Abstract
Steroid hormones induce transcription of their responsive genes by complex mechanisms including synergism between the hormone receptors and other transcription factors. On the mouse mammary tumor virus (MMTV) promoter progesterone induction is mediated by the reciprocal synergism between progesterone receptor (PR) and the ubiquitous transcription factor nuclear factor 1 (NF1). PR binding mediates ATP-dependent displacement of histone H2A and H2B, enabling NF1 access to its target site. In minichromosomes assembled in vitro NF1 binding facilitates access of PR to the hormone-responsive elements (HREs) by precluding reforming of the histone octamer, but the function of NF1 in living cells remains unclear. Here we show that depleting NF1 by small interfering RNAs or mutating the NF1-binding site significantly compromises transcription of the MMTV promoter. The central HREs 2 and 3 are not needed for ATP-dependent H2A/H2B displacement or NF1 binding but are critical for full PR binding and MMTV transactivation. We found that NF1 binding to the MMTV promoter on a H3/H4 histone tetramer particle exposes the central HREs and facilitates their binding by PR, suggesting a possible mechanism for the reciprocal synergism between PR and NF1.
Keywords: Chromatin/Immunoprecipitation/ChIP, Chromatin/Regulation, Chromatin/Remodeling, Chromatin/Structure, Hormones/Steroid, Receptors/Steroid/Thyroid, Transcription/Regulation
Introduction
The basic unit of chromatin, the nucleosome, consists of a flat cylinder formed by an octamer of the four core of histones, around which 146-bp DNA is wrapped in 1.65 left-handed superhelical turns (1). The central core of the histone octamer is formed by a stable symmetrical tetramer of histones H3 and H4 capable of organizing the central 96 bp of nucleosomal DNA (2, 3). The histone octamer is formed by the symmetrical addition of a dimmer of H2A/H2B on each side of the H3/H4 tetramer. A fraction of the nucleosomes in various eukaryotic genomes is positioned relative to the DNA sequence (4). Modulation of the structure and dynamics of nucleosomes is an important regulatory mechanism of all DNA-based processes in eukaryotic cells, such as transcription, DNA replication, and repair.
Steroid hormones regulate gene expression by binding to their intracellular receptors, which activate signal transduction cascades and interact in the cell nucleus with other transcription factors and/or with specific DNA sequences, called hormone-responsive elements (HREs)4 (5). When bound to DNA, the hormone receptors modulate the transcription of associated promoters by recruiting coregulators, among them chromatin-remodeling complexes. The activity of these complexes can result in changes in the position, structure, or dynamics of specific nucleosomes, which may preclude or facilitate loading of transcription factors (6). The SWI/SNF and the RSC complexes are the prototypes of ATP-dependent chromatin remodeling machines described initially in yeast but conserved in all eukaryotes (7–12). Action of these complexes can result in different outcomes, among them transfer, nucleosome sliding, dinucleosome formation, and H2A/H2B displacement (3, 9, 13, 14).
The mouse mammary tumor virus (MMTV) long terminal repeat region encompasses a hormone-dependent promoter with several cis-acting elements, including five HREs and a binding site for nuclear factor 1 (NF1) located immediately downstream. Binding of the progesterone receptor (PR) to the five HREs on free DNA is highly cooperative and precludes binding of NF1 to the adjacent site. In chromatin the MMTV promoter is organized into positioned nucleosomes (15), with a nucleosome located over the promoter covering the five HREs and the NF1-binding site. On this promoter nucleosome, the binding site for NF1 is not accessible, and only two of the five HREs, the strong palindromic HRE1 and the weak half-palindrome HRE4, can be bound by hormone receptors, whereas the central HREs, in particular the palindromic HRE2 and the half-palindrome HRE3, are not accessible for receptor binding (16). Following hormone induction in vivo all HREs and the binding site for NF1 are occupied simultaneously on the surface of a nucleosome-like structure, and a functional synergism is observed between glucocorticoid or progesterone receptor and NF1 (17). Transient transfection experiments have shown that the central HREs 2 and 3 are essential for hormone-activated transcription (18).
There have been many reports indicating a role for SWI/SNF, Brg1, and Brm in glucocorticoid regulation of MMTV transcription (19–24), but the situation with progesterone is less clear. Progesterone treatment of the breast cancer cell line carrying an integrated single copy of an MMTV transgene leads to recruitment of PR, SWI/SNF, and SNF2h-related complexes to the MMTV promoter (3, 25). Recruitment is accompanied by selective displacement of histones H2A and H2B from the nucleosome B (3). Moreover, after 5 min of hormone treatment, the cytoplasmic signaling cascade Src/Ras/Erk is activated via an interaction of PR with the estrogen receptor, which activates Src (26). As a consequence of Erk activation, PR is phosphorylated, Msk1 is activated, and the ternary complex PR-Erk-Msk1 is recruited to nucleosome B (27). Msk1 phosphorylates H3 at serine 10, which is followed by displacement of HP1g and recruitment of Brg1, PCAF, and RNA polymerase II (27). Based on these results, we have proposed a hypothetical model for MMTV promoter activation by progesterone that has been updated as our knowledge of the system increased (25, 27, 28). However, several steps in this model have not been tested. In particular the recruitment of NF1 and whether it can be accomplished in the absence of receptor binding to the central hidden HREs is not known.
To answer these questions we have used cultured breast cancer cells as well as minichromosomes and recombinant mononucleosomes assembled in vitro on either wild type MMTV sequences or on a promoter with point mutations that inactivate HRE2 and HRE3 (HRE 2−/3−). We have also used nucleosomes assembled on a MMTV promoter with the NF1 located outside of the nucleosome (29). Using assembled wild type and HRE 2−/3− MMTV promoters in minichromosomes using Drosophila embryo extracts, we show that the mutation precludes activation of transcription induced by recombinant PR and NF1. Mononucleosomes assembled with recombinant histones and wild type or mutant promoter sequences exhibit equal stability and positioning and can be efficiently remodeled by purified yeast SWI/SNF. In the presence of competitor DNA, PR is needed for recruitment of SWI/SNF, subsequent displacement of H2A/H2B dimers, and binding of NF1 to both wild type and mutant promoter nucleosomes. Moreover, nucleosomes containing the NF1-binding site located in the linker DNA can bind NF1, which does not recruit SWI/SNF in vitro. The complex of PR and NF1 on a tetramer of histones H3 and H4, as obtained with the HRE 2−/3− MMTV promoter, is incompetent for transactivation, demonstrating that binding of PR to central HREs 2 and 3 is essential for full induction. Thus, we have proven some of the predictions of our model.
EXPERIMENTAL PROCEDURES
Cell Culture and Hormone Treatments
T47D-MTVL breast cancer cells carrying one stably integrated copy of the luciferase reporter gene driven by the MMTV promoter (17) were routinely grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. For the experiments, the cells were plated in RPMI medium without phenol red supplemented with 10% dextran-coated charcoal-treated fetal bovine serum, and 48 h later the medium was replaced by medium without serum. After 24 h in serum-free conditions, the cells were incubated with R5020 (10 nm) or vehicle (ethanol) for different times at 37 °C.
