Abstract
Mycobacterial AdnAB exemplifies a family of heterodimeric motor-nucleases involved in processing DNA double strand breaks (DSBs). The AdnA and AdnB subunits are each composed of an N-terminal UvrD-like motor domain and a C-terminal RecB-like nuclease module. Here we conducted a biochemical characterization of the AdnAB motor, using a nuclease-inactivated heterodimer. AdnAB is a vigorous single strand DNA (ssDNA)-dependent ATPase (kcat 415 s−1), and the affinity of the motor for the ssDNA cofactor increases 140-fold as DNA length is extended from 12 to 44 nucleotides. Using a streptavidin displacement assay, we demonstrate that AdnAB is a 3′ → 5′ translocase on ssDNA. AdnAB binds stably to DSB ends. In the presence of ATP, the motor unwinds the DNA duplex without requiring an ssDNA loading strand. We integrate these findings into a model of DSB unwinding in which the “leading” AdnB and “lagging” AdnA motor domains track in tandem, 3′ to 5′, along the same DNA single strand. This contrasts with RecBCD, in which the RecB and RecD motors track in parallel along the two separated DNA single strands. The effects of 5′ and 3′ terminal obstacles on ssDNA cleavage by wild-type AdnAB suggest that the AdnA nuclease receives and processes the displaced 5′ strand, while the AdnB nuclease cleaves the displaced 3′ strand. We present evidence that the distinctive “molecular ruler” function of the ATP-dependent single strand DNase, whereby AdnAB measures the distance from the 5′-end to the sites of incision, reflects directional pumping of the ssDNA through the AdnAB motor into the AdnB nuclease. These and other findings suggest a scenario for the descent of the RecBCD- and AddAB-type DSB-processing machines from an ancestral AdnAB-like enzyme.
Keywords: DNA/Damage, DNA/Helicase, DNA/Nuclease, DNA/Recombination, DNA/Repair, Enzymes/ATPases
Introduction
DNA repair systems play an important role in bacterial pathogenesis, by allowing disease-causing bacteria to withstand DNA-damaging mediators of host innate immune systems (1–6). Mycobacteria, including the agent of human tuberculosis, have two distinct pathways to repair DNA double-strand breaks (DSBs),2 the most lethal form of DNA damage: (i) a RecA-dependent homologous recombination (HR) system and (ii) a non-homologous end-joining (NHEJ) system driven by dedicated DNA ligases and the end-binding protein Ku (7–11). In HR, an intact copy of the broken chromosome segment, typically a newly replicated sister chromatid, serves as a template for DNA synthesis across the break, ultimately yielding a faithfully restored chromosome with no mutations. By contrast, NHEJ protects the bacterial chromosome against DSBs during quiescent states, when there is no sister chromatid available to direct HR (12, 13). The recent finding that dormant mycobacteria develop into spores containing a single chromosome (14) suggests a role for NHEJ in spore viability under conditions of clastogenic stress, as demonstrated for the classic spore-forming bacterium, Bacillus subtilis (15, 16). The mycobacterial NHEJ mechanism, unlike that of HR, is conspicuously mutagenic (9, 11).
Nucleolytic resection of DSB ends is an essential step in HR and also during NHEJ of incompatible ends. In the HR mechanism, one or both of the DSB ends is resected by an exonuclease to leave a 3′ single-stranded tail, which then invades the homologous sister chromatid and primes repair DNA synthesis. The resection of DSB ends in bacteria is a regulated motor-driven process performed by a multisubunit helicase-nuclease complex encoded in an operon-like gene cluster (17). Three distinct clades of bacterial DSB-resecting machines have been identified, exemplified by Escherichia coli RecBCD (18, 19), Bacillus subtilis AddAB (20–22), and Mycobacterium smegmatis AdnAB (23). RecBCD is a heterotrimer composed of two helicase motor subunits, RecB and RecD, that initiate unwinding from a DSB end by translocating with opposite polarities on the two DNA strands (24, 25) and one nuclease module, located at the C terminus of RecB, that incises the single strands displaced by the helicases. AddAB is a heterodimer consisting of a single helicase motor in the N-terminal portion of the AddA subunit, plus two distinct RecB-like nuclease modules, one at the C terminus of AddA and another at the C terminus of AddB, each of which is dedicated to processing one of the single strands displaced by the helicase motor (22). Mycobacterial AdnAB is the founder of a newly recognized family of heterodimeric helicase-nucleases composed of two motors and two nucleases. The AdnA and AdnB subunits are each composed of an N-terminal UvrD-like motor domain and a C-terminal RecB-like nuclease module (23). AdnAB is a signature of the Actinomycetales taxon, including many bacteria that cause human disease (e.g. tuberculosis, diphtheria, nocardiosis, and Whipple disease).
Mycobacterial AdnAB shares certain generic features with the other motor-nuclease complexes. Its NTPase activity is off until triggered by double strand DNA with free ends, after which the energy of NTP hydrolysis is coupled to DSB end-resection by the AdnAB nuclease. The distinctive properties of AdnAB concern its dual single strand DNA (ssDNA) nuclease activities, which comprise an ATP-regulated molecular ruler. Absent ATP, AdnAB incises ssDNA by measuring the distance from the free 5′-end to dictate the sites of cleavage, which are predominantly 5 or 6 nucleotides from the 5′-end. ATP hydrolysis elicits a distal displacement of the cleavage sites 16–17 nucleotides from the 5′ terminus. We demonstrated a strict division of labor between the subunits, whereby mutations in the nuclease active site of the AdnB subunit ablated the ATP-inducible cleavages, while synonymous changes in the AdnA nuclease active site abolished ATP-independent cleavage. By studying the effects of mutations in the AdnA and AdnB phosphohydrolase active sites on the nuclease activities, we showed that ATP hydrolysis by the AdnB motor triggers the AdnB nuclease in cis (23).
Studies of the DNA motor function of mycobacterial AdnAB are stymied by concomitant DNA degradation. This problem can be circumvented, in principle, by simultaneous mutation of the active sites of both subunit nucleases. Such mutations completely suppress DSB end resection and single strand DNase activity. Moreover, the nuclease-dead versions of AdnAB could unwind linear plasmid DNA without degrading the displaced single strands (23).
Here we exploit a nuclease-dead AdnAB to conduct a biochemical characterization of the AdnAB motor, including DNA binding, unwinding, ATP hydrolysis, and directionality of translocation. We integrate the findings into a model of DNA unwinding in which the leading AdnB and lagging AdnA subunit motors track in tandem, 3′ to 5′, along the same DNA single strand. We present evidence that the ruler function of the ATP-dependent single strand DNase reflects directional pumping of the ssDNA through the AdnAB motor into the AdnB nuclease domain. Inspired by structural studies of RecBCD (18, 26–28), we discuss a plausible scenario for the descent of bacterial motor nuclease machines from an ancestral AdnAB-like enzyme with dual motors and dual nucleases.
