Abstract
Spermatogenesis, a fundamental process in the male reproductive system, requires a series of tightly controlled epigenetic and genetic events in germ cells ranging from spermatogonia to spermatozoa. Jmjd1a is a key epigenetic regulator expressed in the testis. It specifically demethylates mono- and di-methylated histone H3 lysine 9 (H3K9me1 and H3K9me2) but not tri-methylated H3K9 (H3K9me3). In this study, we generated a Jmjd1a antibody for immunohistochemistry and found Jmjd1a was specifically produced in pachytene and secondary spermatocytes. Disruption of the Jmjd1a gene in mice significantly increased H3K9me1 and H3K9me2 levels in pachytene spermatocytes and early elongating spermatids without affecting H3K9me3 levels. Concurrently, the levels of histone acetylation were decreased in Jmjd1a knock-out germ cells. This suggests Jmjd1a promotes transcriptional activation by lowering histone methylation and increasing histone acetylation. Interestingly, the altered histone modifications in Jmjd1a-deficient germ cells caused diminished cAMP-response element modulator (Crem) recruitment to chromatin and decreased expression of the Crem coactivator Act and their target genes Tnp1 (transition protein 1), Tnp2, Prm1 (protamine 1), and Prm2, all of which are essential for chromatin condensation in spermatids. In agreement with these findings, Jmjd1a deficiency caused extensive germ cell apoptosis and blocked spermatid elongation, resulting in severe oligozoospermia, small testes, and infertility in male mice. These results indicate that the Jmjd1a-controlled epigenetic histone modifications are crucial for Crem-regulated gene expression and spermatogenesis.
Keywords: Spermatogenesis, Gene Expression, Gene Knockout, Histone Modification, Gene Regulation, CREM, Histone Demethylation, Jmjd1a, Male Infertility, Testis
Introduction
Male reproductive function of mammals relies on normal spermatogenesis within the seminiferous epithelium of the testis (1). During spermatogenesis, the spermatogonia undergo mitosis and differentiate into spermatocytes. The primary spermatocytes process through preleptotene, leptotene, zygotene, pachytene, and diplotene meiotic steps to generate secondary spermatocytes. Subsequently, the secondary spermatocytes enter the spermatid stages and undergo dramatic morphological changes, finally differentiating into mature spermatozoa. During this last phase, acrosomes and tails form, chromatins condense, and spermatid heads elongate (1). Interestingly, in cross-sectioned seminiferous tubules, each type of germ cell appears developmentally synchronized, and a defined grouping of germ cell types at particular developmental steps is constantly associated. In the mouse seminiferous epithelium, 12 groups of cell associations or “stages” (I–XII) can be steadily recognized according to the criteria established previously (1). Many genes critical for spermatogenesis, such as cAMP-response element modulator (Crem),2 Tnp1, Tnp2, Prm, and Prm2, are expressed in a germ cell type-specific manner and/or seminiferous epithelial stage-specific manner (2, 3).
Spermiogenesis requires extensive chromatin condensation. Histone displacement, a process in which histones are initially replaced by Tnp1 and Tnp2 and subsequently by Prm1 and Prm2, is required for chromatin condensation. Genetic ablation of transition proteins or protamines causes defective spermiogenesis (4–7). The transcripts of these genes can be first detected in step 7 round spermatids in mice, and the transcriptional activation of these genes is mainly regulated by the transcription factor Crem (2, 3). Unlike its close family member cAMP-response element-binding protein, which is activated by phosphorylation, the transcriptional activity of Crem is mainly mediated by a coactivator, the activator of Crem in the testis (Act) (8). It has been demonstrated that the Crem-regulated gene expression pathway is essential for normal spermatogenesis (3, 9–11).
Although the mechanisms responsible for histone displacement are unclear, histone variants and histone modifications may play important roles. During spermatogenesis, high levels of general and testis-specific histone variants are expressed and associated with DNA. This association alters nucleosome stability and facilitates histone displacement (12). Accordingly, knock-out of certain histone variants can arrest spermiogenesis at elongation steps and reduce male fertility (13–15). Correct epigenetic modifications of histones may also be important for regulating gene expression essential for spermatogenesis and spermatid chromatin condensation. It has been observed that highly acetylated histone H4 is present only in early elongating spermatids prior to histone displacement and positively correlates with histone condensation (16–18). Histone methylation is also tightly regulated during spermatogenesis, and distinct patterns of histone methylation have been observed at specific stages of spermatogenesis (19–21). In agreement with these observations, histone modification enzymes are robustly and specifically expressed in male germ cells (18, 20, 22–25). Although it is unclear how histone modifications are involved in the regulation of gene expression and histone displacement, it is conceivable that epigenetic modifications of histones may alter chromatin architecture and change gene expression profile required for normal spermatogenesis. Indeed, genetic ablation of specific histone modifiers such as Suv39h, G9a, or Meisetz has been shown to block spermatogenesis at early steps (19, 23, 26).
Jmjd1a, a histone demethylase, is preferentially expressed in the testis (27). Jmjd1a specifically demethylates mono- and di-methyl histone H3 lysine 9 (H3K9me1 and H3K9me2) but not tri-methyl H3 lysine 9 (H3K9me3) (28). Jmjd1a has been suggested as a coactivator for estrogen and androgen receptors (28, 29) and shown to regulate stem cell self-renewal, myocyte development, hypoxia-induced stress response, and energy metabolism (30–36).
In this study, we have generated a Jmjd1a antibody and examined Jmjd1a distribution in the testis. We have found that Jmjd1a is expressed in a germ cell type-specific and developmental step-specific manner. Furthermore, we have generated Jmjd1a knock-out mice and found that Jmjd1a deficiency results in severe oligozoospermia and male infertility. In addition, we also investigated the Jmjd1a-regulated epigenetic and genetic events essential for spermatogenesis, such as activation of Crem target genes.
MATERIALS AND METHODS
Construction of the Jmjd1a Targeting Vector
Genomic DNA was purified from the TC-1 mouse embryonic stem (ES) cells with a 129SvEv/j strain background (37) and used for amplifying the homologous arms of the Jmjd1a targeting vector by using an LA PCR kit (Takara Bio. Inc.). The 5′ arm DNA was amplified by using primers Jmjd1a-5F (cggttaattaactttcctctttaggggcac) and Jmjd1a-5R (aatgcggccgcttgtaaaaccaaccaac). The 3′ arm DNA was amplified by using primers Jmjd1a-3F (aatctcgagtaccatgcgcgtgagtgataaagctac) and Jmjd1a-3R (aaaggatccgcctggtctacagagcacaaactctca). PCR products were subcloned into the pCR2.1-TOPO plasmids using a TOPO TA cloning kit (Invitrogen) and verified by DNA sequencing. The 5′ arm DNA was isolated from the TOPO vector and subcloned into the PacI and NotI sites of the pFRT-LoxP plasmid (38), so the integrated 5′ arm was upstream of a PGK-neo cassette (Fig. 1A). The 3′ arm DNA was transferred from the TOPO vector to the pFRT-LoxP plasmid by XhoI and BamHI digestions, and the integrated 3′ arm was located between the PGK-neo and the HSV-tk (thymidine kinase) cassettes (Fig. 1A). The targeting vector was linearized by PacI digestion.