Chromatin Immunoprecipitation (ChIP) in Cultured Cells
ChIP assays were performed as described (44) using the NF1-specific antibody (a gift from Dr Naoko Tanese), the H2A antibody (a gift from Stefan Dimitrov), anti-PR (H190) and anti-Brg1 (H88) (both from Santa Cruz), and anti-SMARCA2/BRM (ab15597) (from Abcam). Quantification of chromatin immunoprecipitation was performed by real time PCR using a Roche Applied Sciences Lightcycler. The fold enrichment of target sequence in the immunoprecipitated compared with input (Ref) fractions was calculated using the comparative Ct (the number of cycles required to reach a threshold concentration) method with the equation 2Ct(IP)−Ct(Ref). Primers sequences are available on request. For the coimmunoprecipitated BAF-NF1 anti BAF250 antibody from Upstate-Millipore, catalog number 04-080 was used.
RNA Interference Experiments
All of the siRNAs were purchased from Dharmacon and transfected into the T47D-MTVL cells using Lipofectamine 2000 (Invitrogen). After 48 h the medium was replaced by fresh medium without serum. After 1 day in serum-free conditions, the cells were incubated with R5020 (10 nm) or vehicle (ethanol) for different times at 37 °C. The down-regulation of NF1 expression was determined by Western blotting.
RNA Extraction and Reverse Transcription-PCR
Total RNA was prepared and cDNA generated as previously described (27). Quantification of LUC and glyceraldehyde-3-phosphate dehydrogenase gene products was performed by real time PCR. Each value calculated using the standard curve method was corrected by the human glyceraldehyde-3-phosphate dehydrogenase and expressed as relative RNA abundance over time zero. Primer sequences are available on request.
Minichromosome Reconstitution, Transcription, and Immunoprecipitation Using Postblastodermic Drosophila melanogaster Extracts
Extracts were used to assemble chromatin as previously described (45). The plasmid pMMTVCAT, used as transcription template, contains the wild type MMTV promoter from −640 to +126. In vitro transcription reactions with recombinant human PR and NF1 were performed as described (40). Transcription was quantified with Image Gauge package (Fujifilm). For ChIPs experiments, 10 ng of DNA of the reconstituted material was incubated with recombinant PR and NF1 during 30 min and subjected to ChIP assays as previously reported (40).
Mononucleosome Reconstitution and Purification
The 232-bp EcoRI-BamHI fragment containing either the wild type MMTV promoter sequence from −221 to +1, the MMTV HRE 2/3 mutant, or the HRE 1 mutant was used for mononucleosome reconstitution. The +50 construct with the NF1 site located into the linker DNA was obtained and labeled as previously described (29). The histones used for reconstitution experiments were recombinant Xenopus laevis histones expressed in Escherichia coli, and nucleosomes were reconstituted by the salt dialysis technique as described (34). Tetramer particles were reconstituted as previously described (2).
Electrophoretic Mobility Shift Assay
Recombinant human PR, isoform B (PRB), and pig NF1 C2 were expressed in baculovirus and purified as previously described (30). Nucleosomes and naked DNA were incubated with different amounts of PR or NF1 incubated for 20 min at room temperature and analyzed by electrophoresis at 4 °C in a 3.5% acrylamide, 20% glycerol, 0.5% agarose, 0.3× TBE gel. For PR and NF1 binding to tetramer particles, a 3.5% acrylamide gel was used.
Nucleosome Remodeling Assays
The ySWI/SNF was prepared as described (46). The reactions (20 μl) were done in 10 mm HEPES (pH 7.9), 60 mm KCl, 6 mm MgCl2, 60 mm EDTA, 2 mm dithiothreitol, 13% glycerol containing 20 nm of MMTV promoter nucleosomes, and 6 nm of SWI/SNF in the presence of 1 mm ATP. The nucleosomes were incubated for 30 min at 30 °C followed by an additional 30 min with 30 ng/ml poly(dI-dC) as competitor to remove SWI/SNF from the nucleosomes. The different mononucleosome populations were separated by electrophoresis on 5% polyacrylamide gels (acrylamide:bisacrylamide, 60:1) in 0.2× TBE (34).
Restriction Endonuclease Accessibility
Wild type and +50 mononucleosomes were incubated or not with NF1 and/or 1 μg of poly(dI-dC) in TGA buffer (10 mm Tris-HCl (pH 8.0), 0.5 mm EDTA, 5% glycerol, 0.5 mm 2-mercaptoethanol, 90 mm NaCl, 3 μg/μl bovine serum albumin) for 20 min at room temperature before being remodeled by SWI/SNF (described above). Remodeled nucleosomes were digested at 37 °C with 500 units/ml of SacI in a total volume of 160 μl. At the indicated time points, the aliquots of 19 μl were removed and added to 181 μl of stop buffer to give a final concentration of 12.5 mm EDTA and 0.5% SDS. The samples were then extracted with phenol, phenol/chloroform, and chloroform/isoamylic alcohol and precipitated with three volumes of ethanol. After washing with 80% ethanol and drying, the samples were analyzed on 8% denaturing polyacrylamide gels.
Cleavage by Nucleases and Mononucleosome Stability Experiments
DNase I and Exonuclease III digestion of wild type and mutant nucleosomes was performed as described (2). Stability experiments were done as previously reported (47).
Plasmids and Generation of Stable Cell Lines
The pMCBB HRE 2−/3− plasmid harbors mutations of the conserved G at position −112 to a C in HRE 2 and of the C at position −94 to a G in the HRE3. These residues are important for recognition by the hormone receptor (17, 48). The pMCBB NF1− plasmid bears mutations of the G at position −73 to an A and of the C at position −65 to a T (see Fig. 1B). Both pMCBB HRE2/3 mutant and pMCBB NF1 mutant were generated by PCR using primers containing the described mutations and the pMCBB wild type plasmid as template (33).
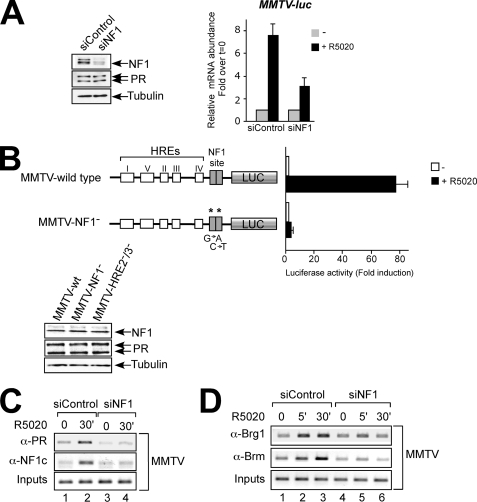
FIGURE 1.