EXPERIMENTAL PROCEDURES
Recombinant AdnAB Proteins
Wild-type AdnAB heterodimer and the mutant AdnA(D934A)-AdnB(D1014A) heterodimer (“nuclease-dead” AdnAB) were produced in E. coli and purified from a soluble extract by nickel affinity and anion exchange chromatography and glycerol gradient sedimentation as described previously (23). A nuclease-dead AdnB heterodimer with an inactivating D285A mutation in the AdnB subunit motor domain (AdnAB*) was produced and purified by the same procedure. The protein concentrations were determined as described previously (23). The figure legends specify the amounts of added enzyme with respect to the AdnB subunit.
DNA Binding Assay
Reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 10% glycerol, 1 mm DTT, 0.1 μm of 5′ 32P-labeled DNA as specified, and 1 pmol or 2 pmol of nuclease-dead AdnAB were incubated for 30 min at 22 °C. The reactions were adjusted to 20% glycerol and then analyzed by electrophoresis through a 15-cm native 6% polyacrylamide gel containing 45 mm Tris borate, 1.25 mm EDTA. The gels were run at 110 V in a cold room for 2–3 h. The gels were then transferred to DEAE paper and dried under vacuum. The free DNA and protein·DNA complexes were visualized by autoradiography.
Helicase Assay
Oligonucleotides were 5′-radiolabeled by using T4 polynucleotide kinase and [γ-32P]ATP and then purified by electrophoresis through a native 18% native polyacrylamide gel. The radiolabeled strand was annealed to a 5-fold excess of an unlabeled complementary DNA strand to form the blunt duplex and tailed duplex substrates shown in Fig. 2. Helicase reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 2 mm MgCl2, 0.9 mm DTT, 100 nm (1 pmol) of 5′ 32P-labeled duplex or tailed duplex DNA as specified and 85 nm (0.85 pmol) of nuclease-dead AdnAB were preincubated for 2 min on ice. The reactions were initiated by adding 2 mm ATP and 20 μm of an unlabeled “trap” 24-mer DNA oligonucleotide (identical to the labeled strand of the helicase substrate). The trap strand prevented the spontaneous reannealing of the unwound 32P-labeled DNA strand. The reaction mixtures were incubated for 15 min at 37 °C and then quenched by adding 2 μl of 200 mm EDTA, 40% glycerol, 2% SDS, 0.3% bromphenol blue. A control reaction mixture containing labeled DNA but no AdnAB was heated at 95 °C for 5 min. The reaction products were analyzed by electrophoresis through a native 18% polyacrylamide gel in 89 mm Tris borate, 2.5 mm EDTA. The products were visualized by autoradiography.
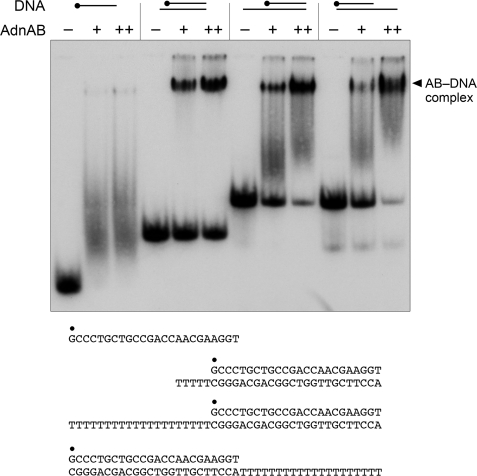
FIGURE 2.
AdnAB is a helicase that requires no loading strand. Reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 0.9 mm DTT, 2 mm MgCl2, 2 mm ATP, 0.1 μm (1 pmol) of 5′ 32P-labeled blunt duplex 24-mer, 3′ dT20-tailed, or 5′ dT20-tailed duplex as specified, and 0.85 pmol of nuclease-dead AdnAB or AdnAB* (a nuclease-dead AdnAB heterodimer with an ATPase-inactivating D285A mutation in the AdnB subunit) were incubated for 15 min at 37 °C. The products were analyzed by electrophoresis through a 15-cm 18% native polyacrylamide gel in 0.5× TBE (45 mm Tris borate, 1.2 mm EDTA) and visualized by autoradiography. AdnAB and AdnAB* were omitted from control reactions (− lanes). DNA-containing reaction mixtures lacking AdnAB that were heat-denatured prior to PAGE are included in lanes marked with ▵.
Streptavidin Displacement Assay of AdnAB Translocation
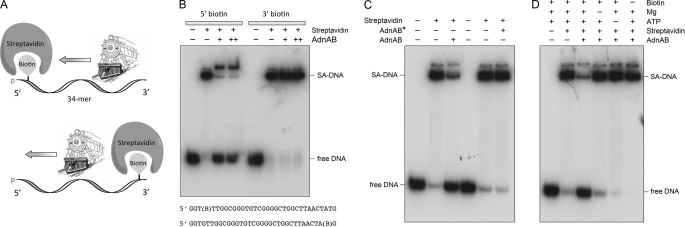
Synthetic 34-mer DNA oligonucleotides of otherwise identical nucleobase sequence containing a Biotin-ON internucleotide spacer either at the fourth position from the 5′ terminus or the second position from the 3′ terminus (Fig. 4) were purchased from Eurofins MWG Operon. The biotinylated ssDNAs were 5′ 32P-labeled by using T4 polynucleotide kinase and [γ-32P]ATP and then purified by electrophoresis through a native 18% native polyacrylamide gel. The DNA·SA complexes were formed by preincubating 50 nm biotinylated [32P]DNA with 2 μm streptavidin (Sigma) in 20 mm Tris-HCl (pH 8.0), 2 mm MgCl2, 2 mm ATP, 1 mm DTT for 10 min at room temperature. The mixtures were supplemented with 20 μm free biotin (Fisher), and the displacement reactions (10 μl) were initiated by adding nuclease-dead AdnAB. After incubation for 15 min at 37 °C, the reactions were quenched by adjustment to 31 mm EDTA, 4% glycerol (and in some experiments 0.1% SDS) and addition of 108 pmol of unlabeled ssDNA (44-mer from supplemental Table S1) to mask any binding of AdnAB to [32P]DNA released from the SA·DNA complex. The reaction products were analyzed by electrophoresis through a 15-cm native 15% polyacrylamide gel containing 89 mm Tris borate, 2.5 mm EDTA. The free 32P-labeled 34-mer DNA and the DNA·SA complexes were visualized by autoradiography.
FIGURE 4.