FIGURE 1.
Generation of Jmjd1a−/− mice. A, gene-targeting strategy used to generate Jmjd1a knock-out allele in ES cells. The relationships among the 3′ region of the Jmjd1a gene, the targeting vector, and the targeted Jmjd1a allele are sketched. The two black bars under the Jmjd1a locus indicate the 5′ and 3′ DNA probes for Southern blot. The gray bars under the Jmjd1a locus and the targeted allele indicate the DNA fragments amplified by PCR for genotyping. B, identification of targeted ES clones by Southern blot. ES cell DNA was digested by XhoI and EcoRV for Southern blot using 5′ probe and by EcoRV for Southern blot using 3′ probe as indicated in A. The 5′ probe detected a 10.9-kb fragment from the WT (+) Jmjd1a allele and a 7.3-kb fragment from the targeted (−) Jmjd1a allele. The 3′ probe detected a 11.4-kb fragment from the WT Jmjd1a allele and a 8.1-kb fragment from the targeted Jmjd1a allele. C, genotype analysis of mice by PCR. PCRs were performed using mouse genomic DNA and allele-specific primer pairs. The upper band (226 bp) and the lower band (120 bp) represent WT and targeted Jmjd1a alleles, respectively. D, Western blot. Testis lysates were prepared from Jmjd1a heterozygous (+/−) and knock-out (−/−) mice. Western blot was performed using Jmjd1a antibody. β-Actin served as a loading control.
Electroporation, Southern Blot, and Generation of Jmjd1a Knock-out Mice
TC-1 ES cells were cultured and electroporated with the targeting vector DNA as described (37). Cells were cultured in selection medium containing 300 μg/ml geneticin and 0.2 μm 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil for 7 days. Surviving clones were isolated and screened by Southern blot analyses using 5′ and 3′ probes located outside the targeting region and a neo probe (Fig. 1A). DNA samples were digested with XhoI and EcoRV for Southern blot (Fig. 1A).
Targeted ES clones were injected into E3.5 blastocysts of C57BL/6 mice, and the injected blastocysts were transferred into the uteri of pseudo-pregnant mice. Resulting chimeric males were mated with C57BL/6 females, and their agouti progenies were genotyped by Southern blot, as described above, and PCR using primers Jmjd1a-ICF (ATCTGATGCAGCCAATGTCA) and Jmjd1a-ICR (GGCTCCTGGCTTCTCTTTTC) for detecting the Jmjd1a wild type (WT) allele and primers KOV1 (gaaagtataggaacttcgtcgacctc) and Jmjd1a33 (ctaagccagggataaggactttca) for detecting the Jmjd1a knock-out allele (Fig. 1A). Heterozygous (Jmjd1a+/−) mice were used in further breeding to produce age-matched WT, Jmjd1a+/−, and homozygous knock-out (Jmjd1a−/−) mice for experiments (Fig. 1C).
Generation of Jmjd1a Antibody
Jmjd1a antibody was generated in rabbits immunized with a recombinant polypeptide of the mouse Jmjd1a. Briefly, RNA was purified from mouse testes, and cDNA was made by reverse transcription. The DNA fragment for residues Asn308–Asn566 of Jmjd1a was amplified by PCR using primers Jmjd1a-30 (catgcgcatatgactccatctagcaaggacccaag) and Jmjd1a-31 (ccatggatccacaggagagtccttctgctggtc). The amplified DNA was subcloned into the NdeI and BamHI sites of the pET-16b plasmid (Novagen). BL21-competent bacteria were transformed with the plasmids and used to produce the Jmjd1a polypeptide. The soluble 32-kDa Jmjd1a polypeptide was purified from bacterial lysates prepared from isopropyl 1-thio-β-d-galactopyranoside-induced bacterial culture by using 1 ml of nickel-nitrilotriacetic acid beads as described (39). The concentration of purified protein was estimated by comparing with serial dilutions of bovine serum albumin standards on an SDS-polyacrylamide gel stained with Coomassie Blue. The purified protein (1 mg/ml) was used as a Jmjd1a antigen, and the rabbit immunization was carried out through a service contract with the Proteintech Group, Inc.
Western Blot
Mouse testes were collected and homogenized in lysis buffer containing 50 mm Tris-HCl (pH 7.4), 0.1% SDS, 2 μm phenylmethylsulfonyl fluoride, and 10 μg/ml leupeptin. Following sonication and centrifugation, protein concentrations in the tissue extracts were measured by using a Bio-Rad protein assay kit. Aliquots containing 10 μg of total protein were separated by SDS-PAGE and transferred to nitrocellulose membranes. Blots were probed with primary antibodies against Jmjd1a, H3K9me1 (ab9045, Abcam), H3K9me2 (07-441, Upstate), H3K9me3 (07-442, Upstate), H3 (ab1791, Abcam), H3K27me2 (9755, Cell Signaling), acetylated H4K5/8/12/16 (06-866, Upstate), acetylated H3K9/14 (06-599, Upstate), acetylated H3K9 (39586, Active Motif), acetylated H3K56 (04-1135, Upstate), Crem (sc-440, Santa Cruz Biotechnology), Tnp1 (gift from Dr. Stephen Kistler, University of South Carolina), and β-actin (Sigma). Appropriate secondary antibodies conjugated with horseradish peroxidase and the ECL chemiluminescent reagents (Amersham Biosciences) were used to detect the bound primary antibodies.
Sperm Count
Sperm count was performed as described (40, 41). Briefly, for each adult mouse, one caudal epididymis was used for histological examination, and the other one was minced in 1 ml of PBS. Sperms were allowed to disperse into solution by incubating for 5 min. A small volume of solution containing the sperm was transferred into a hemocytometer and allowed to settle for 15 min prior to counting. The average number of sperm per epididymis was calculated from three mice for each genotype group.
