Abstract
Changes in membrane morphology and membrane protein dynamics based on its fluidity are critical for cancer metastasis. However, this subject has remained unclear, because the spatial precision of previous in vivo imaging has been limited to the micrometer level and single molecule imaging is impossible. Here, we have imaged the membrane dynamics of tumor cells in mice with a spatial precision of 7–9 nm under a confocal microscope. A metastasis-promoting factor on the cell membrane, protease-activated receptor 1 (PAR1), was labeled with quantum dots conjugated with an anti-PAR1 antibody. Movements of cancer cells and PAR1 during metastasis were clearly observed in vivo. Images used to assess PAR1 dynamics were taken of representative cells for four stages of metastasis; i.e. cancer cells far from blood vessels in tumor, near the vessel, in the bloodstream, and adherent to the inner vascular surface in the normal tissues near tumor were photographed. The diffusion constant of PAR1 in static cells far from tumor blood vessels was smaller than in moving cells near the vessels and in the bloodstream. The diffusion constant of cells adhering to the inner vascular surface in the normal tissues was also very small. Cells formed membrane protrusion during migration. The PAR1 diffusion constant on these pseudopodia was greater than in other membrane regions in the same cell. Thus, the dynamics of PAR1 movement showed that membrane fluidity increases during intravasation, reaches a peak in the vessel, decreases during extravasation, and is also higher at locally formed pseudopodia.
Keywords: Antibodies/Monoclonal, Cancer, Cell/Migration, Methods/Microscopic Imaging, Receptors/Membrane, Tumor/Metastases, Nanotechnology, Quantum Dots
Introduction
During metastasis, cancer cells detach from the parent tumor, invade surrounding connective tissue and blood vessels, are transported in the bloodstream, and invade other organs after extravasation (1, 2). Membrane dynamics are significantly altered in metastatic cancer cells (3). Many studies using cultured cells have suggested that metastatic cancer cells form pseudopodia termed filopodia, lamellipodia, and invadopodia; this process is driven by actin polymerization in the direction of cellular migration and invasion (4–7). Additionally, greater membrane fluidity is thought to enhance the malignancy of cultured cancer cells (8, 9). High membrane fluidity is coupled to increased diffusion speed of membrane proteins. Greater diffusion speed accelerates the reaction rate between receptors and their ligands or adhesion proteins and their extracellular substratums. In this way, the metastatic ability of cancer cells is activated (9). Thus, to elucidate the mechanisms of cancer metastasis, analysis of membrane protein dynamics during metastasis is crucial. In living tumors in vivo, there are blood vessels and three-dimensional communication systems between cells, unlike in cultures in vitro. It is therefore essential that in vivo membrane morphology and fluidity based on membrane protein dynamics are clarified.
Previous studies used imaging of GFP3- or luciferase-expressing cancer cells in vivo to examine the behavior of metastatic cancer cells (10–14). However, because the spatial precision of such imaging is limited to the micrometer level and single molecule imaging is impossible, the details of in vivo dynamics of individual membrane proteins remain unknown. We have been developing single molecule imaging using fluorescent molecules and quantum dots (QDs) with 1 nm precision in vitro and have elucidated the molecular mechanisms of motor proteins, myosin, kinesin, and dynein (15–18). By applying this imaging method in vivo, our previous studies (19, 20) succeeded in tracking an anti-HER2 (human epidermal growth factor receptor 2) monoclonal antibody conjugated with bright QDs (21, 22) in vivo with a spatial precision of 30 nm. However, the size of a typical protein ranges from several nanometers to 20 nm. Therefore, 30 nm precision is not suitable to understand the molecular function-associated dynamics of proteins.
Here, we have further developed a method to image a tumor cell membrane protein with antibody-conjugated QDs. We used this technique to visualize the details of membrane fluidity and morphology during metastasis in living mice with a spatial precision of 7–9 nm under a Nipkow disk confocal microscope. This new nanotechnology would enable us to understand the functional dynamics of proteins and nanometer-scale anticancer agents in vivo.
EXPERIMENTAL PROCEDURES
PAR1 Antibody
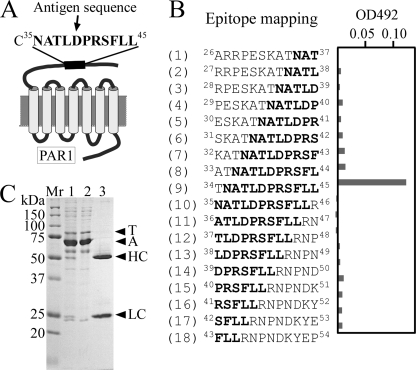
The oligopeptide CNATLDPRSFLL, including a sequence from Asn35 to Leu45 of PAR1, was cross-linked with keyhole limpet hemocyanine, and the cross-linked oligopeptide was used as an antigen to generate an anti-human PAR1 monoclonal antibody. Epitope mapping for the antibody was performed by enzyme-linked immunosorbent assay utilizing the culture supernatant of hybridoma cells for the primary antibody and a horseradish peroxidase-conjugated anti-mouse IgG antibody (Upstate) as the secondary antibody. The absorbance was read at 492 nm to detect substrate reactivity by the horseradish peroxidase-conjugated antibody. To obtain high purity anti-PAR1 antibody, the cloned hybridoma cells were injected into the abdominal cavity of a severe combined immunodeficient (SCID) mouse (Charles River), which is an immunodeficient mouse without immunoglobulins, and ascites were prepared from the mouse. Anti-PAR1 antibody was purified from the ascites using protein G-Sepharose (Amersham Biosciences), and the purified antibody was applied to a preparation of anti-PAR1 antibody-conjugated QDs (anti-PAR1-QDs) using a Qdot® 705 Antibody Conjugation Kit (Invitrogen), where the number indicates the emission wavelength.