Directionality of AdnAB translocation on ssDNA. A, schematic representation of directional 5′ streptavidin displacement by the AdnAB motor acting as a cowcatcher to pry apart the otherwise stable SA-biotin interaction. See text for discussion. B, translocation reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 1 mm DTT, 2 mm MgCl2, 2 mm ATP, 50 nm (0.5 pmol) 5′ 32P-labeled 5′ or 3′ biotinylated 34-mer ssDNA (shown at the bottom, with B signifying the position of the biotin spacer), 2 μm streptavidin (where indicated by +), 20 μm biotin trap, and 0.25 pmol (+), or 0.5 pmol (++) of nuclease-dead AdnAB were incubated for 15 min at 37 °C. The reactions were quenched with EDTA, and the mixtures were analyzed by native PAGE. The radiolabeled DNAs were visualized by autoradiography. The species corresponding to SA·DNA complex and free DNA are indicated on the right. C, reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 1 mm DTT, 2 mm MgCl2, 2 mm ATP, 50 nm (0.5 pmol) of 5′ 32P-labeled 5′ biotinylated 34-mer ssDNA, 2 μm streptavidin (where indicated by +), 20 μm biotin trap, and 0.5 pmol of nuclease-dead AdnAB or AdnAB* (a nuclease-dead AdnAB with an inactive AdnB subunit motor) were incubated for 15 min at 37 °C. The products were resolved by native PAGE. D, reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 1 mm DTT, 50 nm 5′ 32P-labeled 5′ biotinylated 34-mer ssDNA, 2 μm streptavidin (where indicated by +), 0.5 pmol nuclease-dead AdnAB, and 2 mm ATP, 2 mm MgCl2 or 20 μm biotin trap, where specified by +, were incubated for 15 min at 37 °C. The products were resolved by native PAGE.
ATPase Assay
Reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 0.5 mm DTT, 1 mm MgCl2, 1 mm [α-32P]ATP, ssDNA, and AdnAB as specified were incubated at 37 °C. The reactions were quenched by adding 2 μl of 5 m formic acid. Aliquots (2 μl) of the mixtures were applied to PEI-cellulose TLC plates, which were developed with 0.45 m ammonium sulfate. [α-32P]ATP and [α-32P]ADP were quantified by scanning the plates with a Fujix BAS2500 imager.
RESULTS AND DISCUSSION
AdnAB Binds Stably to a DSB End
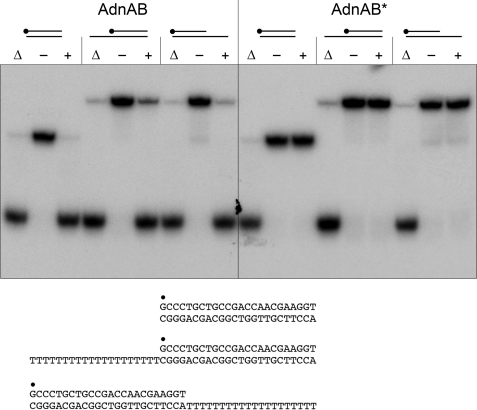
The DNA interactions of AdnAB were studied using the mutant heterodimer AdnA(D934A)-AdnB(D1014A) bearing nuclease-inactivating mutations in both subunits. The nuclease-dead AdnAB was incubated with a 5′ 32P-labeled 24-mer DNA oligonucleotide, either alone as ssDNA, or pre-annealed to complementary unlabeled DNA oligonucleotides to form the 5′ or 3′ tailed duplexes shown in Fig. 1. The binding reaction mixtures were then analyzed by native PAGE, and the radiolabeled DNAs were visualized by autoradiography (Fig. 1). A diffuse smear of shifted ssDNA seen in the presence of AdnAB reflected the formation of a metastable AdnAB-ssDNA complex that dissociated during the electrophoretic separation. By contrast, AdnAB formed a single discrete complex of much slower electrophoretic mobility when presented with a 24-bp duplex ligand in which one end comprised a blunt DSB and the other a DSB with a 4-nucleotide 3′ tail (Fig. 1). Tailed duplexes with longer (20-nucleotide) 5′ or 3′ tails yielded a mixed gel-shift pattern comprising the single discrete AdnAB·DNA complex plus a trailing diffuse smear, the latter again indicating the dissociable complex of AdnAB on the 20-mer ssDNA tail (Fig. 1). We conclude from the native gel shift experiment that AdnAB binds stably to a DSB end (either blunt or with a short overhang). This is a property shared with E. coli RecBCD (29, 30) and Bacillus AddAB (22).
FIGURE 1.
AdnAB binds stably to DSB ends. Reaction mixtures containing 0.1 μm (1 pmol) of 5′ 32P-labeled ssDNA, 3′-tailed, or 5′-tailed DNA as specified, and 1 pmol (indicated by +) or 2 pmol (indicated by ++) of nuclease-dead AdnAB were incubated for 30 min at 22 °C. AdnAB was omitted from control reactions (− lanes). The products were analyzed by native PAGE. The free 32P-labeled DNAs and slower migrating protein·[32P]DNA complexes (annotated on the right) were visualized by autoradiography. The DNAs are shown at the bottom, with the 5′ 32P label denoted by ●.
AdnAB Unwinds Duplex DNA without Requiring a Loading Single Strand
The helicase activity of the nuclease-dead AdnAB complex was assayed with three different substrates: (i) a 3′-tailed duplex substrate consisting of a 24-bp duplex with a 3′ T20 tail to serve as a potential “loading strand”; (ii) a 5′-tailed duplex substrate consisting of the identical 24-bp duplex segment, but with a 5′ T20 tail as loading strand; and (iii) a blunt-ended 24-bp duplex that lacks a loading strand (Fig. 2). Tailed duplexes like these are used routinely to infer helicase directionality. For example, the mycobacterial 3′ to 5′ helicases UvrD1 and UvrD2 that initiate unwinding when bound to a loading strand readily displace the 32P-labeled 24-mer from the 3′-tailed duplex, but are unable to unwind the 5′-tailed substrate (31).
The helicase assay format we used entailed preincubation of AdnAB and labeled DNA, followed by initiation of unwinding by addition of ATP, with simultaneous addition of a “trap” of excess unlabeled displaced strand that: (i) minimizes reannealing of any 32P-labeled 24-mer that was unwound by AdnAB and (ii) competes with the loading strand for binding to any free AdnAB or AdnAB that dissociated from the labeled DNA without unwinding it. Consequently, the assay predominantly gauges a single round of strand displacement by AdnAB bound to the labeled duplex prior to the onset of ATP hydrolysis. We found that AdnAB unwound all three DNA substrates to yield a radiolabeled free single strand that migrated faster than the input tailed duplex during native PAGE; the helicase reaction product comigrated with free 24-mer generated by thermal denaturation of the substrate (Fig. 2). These results indicate that AdnAB can initiate unwinding from a blunt DSB end. The fact that AdnAB does not require a loading single strand precludes any inferences about helicase directionality from this experiment.