Measurement of Testosterone
Blood samples were collected from 10-week-old male WT and Jmjd1a−/− mice by periorbital puncture. Serum samples were prepared as described (42). The testosterone in the testis was extracted as described (42). Briefly, one testis from each mouse was homogenized in 250 μl of PBS at room temperature. Homogenates were first extracted with 5 ml of diethyl ether and then extracted with 3 ml of diethyl ether again. The combined diethyl ether extracts were evaporated at room temperature in a fume hood and then resuspended in 1 ml of PBS. The concentrations of testosterone in the serum samples and the testis extracts were measured by using an ELISA kit (DSL-10-4000, Diagnostic Systems Laboratories).
Histological Examination
Testes were fixed in modified Davidson's fluid overnight at 4 °C (43). Fixed tissues were washed in PBS, dehydrated in ethanol, incubated in xylene, and then embedded in paraffin. Tissue sections were cut to a thickness of 5 μm and stained with hematoxylin and eosin or with a periodic acid-Schiff and hematoxylin kit (395B, Sigma). The specific steps of the germ cell development and differentiation during spermatogenesis and the stages of seminiferous epithelium were determined according to the well established criteria described previously (1). For preparation of semi-thin sections, testes were perfused with 2.5% glutaraldehyde and 2.0% formaldehyde and fixed at 4 °C overnight. The fixed samples were dehydrated and embedded in Spurr's low viscosity resin (EMS, Hatfield, PA). Semi-thin sections 1 μm in thickness were prepared by using an RMC MT6000-XL ultramicrotome and were stained with toluidine blue for microscopy.
Immunohistochemistry (IHC)
Deparaffinized and hydrated tissue sections were incubated in a 10 μm sodium citrate buffer at 95 °C for 10 min for retrieving antigens and then treated in methanol containing 3% H2O2 for quenching endogenous peroxidase. After incubating in PBS containing 10% goat serum, 1% bovine serum albumin, and 0.1% Triton X-100, tissue sections were incubated at 4 °C overnight with a primary antibody against Jmjd1a, H3K9me1, H3K9me2, or H3K9me3. Subsequently, the washed sections were incubated with a biotin-conjugated secondary antibody and the avidin-conjugated horseradish peroxidase in the Vectastain ABC-AP kit (AK-5200, Vector Laboratories). The horseradish peroxidase activities were visualized by using a DAB substrate kit (SK-4100, Vector Laboratories). The sections were counterstained with hematoxylin or periodic acid-Schiff and hematoxylin and sealed in Permount for microscopy and imaging.
Testicular Chromosome Spread and Immunofluorescence
Seminiferous tubules were isolated from wild type and knock- out mice. After incubating in 1% sodium citrate for 30 min, the tubules were fixed in Carnoy's solution (75% methanol and 25% acetic acid) overnight at 4 °C. The fixed tubules were then incubated in 60% acetic acid for 2 min. Single cell suspension was obtained by repeated pipetting. Cell suspension was then spread on pre-heated slides at 60 °C and dried (26). Immunofluorescence was done as described above except that the TSA kit (Invitrogen, T-20911) was used to amplify Jmjd1a signals. Propidium iodide (1 μg/ml) was used to stain chromosomes.
Real Time RT-PCR
Total RNA was purified from the testes of 10-week-old WT and Jmjd1a−/− mice using TRIzol reagent (Invitrogen). The real time RT-PCR was performed by using the reverse transcriptase core kit (Eurogentec) and the Universal Mouse Probe Sets for TaqMan PCR (Roche Applied Science). The mRNA-specific primer pairs were designed, and the specific match of universal mouse probe sets was determined by running the software at Universal Probe Library Design Center. Relative mRNA concentrations were obtained by normalizing to the endogenous levels of 18 S RNA.
Chromatin Immunoprecipitation (ChIP)
Cells were isolated from the testes of adult WT and Jmjd1a−/− mice as described (44). Briefly, the tunica albuginea of mouse testes was removed. The remaining tissues were digested by 0.5 mg/ml collagenase in Dulbecco's modified Eagle's medium with 0.5% bovine serum albumin at 32 °C for 15 min, washed in Dulbecco's modified Eagle's medium, and further digested by 0.5 mg/ml trypsin and 1 μg/ml DNase in Dulbecco's modified Eagle's medium with 0.5% bovine serum albumin. After pipetting 10–12 times and washing with Dulbecco's modified Eagle's medium, cells were filtered through a 40-micron filter and diluted to 2 × 106/ml. The cells were fixed with 1% formaldehyde for 10 min at room temperature, washed in cold PBS, and incubated in fixation-stopping solution (100 mm Tris-HCl (pH 9.4) and 10 mm dithiothreitol). ChIP assay was performed as described previously (45) by using antibodies against H3K9me1, H3K9me2, H3K9me3, and Crem. 2 × 106 cells were used for each of H3K9Me1, H3K9Me2, and H3K9Me3 ChIP assays. 2 × 107 cells were used for each Crem ChIP assay. Eluted DNA samples were processed and subjected to PCR analyses of the proximal regions of Tnp1 and Odf1 gene promoters containing functional Crem-binding sites. In PCR, primers Tnp1-ChIP3F (gtccttttggctggtatgga) and Tnp1-ChIP3R (cttagccaaagctggtggag) were used to amplify fragment a. Primers Tnp1-ChIP2F (aatgaccacggctaaagcac) and Tnp1-ChIP2R (gccgggagtccatgtattta) were for fragment b amplification. Primers Odf1-ChIP5F (gggtctcaggggaccataac) and Odf1-ChIP5R (ctcttctcagaggcctccttt) were for fragment c amplification.
Apoptosis Analysis
Paraffin sections prepared from the testes of 10-week-old WT and Jmjd1a−/− mice were subjected to apoptosis analysis. The analysis was performed by using the ApopTag peroxidase in situ apoptosis detection kit (Chemicon). Digital images were recorded for each section. Two nonadjacent sections for each mouse and three mice per group were analyzed. For each section, the number of apoptotic cells in about 100 cross-sectioned seminiferous tubules was counted using the University of Texas Health Science Center, San Antonio, ImageTool software. The apoptotic cells were divided into three groups as follows: the pre-pachytene germ cells that attached to the basal membrane; the pachytene, diplotene, and secondary spermatocytes; and the spermatids.