DNA Constructs
To make a construct of the PAR1-GFP gene that would be stably expressed in cultured cells, the open reading frame of human PAR1 cDNA was cloned into the HindIII and BamHI sites of the pEGFP vector (Clontech). The PAR1-GFP gene was excised from the plasmid at the HindIII and NotI sites and cloned into the HindIII and NotI sites of the pLNCX2 retroviral vector (BD Bioscience). The sequences of these DNA constructs were checked according to the ABI DNA sequencing system protocol.
Cell Culture
The human KPL-4 (KPL) breast cancer cell line (23), non-metastatic in SCID mice, was kindly provided by Dr. J. Kurebayashi (Kawasaki Medical School, Japan). The KPL cells were transformed into the metastatic cancer cell PAR1-GFP-expressing KPL (PAR1-KPL) by transduction with the pLNCX2 retroviral vector system containing the PAR1-GFP gene as the insert, and the cells were then cloned. KPL and PAR1-KPL cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum. PAR1-KPL cells were grown in the presence of 400 μg/ml G418.
Optical System with Confocal Microscope
The optical system for observation of the fluorescence of GFP or QDs consisted primarily of an epifluorescence microscope (IX-71, Olympus) with modifications, a Nipkow disk-type confocal unit (CSU10, Yokogawa), and an electron multiplier type charge-coupled device camera (EM-CCD, Ixon DV887, Andor Technology). A UPlanFL N (×100, 1.30 numerical aperture, Olympus) objective lens was used for in vitro imaging and a PlanApo (×60, 1.40 numerical aperture, Olympus) objective lens was used for in vivo imaging. GFP was illuminated by a blue laser (488 nm wavelength, Furukawa Electric), and QDs were illuminated by a green laser (532 nm wavelength, CrystaLaser). The laser-excited fluorescence was filtered with a 500–550 nm bandpass filter for GFP, a 685–725 nm bandpass filter for QDs, and a >580 nm long-pass filter for imaging QDs and autofluorescence of red blood cells. Images were taken at a rate of 5–10 frames per second. For in vivo imaging, to remove the oscillation of heartbeat and respiration in observations, an aluminum stage was developed for this study and attached to the above microscopy system.
In Vitro Imaging
To investigate the specificity of the PAR1 antibody, KPL and PAR1-KPL cells were mixed with 40 nm anti-PAR1-QDs in serum-free L-15 medium (Invitrogen) for 30 min at 37 °C. After washing with L-15 medium, these cells were incubated with L-15 containing 0.5% fetal bovine serum in a glass-bottomed dish and then observed. The captured images were converted to autovideo interleaving files, and fluorescence intensities of QDs in the files were calculated as gray values using ImageJ software. To track PAR1 movements, PAR1-KPL cells were mixed with 2.5 nm anti-PAR1-QDs in serum-free L-15 medium for 30 min at 37 °C. After washing with L-15, these cells were incubated with L-15 containing 0.5% fetal bovine serum in a glass-bottomed dish and then observed. The position of QDs on the cell membrane was tracked using a previously described single molecule tracking method (18).
In Vivo Imaging
PAR1-KPL cells (1 × 106) were suspended in 100 μl of L-15 medium containing 10% fetal bovine serum and transplanted subcutaneously into the skin of female SCID mice at 5–7 weeks of age. 5–10 weeks after transplantation, anti-PAR1-QDs were injected into the tail vein of the mice. The probe concentration in the blood was 5 nm. This concentration does not inhibit migration and invasion of PAR1-KPL cells in vitro. The mice labeled with the probe were placed under anesthesia with ketamine and xylazine, and the anesthetized condition was maintained for the course of the imaging session. Minimal surgery was performed to expose the living tumor by removing the skin with as little damage to the surrounding blood vessels as possible. The stripped tumor was turned over and fixed with thread. By this surgical procedure, the area near the large vessel in the lower section of the tumor was exposed. A polyvinyl chloride plate (0.5 mm thickness) containing a small window (10 mm × 10 mm) was mounted on the exposed tumor, and then the skin around the tumor was bonded to the plate with Aronalpha instant adhesive (Toagosei Co. Ltd.). Our previous study used a suture thread to connect the polyvinyl chloride plate to the skin around tumor (20). In this study, use of Aronalpha made both connections stiffer without damaging the tumor cells, allowing removal of oscillations derived from the heartbeat and respiration during observation. Tumor-bearing mice with a mounted plate were fixed to a handmade aluminum stage designed to stabilize the plate with screws. The position of QDs on the tumor cell membrane was tracked using a previously described single molecule tracking method (18). For cells in vessels, the trajectories of QDs were calculated from the position of the QD relative to that of the cell. Animals were used in accordance with guidelines approved by the committee on animal experiments of Tohoku University.
Mean Square Displacement Analysis of QD Movement
To investigate the dynamic behavior of QDs, mean square displacement (MSD) analysis was carried out as previously described (24, 25). When MSD is fitted against time, the linear plot of MSD produced is assumed to represent random Brownian movement.