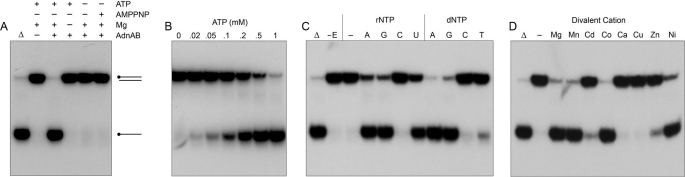
The helicase activity of nuclease-dead AdnAB was evaluated further using the blunt 24-mer substrate. Strand displacement was abolished by omission of either ATP or magnesium, and by substitution of AMPPNP for ATP (Fig. 3A). The extent of unwinding was proportional to ATP concentration, being virtually complete at 1 mm ATP and attaining a half-maximum at ∼0.2 mm ATP (Fig. 3B). The NTP substrate specificity of the AdnAB helicase was investigated by comparing the extents of unwinding in the presence of 1 mm ATP, GTP, CTP, UTP, dATP, dGTP, dCTP, and dTTP (Fig. 3C). All of the purine rNTPs and dNTPs supported efficient unwinding. By contrast, AdnAB displayed poor helicase activity with the pyrimidine NTPs, with UTP being more effective than TTP, whereas CTP and dCTP were virtually inert. The ability of the various NTPs to support the AdnAB helicase correlated with their relative efficacy in NTP hydrolysis (23). The divalent cation requirement for the helicase reaction in the presence of 2 mm ATP was satisfied by 2 mm magnesium, manganese, cobalt, or nickel (Fig. 3D). By contrast, calcium and copper were inert, whereas cadmium and zinc supported a feeble unwinding activity (Fig. 3D).
FIGURE 3.
Properties of the AdnAB helicase. A, reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 0.9 mm DTT, 0.1 μm (1 pmol) 5′ 32P-labeled 24-bp blunt duplex DNA, 0.85 pmol of nuclease-dead AdnAB, and 2 mm MgCl2, 2 mm ATP, or 2 mm AMPPNP (where specified by +) were incubated for 15 min at 37 °C. B, ATP titration. Reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 2 mm MgCl2, 0.9 mm DTT, 0.1 μm (1 pmol) 5′ 32P-labeled 24-bp blunt duplex DNA, 0.85 pmol of nuclease-dead AdnAB, and increasing amounts of ATP as specified were incubated for 15 min at 37 °C. C, nucleotide specificity. Reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 2 mm MgCl2, 0.9 mm DTT, 0.1 μm (1 pmol) 5′ 32P-labeled 24-bp blunt duplex DNA, 0.85 of pmol nuclease-dead AdnAB, and either no nucleotide (− lane) or 1 mm of the indicated NTP or dNTP were incubated for 15 min at 37 °C. Enzyme was omitted from the control reaction in lane -E. D, divalent cation specificity. Reaction mixtures containing 20 mm Tris-HCl (pH 8.0), 2 mm ATP, 0.1 μm (1 pmol) 5′ 32P-labeled 24-bp blunt duplex DNA, 0.85 pmol of nuclease-dead AdnAB, and 2 mm of the specified divalent cation were incubated for 15 min at 37 °C. The reaction products were analyzed by native PAGE and visualized by autoradiography. DNA-containing reaction mixtures lacking AdnAB that were heat-denatured prior to PAGE are included in lanes marked by ▵.
Helicase Activity Depends on the AdnB Motor
Previous studies suggested that the AdnB subunit provides the principal or “dominant” ATPase motor function of the AdnAB complex, based on the observations that ATPase activity is abolished by mutations in the phosphohydrolase active site of the AdnB subunit but impacted only modestly by synonymous mutations in the AdnA phosphohydrolase active site (23). We invoked a scenario in which ATP hydrolysis by AdnB was required to trigger the AdnA ATPase, which can be thought of as the “dependent” motor of a two-motor complex. In keeping with this idea, we found that AdnAB digestion of linear plasmid DNA (a combination of helicase and nuclease activities) was ablated by AdnB motor mutations, but was either unaffected of partially reduced by AdnA motor mutations (23). Here we verified the dominance of the AdnB motor by introducing the ATPase-inactivating AdnB mutation D285A (designated AdnB*) into the nuclease-dead AdnAB complex. The AdnAB* heterodimer was inert in unwinding blunt, 5′-tailed, or 3′-tailed duplex substrates (Fig. 2).
Activation of the AdnAB Motor by Single-stranded DNAs
Single-stranded DNA can serve as an ATPase activator for wild-type AdnAB but is degraded in the process (23). Here we used the nuclease-dead AdnAB mutant to probe the responsiveness of the AdnAB motor to single-stranded DNAs without the complicating factor of DNA decay. We surveyed a series of single-stranded DNA oligonucleotides of varying chain lengths, from 6 to 44 nucleotides. ATP hydrolysis was assayed by the conversion of [α-32P]ATP to [α-32P]ADP. The extent of ADP formation displayed a hyperbolic dependence on the concentration of each of the DNA oligonucleotides, except for the 6-mer, which was effectively inert up to 1 μm DNA (supplemental Fig. S1). Kinetic parameters were calculated by nonlinear regression curve fitting of the data to the Michaelis-Menten equation. The Km and kcat values are compiled in Table 1. (Note that because the ATPase assays in these experiments are single time point measurements, rather than velocities, the turnover numbers are slightly underestimated; see below.)
TABLE 1.
Effects of ssDNA length on ATPase activity
| ssDNA length | Km DNA | kcat | kcat/Km |
|---|---|---|---|
| nt | nm | s−1 | |
| 44 | 5.3 | 237 | 44.7 |
| 36 | 14.3 | 215 | 15.0 |
| 30 | 14.9 | 267 | 17.9 |
| 24 | 20.4 | 191 | 9.4 |
| 18 | 88.9 | 264 | 3.0 |
| 12 | 731 | 225 | 0.31 |
The instructive findings were that the affinity of single-stranded DNA for the putative “activator site” on the AdnAB motor was very much dependent on polynucleotide size, such that the apparent Km for DNA decreased progressively from 731 nm to 5.3 nm as the DNA was lengthened from 12 to 44 nucleotides (Table 1). The biggest incremental gains in affinity occurred in the transition from 12-mer to 18-mer (8-fold) and 18-mer to 24-mer (4-fold). By contrast, there was relatively little variation in the apparent turnover number. The net increase in kcat/Km was 140-fold in the transition from 12-mer to 44-mer ssDNA cofactor. Note that the lowest observed value for DNA Km (5.3 nm) is nearly equal to the concentration of AdnAB in the ATPase reaction mixture (4.2 nm).
We extended the kinetic analysis by measuring the velocity of ATP hydrolysis as a function of ATP concentration in the presence of 1 μm 44-mer ssDNA cofactor (supplemental Fig. S2). From these data, we calculated that the AdnAB motor has a Km of 87 μm ATP and a kcat of 415 s−1. These values are similar to those reported for ATP hydrolysis by E. coli RecBCD in the presence of double-stranded DNA (32). Bacillus AddAB has a similar ATPase kcat in the presence of double-stranded DNA (247 s−1), but the AddAB Km for ATP is 10-fold higher than that of mycobacterium AdnAB (21). The key point is that the three clades of bacterial DSB-processing motors are comparably powered with respect to nucleotide hydrolysis.