RESULTS
Generation of Jmjd1a Null Mice
The mouse Jmjd1a gene spans about 43 kb and contains 26 exons that encode multiple alternative splicing variants. To inactivate the enzyme activity in all splicing isoforms, we constructed a gene-targeting vector containing a 6-kb 5′ arm from intron 12 to intron 16 and a 4.8-kb 3′ arm containing exon 26 (Fig. 1A). After electroporating the targeting vector DNA, we cultured ES cells in a selection medium with neomycin and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil and then performed Southern blot to identify clones with correct homologous recombination. After the DNA samples were digested with XhoI and EcoRV restriction enzymes, the 5′ probe detected a 7.3-kb fragment from the targeted allele and a 10.9-kb fragment from the WT allele, whereas the 3′ probe detected a 8.1-kb fragment from the targeted allele and a 11.4-kb fragment from the WT allele (Fig. 1, A and B). Further Southern blot analysis using a neo probe confirmed that the genome of all three targeted clones had no detectable random insertion of the vector DNA, indicating that only the Jmjd1a allele is mutated in these clones (data not shown). Because the deleted region in the targeted allele contains exons 17–25 for the demethylase domain of Jmjd1a, the targeted Jmjd1a allele is a null allele in terms of its enzymatic activity (Fig. 1A).
Three lines of targeted ES cells were injected into C57BL/6 morula followed by embryo transplantation into pseudopregnant foster mothers. Fourteen male chimeric mice were produced from two of the three ES cell lines. Chimeric mice with germ line transmission were identified by crossing with WT C57BL/6 females. Heterozygous (Jmjd1a+/−) and homozygous (Jmjd1a−/−) mice were subsequently generated and identified by PCR-based genotyping analysis (Fig. 1C). Western blot using testis extracts of Jmjd1a+/− and Jmjd1a−/− mice and an antibody against the N terminus of Jmjd1a confirmed the absence of the 150-kDa Jmjd1a protein in Jmjd1a−/− mice (Fig. 1D). In addition, a weak 90-kDa band was only detected in Jmjd1a−/− mice, likely representing a demethylase-deficient N-terminal polypeptide created by the gene-targeting event.
From an examined batch of Jmjd1a+/− breeding pairs, 102 WT, 213 Jmjd1a+/−, and 101 Jmjd1a−/− pups were obtained, indicating a normal Mendelian ratio (1:2:1). All of these mice exhibited normal body growth. These results suggest that Jmjd1a is not essential for embryonic development, postnatal survival, or somatic growth.
Disruption of the Jmjd1a Gene Causes Severe Oligozoospermia and Male Infertility
Female Jmjd1a−/− and Jmjd1a+/− mice and male Jmjd1a+/− mice exhibited normal reproductive function. When sexually mature male Jmjd1a−/− mice were paired with young adult WT females, they were able to successfully mount the females and ejaculate to produce coital plugs in the vagina, indicating that Jmjd1a is not required for male sexual behavior. However, no progeny was produced from a total of five breeding pairs of male Jmjd1a−/− and normal female mice in a period of 6 months. These results indicate that Jmjd1a is absolutely essential for male reproductive function.
To investigate why male Jmjd1a−/− mice were infertile, we examined their testosterone levels, testis weights, and sperm numbers. We found no significant differences in serum testosterone levels and body weights between male Jmjd1a−/− and WT mice at 12 weeks of age (data not shown). However, the testes of Jmjd1a−/− mice were much smaller than those of their WT littermates (Fig. 2A). The ratio of average testis weight to body weight for Jmjd1a−/− mice was reduced 40% versus WT mice (Fig. 2A). Surprisingly, in contrast to the numerous mature sperm found in the epididymal lumen of WT mice, very few sperm could be observed in the epididymal lumen of Jmjd1a−/− mice (Fig. 2B). Sperm counting further revealed that the average number of total epididymal sperm in Jmjd1a−/− mice was only 1‰ of that in WT mice (Fig. 2B). These results clearly demonstrate that disruption of the Jmjd1a gene results in a severe oligozoospermia syndrome responsible for the sterile phenotype of male Jmjd1a−/− mice.
FIGURE 2.
Testis weight, sperm count, and Jmjd1a distribution in the testis. A, photographs of testes and the ratios of testis weight (T.W.) to body weight (B.W.) obtained from 12-week-old WT and Jmjd1a−/− (KO) mice. S.D., standard deviation; N., number; p value (<0.01), calculated by unpaired t test. B, hematoxylin and eosin-stained sections of the caudal epididymides prepared from 12-week-old WT and Jmjd1a−/− (KO) mice and the average sperm number collected from each epididymis in WT and KO mice. The statistical analysis was performed with unpaired t test. S, spermatozoa; L, lumen. C, Jmjd1a IHC. Paraffin sections were prepared from the testes of 7-week-old mice and used for IHC. Brown color indicates Jmjd1a immunoreactivity. The sections were counterstained with periodic acid-Schiff and hematoxylin. The section from a Jmjd1a−/− (KO) mouse served as a negative control. The seminiferous stages were labeled from I to XII. Sg, spermatogonia; Pl, preleptotene; L, leptotene; Z, zygotene; P, pachytene; 2nd, secondary spermatocyte; RS, round spermatid; ES, elongating spermatid; SC, Sertoli cell; LC, Leydig cell. D, immunofluorescence staining of chromosomes in a pachytene spermatocyte with Jmjd1a antibodies and propidium iodide (PI).
Jmjd1a Is Mainly Expressed in Pachytene and 2nd Spermatocytes during Spermatogenesis
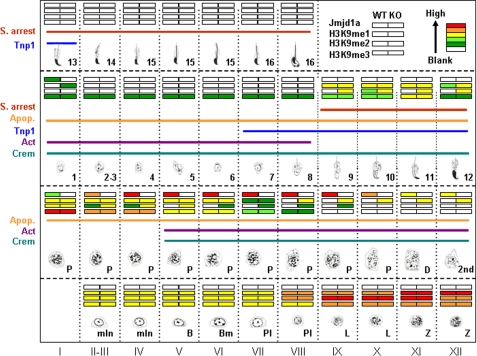
To assess the cell type-specific function of Jmjd1a in the testis, it is essential to determine what cell types express Jmjd1a protein during spermatogenesis. For this purpose, we generated a recombinant Jmjd1a polypeptide (Asn308–Asn522) as an antigen and produced Jmjd1a polyclonal antibody. We performed IHC for Jmjd1a with this antibody and periodic acid-Schiff and hematoxylin staining for the acrosome to distinguish developmental steps of spermatids and morphological stages of the germinal epithelial cycle of seminiferous tubules. We detected a Jmjd1a protein in the nuclei of stages I–X pachytene spermatocytes. Jmjd1a immunostaining signals were much stronger in stages IV–IX pachytene cells versus stages II, III, and X pachytene cells (Fig. 2C). Relatively weak signals were observed in the diplotene and 2nd spermatocytes (Fig. 2C and Fig. 7). In addition, very weak Jmjd1a immunostaining signals were detected in step 1 round spermatids (Fig. 2C). In contrast, Jmjd1a immunoreactivity was absent in stages II–VI spermatogonia and in stages VII and VIII preleptotene, stages IX and X leptotene, and stages XI and XII zygotene spermatocytes. Jmjd1a was also undetectable in stages II–XII round spermatids and all stages elongating spermatids (Fig. 2C). In addition, Jmjd1a immunostaining signals were not found in Sertoli cells and Leydig cells (Fig. 2C). Surprisingly, strong Jmjd1a immunofluorescence signals were scattered on the chromosomes of pachytene spermatocytes (Fig. 2D), suggesting that Jmjd1a is associated with the chromosome for histone modification. Taken together, these results demonstrate that Jmjd1a is specifically expressed in pachytene, diplotene, and 2nd spermatocytes and step 1 round spermatids. This interesting spatial and temporal expression pattern suggests that Jmjd1a should function in a germ cell type- and stage-specific manner during spermatogenesis.