Labeling of Blood Cells in Mice with Anti-PAR1-QDs
Anti-PAR1-QDs were injected into the tail vein of female SCID mice to test whether QDs would bind to blood cells. The probe concentration in the blood was 5 nm. 2 h after probe injection, blood was taken from the heart under anesthesia with ketamine and xylazine. The blood cells were separated into two fractions, whole blood cells, and leukocytes and then examined. Blood cells were collected from whole blood by centrifugation at 400 × g for 5 min, washed with phosphate-buffered saline ten times, and observed in a glass-bottomed dish with the same optical system used for in vivo imaging. To isolate leukocytes, blood cells were washed with phosphate-buffered saline five times and then treated with Red Blood Cell Lysing Buffer (Sigma) according to the manufacturer's instructions. The treatments were repeated 3–5 times and observed as in whole blood cells.
RESULTS
Development of in Vivo Nano-imaging Method
To image the membrane dynamics of metastatic cancer cells with a high spatial precision, we improved three points in our previous imaging method: the targeted protein, the type of cancer cell, and the method of fixing tumors on a microscope stage. First, a metastasis-promoting factor on the cell membrane, protease-activated receptor 1 (PAR1), was targeted. PAR1 is a G-protein-coupled receptor that plays a critical role in metastatic processes of various cancers of the breast, colon, lung, pancreas, and prostate (26–28). PAR1 expression correlated strongly with cellular metastatic capability in breast cancer (29), whereas HER2 did not. Matrix metalloprotease 1 activates PAR1 by cleaving its exodomain at the Arg41–Ser42 peptide bond, and this activation promotes cell migration and invasion. We prepared anti-PAR1 antibody that specifically binds to a human PAR1 sequence, 35NATLDPRSFLL45 (Fig. 1, A–C), which differs from mouse PAR1 by three amino acids (letters in italic), 35DATVNPRSFLL45 (30).
FIGURE 1.
Preparation and characterization of the anti-PAR1 monoclonal antibody. A, frame format of molecular structure of PAR1. Matrix metalloprotease 1 activates PAR1 by cleaving its exodomain at the Arg41–Ser42 peptide bond. This activation promotes PAR1-dependent Ca2+ signaling following cancer cell migration and invasion. The peptide CNATLDPRSFLL, including a sequence from Asn35 to Leu45 of PAR1, was synthesized and used as an antigen to make an anti-PAR1 antibody capable of functioning as both a cancer cell tracer and a PAR1-inhibitor (anticancer agent). B, epitope mapping for the PAR1 antibody. (1)-(18) represent peptides of 12 amino acids, including part or all of the 35NATLDPRSFLL45 sequence, were used for epitope mapping by enzyme-linked immunosorbent assay. OD492 indicates the absorbance of peroxidase-conjugated antibodies at 492 nm. The PAR1 antibody specifically reacted with the ninth peptide, TNATLDPRSFLL, but not with the tenth peptide, NATLDPRSFLLR, despite the fact that both peptides included the same NATLDPRSFLL sequence. The reason for this might be that an amino group from Asn in the tenth peptide inhibited the binding of the anti-PAR1 antibody because another peptide, Cys-bound NATLDPRSFLL, was used for the antigen (A). Additionally, Arg46 in the tenth peptide might inhibit the binding of antibody and peptide. The results indicate that the epitope for the antibody is 35NATLDPRSFLL45. C, purification of PAR1 antibody. To obtain high purity PAR1 antibody, the PAR1 antibody was purified from the ascites with protein G beads and checked using 10% SDS-PAGE. Lane 1 shows the pattern of a crude sample of the ascites. Lanes 2 and 3 show the patterns of samples unbound by Protein G and released from Protein G after binding, respectively. T, A, HC, and LC show the transferrin, albumin, heavy chain of IgG, and light chain of IgG, respectively. The sample of purified PAR1 antibody contained no transferrin or albumin (lane 3).
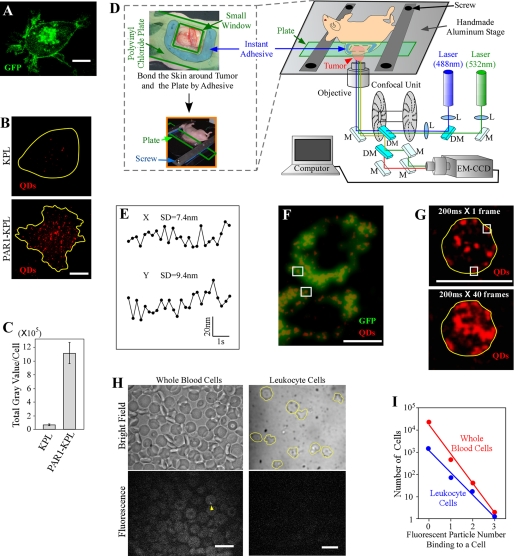
In our previous imaging studies, we prepared tumor-bearing mice by transplantation of human KPL-4 (KPL) breast cancer cells (23), which express HER2 at a high level but PAR1 at low levels. Thus, KPL cells are non-metastatic in mice. To examine the metastatic process, KPL cells were transformed into a PAR1-GFP-expressing KPL (PAR1-KPL) cell line by transduction of a PAR1-GFP gene (Fig. 2A). PAR1 expression induced metastasis of KPL cells (supplemental Fig. S1). To examine the specificity of anti-PAR1 antibodies, we performed immunostaining with anti-PAR1 antibody-conjugated QDs (anti-PAR1-QDs) in vitro. Anti-PAR1-QDs specifically reacted to human PAR1 on the cell membranes of PAR1-KPL cells, whereas its fluorescence intensity on KPL cells was only 1/17 of that on PAR1-KPL (Fig. 2, B and C). PAR1 antibody at a concentration of 10 nm did not prevent the endocytosis of PAR1 (supplemental Fig. S2) and the metastatic activity of PAR1-KPL cells activated by matrix metalloprotease 1. At a high concentration of 10 μm, antibody decreased the activity to ∼40% (data not shown). These results demonstrate that the anti-PAR1 antibody could function as a specific tracer against PAR1-expressing metastatic cancer cells and an effective anticancer agent.