The AdnAB Motor Translocates Unidirectionally on Single-stranded DNA
RecBCD and AdnAB each contain two motor domains in separate subunits, but they appear to be organized in fundamentally distinct ways. In RecBCD, the RecB and RecD motors act in parallel. The RecB subunit engages and translocates along the 3′ DNA strand during duplex unwinding while the RecD subunit translocates along the 5′ DNA strand (24, 25). By analogy to an electrical circuit, parallel motors hydrolyze ATP independently, so that mutation of the RecD phosphohydrolase active site does not preclude ATP hydrolysis and DNA translocation by RecB; in turn, mutation of the RecB phosphohydrolase site does not prevent ATP hydrolysis and DNA translocation by RecD (33, 34). By contrast, mycobacterial AdnAB seems to behave like a serial motor, in which ATP hydrolysis by the leading (dominant) AdnB subunit precedes ATP hydrolysis by the lagging (dependent) AdnA subunit. Although this conceptual framework predicts correctly that mutating the lead motor abolishes all ATP hydrolysis and DNA unwinding, it lacks a connection to DNA dynamics.
To flesh out the model of motor dependence, we envision that the AdnB subunit uses ATP hydrolysis to pump single-stranded DNA through its N-terminal domain in a path that feeds the polynucleotide into the N-terminal motor domain of the AdnA subunit, where it can then trigger the AdnA phosphohydrolase activity to aid in further translocation of the AdnAB complex along the ssDNA (supplemental Fig. S3A). The model explains the obligate role of the AdnB motor and the persistence of ATP hydrolysis (and DSB processing) when the AdnA motor is off, presumably because the AdnB motor suffices to pump ssDNA along the track through the AdnA subunit.
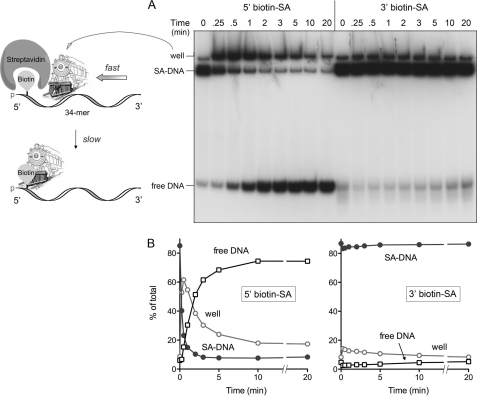
For the model to be plausible, its central tenets need to be interrogated. First, it must be established that AdnAB translocates along single-stranded DNA. Second, the tandem motor model predicts that translocation is unidirectional, albeit without prejudice as to which direction. To address these issues, we employed the streptavidin displacement assay developed by Kevin Raney and colleagues (35–37). 5′ 32P-labeled 34-mer DNA oligonucleotides containing a single biotin moiety at the fourth internucleotide from the 5′-end or the second internucleotide from the 3′-end were preincubated with excess streptavidin (SA) to form a stable SA·DNA complex that was easily resolved from the free biotinylated 34-mer DNA during native PAGE (Fig. 4B). The translocation assay scores the motor- dependent displacement of SA from the DNA in the presence of ATP and excess free biotin, which instantly binds to free SA and precludes SA rebinding to the labeled DNA. The rationale of the assay is that directional tracking of the motor along the DNA single strand will displace SA from one DNA end, but not the other. As depicted in Fig. 4A, the enzyme acts like a “cowcatcher” on a locomotive engine. When moving 3′ to 5′, it can displace SA as it collides with the 5′-biotin·SA. By contrast, a 3′-biotin·SA is not expected to be displaced by a 3′ → 5′ translocase, because the motor moves away from the SA and simply falls off the free 5′-end. The converse outcomes apply to a 5′ → 3′ translocase; it displaces a 3′ SA, but not a 5′ SA.
The instructive finding was that nuclease-dead AdnAB readily displaced SA from a 5′ biotin·SA complex on the 34-mer single-stranded DNA to yield the free 32P-labeled 34-mer strand, but was unable to displace SA from a 3′ biotin·SA complex tested in parallel (Fig. 4B). Stripping of the 5′ biotin·SA complex by AdnAB to liberate free DNA depended on magnesium and ATP (Fig. 4D). Inclusion of the excess biotin trap was necessary to detect the free DNA (Fig. 4D). Moreover, the 3′ → 5′ translocation activity was dependent on the AdnB motor subunit, insofar as the AdnAB* mutant heterodimer was unable to displace the 5′ SA (Fig. 4C). Thus, AdnAB meets the criteria for a unidirectional 3′ → 5′ ssDNA translocase. These results engender a plausible model for the action of the AdnAB motor during DSB end processing, whereby the leading AdnB and lagging AdnA subunit motors translocate in tandem in the 5′ direction along the displaced 3′ strand of the duplex DNA substrate (supplemental Fig. S3B).