NF1 is necessary for full recruitment of PR and BAF to the MMTV promoter. A, T47D-MTVL cells were transfected with control and NF1 siRNAs and treated with 10 nm R5020. Left panel, NF1 levels were analyzed by Western blotting. Right panel, MMTV promoter transcription was analyzed by real time PCR. The values represent the means ± S.D. of three experiments performed in duplicate. Panel B, T47D cells stably expressing luciferase reporter genes driven by the wild type MMTV promoter or by NF1− mutant promoters were treated with 10 nm R5020 or vehicle for 16 h and lysed, and luciferase activity was measured. The values represent the means ± S.D. from two experiments performed in duplicate. Lower panel, NF1, PR, and tubulin levels were analyzed by Western blotting in wild type, NF1−, and HRE 2−/3− mutant cell lines. Panel C, T47D-MTVL cells transfected with the indicated siRNAs were treated with hormone as indicated and subjected to ChIP assays with α-PR and α-NF1. The precipitated DNA fragments were analyzed by PCR for sequences corresponding to the MMTV nucleosome B. A representative of three independent experiments is shown. D, T47D-MTVL cells transfected with the indicated siRNAs were treated with hormone as indicated and subjected to ChIP assays with α-Brg1 and α-Brm. The precipitated DNA fragments were analyzed by PCR for sequences corresponding to the MMTV nucleosome B. A representative of three independent experiments is shown.
For generation of stable cell lines, the 1.5-kb DNA sequences of the MMTV promoter from the pMCBB wild type, HRE 2−/3− and NF1− were cloned into the pAGE5 plasmid generating the pAMB wild type, pAMB HRE 2−/3−, and pAMB NF1− plasmids containing a neomycin resistance. 1 μg of these plasmids were linearized with PvuI and used to stably transfect T47D wild type cells using Lipofectamine 2000 reagent (Invitrogen). After selective growth in Dulbecco's modified Eagle's medium, 10% fetal bovine serum containing 0.5 mg/ml G418 (Invitrogen), resistant foci were pooled.
RESULTS
NF1 Is Necessary for Full MMTV Induction and Efficient Recruitment of PR and BAF
To analyze the role of NF1 in progesterone activation of the MMTV promoter, we first studied the effect of down-regulation of the NF1 levels in the breast cancer cell line T47D-MTVL that carries an integrated copy of the MMTV promoter driving the luciferase gene (17). Transfection with NF1C siRNA resulted in significant reduction (60%) of NF1C protein levels without affecting the cellular levels of PR (Fig. 1A, left panel). Depletion of NF1C compromised hormonal MMTV transactivation by 50% (Fig. 1A, right panel), confirming the essential role of NF1C in MMTV promoter induction reported in minichromosomes assembled in Drosophila embryo extracts (30).
Hormonal induction was also compromised in T47D cells stably transfected with a MMTV promoter carrying point mutations in each half of the palindromic NF1-binding site that precluded NF1 binding (Fig. 1B, second row, and data not shown), whereas no change in PR and NF1 protein levels was observed (Fig. 1B, lower panel). This demonstrates that the five intact HREs are not sufficient for efficient progesterone induction and that binding of NF1 to its target sequences in the MMTV promoter is required.
To study the mechanism of this NF1 requirement, we performed chromatin immunoprecipitation experiments in cells depleted of NF1. As expected we found diminished hormone-dependent NF1C recruitment to the MMTV promoter (Fig. 1C, second row). But we also found that the levels of PR bound to the promoter upon hormone treatment were reduced in NF1-depleted cells (Fig. 1C, first row). This finding confirms previous results with in vitro assembled minichromosomes (30) and shows that in nuclear chromatin, NF1 binding also facilitates full loading of PR on MMTV promoter chromatin.
Because PR recruits BAF complexes to the promoter upon hormone addition, we tested whether NF1 depletion affected binding of BAF subunits to the MMTV promoter. We found that both Brg1 and in particular Brm, the two ATPases of the BAF complex, were reduced in hormone-stimulated cells depleted of NF1C (Fig. 1D, lanes 5 and 6).
If NF1 could interact with components of the BAF complex as previously described in other systems (31, 32), the reduction in BAF loading observed when knocking down NF1 could be explained by a decrease in NF1 recruitment (Fig. 1D). To clarify this point, we tested the interaction between NF1 and BAF in T47D-MTVL cells (supplemental Fig. S1). Coimmunoprecipitation experiments showed no interaction between NF1 and the BAF-specific subunit BAF250 before and after hormone addition (supplemental Fig. S1, compare lane 2 with lane 3). These results suggest that NF1 binding is required for full loading of PR and BAF molecules, as necessary for optimal MMTV induction.
The Central HREs 2 and 3 Are Essential for MMTV Activation
The central HREs 2 and 3 are critical for hormonal induction in transient transfection experiments (18), although their major grooves are initially nonaccessible for PR binding when the promoter is organized in nucleosomes (16, 17). To test the function of these hidden HREs when integrated in nuclear chromatin, we studied the effects of mutations in HREs 2 and 3 in stably transfected T47D cells. Although a control wild type MMTV promoter was induced 60-fold in response to overnight exposure to hormone, induction of the HRE 2−/3− mutant promoter was less than 6-fold (Fig. 2A, second row), indicating that binding of PR to these central HREs is critical for transcriptional activation by progesterone. No change in PR and NF1 protein levels was observed in wild type and HRE 2−/3− mutant cells (Fig. 1B, lower panel).
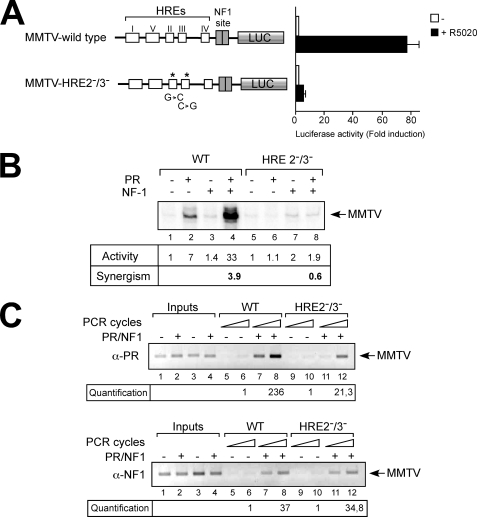
FIGURE 2.
The central HREs 2 and 3 are critical for transcriptional induction of the MMTV promoter. A, T47D cell stably expressing luciferase reporter genes driven by the wild type MMTV promoter or by HRE 2−/3− mutant promoters were treated with 10 nm R5020 or vehicle for 16 h and lysed, and luciferase activity was measured. The values represent the means ± S.D. from two experiments performed in duplicate. B, wild type (WT) and HRE 2−/3− mutant MMTV minichromosomes (25 ng of DNA in each reaction) assembled on post-blastodermic Drosophila embryo extracts were incubated with purified recombinant activated PR and NF1 and transcribed with HeLa nuclear extract (30). The products were visualized by primer extension analysis and sequencing gel electrophoresis. The position of the product from the wild type MMTV promoter is indicated. A representative of three independent experiments is shown. C, wild type and HRE 2−/3− mutant MMTV minichromosomes were incubated with purified recombinant activated PR and NF1 and subjected to ChIP assays as described previously (30) using specific antibodies against PR (upper panel) and NF1 (lower panel). The precipitated DNA fragments were subjected to PCR analysis at two different cycle numbers, to test for the presence of the MMTV nucleosome B sequence. Input material (5%) is shown for comparison. Lanes 1 and 2, wild type promoter; lanes 3 and 4, HRE 2−/3− promoter. Quantification was done by real time PCR as previously described (27). A representative of three independent experiments is shown.