FIGURE 7.
Temporal and spatial distributions of Jmjd1a, H3K9me1, H3K9me2, H3K9me3, Crem, Act, and Tnp1 in WT and Jmjd1a−/− mice during spermatogenesis and their relationships relevant to germ cell apoptosis (Apop.) and spermatid-elongating arrest (S. arrest) in Jmjd1a−/− mice. The 12 stages of seminiferous epithelium and the developmental steps of spermatogenesis are listed. The levels of Jmjd1a, H3K9me1, H3K9me2, and H3K9me3 in the testes of WT and Jmjd1a−/− (KO) mice are presented in color codes as indicated (upper right corner). The steps with Crem and Act proteins and Tnp1 mRNA expression are indicated. The steps with germ cell apoptosis and developmental arrest of elongating spermatids in Jmjd1a−/− mice are also indicated. mIn, mitotic intermediate spermatogonia; B, type B spermatogonia; Bm, mitotic type B spermatogonia; Pl, preleptotene cells; L, leptotene cells; Z, zygotene cells; P, pachytene cells; 2nd, secondary spermatocytes; 1–16, steps 1–16 spermatids.
Disruption of Jmjd1a Significantly Increases H3K9me1 in Pachytene Spermatocytes
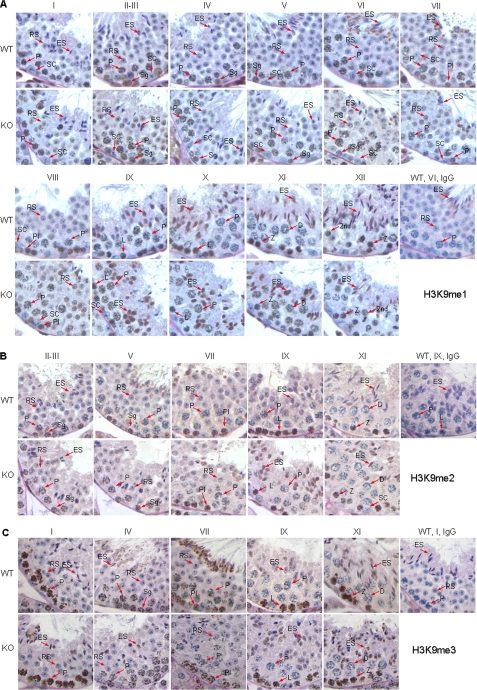
Jmjd1a demethylates H3K9me1 in biochemical reactions (28). To examine the H3K9me1 pattern and the impact of Jmjd1a knock-out on H3K9me1 status in the germinal epithelium of seminiferous tubules, we performed IHC with antibodies specific to H3K9me1. In the testes of adult WT mice, strong H3K9me1 immunostaining signals were observed in the nuclei of all fist layer germ cells of the stage II–XII germinal epithelia, including spermatogonia, preleptotene, leptotene, and zygotene cells, whereas no signals were seen in the nuclei of Sertoli cells (Fig. 3A). Moderate levels of H3K9me1 immunoreactivity were detected in the nuclei of stages I–IV pachytene spermatocytes, and a weak signal was observed in the nuclei of stage VII pachytene spermatocytes (Fig. 3A). Interestingly, H3K9me1 was undetectable in stages VIII–X pachytene and stage XI diplotene spermatocytes (Fig. 3A). H3K9me1 signal remained very low in 2nd spermatocytes and steps 1–8 round spermatids, but it became much stronger in steps 9–12 spermatids (Fig. 3A). H3K9me1 signal was absent in the nuclei of steps 13–16 spermatids (Fig. 3A). These results indicate that the degree of H3K9me1 is dynamically regulated in a spermatogenic step-specific manner.
FIGURE 3.
Stage-specific H3K9 methylation in the testes of WT mice and increased H3K9me1 and H3K9me2 in the testes of Jmjd1a−/− (KO) mice. Testis sections were prepared from 7-week-old WT and Jmjd1a−/− (KO) mice. The stages of seminiferous epithelium are indicated. Refer to the legend for Fig. 2 for abbreviations of cell type nomenclatures. Substitution of nonimmune IgG for specific antibodies was performed as negative controls. A, H3K9me1 IHC (brown color) in the testes of WT and KO mice. B, H3K9me2 IHC (brown color) in the testes of WT and KO mice. C, H3K9me3 IHC (brown color) in the testes of WT and KO mice.
In the testes of adult Jmjd1a−/− mice, H3K9me1 levels in spermatogonia and stages I–VII pachytene spermatocytes were unchanged. However, H3K9me1 levels in Jmjd1a−/− testes were markedly increased in late step pachytene, diplotene, and 2nd spermatocytes in stages VIII–XII germinal epithelia versus WT (Fig. 3A). Furthermore, H3K9me1 immunostaining signals remained higher in step 1 round spermatids in Jmjd1a−/− testes versus WT. Similar to WT testes, the H3K9me1 signal was undetectable in steps 13–16 spermatids in Jmjd1a−/− testes (Fig. 3A). Western blot with testis lysates confirmed that H3K9me1 was significantly increased in the testes of all examined Jmjd1a−/− mice, and it was also increased in the testes of two of the three examined Jmjd1a+/− mice (Fig. 4A). These results demonstrate that Jmjd1a is required for demethylating H3K9me1 in late step pachytene spermatocytes and early step round spermatids.
FIGURE 4.
