar]
- 23.Blobel G. A. (2002) J. Leukocyte Biol. 71, 545–556 [PubMed] [Google Scholar]
- 24.Dames S. A., Martinez-Yamout M., De Guzman R. N., Dyson H. J., Wright P. E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5271–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freedman S. J., Sun Z. Y., Poy F., Kung A. L., Livingston D. M., Wagner G., Eck M. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5367–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu J., Milligan J., Huang L. E. (2001) J. Biol. Chem. 276, 3550–3554 [DOI] [PubMed] [Google Scholar]
- 27.Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. (2002) Science 295, 858–861 [DOI] [PubMed] [Google Scholar]
- 28.Mahon P. C., Hirota K., Semenza G. L. (2001) Genes Dev. 15, 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu Y. Z., Moran S. M., Hogenesch J. B., Wartman L., Bradfield C. A. (1998) Gene Expr. 7, 205–213 [PMC free article] [PubMed] [Google Scholar]
- 31.Kasper L. H., Boussouar F., Boyd K., Xu W., Biesen M., Rehg J., Baudino T. A., Cleveland J. L., Brindle P. K. (2005) EMBO J. 24, 3846–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira T., Zheng X., Ruas J. L., Tanimoto K., Poellinger L. (2003) J. Biol. Chem. 278, 6816–6823 [DOI] [PubMed] [Google Scholar]
- 33.Gu W., Shi X. L., Roeder R. G. (1997) Nature 387, 819–823 [DOI] [PubMed] [Google Scholar]
- 34.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berchner-Pfannschmidt U., Petrat F., Doege K., Trinidad B., Freitag P., Metzen E., de Groot H., Fandrey J. (2004) J. Biol. Chem. 279, 44976–44986 [DOI] [PubMed] [Google Scholar]
- 36.Liu Q., Berchner-Pfannschmidt U., Möller U., Brecht M., Wotzlaw C., Acker H., Jungermann K., Kietzmann T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4302–4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metzen E., Berchner-Pfannschmidt U., Stengel P., Marxsen J. H., Stolze I., Klinger M., Huang W. Q., Wotzlaw C., Hellwig-Bürgel T., Jelkmann W., Acker H., Fandrey J. (2003) J. Cell Sci. 116, 1319–1326 [DOI] [PubMed] [Google Scholar]
- 38.Siegel R. M., Chan F. K., Zacharias D. A., Swofford R., Holmes K. L., Tsien R. Y., Lenardo M. J. (2000) Sci. STKE PL1. [DOI] [PubMed] [Google Scholar]
- 39.Frisch S. M., Mymryk J. S. (2002) Nat. Rev. Mol. Cell Biol. 3, 441–452 [DOI] [PubMed] [Google Scholar]
- 40.Kurokawa R., Kalafus D., Ogliastro M. H., Kioussi C., Xu L., Torchia J., Rosenfeld M. G., Glass C. K. (1998) Science 279, 700–703 [DOI] [PubMed] [Google Scholar]
- 41.Chakravarti D., Ogryzko V., Kao H. Y., Nash A., Chen H., Nakatani Y., Evans R. M. (1999) Cell 96, 393–403 [DOI] [PubMed] [Google Scholar]
- 42.Hamamori Y., Sartorelli V., Ogryzko V., Puri P. L., Wu H. Y., Wang J. Y., Nakatani Y., Kedes L. (1999) Cell 96, 405–413 [DOI] [PubMed] [Google Scholar]
- 43.Sang N., Fang J., Srinivas V., Leshchinsky I., Caro J. (2002) Mol. Cell. Biol. 22, 2984–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avantaggiati M. L., Ogryzko V., Gardner K., Giordano A., Levine A. S., Kelly K. (1997) Cell 89, 1175–1184 [DOI] [PubMed] [Google Scholar]
- 45.Grossman S. R., Perez M., Kung A. L., Joseph M., Mansur C., Xiao Z. X., Kumar S., Howley P. M., Livingston D. M. (1998) Mol. Cell 2, 405–415 [DOI] [PubMed] [Google Scholar]
- 46.Wadgaonkar R., Collins T. (1999) J. Biol. Chem. 274, 13760–13767 [DOI] [PubMed] [Google Scholar]
- 47.Pan Y., Oprysko P. R., Asham A. M., Koch C. J., Simon M. C. (2004) Oncogene 23, 4975–4983 [DOI] [PubMed] [Google Scholar]
- 48.Ashcroft M., Taya Y., Vousden K. H. (2000) Mol. Cell. Biol. 20, 3224–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmid T., Zhou J., Köhl R., Brüne B. (2004) Biochem. J. 380, 289–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blagosklonny M. V., An W. G., Romanova L. Y., Trepel J., Fojo T., Neckers L. (1998) J. Biol. Chem. 273, 11995–11998 [DOI] [PubMed] [Google Scholar]
- 51.Ravi R., Mookerjee B., Bhujwalla Z. M., Sutter C. H., Artemov D., Zeng Q., Dillehay L. E., Madan A., Semenza G. L., Bedi A. (2000) Genes Dev. 14, 34–44 [PMC free article] [PubMed] [Google Scholar]
- 52.Kaluzová M., Kaluz S., Lerman M. I., Stanbridge E. J. (2004) Mol. Cell. Biol. 24, 5757–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koumenis C., Alarcon R., Hammond E., Sutphin P., Hoffman W., Murphy M., Derr J., Taya Y., Lowe S. W., Kastan M., Giaccia A. (2001) Mol. Cell. Biol. 21, 1297–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammond E. M., Mandell D. J., Salim A., Krieg A. J., Johnson T. M., Shirazi H. A., Attardi L. D., Giaccia A. J. (2006) Mol. Cell. Biol. 26, 3492–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grossman S. R., Deato M. E., Brignone C., Chan H. M., Kung A. L., Tagami H., Nakatani Y., Livingston D. M. (2003) Science 300, 342–344 [DOI] [PubMed] [Google Scholar]
- 56.Espinosa J. M., Emerson B. M. (2001) Mol. Cell 8, 57–69 [DOI] [PubMed] [Google Scholar]
- 57.Barlev N. A., Liu L., Chehab N. H., Mansfield K., Harris K. G., Halazonetis T. D., Berger S. L. (2001) Mol. Cell 8, 1243–1254 [DOI] [PubMed] [Google Scholar]
- 58.Kim W. Y., Kaelin W. G., Jr. (2006) Semin. Oncol. 33, 588–595 [DOI] [PubMed] [Google Scholar]
- 59.Filippi S., Latini P., Frontini M., Palitti F., Egly J. M., Proietti-De-Santis L. (2008) EMBO J. 27, 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holmquist-Mengelbier L., Fredlund E., Löfstedt T., Noguera R., Navarro S., Nilsson H., Pietras A., Vallon-Christersson J., Borg A., Gradin K., Poellinger L., Påhlman S. (2006) Cancer Cell 10, 413–423 [DOI] [PubMed] [Google Scholar]