We then tested whether the relevance of HREs 2 and 3 can be reproduced on MMTV promoter constructs assembled in minichromosomes using Drosophila embryo extracts (30, 33). The addition of recombinant PR or NF1 separately had weak effects on transcription of either wild type or HREs 2−/3− mutant MMTV minichromosomes (Fig. 2B, lanes 2, 3, 6, and 7, respectively; quantification below each lane). However, when both PR and NF1 were present, a synergistic transcriptional activation was observed on wild type MMTV but not on the HREs 2−/3− mutant promoter (Fig. 2B, lanes 4 and 8). Therefore, the synergism between PR and NF1 depends on PR binding to the central HREs 2 and 3.
We next investigated the binding of the two transcription factors to the wild type and mutant MMTV promoters. In ChIP experiments performed in the simultaneous presence of PR and NF1 (Fig. 2C), we detected binding of PR and NF1 to wild type promoter, as previously reported (Fig. 2C, lanes 7 and 8, upper and lower panel, respectively). In contrast, the HRE 2−/3− mutant promoter bound 11-fold less PR than the wild type promoter (Fig. 2C, upper panel, compare lanes 7 and 8 versus lanes 11 and 12 and quantification by real time PCR below) but equal amounts of NF1 (Fig. 2C, lower panel, compare lanes 7 and 8 versus lanes 11 and 12 and quantification by Real Time PCR below). These results indicate that the remodeling initiated via PR bound to HRE1 is sufficient for full NF1 access to the MMTV promoter in the absence of PR bound to the central HREs 2 and 3.
Binding of Factors to the Wild Type and HRE2−/3− Promoter Nucleosomes and Effect of Nucleosome Remodeling
To perform mechanistic studies short end-labeled DNA fragments containing either the wild type or the HRE 2−/3− MMTV promoter were used for protein binding assays. At low concentrations of PR, a retarded band was formed on wild type and mutant promoter DNAs that corresponds to complexes with a single PR dimer bound to the distal HRE1 (Fig. 3A, lanes 2–5). At higher concentrations of PR, more slowly migrating bands appeared with the wild type promoter but not with the mutant promoter (Fig. 3A, compare lanes 6 and 7 with lanes 9–13), indicating that HREs 2 and 3 were needed for the appearance of these larger complexes.
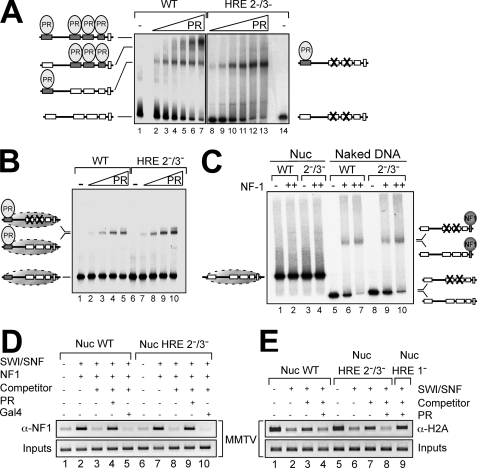
FIGURE 3.
PR dependent recruitment of SWI/SNF promotes NF1 binding and H2A displacement in wild type and HRE 2−/3− mutant MMTV mononucleosomes. A, end-labeled wild type (WT) MMTV promoter DNA (lanes 1–7) or the corresponding HRE 2−/3− mutant DNA (lanes 8–14) were incubated at room temperature for 20 min with increasing amounts of recombinant PR in 20-μl reactions, and the samples were analyzed by electrophoresis on a 3.5% acrylamide, 0.5% agarose, 20% glycerol, 0.3× TBE gel followed by autoradiography. A representative of three independent experiments is shown. B, end-labeled wild type and HRE 2−/3− mutant DNA assembled into nucleosome core particles were incubated at room temperature for 20 min with increasing amounts of recombinant PR in 20-μl reactions, and the samples were analyzed as in A. C, reconstituted wild type and HRE 2−/3− mutant nucleosomes (Nuc) and naked DNA were incubated at room temperature for 20 min with increasing amounts of recombinant NF1 in 20-μl reactions, and the samples were analyzed as in A. D, reconstituted wild type and HRE 2−/3− mutant nucleosomes were treated with SWI/SNF in the presence of ATP, competitor DNA, NF1, PR, and GAL4 as indicated. Following incubation at 30 °C for 30 min, the nucleosomes were cross-linked in 0.25% formaldehyde and immunoprecipitated with an antibody against NF1. The precipitated DNA fragments were subjected to PCR analysis (20 cycles) with oligonucleotides corresponding to the MMTV promoter nucleosome B. 5% of the input DNA was used as loading control. A representative of three independent experiments is shown. E, wild type, HRE 2−/3−, and HRE 1− MMTV mononucleosomes were treated with SWI/SNF in the presence of ATP, competitor DNA, and PR as indicated. Following incubation at 30 °C for 30 min, the nucleosomes were cross-linked in 0.25% formaldehyde and immunoprecipitated with an antibody against H2A. The precipitated DNA fragments were subjected to PCR analysis (20 cycles) with oligonucleotides corresponding to the MMTV promoter nucleosome B. 5% of the input DNA was used as loading control. A representative of three independent experiments is shown.
When the MMTV promoter DNA fragments were assembled into nucleosomes using recombinant histone octamers and the salt dialysis protocol (3), digestion with exonuclease III, DNase I, and restriction enzymes indicated that the wild type and the HRE 2−/3− mutant nucleosomes have similar structure and stability (supplemental Fig. S2 and S3). In band shift assays nucleosomes assembled with wild type or HRE 2−/3− mutant DNA behaved similarly in terms of PR binding. Even at high PR concentrations a single retarded complex corresponding to PR bound to the HRE1 was detected (Fig. 3B, lanes 4 and 5 versus lanes 9 and 10). Nucleosomes assembled on a DNA fragment carrying a mutated HRE1 did not yield this retarded complex (data not shown and (34), confirming that the HRE1 is the only element exposed on the surface of the nucleosome for binding of PR. NF1 cannot bind to its cognate sequences on wild type or HRE 2−/3− mutant promoter nucleosomes as it does on DNA (Fig. 3C, lanes 1 and 2 versus lanes 5 and 7 and lanes 3 and 4 versus lanes 8 and 10), confirming the inaccessibility of the NF1-binding site in nucleosomes (29).