Disposition of the Nuclease Modules in AdnAB: Insights from the Structure of RecBC
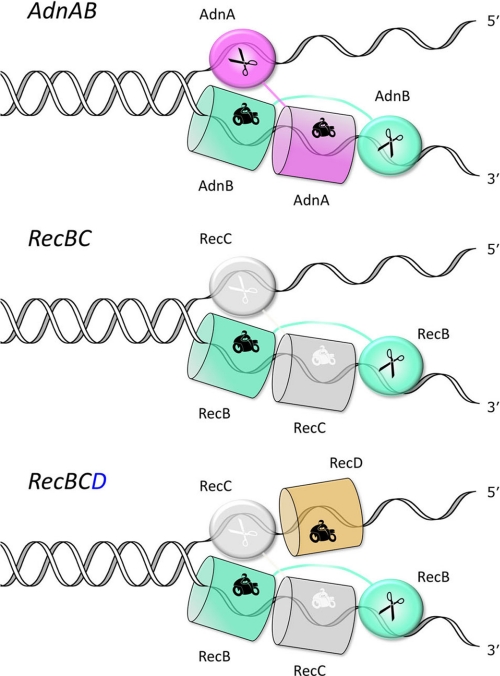
The tandem motor model leaves open the issue of how the nuclease modules of the AdnA and AdnB subunits are disposed with respect to the displaced DNA strands. Our previous findings that both nucleases measure the distance from the 5′-end of single-stranded DNA substrates (23) raised the prospect that DNA degradation might be focused on the 5′-displaced strand, an idea that makes sense given the HR goal of providing a 3′-OH single strand substrate for a RecA-mediated homology search. However, we thought this scenario unlikely in view of current functional and structural understanding of the RecBC and AddAB motor nuclease complexes. In Bacillus AddAB, there appears to be a division of nuclease labor during DSB resection, whereby the RecB-like AddA subunit nuclease (the homolog of mycobacterial AdnB nuclease) is dedicated to degrading the 3′-displaced strand, whereas the AddB subunit nuclease (the homolog of the mycobacterial AdnA nuclease) degrades the 5′-displaced strand (22). Positing a similar functional specialization in AdnAB, we envisioned a model for the domain organization of the AdnAB motor and nuclease units as shown in Fig. 5 (top panel). The key features are: (i) the placement of the AdnB nuclease behind the tandem motor on the displaced 3′ strand, so that access of the 3′ strand to the nuclease active site requires pumping of the DNA through the tandem motor and (ii) the assignment of the AdnA nuclease to receive the displaced 5′ strand directly at the “Y-fork” of the double strand DNA-ssDNA junction. This model is indebted to the available crystal structures of the RecBC heterodimer portion of RecBCD bound to Y-fork DNAs (18, 27), which reveal the separate paths of the displaced strands through the RecB and RecC subunits (Fig. 5, middle panel). In RecBC, the RecB N-terminal motor domain drives strand separation by translocating 3′ to 5′ along the displaced 3′ strand. The 3′ DNA is pumped by RecB through the N-terminal domain of the RecC subunit, and then into the C-terminal RecB nuclease domain, located behind RecC and tethered to the RecB motor domain by an extended linker peptide. A key insight from the crystal structure was the realization that the RecC N-terminal domain is a degenerated motor, retaining the folds of the subdomains that comprise the SF1 helicases (e.g. PcrA and UvrD), but lacking the specific amino acids responsible for NTP phosphohydrolase and DNA translocase activities (18). Ridgen (26) has since reported that a segment of ∼200 amino acids of the RecC C-terminal domain (through which the 5′-displaced strand passes) is a degenerated RecB-like nuclease that retains the nuclease fold but lacks the metal-binding site and catalytic amino acids. If RecC exemplifies a late stage of erosion of the catalytic capacity of a bifunctional motor-nuclease subunit within a heterodimeric motor nuclease complex, then we view mycobacterial AdnA as an extant and functional representative of the ancestral state from which RecC might have devolved.
FIGURE 5.
AdnAB domain model based on the structure of DNA-bound RecBC.
Testing the Model and Accounting for the Nuclease “ATP Ruler” Function
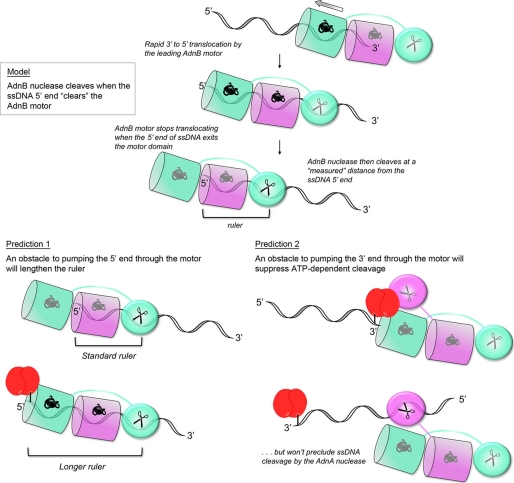
How can the RecBC-based model of AdnAB rationalize the distinctive ATP-dependent molecular ruler function displayed by AdnAB as it cleaves ssDNA? As illustrated in Fig. 6, we posit that: (i) the requirement for NTP hydrolysis to trigger ssDNA incision by the AdnB nuclease module simply reflects the action of the lead AdnB motor in pumping the DNA 3′-end through the tandem motor unit until the ssDNA enters the AdnB nuclease active site; (ii) the rate of 3′-to-5′ translocation is much faster than the rate of ssDNA cleavage; (iii) the lead AdnB motor stops translocating when the 5′-end of the ssDNA clears the lead motor domain (just as cargo stops moving at the end of a conveyor belt), at which time the AdnB nuclease can incise at a “measured” distance from the ssDNA 5′-end. Thus, the length of the ruler reflects the distance along the ssDNA track from the end of the motor to the nuclease active site (Fig. 6, upper panel). This value is 16–17 nucleotides for the series of ssDNA substrates tested previously (23). (Note that, for simplicity, the model assumes that DNA translocation by the lagging AdnA motor is either coupled to the lead AdnB motor or else translocates much slower than the lead motor.)
FIGURE 6.
Model of the AdnAB nuclease ruler and predictions of experimental testing.
The model begets several predictions regarding the effects of a physical obstacle to DNA pumping on the outcomes of the ssDNA cleavage reactions. First, it should be the case that an obstacle to pumping the 5′ ssDNA end through the motor will lengthen the ruler, by arresting translocation before the DNA 5′-end clears the lead motor (Fig. 6, lower left panel). Second, an obstacle to pumping the 3′ ssDNA end through the motor ought to suppress ATP-dependent DNA cleavage, but not preclude ATP-independent ssDNA cleavage by the AdnA subunit nuclease at sites close to the DNA 5′-end (Fig. 6, lower left panel).
We endeavored to test these predictions by surveying the effects of bulky 5′ and 3′ biotin-streptavidin adducts (serving as the putative obstacles) on the ssDNA nuclease activities of wild-type AdnAB. At first glance, this strategy might seem off-base, given our demonstration above that the AdnAB motor can displace a 5′ SA adduct from 5′ (bio)ssDNA. However, the enabling quality of the 5′ biotin·SA obstacle was revealed by an analysis of the kinetics of streptavidin displacement from the radiolabeled 34-mer ssDNA in the presence of a free biotin trap (Fig. 7). Aliquots were removed at various times, and the ATPase reaction was quenched immediately with EDTA. The products were then analyzed by native PAGE (Fig. 7A). We observed that addition of nuclease-dead AdnAB resulted in the rapid conversion of the SA·DNA complex into a more slowly migrating species that barely entered the gel at the sample well. This “well-shift” complex comprised the majority of the radiolabeled DNA at 0.25, 0.5, and 1 min, before declining steadily between 1 and 10 min, concomitant with the accumulation of free DNA (Fig. 7, A and B); this pattern is consistent with a precursor-product relationship of the well-shift complex and the free DNA. We surmise that SA displacement by the AdnAB motor is a biphasic process, entailing rapid translocation of AdnAB (the locomotive) along the ssDNA until it collides with the 5′ biotin·SA without actually displacing it (thereby yielding the larger AdnAB·SA·DNA complex in the well), followed by a slow step of SA displacement by the cowcatcher (Fig. 7A). By contrast, we did not see significant accumulation of a well-shifted complex or a significant increase in free DNA when the AdnAB motor was reacted with a 34-mer ssDNA with a 3′ biotin·SA adduct (Fig. 7, A and B). The physical details of how SA is pried loose by DNA helicases or translocases are unknown (35), but the kinetic delay seen here could signify multiple attempts by stalled AdnAB at a force-dependent alteration in the biotin·SA interface. In any event, this experiment establishes that 5′ biotin·SA is a transient physical obstacle to the movement of ssDNA though the AdnAB motor.