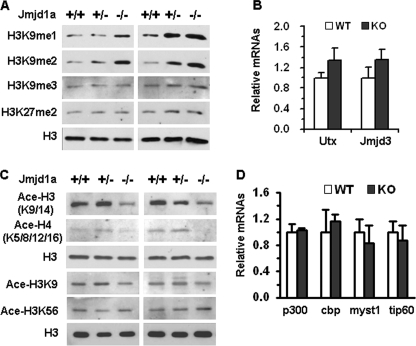
Jmjd1a deficiency increases histone methylation and decreases histone acetylation in the testis. A, Western blot analyses of H3K9me1, H3K9me2, H3K9me3, H3K27me2, and total H3 in the testes of 12-week-old WT (+/+), heterozygous (+/−), and KO (−/−) Jmjd1a mice. Two independent sets of samples were assayed. Total H3 levels served as loading controls. B, Utx and Jmjd3 mRNA levels measured by real time RT-PCR. Total RNA samples were prepared from the testes of 12-week-old WT (n = 3) and KO (n = 3) mice. Each sample was assayed twice. The level of mRNA was normalized to 18 S level of the same sample. Data have no statistical difference and are presented as mean ± S.D. C, Western blot analyses of acetylated H3 (Ace-H3K9/14), H4 (Ace-H4K5/8/12/16), H3K9, and H3K56. Total H3 served as loading control. Two independent sets of samples were assayed. The first three testis samples used in A and C were the same, so the total H3 was the same. D, real time RT-PCR measurements of p300, Cbp, Myst1, and Tip60 mRNA levels. RNA samples and analytical methods were described in B.
Inactivation of Jmjd1a Increases H3K9me2 in Pachytene Spermatocytes and Step 9–10 Spermatids without Affecting H3K9me3 Levels
In biochemical reactions, in addition to H3K9me1, Jmjd1a also demethylates H3K9me2 but not H3K9me3 (28). We used IHC to define H3K9me2 patterns and assessed the effects of Jmjd1a knock-out on H3K9me2 levels. In WT germinal epithelium, the H3K9me2 was high in spermatogonia (stages II–VI), preleptotene (stages VII and VIII), leptotene (stages IX and X), zygotene (stages XI and XII), and stage I pachytene spermatocytes (Fig. 3B). H3K9me2 levels were reduced in stages II–X pachytene, stage XI diplotene, and stage XII 2nd spermatocytes. H3K9me2 levels remained low in steps 1–9 spermatids. Interestingly, H3K9me2 immunoreactivity was transiently increased in step 10 elongating spermatids and then decreased to low levels again in steps 11–16 spermatids (Fig. 3B).
Ablation of Jmjd1a in mice did not alter the H3K9me2 levels in spermatogonia, preleptotene, leptotene, zygotene, and the first step pachytene spermatocytes but increased H3K9me2 levels in stages II–IX pachytene spermatocytes (Fig. 3B). H3K9me2 immunoreactivity was similar in stage X pachytene, stage XI diplotene, and stage XII 2nd spermatocytes and in steps 1–8 spermatids in the germinal epithelia of Jmjd1a−/− and WT mice (Fig. 3B). However, H3K9me2 levels were markedly elevated in steps 9–12 spermatids in Jmjd1a−/− germinal epithelia before declining to low levels in steps 13–16 spermatids (Fig. 3B). Western blot clearly revealed that the H3K9me2 level increased 2–3-fold in Jmjd1a−/− testes versus WT testes, and it also increased in two of the three examined Jmjd1a+/− mice (Fig. 4A). These results indicate that the H3K9me2 level is stringently regulated in a spermatogenic step-specific manner, and Jmjd1a is required to maintain relatively low levels of H3K9me2 in pachytene spermatocytes and step 9–10 spermatids.
We also examined H3K9me3 by IHC. Moderate H3K9me3 immunoreactivity was detected in spermatogonia and preleptotene spermatocytes. H3K9me3 immunostaining signals began to increase in leptotene spermatocytes and reached the peak level in stage I pachytene spermatocytes (Fig. 3C). H3K9me3 immunostaining signals were gradually reduced in stages II–IX pachytene spermatocytes and became undetectable in stage X pachytene spermatocytes. In round spermatids, H3K9me3 signals were observed in central regions of the nuclei (Fig. 3C). Disruption of the Jmjd1a gene did not change H3K9me3 levels throughout the spermatogenic cycle in Jmjd1a−/− mice (Fig. 3C). Western blot also confirmed that Jmjd1a deficiency had no effect on H3K9me3 levels in the testis (Fig. 4A). These data agree with in vitro assays that found H3K9me3 was not a substrate of Jmjd1a (28). In addition, Jmjd1a deficiency also did not alter H3K27me2 levels in the testis (Fig. 4A). This was consistent with the similar expression levels of H3K27-specific histone demethylases, Utx and Jmjd3 (46), in the testes of Jmjd1a−/− and WT mice (Fig. 4B). These results demonstrate that the H3K9me3 levels vary during spermatogenesis, and Jmjd1a does not modulate either H3K9me3 or H3K27me2 in the testis. Therefore, in the testis, Jmjd1a functions as an H3K9me1/H3K9me2-specific demethylase.
Increase in H3K9me1 and H3K9me2 Is Associated with a Decrease in Histone Acetylation in the Testes of Jmjd1a−/− Mice
Because methylation and acetylation are mutually exclusive at histone H3K9 (47), we examined histone acetylation by Western blot. The acetylation levels of both histones H3 and H4 were significantly reduced in the testes of Jmjd1a−/− mice versus WT mice as assayed by the antibodies against acetylated H3K9/14 and H4K5/8/12/16 (Fig. 4C). Further analysis with an antibody specific to acetylated H3K9 confirmed a significant decrease in H3K9 acetylation in Jmjd1a−/− testis. In contrast, the levels of H3K56 acetylation remained unchanged in Jmjd1a−/− testis (Fig. 4C). Real time RT-PCR analyses revealed that the broadly expressed histone acetyltransferases cAMP-response element-binding protein-binding protein and p300 and the testis-specific histone acetyltransferases Myst1 and Tip60 (48) were equally expressed in the testes of Jmjd1a−/− and WT mice (Fig. 4D). These results suggest that the reduced histone acetylation in the testes of Jmjd1a−/− mice may be directly associated with the increased histone methylation and is not due to any change of histone acetyltransferase expression. These results also suggest that Jmjd1a may serve as a transcriptional activator in the testis through indirectly enhancing histone acetylation.