We next explored the effect of remodeling by SWI/SNF on NF1 binding to nucleosomes using electrophoretic mobility shift assays (35, 36). In 5% acrylamide gels, wild type, HRE1− and HRE 2−/3− mutant nucleosomes migrated as a main band (supplemental Fig. S4, lanes 1, 3, and 5, black arrows), representing a mixture of the two main populations with the dyad axis at −107 and −127 and weaker, more fast migrating bands (supplemental Fig. S4, lanes 1, 3, and 5, gray arrow), corresponding to nucleosomes with the histone octamer localized at the ends of the DNA fragment (2). Incubation with SWI/SNF and ATP resulted in a faster mobility of a larger fraction of wild type and mutant nucleosomes (supplemental Fig. S4, lanes 2, 4, and 6). Moreover, SacI accessibility experiments performed after SWI/SNF remodeling of wild type and HRE 2−/3− mutant nucleosomes showed the same percentages of digestion (37.4 and 38,8%, respectively). Both types of nucleosomes bound NF1 upon remodeling (Fig. 3D, lanes 1 and 2 and lanes 6 and 7), but the addition of an excess of competitor DNA eliminated remodeling, as indicated by the low cleavage efficiency of SacI (3) and the lack of NF1 binding (Fig. 3D, lanes 3 and 8). However, when the nucleosomes were preincubated with PR and treated with SWI/SNF and ATP in the presence of competitor DNA, NF1 bound to wild type and to HRE 2−/3− mutant nucleosomes (Fig. 3D, lanes 3 and 4 versus lanes 8 and 9). Along with the appropriate controls (Fig. 3D, lanes 5 and 10), these results indicate that SWI/SNF recruited to MMTV nucleosomes by PR bound to the exposed HRE1 mediates binding of NF1 in the absence of PR binding to the central HREs 2 and 3. Similar to NF1 binding, H2A displacement was catalyzed by SWI/SNF in a PR-dependent manner both in wild type and HRE 2−/3− mutant promoter nucleosomes (Fig. 3E, lanes 1–8), whereas with HRE1− mutant promoter nucleosomes, no histone H2A displacement was observed (Fig. 3E, lane 9).
To test whether NF1 can recruit SWI/SNF to MMTV nucleosomes, we used a mutant MMTV promoter containing an insertion of 50 bp downstream of the HREs that displace the NF1 site to the linker DNA (+50) (29). NF1 bound very efficiently to +50 mutant nucleosomes, forming a single retarded complex (supplemental Fig. S5A, lanes 6–8). Incubation with SWI/SNF and ATP in the absence of competitor DNA and in the absence or presence of NF1 made the SacI restriction site more accessible for cleavage in wild type and +50 nucleosomes (supplemental Fig. S5B, compare lanes 1 with lanes 5 and 9 for wild type nucleosome and lane 3 with lanes 7 and 10 for mutant), whereas no remodeling was observed in the presence of an excess of competitor DNA (supplemental Fig. S5B, compare lane 5 with lane 2 for the wild type and lane 7 with lane 4 for the mutant), even when the nucleosomes are preincubated with NF1 (supplemental Fig. S5B, compare lane 2 with lane 6 for the wild type and lane 4 with lane 8 for the mutant), indicating that NF1 cannot recruit SWI/SNF to MMTV nucleosomes. Similar results were obtained with the +30 insertion mutant (data not shown).
MMTV Promoter Sequences on H3/H4 Tetramer Particles Bind NF1 and PR Synergistically
A tetramer of histones H3/H4 is known to position MMTV promoter sequences, and the resulting particles bind NF1 with relatively high affinity (2). We have used a band shift assay that detects very large complexes to test whether PR and NF1 can bind simultaneously to MMTV sequences organized around a H3/H4 tetramer. Band shift experiments show that recombinant PR and NF1 can bind individually to H3/H4 tetramers (Fig. 4A, lanes 2 and 3). Incubation of tetramer particles with NF1 together with increasing concentrations of PR produced two slow migrating complexes containing both proteins (Fig. 4A, lanes 4 and 5). The faster complex corresponds to tetramers carrying NF1 and one molecule of PR (Fig. 4A, lane 4), whereas the slowest migrating complexes, observed at higher concentrations of PR, correspond to tetramers with NF1 and multiple PRs (Fig. 4A, lane 5). The same slow migrating complexes were observed when either PR or NF1 were prebound to the tetramer, and the second transcription factor was added later (Fig. 4A, lanes 7 and 8).
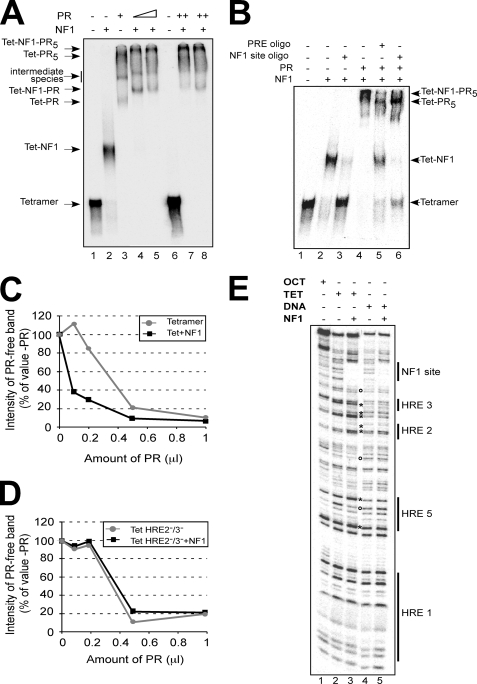
FIGURE 4.
PR and NF1 bind cooperatively to the MMTV promoter assembled on a tetramer of histones H3 and H4. A, end-labeled reconstituted MMTV tetramer particles were incubated with PR and NF1 as indicated. The established complexes were analyzed by electrophoresis and autoradiography. The identity of the main bands is indicated on the left margin. B, end-labeled reconstituted MMTV tetramer particles were incubated with PR, NF1, and 100-fold molar excess of an oligonucleotide (oligo) containing either a progesterone-responsive element (PRE) or a NF1 consensus site, as indicated. After incubation the established complexes were analyzed as described for A. The identity of the main bands is indicated on the right margin. C, end-labeled reconstituted MMTV tetramer particles were incubated with NF1 and increasing amounts of PR, and the samples were analyzed as in A. The bands corresponding to free TET and free TET-NF1 were quantified and are plotted, as percentages of the values in the absence of added PR, against the amount of added PR. D, end-labeled reconstituted HRE 2−/3− MMTV tetramer particles were incubated with NF1 and increasing amounts of PR, and the samples were analyzed and plotted as in C. E, end-labeled reconstituted MMTV octamer and tetramer particles were incubated with NF1 and digested with DNase I as previously described (34). End-labeled free MMTV DNA was used as a control (lanes 4 and 5). The asterisks and circles indicate the NF1-dependent DNase I-hypersensitive and protected sites, respectively. A representative of two independent experiments is shown.