FIGURE 2.
Materials and methods for in vivo nano-imaging. A, fluorescent image of PAR1-KPL cells. Fluorescence of PAR1-GFP was clearly observed in the cell membrane and endocytotic vesicles in PAR1-KPL cells (excitation, 488 nm; emission, 500–550 nm; and exposure time, 3 s). B, imaging of KPL and PAR1-KPL cells with anti-PAR1-QDs (40 nm). QDs on the cell membrane located near the glass surface were visualized in vitro. Yellow lines show the cellular outline delineated using phase-contrast images. Excitation, 532 nm; emission, 685–725 nm; and exposure time, 0.1 s. C, fluorescence intensity of QDs measured on the cell membrane of KPL and PAR1-KPL cells in vitro. Error bars indicate ±S.E. KPL cells, n = 13. PAR1-KPL cells, n = 11. D, schematic drawing of in vivo nano-imaging. Tumors were stripped, turned over, and fixed with thread (see supplemental Fig. S2). A polyvinyl chloride plate containing a small window (10 mm × 10 mm) was mounted on the exposed tumor, and then the skin around the tumor and the plate were bonded by instant adhesive. The plate was fixed to a handmade aluminum stage designed to stabilize the plate with screws. In the optical schematic for in vivo nano-imaging, L, M, and DM show the lens, mirror, and dichroic mirror, respectively. An example of a tumor-bearing nude mouse is provided. E, spatial precision of in vivo nano-imaging. Immobile QDs were tracked in tumor stroma. The standard deviation of the position of QDs was 7.4 nm in the x-axis and 9.4 nm in the y-axis. F, fluorescent imaging of PAR1-GFP and anti-PAR1-QDs on the edge of tumor cells in mice. Fluorescence of anti-PAR1-QDs was compared with that of PAR1-GFP on tumor cell membrane. Only the QDs that colocalized with PAR1-GFP on the cell edge (white squares) were analyzed. The laser-excited fluorescence of PAR1-GFP and anti-PAR1-QDs were filtered with a 500–550 nm bandpass filter and a 685–725 nm bandpass filter, respectively. G, determination of the cellular outline by superimposed images of anti-PAR1-QDs fluorescence on tumor cells in mice. An outline of a cell (yellow lines) determined by superimposed images (bottom panel) was applied to a cell image in a single frame (top panel). QDs on cell edges (white squares) were analyzed. H, labeling of blood cells in SCID mice with anti-PAR1-QDs. Bright field and fluorescent images of whole blood cells and leukocyte cells are shown. In whole blood cells, most cells were red blood cells that were autofluorescent. Yellow lines show the outline of leukocytes. Although there was low level binding of a few QDs per cell (yellow arrowhead), high affinity binding of QDs as seen in Figs. 3 and 4 was not observed. Excitation, 532 nm; emission, >580 nm; and exposure time, 0.2 s. I, graphic representation of fluorescent images of whole blood cells (red symbol) and leukocyte cells (blue symbol). The number of the cells binding QDs was fitted to an exponential function. Approximation formulas for the fitting were 16,000 × e−3x (red symbols) and 1,100 × e−2.3x (blue symbols), where x is the number of attached particles. Bars, 10 μm.
Exposed mouse tumors (supplemental Fig. S3, A–D) were bonded on a polyvinyl chloride plate containing a small window using Aronalpha instant adhesive, which features low osmolarity and fluidity, without damaging tumor cells (Fig. 2D). Aronalpha minimized oscillations derived from heartbeat and respiration, in contrast to larger oscillations observed in the previous method using thread (20). The plate was fixed tightly to a handmade aluminum stage using screws (Fig. 2D). After injection of QDs into the tail vein of tumor-bearing mice, QDs bound to the tumor stroma were imaged under a confocal microscope (Fig. 2D). The center of the QD image was calculated by fitting the fluorescence intensity profiles of QDs to two-dimensional Gaussian curves (18). We measured values of ∼7 nm for the X-axis and ∼9 nm for the Y-axis (Fig. 2E), indicating that we can track the movement of PAR1 labeled with anti-PAR1-QDs with a spatial precision of 7–9 nm.
To confirm whether the QDs bound to the surface of targeted tumor cells, two methods were employed after injection of anti-PAR1-QDs into tumor-bearing SCID mice. First, the fluorescence positions of anti-PAR1-QDs were compared with those of PAR1-GFP on tumor cell membranes. We analyzed the QDs that colocalized with PAR1-GFP on cell membrane (Fig. 2F). The second method involved superimposed imaging of anti-PAR1-QD fluorescence over 40 frames, showing the position of PAR1 on the cell membrane. Membrane proteins move randomly along the cell membrane, and the traces of QDs show the outline of the membrane (Fig. 2G).
We confirmed that anti-PAR1-QDs labeled targeted PAR1-KPL cells but not the blood cells in SCID mice. We injected anti-PAR1-QDs into the tail vein of mice that were not transplanted with PAR1-KPL cells and examined the QDs bound to blood cells. Most of the cells (>99.99% for whole blood cells and >99.9% for leukocytes) were labeled with three or fewer QDs (Fig. 2, H and I), but many anti-PAR1-QDs (typically >10 QDs) bound to PAR1-KPL cells (Figs. 3 and 4). The probability of a cell being bound by more than ten QDs is extremely low. Therefore, in tumor-bearing mice, cells labeled with dozens of QDs represent metastatic cancer cells.