Jmjd1a Deficiency Diminishes Crem Recruitment, Act Expression, and Transcriptional Activation of Their Target Genes Required for Chromatin Condensation in Spermatids
The grades of histone acetylation and methylation are usually associated with recruitment of transcription factors to the chromatin and transcriptional activation of certain gene promoters. Crem is an essential transcription factor that works with its coactivator Act in germ cells to mediate expression of multiple genes required for spermatid elongation, including Tnp1, Tnp2, Prm1, and Prm2 (3, 8). Because Jmjd1a deficiency increased H3K9me1 and H3K9me2 and decreased histone H3 acetylation, we examined Crem and Act expression as well as their target gene expression in the testes of Jmjd1a−/− mice. Although the Crem expression level was unaltered, the level of Act expression decreased more than 70% in the testes of Jmjd1a−/− mice versus WT mice, suggesting that the histone modification altered by Jmjd1a deficiency selectively affects some but not all gene expression (Fig. 5A). There are two known Crem-binding sites (a and b in Fig. 5B) in the proximal region of the Tnp1 promoter and the recruitment of Crem to these sites activates Tnp1 transcription (49). Importantly, ChIP assays revealed that the recruitment of Crem to both a and b sites was significantly reduced in the testes of Jmjd1a−/− mice versus WT mice (a and b, Fig. 5C). Similarly, the recruitment of Crem to a functional binding site in the promoter of another direct target gene Odf1 was also reduced (Fig. 5, B and region c in C). Furthermore, ChIP assays demonstrated that Jmjd1a deficiency increased H3K9me1 levels at all three Crem-binding regions in both promoters and H3K9me2 levels at Crem-binding regions a in Tnp1 promoter and c in Odf1 promoter, although it had little effect on H3K9me3 levels at these Crem-binding regions (Fig. 5C). These results suggest that Jmjd1a-regulated histone codes are required for Act expression and Crem recruitment to the functional binding sites of its target genes.
FIGURE 5.
Jmjd1a deficiency decreases Act expression and diminishes Crem-mediated gene expression. A, relative Crem and Act mRNA expression levels in the testes of 12-week-old WT and Jmjd1a−/− (KO) mice (n = 3 for each group). **, p < 0.01 by unpaired t test. B, cAMP-response elements (CRE) for Crem binding in the proximal regions of the Tnp1 and Odf1 promoters are indicated. Exon 1 and exon 2 (E2) also are indicated. The DNA fragments (a–c) that were amplified by PCR in ChIP assays are marked. C, ChIP assays for regions a and b of the Tnp1 promoter and c of the Odf1 promoter (B). Germ cells were isolated from the testes of WT and Jmjd1a−/− (KO) mice. Input represents 20% of materials used for Crem ChIP. ChIP assays were performed using antibodies against H3K9me1, H3K9me2, H3K9me3, and Crem as indicated. Substitution of nonimmune IgG for specific antibodies served as negative controls. D, expression levels of Crem and Act target genes in the testes of 12-week-old WT and Jmjd1a−/− (KO) mice (n = 3 for each group). Data are presented as mean ± S.D. *, p < 0.05 by unpaired t test. E, Western blot analysis of Tnp1 in the testes of 12-week-old WT (+/+), Jmjd1a+/−, and Jmjd1a−/− mice. β-Actin served as a loading control.
In agreement with reduced Crem recruitment to its target promoter and reduced Act expression in the testes of Jmjd1a−/− mice, the expression levels of all examined Crem and Act target genes, including Tnp1, Tnp2, Prm1, Prm2, Odf1, and Gsg3, were significantly reduced in the testes of Jmjd1a−/− mice, and especially the mRNA level of Tnp1 was reduced 90% when compared with that in the testes of WT mice (Fig. 5D). Western blot using the Tnp1 antibody also revealed a remarkable decrease in Tnp1 in the testes of Jmjd1a−/− mice versus WT mice (Fig. 5E). These results indicate that ablation of Jmjd1a significantly inhibits Crem- and Act-mediated target gene expression in the testes due to, at least in part, the failed Crem recruitment to chromatin and decreased Act expression.
Disruption of the Jmjd1a Gene Blocks Spermatid Elongation
To characterize the physiological significance of Jmjd1a-regulated histone modifications and gene expression during spermatogenesis, we carefully examined the cell morphology and the numbers of each cell type associated with specific stages of the seminiferous epithelia in Jmjd1a−/− mice. No differences were observed in the morphology of Sertoli cells in the testes of Jmjd1a−/− mice versus WT mice. This is consistent with the absence of Jmjd1a in Sertoli cells. In the germ cell lineage, Jmjd1a deficiency also did not affect the morphology of spermatogonia, spermatocytes, and steps 1–8 round spermatids (Fig. 6A).
FIGURE 6.
Ablation of Jmjd1a arrests spermiogenesis and causes apoptosis of developing germ cells. A, Jmjd1a deficiency blocked spermiogenesis at spermatid-elongating stages. Testis sections were prepared from 12-week-old WT and Jmjd1a−/− (KO) mice and stained with periodic acid-Schiff and hematoxylin. The stages of seminiferous epithelium are indicated from I to XII. Note the abnormal head morphologies of elongating spermatids at stages X–XII and stages I–IV and the drastically reduced number of elongated spermatids at stages X–VII in KO testes (green boxes). Sg, spermatogonia; Pl, preleptotene; L, leptotene; Z, zygotene; P, pachytene; 2nd, secondary spermatocyte; RS, round spermatid; ES, elongating spermatid; SC, Sertoli cell; LC, Leydig cell. B, toluidine blue-stained semi-thin sections of WT and Jmjd1a−/− (KO) mouse testes. Developmental steps of spermatids are indicated. acr, acrosome; DS, dorsal side; VS, ventral side. C, detection of apoptotic cells in the testis sections of 12-week-old WT and KO mice by terminal deoxynucleotidyltransferase nick end-labeling assay. Apoptotic cells exhibit brown nuclei in this assay. The lower panels are the enlarged pictures of the boxed regions in the upper panels. The asterisk indicates the small apoptotic nuclei of the elongating spermatids. D, quantitative analysis of apoptotic germ cells in the seminiferous epithelium of WT and KO testes. Three 12-week-old mice per genotype group and two sections per testis were analyzed. Apoptotic cells in group a (pre-pachytene germ cells), group b (pachytene, diplotene and secondary spermatocytes), and group c (spermatids) were counted and recorded separately. Data are presented as the average number (No.) of apoptotic cells per cross-sectioned seminiferous tubule. The Total bar graphs represent the sum of groups a–c. *, p < 0.05 by unpaired t test.