The nature of the observed complexes was confirmed by adding an excess of nonradioactive oligonucleotides. An excess of NF1 oligonucleotide competed for the TET-NF1 complex (Fig. 4B, lane 3) and also for the very slow migrating complex of TET-NF1-PR5 (Fig. 4B, compare lanes 4 and 6). A progesterone-responsive element oligonucleotide competes for this slow migrating complex and generates the TET-NF1 complex (Fig. 4B, compare lanes 4 and 5). Along with the results obtained either with histone octamers (Fig. 3, D and E) or in cultured cells (3, 37), these data indicate that the product of the MMTV nucleosome remodeling generated by SWI/SNF, namely the tetramer particle, can accommodate the full loading of the promoter with PR and NF1 as observed in cells after hormone induction (Fig. 1C and Ref. 17).
Finally we tested whether binding of NF1 to an MMTV promoter assembled on a H3/H4 tetramer facilitates binding of PR. We incubated the MMTV tetramers with limiting amounts of NF1 to generate a mixture of free H3/H4 tetramer and the TET-NF1 complex (supplemental Fig. S6). To this mixture we added increasing amounts of PR. The relative affinity of PR for free tetramer and TET-NF1 was determined by measuring the progressive disappearance of the corresponding bands as the concentration of PR increases (supplemental Fig. S6). A comparison of the amount of PR needed to reach 50% disappearance of the corresponding band indicates that the TET-NF1 complex has a 6-fold higher affinity for PR than the free tetramer particle (Fig. 4C). This difference is dependent on the internal HREs 2 and 3, because displacement curves performed with HRE 2−/3− mutant tetramers showed no effect of NF1 on PR binding affinity (Fig. 4D). This is not due to a protein-protein interaction between PR and NF1 as assayed by in vitro coimmunoprecipitation experiments (supplemental Fig. S7). Therefore, we conclude that NF1 synergizes with PR on hormonal induction of MMTV not only by stabilizing the tetramer particle (30) but also by helping PR to bind the central HREs on the remodeled histone H3/H4 tetramer.
NF1 Binding to MMTV Promoter on a H3/H4 Tetramer Exposes the Central HREs
Next, we tested whether NF1 enhanced PR binding to the HREs 2 and 3 by changing the path on the DNA helix on the H3/H4 tetramer. For this we compared the effect of NF1 binding on the DNase I digestion patterns of free MMTV DNA and DNA assembled on tetramer particles (Fig. 4E). A comparison of the DNase I cleavage patterns of octamers, tetramers, and free DNA (lanes 1, 2, and 4, respectively) showed that the pattern of the tetramer particles is intermediate between those of octamer particles and those of free DNA (2). With both free DNA and tetramer particles, we observed a clear NF1-dependent protection against nuclease over the NF1 site, indicating specific protein binding (Fig. 4E, lanes 3 and 5). Moreover, in MMTV tetramer particles, we observed NF1-dependent changes in the DNase I cleavage pattern, mostly in form of hypersensitive sites, localized in the region covering the HREs 2 and 3 (Fig. 4E, compare lanes 2 with lane 3, marked with asterisks). Additional changes in the nuclease cleavage pattern were observed in a region including and flanking the HRE5. These changes were not observed when free MMTV DNA was used as substrate for NF1 binding and DNase I digestion (Fig. 4E, lanes 4 and 5). Thus, NF1 by binding to its site on the surface of a H3/H4 tetramer alters the path of the DNA helix, making it more accessible for nuclease over the central HREs. This finding reveals a previously unknown role for NF1 during the activation of the MMTV promoter that could be critical for the observed functional synergism between PR and NF1.
DISCUSSION
The results presented in this paper show that NF1 binding can take place in the absence of PR binding to the central HREs 2 and 3 and only requires the interaction of PR with the accessible HRE1. However, NF1 binding is essential for MMTV activation by facilitating full loading of PR and associated BAF to the central HREs 2 and 3. Moreover, we show that PR and NF1 can bind synergistically to MMTV promoter sequences assembled on a tetramer of histones H3 and H4, a finding compatible with the idea that the hormone-induced remodeling of MMTV chromatin generates a tetramer of H3/H4 as a platform on which the activated PR can orchestrate the assembly of coactivators and the basic transcriptional machinery.
NF1 Is Needed for Full PR Binding and Efficient MMTV Induction
We have postulated that NF1 binding is essential for stabilizing the open nucleosome conformation and for facilitating binding of PR to the central HREs (30). In agreement with this prediction, we show here that NF1C depletion leads to decreased accumulation of PR and BAF at the MMTV promoter and compromises transactivation. Inactivation of the NF1-binding site by point mutations also compromised MMTV induction in stably transfected cells.
In MMTV minichromosomes the synergistic effect of NF1 on PR binding and promoter activation is maintained when only the NF1 DNA-binding domain is used instead of the complete protein (30), consistent with a role of DNA binding rather than transactivation by NF1 in the context of the MMTV promoter. On the other hand, an association of the BAF complex with the CSF1 promoter has been reported to require intact NF1/CTF-binding sites, suggesting that prebound NF1 could target the BAF complex to the CSF1 promoter (32). However, experiments with MMTV nucleosomes mutated to place the NF1-binding site in the accessible linker DNA show that, in contrast to PR, NF1C does not recruit yeast SWI/SNF to the promoter. Moreover, no interaction between NF1 and BAF complex has been observed before and after hormone addition in T47DMTVL cells (supplemental Fig. S1). These results support the notion that NF1 binding does not enhance binding of PR by contributing to remodeling of MMTV chromatin.
The HREs 2 and 3 Are Essential for Hormonal Activation but Are Not Needed for Initial BAF-dependent Remodeling and 0NF1 Binding
The central HREs 2 and 3 have been shown to be important for hormone induction of the MMTV promoter in transient transfection experiments (18) but are not accessible for PR binding on in vitro assembled MMTV promoter nucleosomes (16). Here we confirm the importance of these HREs for hormone induction in stably transfected cells and show that the HRE 2−/3− mutant MMTV mononucleosomes are remodeled as efficiently as the wild type nucleosomes, resulting in similar binding of NF1. NF1 binding is important for PR occupancy of HREs 2 and 3, as shown in the NF1 knockdown experiments. These results suggest that PR binds to the central HREs as part of a complex with the activated Erk and Msk1 kinases and possibly PCAF, leading to enhanced phosphoacetylation of histone H3 and further recruitment of BAF (37). This is a prerequisite for subsequent steps in promoter activation such as recruitment of coactivators and RNA polymerase.