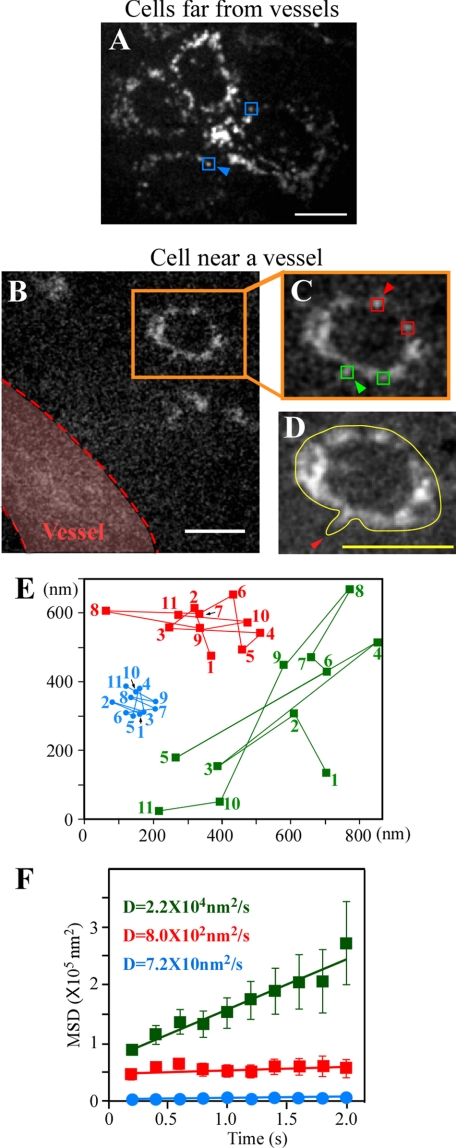
FIGURE 3.
Membrane dynamics in metastatic cancer cells outside of vessels. A, QD fluorescent image of cells far from tumor vessels. The PAR1 diffusion constant as represented by blue squares was analyzed in E and F. B, QD fluorescent image of a cell near a vessel. Orange squares show a cancer cell forming membrane protrusions in the direction of the vessel. Red dotted lines represent the outline of a vessel determined by superimposed images of auto-fluorescence of blood cells. C, magnified view of B. QDs on the membrane facing the vessel, represented by green squares, and QDs on the membrane on the opposite side of the cell, represented by red squares, were differentially selected and analyzed in E and F. D, cell image superimposed for 3.6 s. The yellow line shows an outline of the cell. The red arrowhead indicates a clearly imaged membrane protrusion. E, traces of blue, orange, and green squares as shown with arrowheads in A and C. Numbers show the tracking order. F, MSD plots of QDs on membrane in A (blue), on the membrane facing the vessel in C (green), and on the membrane in the tail of the cell in C (red). D, diffusion constant. Error bars indicate ±S.E. Blue data, n = 24 (6 trajectories/cell × 4 cells). Red data, n = 39 (13 trajectories/cell × 3 cells). Green data, n = 39 (13 trajectories/cell × 3 cells). Squares in A and C show typical QDs on the edges of cells. Excitation, 532 nm; emission, >580 nm. The exposure time in A–C was 0.2 s. Bars, 10 μm.
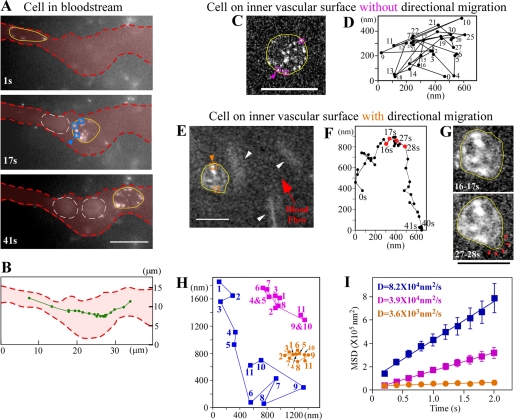
FIGURE 4.
Membrane dynamics in metastatic cancer cells in vessels. A, imaging of cells in the bloodstream. Cells are shown after 1 s, 17 s, and 41 s. Yellow lines show outlines of cancer cells. Red dotted lines show outlines of vessels determined by superimposed images of autofluorescent blood cells. White dotted lines indicate outlines of red blood cells. B, trajectory of the barycentric position of the cell in A at every 2 s (green line). C, fluorescent image of a cell adhering to the inner vascular surface without directional movement. The yellow line shows an outline of the cancer cell. D, trajectory of the barycentric position of the cell in C at every second. Numbers show the tracking order. E, imaging of directional cell migration on the inner vascular surface. The yellow line represents an outline of the cancer cell. White arrowheads show red blood cells with a comet-like configuration. F, trajectory of the barycentric position of the cell in E at every second. G, cells in E superimposed for 16–17 s and 27–28 s. Yellow lines show outlines of cancer cells. Red arrowheads represent lamellipodia-like structures. H, traces of blue, purple, and orange squares, as shown with arrowheads in A, C, and E. Numbers show the tracking order. I, MSD plots of QDs on membranes of cells in the bloodstream (blue), on the inner vascular surface without directional migration (purple), and on the inner vascular surface with migration (orange). D, diffusion constant. Error bars indicate ±S.E. Blue data, n = 88 (22 trajectories/cell × 4 cells). Purple data, n = 115 (23 trajectories/cell × 5 cells). Orange data, n = 78 (26 trajectories/cell × 3 cells). Squares in A, C, and E show typical QDs on the edge of cells. Excitation, 532 nm; emission, >580 nm; exposure time, 0.2 s. Bars, 10 μm.