FIGURE 7.
A 5′ biotin-streptavidin adduct is a kinetic obstacle to AdnAB translocation. Reaction mixtures (90 μl) containing 20 mm Tris-HCl (pH 8.0), 1 mm DTT, 2 mm MgCl2, 2 mm ATP, 2 μm streptavidin, 50 nm (4.5 pmol) 5′ 32P-labeled 5′ or 3′ biotinylated 34-mer ssDNA, 20 μm biotin, and 4.6 pmol of nuclease-dead AdnAB were incubated at 37 °C. Aliquots (10 μl) were withdrawn at the times specified and quenched with EDTA. The mixtures were analyzed by native PAGE, and the radiolabeled DNA was visualized by autoradiography (A). The distributions of radiolabeled DNA between DNA·SA complex, free DNA, and the well-shift complex were quantified by scanning the gel and are plotted as a function of time in B. The results are consistent with a biphasic process of 5′ SA adduct displacement, depicted in schematic form in A, whereby the AdnAB motor rapidly translocates to abut the 5′ SA complex, followed by a slow step in which SA is pried off the DNA by the AdnAB cowcatcher.
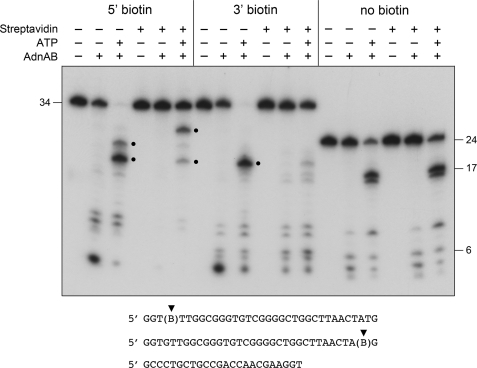
The experiment interrogating the nuclease ruler model is shown in Fig. 8. Wild-type AdnAB was incubated with 5′ 32P-labeled 34-mer DNA with a 5′ or 3′ biotin, with or without SA, in the absence or presence of ATP. The reactions were quenched with EDTA, and the products were analyzed by denaturing PAGE. A non-biotinylated 5′ 32P-labeled 24-mer DNA served as a control to demonstrate the nuclease ruler phenomenon, whereby AdnA nuclease cleaves close to the 5′-end in the absence of ATP to generate a cluster of small (4-, 5-, 6-, and 8-nucleotide) products, and the addition of ATP triggers cleavage by AdnB nuclease at distal sites to yield a 16-mer/17-mer doublet (Fig. 8). The inclusion of SA had no apparent effect on cleavage of the non-biotinylated 24-mer DNA (Fig. 8). The ATP ruler effect was also evident for the 5′- and 3′-biotinylated 34-mer oligonucleotides when the nuclease assays were performed in the absence of SA, i.e. a cluster of smaller 5′ 32P-labeled cleavage products was generated when ATP was omitted, and this cleavage pattern shifted distally in the presence of ATP (Fig. 8).
FIGURE 8.
Testing the ATP ruler model: effect of 5′ and 3 streptavidin obstacles on the outcomes of ssDNA cleavage. Reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 8.0), 1 mm DTT, 2 mm MgCl2, 50 nm (0.5 pmol) 5′ 32P-labeled 5′ or 3′ biotinylated 34-mer ssDNA or non-biotinylated 24-mer DNA, and 1 mm ATP, 2 μm streptavidin, or 0.26 pmol of wild-type AdnAB (where indicated by +) were incubated for 15 min at 37 °C. The reactions were quenched with formamide/EDTA, heated for 5 min at 95 °C, and then analyzed by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 m urea, 45 mm Tris borate, 1.25 mm EDTA. An autoradiograph of the gel is shown. The ATP-induced cleavage products of the biotinylated DNAs are highlighted by ●.
The model's predictions concerning the effects of an SA adduct on the cleavage patterns of the biotinylated ssDNAs were fulfilled by the experiment. ATP-independent cleavage of the 5′ biotin ssDNA was abolished by SA. By contrast, SA did not prevent ATP-independent incision of the 3′ biotin ssDNA. These results signify that a free 5′-end is needed to access the AdnA nuclease. Opposite SA effects were noted for the ATP-dependent incisions at 5′ distal sites (denoted by dots in Fig. 8). First, the addition of SA suppressed ATP-dependent cleavage of the 3′ biotin ssDNA, consistent with the proposed directional pumping of the 3′ strand through the tandem motor to access the AdnB nuclease site. Second, SA addition to the 5′ biotin ssDNA elicited a distal shift in the site of ATP-dependent incision to yield a new cleavage product ∼8 nucleotides longer than the major product formed by AdnB nuclease in the absence of SA (Fig. 8). This interval defines the increase in the size of the ruler caused by the 5′ SA obstacle to translocation by the motor. An additional subtlety of the experiment was the effect of the 5′ biotin itself on the pattern of AdnB nuclease cleavage, revealed in the pairwise comparison of ATP-dependent cleavage of the 5′- versus 3′-biotinylated ssDNA. Whereas ATP triggered cleavage of the 3′ biotin ssDNA at a single predominant site, we detected an additional incision of the 5′ biotin ssDNA 3 nucleotides downstream (Fig. 8). (This 5′ biotin effect was seen in multiple iterations of the experiment.) We surmise that the biotin spacer (which maintains the natural distance between flanking phosphodiesters) is itself capable of exerting a slight impediment to the AdnAB motor and, thereby, lengthening the ruler for at least some of the cleavages. In sum, the results weigh in favor of the AdnAB model depicted in Fig. 5.
Evolution of the Bacterial Motor-Nuclease Machines
The present study, plus structural and phylogenetic analyses (17, 18, 23, 26), suggest an evolutionary path connecting the three known clades of bacterial DSB resecting motor nucleases: AdnAB, AddAB, and RecBCD. An ancestral AdnAB-like machine with two motors and two nucleases (Fig. 5) could devolve into an AddAB-like state by degeneration of the helicase active site of the lagging motor subunit. In Bacillus AddAB, this process entailed loss of all the helicase motifs but one (22). Ridgen (26) has invoked an AddAB-type enzyme (one motor, two nucleases) as an intermediate stage in emergence of a RecBC-like heterodimer (itself a functional motor-nuclease) by effacement of the nuclease active site in the lagging subunit (Fig. 5). The recruitment of a new motor subunit, RecD, with directionality opposite to that of the lead RecB motor, and its incorporation into the RecBCD complex on the 5′-displaced strand behind the RecC C-domain (18, 27, 28), appears to be a late innovation. The circumstances that selected for these changes, and the potential biological advantages of one flavor of motor nuclease versus the other, are entirely unclear at this point. Nonetheless, a distinctive feature of mycobacteria (and several other Actinomycetales species) is that they retain both AdnAB-type and RecBCD-type motor nucleases (23), suggesting that there might be a division of labor among these DSB-processing machines.