MMTV Promoter Sequences Assembled on a H3/H4 Tetramer Bind PR and NF1 Synergistically
SWI/SNF-treated MMTV nucleosomes yield a pattern of DNase I digestion indistinguishable from that of a H3/H4 tetramer particle, and incubation of tetramer particles with SWI/SNF does not change the digestion pattern (2). Therefore, it seems that SWI/SNF promotes ATP-dependent remodeling of octamer particles into tetramers but does not use tetramer particles as substrate. The idea that histones H2A and H2B are necessary for SWI/SNF-mediated remodeling is consistent with a previous report showing that arrays of histone tetramers are poor substrate for SWI/SNF remodeling (38). Consistent with this idea, the acidic N terminus of the Swi3p subunit of yeast SWI/SNF was identified as a novel H2A-H2B-binding domain required for ATP-dependent H2A/H2B dimer displacement (39).
A tetramer of histones H3 and H4 positions MMTV promoter sequences in a similar way as histone octamers, but NF1 can bind to a H3/H4 tetramer particle with relatively high affinity (2). Here we show that PR can access all HREs in MMTV sequences positioned around an H3/H4 tetramer. More important, such particles can bind PR and NF1 simultaneously, reminiscent of what is observed in cells carrying a single copy of the MMTV promoter integrated in chromatin (Fig. 1C and Ref. 17). Binding of PR to a wild type MMTV tetramer particle is enhanced if NF1 is bound to the particle, indicating that binding is cooperative. Mutation of the HRE 2 and 3 eliminates this synergistic binding of NF1 and PR. The nature of this synergism is unknown, but it is unlikely to result from a direct interaction between NF1 and PR, because no interaction between the two proteins was observed (supplemental Fig. S7). One alternative possibility is that the deformation of the DNA double helix imposed by NF1 binding weakens the interaction of the DNA with the H3/H4 tetramer, thus facilitating access of PR to the HREs 2 and 3. This idea is consistent with our finding of a higher accessibility for DNase I cleavage over the HREs 2 and 3 when MMTV DNA wrapped around a histone H3/H4 tetramer is bound by NF1. It is reasonable to assume that the exposure of the HREs 2 and 3 detected by DNase I contributes to a better binding of PR to these sites. Thus, a tetramer of histones H3 and H4 is a plausible structure for the “open” nucleosome conformation detected upon hormone induction in cultured cells, which results in full loading of all promoter cis elements with five PR homodimers and one NF1 homodimer.
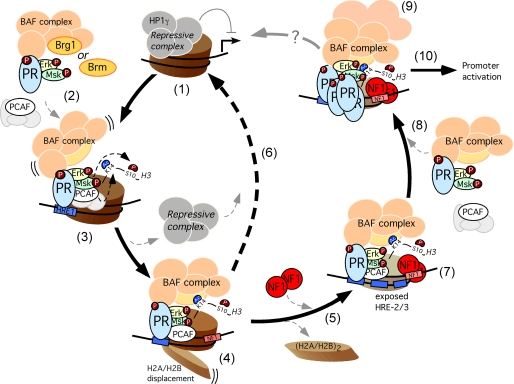
Our present model for activation of the MMTV promoter is shown in Fig. 5. Before hormone addition the MMTV promoter is silent because of its interaction with a repressive complex that includes HP1g (27). Very rapidly after hormone addition (5–10 min), the activated hormone receptor binds to the exposed HRE1 as part of a complex with activated Erk, activated Msk1, and likely PCAF and the BAF complex (37). This complex phosphoacetylates H3, leading to H3S10phK14ac, a modification that displaces the repressive complex and anchors the BAF complex (27, 37). BAF catalyzes the ATP-dependent H2A/H2B displacement that facilitates NF1 binding (3). We hypothesize that in the absence of NF1 the histone octamer particle is reformed, and the HP1g-containing repressive complex brings the promoter to the initial silenced state, thus preventing efficient activation. Whether phosphatases, histone deacetylases, or protein degradation events are involved in this cycle remains to be studied. In the presence of NF1 bound to the H3/H4 tetramer particles, the reassociation of H2A/H2B dimers is prevented, and binding of further PR molecules and BAF complexes to the hidden HREs 2 and 3 is facilitated, leading to full promoter activation. How this full loaded promoter on a H3/H4 tetramer is further converted into a preinitiation complex remains to be established. We also do not know under which conditions the activated tetramer particle reverts to the inactive octamer state.
FIGURE 5.
Model for the initial steps of MMTV promoter activation. Before hormone addition the MMTV promoter is silent and associated with a repressive complex that includes HP1g (step 1). After hormone addition the activated complex of pPR-pErk-pMsk, and likely PCAF and BAF, is recruited to the MMTV promoter (step 2). For simplicity PR is shown as a monomer, although the active form is a homodimer. This complex phosphoacetylates H3 leading to H3S10phK14ac, a modification that displaces the repressive complex (step 3) and anchors the BAF complex leading to ATP-dependent H2A/H2B displacement (step 4). The nucleosome opening facilitates NF1 binding (step 5). In the absence of NF1, the open conformation reverts to the repressed state (step 6), whereas NF1 binding to its cognate site maintains the open H3/H4 tetramer conformation (step 7) and facilitates binding of further PR and associated factors to the previously inaccessible HREs (step 8), thus promoting the recruitment of coactivators and the general transcriptional machinery (step 9). How this fully loaded promoter on a H3/H4 tetramer is further converted into a preinitiation complex (step 10) is unknown.
Finally, it is worth noting that we have not mentioned in these studies the possible role of linker histones. Our previous results (40) and those of other groups (41–43) suggest that changes in histone H1 stoichiometry and phosphorylation by CyclinA/Cdk2 take place at different time points during the hormonal induction and are important for transcriptional activation. Future studies will be required to clarify the relationship of these changes to those reported in this study.
Supplementary Material
Acknowledgments
We thank Dr. Stefan Dimitrov, CNR Grenoble, France for histone H2A antibody and Naoko Tanese, New York University, New York, NY.
This work was supported by grants from the Department d'Innovació Universitat i Empresa, Ministerio de Ciencia e Innovación Grant BMC 2003-02902, Consolider (CSD2006-00049), and Fondo de Investigación Sanitaria Grant PI0411605 and CP04/00087, as well as by European Union Integrated Project HEROIC.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1–S7.