Membrane Dynamics in Metastatic Cancer Cells Outside of Vessels
We observed the in vivo membrane dynamics of metastatic cancer cells in four regions: far from the blood vessel in tumors, near the vessel, in the bloodstream, and adherent to the inner vascular surface in normal tissues near tumors. These locations represent the process of cancer metastasis (Fig. 5). First, the cancer cells outside vessels, cells far from tumor blood vessels, and cells near the vessels were visualized. Anti-PAR1-QDs were bound to the surface of cells in colonies far from tumor blood vessels (Fig. 3A and supplemental video 1). In these cells, mobile edges, such as pseudopodia, were not seen, suggesting that the cells were static. In contrast, cancer cells near tumor blood vessels formed membrane protrusions oriented toward the vessel (Fig. 3, B–D, and supplemental video 2), although actual migration of the cells was not observed. To analyze the diffusional movement of anti-PAR1-QDs quantitatively, the MSD of the QD position was analyzed (24, 25). The MSD plots of anti-PAR1-QDs for cell membranes of cancer cells outside of vessels were fitted to linear functions representing the diffusion coefficient for random diffusion (Fig. 3F). The diffusion constant of PAR1 on cells far from tumor vessels (blue squares in Fig. 3A) was low, at 72 nm2/s (Fig. 3, E and F). This low diffusion constant was not due to systematic noise, because the spatial precision of imaging was ∼7 nm for the x-axis and ∼9 nm for the y-axis at an exposure time of 0.2 s (Fig. 2E), and their calculated diffusion constant, 10.4 nm2/s, was much smaller than that of PAR1. In cells near blood vessels, the QDs on the cell edge facing the vessel (green symbols in Fig. 3, C and E) moved farther along the membrane than those in the edge on the opposite side of the cell (red symbols in Fig. 3, C and E). The diffusion constant of PAR1 facing the vessel (2.2 × 104 nm2/s; green symbols in Fig. 3F), was 28-fold greater than that in the tail of the same cell (8.0 × 102 nm2/s; red symbols in Fig. 3F) and 300-fold greater than that of cells far from the vessel (72 nm2/s; blue symbols in Fig. 3F). These results suggest that the diffusion of PAR1 on vessel-facing membranes of cells near blood vessels might be accelerated by an attractant released from the vessel (31).
FIGURE 5.
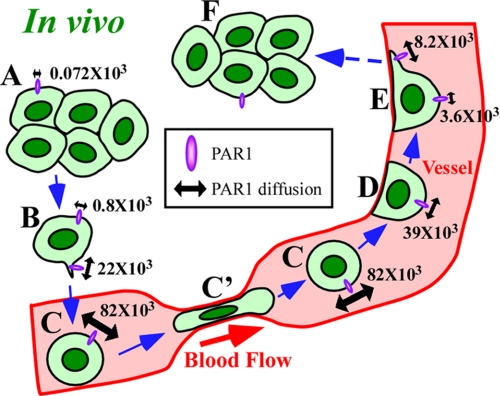
A model for membrane dynamics in metastatic cancer cells. A–F, cells in vivo during metastasis. A, cells far from vessels. B, cell near vessels. C, cell in the bloodstream. C′, cells in narrow vessels. D, cells adhering to the inner vascular surface without directional migration. E, cells migrating directionally on the surface. F, cells after extravasation. Numerical values show the diffusion (nm2/s) of PAR1 labeled with anti-PAR1-QDs. See text for details.
Membrane Dynamics in Metastatic Cancer Cells in Vessels
Next, we visualized cancer cells in vessels, cells in the bloodstream within tumors, and cells adhering to the inner vascular surface in normal tissues near tumors. The cells in the bloodstream moved at a non-uniform velocity (Fig. 4, A and B, and supplemental video 3). The round shape of the cells flowing in a wide vessel changed to a long and thin shape when the cells passed through a narrow region (∼2.5 μm wide) (top panel in Fig. 4A and supplemental Fig. S4). Fig. 4C shows a cell adhering to the inner vascular surface in normal tissues near the tumor (supplemental video 4). The barycentric position of the cell moved randomly without directional migration (Fig. 4D). After adhesion, cancer cells moved with a speed of ∼70 nm/s in the forward (Fig. 4E and during 0–15 s in Fig. 4F) and backward directions (Fig. 4E and during 27–39 s in Fig. 4F) relative to the direction of blood flow (supplemental video 5). We imaged cells on the inner vascular surface; these cells formed a lamellipodia-like structure in the direction of migration (27–28 s in Fig. 4G). In vessels, the trajectories of PAR1 molecules in cells were calculated from the position of the QDs relative to that of the centroid of the cell outlines (Fig. 4H), because the cells were mobile (Fig. 4, B, D, and F). The relative positions of QDs on the cell membrane in the bloodstream moved along the cell edge (Fig. 4, A and H), and the diffusion constant was 8.2 × 104 nm2/s, which was 1100-fold greater than that observed for cells far from a tumor vessel (Fig. 3F and blue symbols in Fig. 4I). In the cells adhering to the inner vascular surface without directional movement, the QDs moved along the cell edge, and the diffusion constant of PAR1 was 3.9 × 104 nm2/s; this was 2-fold lower than in the bloodstream (purple symbols in Fig. 4, C, H, and I). Moreover, in the cells on the inner vascular surface showing directional migration, the diffusion constant of PAR1 (3.6 × 103 nm2/s) was 23-fold less than that in cells in the bloodstream (orange symbols in Fig. 4, E, H, and I). In addition, the PAR1 diffusion constant on the lamellipodia (8.2 × 103 nm2/s) was 2-fold greater than that of other regions in the migrating cells.