Supplementary Material
Acknowledgment
We are grateful to Dale Wigley for illumination discussions of the RecBCD structure, especially the vestigial motor and nuclease domain folds of the RecC subunit.
This work was supported, in whole or in part, by National Institutes of Health Grant AI64693.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- DSB
- double strand break
- HR
- homologous recombination
- NHEJ
- non-homologous end-joining
- ssDNA
- single strand DNA
- AMPPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- AdnAB*
- nuclease-dead AdnB heterodimer with an inactivating D285A mutation in the AdnB subunit motor domain
- DTT
- dithiothreitol
- SA
- streptavidin.
REFERENCES
- 1.Buchmeier N. A., Lipps C. J., So M. Y., Heffron F. (1993) Mol. Microbiol. 7, 933–936 [DOI] [PubMed] [Google Scholar]
- 2.Buchmeier N. A., Libby S. J., Xu Y., Loewen P. C., Switala J., Guiney D. G., Fang F. C. (1995) J. Clin. Investig. 95, 1047–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cano D. A., Pucciarelli M. G., García-del Portillo F., Casadesús J. (2002) J. Bacteriol. 184, 592–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boshoff H. I., Reed M. B., Barry C. E., 3rd., Mizrahi V. (2003) Cell 113, 183–193 [DOI] [PubMed] [Google Scholar]
- 5.Darwin K. H., Nathan C. F. (2005) Infect. Immunol. 73, 4581–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amundsen S. K., Fero J., Hansen L. M., Cromie G. A., Solnick J. V., Smith G. R., Salama N. R. (2008) Mol. Microbiol. 69, 994–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitcher R. S., Green A. J., Brzostek A., Korycka-Machala M., Dziadek J., Doherty A. J. (2007) DNA Repair 6, 1271–1276 [DOI] [PubMed] [Google Scholar]
- 8.Stephanou N. C., Gao F., Bongiorno P., Ehrt S., Schnappinger D., Shuman S., Glickman M. S. (2007) J. Bacteriol. 189, 5237–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong C., Bongiorno P., Martins A., Stephanou N. C., Zhu H., Shuman S., Glickman M. S. (2005) Nat. Struct. Mol. Biol. 12, 304–312 [DOI] [PubMed] [Google Scholar]
- 10.Pitcher R. S., Tonkin L. M., Daley J. M., Palmbos P. L., Green A. J., Velting T. L., Brzostek A., Korycka-Machala M., Cresawn S., Dziadek J., Hatfull G. F., Wilson T. E., Doherty A. J. (2006) Mol. Cell 23, 743–748 [DOI] [PubMed] [Google Scholar]
- 11.Aniukwu J., Glickman M. S., Shuman S. (2008) Genes Dev. 22, 512–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuman S., Glickman M. S. (2007) Nat. Rev. Microbiol. 5, 852–861 [DOI] [PubMed] [Google Scholar]
- 13.Pitcher R. S., Brissett N. C., Doherty A. J. (2007) Annu. Rev. Microbiol. 61, 259–282 [DOI] [PubMed] [Google Scholar]
- 14.Ghosh J., Larsson P., Singh B., Pettersson B. M., Islam N. M., Sarkar S. N., Dasgupta S., Kirsebom L. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10781–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S. T., Setlow B., Conlon E. M., Lyon J. L., Imamura D., Sato T., Setlow P., Losick R., Eichenberger P. (2006) J. Mol. Biol. 358, 16–37 [DOI] [PubMed] [Google Scholar]
- 16.Moeller R., Stackebrandt E., Reitz G., Berger T., Rettberg P., Doherty A. J., Horneck G., Nicholson W. L. (2007) J. Bacteriol. 189, 3306–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cromie G. A. (2009) J. Bacteriol. 191, 5076–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singleton M. R., Dillingham M. S., Gaudier M., Kowalczykowski S. C., Wigley D. B. (2004) Nature 432, 187–193 [DOI] [PubMed] [Google Scholar]
- 19.Dillingham M. S., Kowalczykowski S. C. (2008) Microbiol. Mol. Biol. Rev. 72, 642–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haijema B. J., Venema G., Kooistra J. (1996) J. Bacteriol. 178, 5086–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haijema B. J., Meima R., Kooistra J., Venema G. (1996) J. Bacteriol. 178, 5130–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeeles J. T., Dillingham M. S. (2007) J. Mol. Biol. 371, 66–78 [DOI] [PubMed] [Google Scholar]
- 23.Sinha K. M., Unciuleac M. C., Glickman M. S., Shuman S. (2009) Genes Dev. 23, 1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dillingham M. S., Spies M., Kowalczykowski S. C. (2003) Nature 423, 893–897 [DOI] [PubMed] [Google Scholar]
- 25.Taylor A. F., Smith G. R. (2003) Nature 423, 889–893 [DOI] [PubMed] [Google Scholar]
- 26.Rigden D. J. (2005) BMC Struct. Biol. 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saikrishnan K., Griffiths S. P., Cook N., Court R., Wigley D. B. (2008) EMBO J. 27, 2222–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saikrishnan K., Powell B., Cook N. J., Webb M. R., Wigley D. B. (2009) Cell 137, 849–859 [DOI] [PubMed] [Google Scholar]
- 29.Taylor A. F., Smith G. R. (1995) J. Biol. Chem. 270, 24451–24458 [DOI] [PubMed] [Google Scholar]
- 30.Farah J. A., Smith G. R. (1997) J. Mol. Biol. 272, 699–715 [DOI] [PubMed] [Google Scholar]
- 31.Sinha K. M., Stephanou N. C., Gao F., Glickman M. S., Shuman S. (2007) J. Biol. Chem. 282, 15114–15125 [DOI] [PubMed] [Google Scholar]
- 32.Roman L. J., Kowalczykowski S. C. (1989) Biochemistry 28, 2873–2881 [DOI] [PubMed] [Google Scholar]
- 33.Dillingham M. S., Webb M. R., Kowalczykowski S. C. (2005) J. Biol. Chem. 280, 37069–37077 [DOI] [PubMed] [Google Scholar]
- 34.Spies M., Dillingham M. S., Kowalczykowski S. C. (2005) J. Biol. Chem. 280, 37078–37087 [DOI] [PubMed] [Google Scholar]
- 35.Morris P. D., Raney K. D. (1999) Biochemistry 38, 5164–5171 [DOI] [PubMed] [Google Scholar]
- 36.Morris P. D., Byrd A. K., Tackett A. J., Cameron C. E., Tanega P., Ott R., Fanning E., Raney K. D. (2002) Biochemistry 41, 2372–2378 [DOI] [PubMed] [Google Scholar]
- 37.Byrd A. K., Raney K. D. (2004) Nat. Struct. Mol. Biol. 11, 531–538 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.