- HRE
- hormone-responsive element
- MMTV
- mouse mammary tumor virus
- PR
- progesterone receptor
- ChIP
- chromatin immunoprecipitation
- NF
- nuclear factor
- Erk
- extracellular signal-regulated kinase
- siRNA
- small interfering RNA
- PCAF
- P300/CBP-associated factor
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- BAF
- Brg1/hBrm-associated factor.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2.Spangenberg C., Eisfeld K., Stünkel W., Luger K., Flaus A., Richmond T. J., Truss M., Beato M. (1998) J. Mol. Biol. 278, 725–739 [DOI] [PubMed] [Google Scholar]
- 3.Vicent G. P., Nacht A. S., Smith C. L., Peterson C. L., Dimitrov S., Beato M. (2004) Mol. Cell 16, 439–452 [DOI] [PubMed] [Google Scholar]
- 4.Segal E., Fondufe-Mittendorf Y., Chen L., Thåström A., Field Y., Moore I. K., Wang J. P., Widom J. (2006) Nature 442, 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beato M., Herrlich P., Schütz G. (1995) Cell 83, 851–857 [DOI] [PubMed] [Google Scholar]
- 6.Aalfs J. D., Kingston R. E. (2000) Trends Biochem. Sci. 25, 548–555 [DOI] [PubMed] [Google Scholar]
- 7.Wang W. (2003) Curr. Top Microbiol. Immunol. 274, 143–169 [DOI] [PubMed] [Google Scholar]
- 8.Roberts C. W., Orkin S. H. (2004) Nat. Rev. Cancer 4, 133–142 [DOI] [PubMed] [Google Scholar]
- 9.Narlikar G. J., Fan H. Y., Kingston R. E. (2002) Cell 108, 475–487 [DOI] [PubMed] [Google Scholar]
- 10.Martens J. A., Winston F. (2003) Curr. Opin. Genet. Dev. 13, 136–142 [DOI] [PubMed] [Google Scholar]
- 11.Cairns B. R., Schlichter A., Erdjument-Bromage H., Tempst P., Kornberg R. D., Winston F. (1999) Mol. Cell 4, 715–723 [DOI] [PubMed] [Google Scholar]
- 12.Cairns B. R., Kim Y. J., Sayre M. H., Laurent B. C., Kornberg R. D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1950–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaus A., Owen-Hughes T. (2004) Curr. Opin. Genet. Dev. 14, 165–173 [DOI] [PubMed] [Google Scholar]
- 14.Bruno M., Flaus A., Stockdale C., Rencurel C., Ferreira H., Owen-Hughes T. (2003) Mol. Cell 12, 1599–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richard-Foy H., Hager G. L. (1987) EMBO J. 6, 2321–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piña B., Brüggemeier U., Beato M. (1990) Cell 60, 719–731 [DOI] [PubMed] [Google Scholar]
- 17.Truss M., Bartsch J., Schelbert A., Haché R. J., Beato M. (1995) EMBO J. 14, 1737–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalepakis G., Arnemann J., Slater E., Brüller H. J., Gross B., Beato M. (1988) Cell 53, 371–382 [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharjee R. N., Archer T. K. (2006) Virology 346, 1–6 [DOI] [PubMed] [Google Scholar]
- 20.Cairns B. R., Levinson R. S., Yamamoto K. R., Kornberg R. D. (1996) Genes Dev. 10, 2131–2144 [DOI] [PubMed] [Google Scholar]
- 21.Hebbar P. B., Archer T. K. (2003) Mol. Cell. Biol. 23, 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsiao P. W., Fryer C. J., Trotter K. W., Wang W., Archer T. K. (2003) Mol. Cell. Biol. 23, 6210–6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muchardt C., Yaniv M. (1993) EMBO J. 12, 4279–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie Z., Xue Y., Yang D., Zhou S., Deroo B. J., Archer T. K., Wang W. (2000) Mol. Cell. Biol. 20, 8879–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vicent G. P., Ballaré C., Zaurin R., Saragüeta P., Beato M. (2006) Ann. N. Y. Acad. Sci. 1089, 59–72 [DOI] [PubMed] [Google Scholar]
- 26.Migliaccio A., Piccolo D., Castoria G., Di Domenico M., Bilancio A., Lombardi M., Gong W., Beato M., Auricchio F. (1998) EMBO J. 17, 2008–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vicent G. P., Ballaré C., Nacht A. S., Clausell J., Subtil-Rodríguez A., Quiles I., Jordan A., Beato M. (2006) Mol. Cell 24, 367–381 [DOI] [PubMed] [Google Scholar]
- 28.Vicent G. P., Koop R., Beato M. (2002) J. Steroid Biochem. Mol. Biol. 83, 15–23 [DOI] [PubMed] [Google Scholar]
- 29.Eisfeld K., Candau R., Truss M., Beato M. (1997) Nucleic Acids Res. 25, 3733–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Croce L., Koop R., Venditti P., Westphal H. M., Nightingale K. P., Corona D. F., Becker P. B., Beato M. (1999) Mol. Cell 4, 45–54 [DOI] [PubMed] [Google Scholar]
- 31.Zhao L. H., Ba X. Q., Wang X. G., Zhu X. J., Wang L., Zeng X. L. (2005) Acta Biochim. Biophys. Sin. 37, 440–446 [DOI] [PubMed] [Google Scholar]
- 32.Liu R., Liu H., Chen X., Kirby M., Brown P. O., Zhao K. (2001) Cell 106, 309–318 [DOI] [PubMed] [Google Scholar]
- 33.Venditti P., Di Croce L., Kauer M., Blank T., Becker P. B., Beato M. (1998) Nucleic Acids Res. 26, 3657–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicent G. P., Meliá M. J., Beato M. (2002) J. Mol. Biol. 324, 501–517 [DOI] [PubMed] [Google Scholar]
- 35.Corona D. F., Längst G., Clapier C. R., Bonte E. J., Ferrari S., Tamkun J. W., Becker P. B. (1999) Mol. Cell 3, 239–245 [DOI] [PubMed] [Google Scholar]
- 36.Längst G., Bonte E. J., Corona D. F., Becker P. B. (1999) Cell 97, 843–852 [DOI] [PubMed] [Google Scholar]
- 37.Vicent G. P., Zaurin R., Nacht A. S., Li A., Font-Mateu J., Le Dily F., Vermeulen M., Mann M., Beato M. (2009) PLoS Genet. 5, e1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyer L. A., Shao X., Ebright R. H., Peterson C. L. (2000) J. Biol. Chem. 275, 11545–11552 [DOI] [PubMed] [Google Scholar]
- 39.Yang X., Zaurin R., Beato M., Peterson C. L. (2007) Nat. Struct. Mol. Biol. 14, 540–547 [DOI] [PubMed] [Google Scholar]
- 40.Koop R., Di Croce L., Beato M. (2003) EMBO J. 22, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharjee R. N., Banks G. C., Trotter K. W., Lee H. L., Archer T. K. (2001) Mol. Cell. Biol. 21, 5417–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narayanan R., Adigun A. A., Edwards D. P., Weigel N. L. (2005) Mol. Cell. Biol. 25, 264–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bresnick E. H., Bustin M., Marsaud V., Richard-Foy H., Hager G. L. (1992) Nucleic Acids Res. 20, 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strutt H., Paro R. (1999) Methods Mol. Biol. 119, 455–467 [DOI] [PubMed] [Google Scholar]
- 45.Krajewski W. A., Becker P. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1540–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Logie C., Peterson C. L. (1999) Methods Enzymol. 304, 726–741 [DOI] [PubMed] [Google Scholar]
- 47.Candau R., Chávez S., Beato M. (1996) J. Steroid Biochem. Mol. Biol. 57, 19–31 [DOI] [PubMed] [Google Scholar]
- 48.Scheidereit C., Beato M. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 3029–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.