DISCUSSION
PAR1 diffusion at the mobile edge was greater than at the immobile edge in vivo (Fig. 5). As membrane protein movement is coupled with actin filament density underneath the cell membrane (24, 32), membrane fluidity and actin dynamics might synergistically activate local pseudopodial formation. In mice, cancer cells in colonies far from tumor blood vessels did not show active membrane dynamics (Fig. 5A). This might have been due to low local concentrations of signaling factors derived from blood components (31). The cells near blood vessels are likely attracted to the vessels by signaling factors, resulting in the formation of membrane protrusions (Fig. 5B). The concentration gradient of such signaling factors through the extracellular matrix might also result in increased membrane fluidity and actin dynamics within individual cells on sides facing the blood vessel (31); such directionality might permit the cells to extend membrane protrusion and migrate in the direction of the vessel. These protrusions might be a kind of invadopodia. In the bloodstream, the membrane fluidity of cells was 1100-fold higher than that of cells far from vessels (Fig. 5C). The high membrane fluidity was observed similarly in both wide and narrow vessels. This increased diffusion was likely caused by the existence of fewer actin filaments underneath the cell membrane due to a lack of adhesion between cells and the extracellular substratum (33, 34). This idea is supported by the observation that these cells can use their membrane flexibility to change their shape to pass through narrow vessels (Fig. 5C′) (35). Additionally, greater diffusion of membrane protein in vessels might enhance cancer metastasis to another organ by accelerating the reaction rate between receptors and their ligands or adhesion proteins and their extracellular substratums. After adhering to the inner vascular surface, diffusion decreased (Fig. 5D). The reformation of the actin cytoskeleton underneath the cell membrane likely slowed the diffusion of PAR1. Cells subsequently moved slowly and directionally on the inner vascular surface (Fig. 5E). The network of actin filaments might have become more developed to permit directional cellular migration (3, 4, 6). The membrane fluidity of the lamellipodia-like structure was 2-fold higher than that of other regions (Fig. 5E). Similar results were also obtained in cultured cells. The diffusion constant of QDs on mobile lamellipodia (1.2 × 105 nm2/s) in vitro was 5-fold greater than that on immobile cell edges (2.2 × 104 nm2/s) (supplemental Fig. S5, A–D). These results suggest that the difference in membrane fluidity between the mobile and immobile edge is important for directional migration. Additionally, it was previously reported that the diffusion constant of transferrin receptor without GFP fusion on the cultured cell membrane was ∼105 nm2/s (36), which is very similar to the diffusion constant of PAR1-GFP, suggesting that GFP does not interfere the diffusion of PAR1.
In conclusion, the membrane fluidity of metastasizing tumor cells increases at intravasation, peaks in blood vessels, decreases at extravasation, and is higher in locally formed pseudopodia. Such dramatic changes in membrane fluidity and morphology can enable cancer cells to metastasize. This new method of imaging protein dynamics with high spatial precision could make useful contributions to understanding mechanisms of cancer progression as well as the antitumor effects of nanometer-scale anticancer agents in vivo (37, 38).
Supplementary Material
Acknowledgments
We thank T. Kambara, T. Shimozawa, M. Kaya, H. Tada, M. Kawai, and M. Takeda for helpful discussion and S. Inomata and S. Higuchi for technical assistance.
This work was supported by Grants-in-Aid for Scientific Research in Priority Areas from the Japan MEXT (to H. H. and K. G.), a Grants-in-aid for Research Project, Promotion of Advanced Medical Technology (H18-Nano-001) from the Ministry of Health, Labor, and Welfare of Japan (to N. O.), Special Coordination Funds for Promoting Science and Technology of JST (to K. G., T. M. W., and H. H.), Research for Promoting Technological Seeds of JST (to K. G.), and CREST of JST (to H. H.). We also acknowledge the support of the Biomedical Research Core of Tohoku University Graduate School of Medicine, the Tohoku University Global COE Program “Global Nano-Biomedical Engineering Education and Research Network Center,” and Konica Minolta Medical & Graphic, Inc.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Videos 1–5.
- GFP
- green fluorescent protein
- anti-PAR1-QDs
- anti-PAR1 antibody-conjugated QDs
- HER2
- human epidermal growth factor receptor 2
- IgG
- immunoglobulin G
- MSD
- mean square displacement
- PAR1
- protease-activated receptor 1
- PAR1-KPL
- PAR1-GFP-expressing KPL
- QD
- quantum dot
- SCID
- severe combined immunodeficient.
REFERENCES
- 1.Chambers A. F., Groom A. C., MacDonald I. C. (2002) Nat. Rev. Cancer 2, 563–572 [DOI] [PubMed] [Google Scholar]
- 2.Schedin P. (2006) Nat. Rev. Cancer 6, 281–291 [DOI] [PubMed] [Google Scholar]
- 3.Sahai E. (2007) Nat. Rev. Cancer 7, 737–749 [DOI] [PubMed] [Google Scholar]
- 4.Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., Condeelis J. (2005) J. Cell Biol. 168, 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattila P. K., Lappalainen P. (2008) Nat. Rev. Mol. Cell Biol. 9, 446–454 [DOI] [PubMed] [Google Scholar]
- 7.Sarmiento C., Wang W., Dovas A., Yamaguchi H., Sidani M., El-Sibai M., Desmarais V., Holman H. A., Kitchen S., Backer J. M., Alberts A., Condeelis J. (2008) J. Cell Biol. 180, 1245–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maxfield F. R., Tabas I. (2005) Nature 438, 612–621 [DOI] [PubMed] [Google Scholar]
- 9.Zeisig R., Koklic T., Wiesner B., Fichtner I., Sentjurc M. (2007) Arch. Biochem. Biophys. 459, 98–106 [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Wyckoff J. B., Frohlich V. C., Oleynikov Y., Hüttelmaier S., Zavadil J., Cermak L., Bottinger E. P., Singer R. H., White J. G., Segall J. E., Condeelis J. S. (2002) Cancer Res. 62, 6278–6288 [PubMed] [Google Scholar]
- 11.Condeelis J., Segall J. E. (2003) Nat. Rev. Cancer 3, 921–930 [DOI] [PubMed] [Google Scholar]
- 12.Hoffman R. M. (2005) Nat. Rev. Cancer 5, 796–806 [DOI] [PubMed] [Google Scholar]
- 13.Sahai E., Wyckoff J., Philippar U., Segall J. E., Gertler F., Condeelis J. (2005) BMC Biotechnol. 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins D. E., Hornig Y. S., Oei Y., Dusich J., Purchio T. (2005) Breast Cancer Res. 7, R444–R454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishijima A., Kojima H., Funatsu T., Tokunaga M., Higuchi H., Tanaka H., Yanagida T. (1998) Cell 92, 161–171 [DOI] [PubMed] [Google Scholar]
- 16.Nguyen V. T., Kamio Y., Higuchi H. (2003) EMBO J. 22, 4968–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toba S., Watanabe T. M., Yamaguchi-Okimoto L., Toyoshima Y. Y., Higuchi H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5741–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe T. M., Higuchi H. (2007) Biophys. J. 92, 4109–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li-Shishido S., Watanabe T. M., Tada H., Higuchi H., Ohuchi N. (2006) Biochem. Biophys. Res. Commun. 351, 7–13 [DOI] [PubMed] [Google Scholar]
- 20.Tada H., Higuchi H., Wanatabe T. M., Ohuchi N. (2007) Cancer Res. 67, 1138–1144 [DOI] [PubMed] [Google Scholar]
- 21.Chan W. C., Nie S. (1998) Science 281, 2016–2018 [DOI] [PubMed] [Google Scholar]
- 22.Lidke D. S., Nagy P., Heintzmann R., Arndt-Jovin D. J., Post J. N., Grecco H. E., Jares-Erijman E. A., Jovin T. M. (2004) Nat. Biotechnol. 22, 198–203 [DOI] [PubMed] [Google Scholar]
- 23.Kurebayashi J., Otsuki T., Tang C. K., Kurosumi M., Yamamoto S., Tanaka K., Mochizuki M., Nakamura H., Sonoo H. (1999) Br. J. Cancer 79, 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R. S., Kondo J., Fujiwara T. (2005) Annu. Rev. Biophys. Biomol. Struct. 34, 351–378 [DOI] [PubMed] [Google Scholar]
- 25.Yoo J., Kambara T., Gonda K., Higuchi H. (2008) Exp. Cell Res. 314, 3563–3569 [DOI] [PubMed] [Google Scholar]
- 26.Even-Ram S., Uziely B., Cohen P., Grisaru-Granovsky S., Maoz M., Ginzburg Y., Reich R., Vlodavsky I., Bar-Shavit R. (1998) Nat. Med. 4, 909–914 [DOI] [PubMed] [Google Scholar]
- 27.Boire A., Covic L., Agarwal A., Jacques S., Sherifi S., Kuliopulos A. (2005) Cell 120, 303–313 [DOI] [PubMed] [Google Scholar]
- 28.Arora P., Ricks T. K., Trejo J. (2007) J. Cell Sci. 120, 921–928 [DOI] [PubMed] [Google Scholar]
- 29.Booden M. A., Eckert L. B., Der C. J., Trejo J. (2004) Mol. Cell. Biol. 24, 1990–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeley S., Covic L., Jacques S. L., Sudmeier J., Baleja J. D., Kuliopulos A. (2003) Chem. Biol. 10, 1033–1041 [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi H., Wyckoff J., Condeelis J. (2005) Curr. Opin. Cell Biol. 17, 559–564 [DOI] [PubMed] [Google Scholar]
- 32.Morone N., Fujiwara T., Murase K., Kasai R. S., Ike H., Yuasa S., Usukura J., Kusumi A. (2006) J. Cell Biol. 174, 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiger B., Bershadsky A., Pankov R., Yamada K. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 793–805 [DOI] [PubMed] [Google Scholar]
- 34.Guo W., Giancotti F. G. (2004) Nat. Rev. Mol. Cell Biol. 5, 816–826 [DOI] [PubMed] [Google Scholar]
- 35.Li Q. S., Lee G. Y., Ong C. N., Lim C. T. (2008) Biochem. Biophys. Res. Commun. 374, 609–613 [DOI] [PubMed] [Google Scholar]
- 36.Sako Y., Kusumi A. (1995) J. Cell Biol. 129, 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melero I., Hervas-Stubbs S., Glennie M., Pardoll D. M., Chen L. (2007) Nat. Rev. Cancer 7, 95–106 [DOI] [PubMed] [Google Scholar]
- 38.Davis M. E., Chen Z. G., Shin D. M. (2008) Nat. Rev. Drug Discov. 7, 771–782 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.