Importantly, abnormal morphology began to be observed in step 9 Jmjd1a−/− spermatids (stage IX). For normal spermiogenesis in WT mice, step 9 spermatids started shaping their nuclear heads by developing ventral and dorsal surfaces. At steps 10–12 (stages X–XII), most normal spermatids formed an apex at the junction of dorsal and ventral surfaces. From steps 13 to 15 (stages I–VI), all normal WT spermatids developed an elongated, dense head. At step 16 (stages VII and VIII), normal mature spermatids had moved to luminal layers of the seminiferous epithelium and were ready to be released or already released into the lumen. As a result, very few spermatozoa could be seen in stage VIII seminiferous epithelium (Fig. 6A). However, although step 9 spermatids of Jmjd1a−/− mice initiated their head-shaping process, some of these spermatids could not form normal ventral and dorsal surfaces. At steps 10–12, most Jmjd1a−/− spermatids could not develop normal apexes at the junction of dorsal and ventral surfaces, and they displayed short, irregular and/or polygonal head morphologies (Fig. 6A). At steps 13–15 (stages I–IV), the head morphologies of the Jmjd1a−/− spermatids remained short, irregular, and polygonal. In stages V and VI seminiferous epithelia of Jmjd1a−/− mice, the spermiogenesis toward mature spermatozoa was basically arrested, and the total number of step 15 elongated spermatids was drastically reduced compared with the number observed in stages I–IV seminiferous epithelia (Fig. 6A). As a result, in stages VII and VIII seminiferous epithelia of Jmjd1a−/− mice, very few step 16 elongated spermatids and almost no mature spermatozoa could be observed either in the apical crypts of Sertoli cells or in the lumen of seminiferous tubules (Fig. 6A). Taken together, disruption of the Jmjd1a preferentially restricts the process of spermatid elongation starting at step 9 and blocks spermiogenesis, resulting in severe oligozoospermia and a sterile phenotype of male Jmjd1a−/− mice.
Defective Spermatogenesis Is Associated with Extensive Apoptosis of Germ Cells
We have shown that Jmjd1a deficiency blocked spermiogenesis. To investigate the fate of germ cells in the testes of Jmjd1a−/− mice, we performed apoptosis assays and counted the number of apoptotic cells in three groups. Group A included those germ cells attached to the basement membrane of seminiferous tubules, which were spermatogonia and pre-pachytene spermatocytes; group B mainly included pachytene, diplotene, and secondary spermatocytes; and group C contained both round and elongating spermatids. The average number of group A apoptotic germ cells showed no significant difference in the testes of WT and Jmjd1a−/− mice. However, the average number of apoptotic germ cells in groups B and C were, respectively, 5- and 10-fold higher in Jmjd1a−/− mice versus WT mice (Fig. 6, C and D). These results indicate that Jmjd1a is required for the viability of pachytene, diplotene, and secondary spermatocytes and round and elongating spermatids.
DISCUSSION
Post-translational modifications of the histone N termini play important roles in determining chromatin topology and transcriptional regulation. These modifications mainly include methylation and acetylation of lysine and arginine residues, phosphorylation of serine and threonine residues, and ubiquitylation and sumoylation of lysine residues. Furthermore, lysine residues can be modified by one, two, or three methyl groups. Although histone acetylations usually enhance transcriptional activation, histone methylations may positively or negatively regulate gene expression, depending on which lysine residue is modified and how many methyl groups are added (50). Methylation of H3K9 usually suppresses transcription (50). Importantly, histone methylation is a reversible process, and several classes of histone demethylases have been discovered (51, 52). Recently, a large family of proteins that contain a jumonji C-terminal (JmjC) domain has been identified to have histone demethylase activity (52). As a group, these demethylases can demethylate mono-, di-, and/or tri-methylated lysine residues and methylated arginine residues, whereas individual enzymes exhibit different substrate specificities (46, 53, 54). Jmjd1a is a member of this family and selectively removes mono- and di-methyl groups from H3K9 (28). Although biochemical features of these histone demethylases have been characterized, very few studies have been carried out to understand their physiological significance.
In this study, we have found that Jmjd1a is specifically expressed in the pachytene and secondary spermatocytes and step 1 round spermatids during spermatogenesis (Fig. 7). High levels of H3K9me1 are present in stages I–VI pre-pachytene germ cells and stages I–VI pachytene spermatocytes. H3K9me1 levels are low in stages VII–IX pachytene spermatocytes and absent in late stage pachytene spermatocytes and all round spermatids. However, H3K9me1 reappears in steps 9–11 elongating spermatids (Fig. 7). High levels of H3K9me2 are also present in pre-pachytene germ cells and stage I pachytene spermatocytes; however, it is undetectable at later stages (Fig. 7). Importantly, knock-out of Jmjd1a significantly increases H3K9me1 and H3K9me2 in the pachytene and secondary spermatocytes, round spermatids, and early elongating spermatids. These results indicate that Jmjd1a expressed in the pachytene and secondary spermatocytes and early round spermatids plays an important role in demethylating H3K9me1 and H3K9me2 in these germ cells and maintaining a hypomethylation status of H3K9 in later step round and elongating spermatids. Additionally, low levels of H3K9me3 are observed in spermatogonia and stages II–IX pachytene spermatocytes, whereas higher levels appear in preleptotene, leptotene, zygotene, and stage I pachytene spermatocytes (Fig. 7). In agreement with a previous report that H3K9me3 is not a substrate of Jmjd1a (28), knock-out of Jmjd1a does not alter H3K9me3 patterns in the germ cells. These results suggest that the dynamic levels of H3K9me3 in germ cells may be regulated by other specific histone demethylase(s).
Interestingly, we found that the increase in H3K9me1 and H3K9me2 correlated with a decrease in H3 and H4 acetylations in the germ cells of Jmjd1a−/− mice. This may be partially explained by the exclusive relationship between methylation and acetylation on common lysine residues. H4 hyperacetylation happens just before histone replacement and is observed only in species in which spermatogenesis requires histone replacement (16). Therefore, the reduced H4 acetylation in the germ cells of Jmjd1a−/− mice may affect histone replacement in elongating spermatids during spermiogenesis and lead to defective elongating spermatids. However, the direct link between H4 acetylation and H3K9 methylation is currently unknown (47).
Presumably, changes in histone methylation and acetylation in testes should influence regulation of gene expression important for spermatogenesis. Androgen receptor (AR) is expressed in Sertoli cells but not in germ cells, and it regulates gene expression required for maintaining spermatocyte viability and spermatid maturation (55). Because Jmjd1a has been shown to serve as an AR coactivator (28), it is conceivable that Jmjd1a works with AR to support spermatogenesis. Our results demonstrate that Jmjd1a is not present in Sertoli cells but in the AR-negative germ cells, excluding the direct interaction between AR and Jmjd1a in the germinal epithelium. Another essential regulatory pathway for spermatogenesis is Crem-mediated gene expression (3, 49). Crem and its coactivator Act are initially expressed in pachytene spermatocytes (2, 56). They then work together to activate Tnp1, Tnp2, Prm1, and Prm2 expression in step 7 spermatids (2, 49). These gene products are required for histone replacement and spermatid elongation during spermiogenesis (4–7). We found that Jmjd1a deficiency markedly reduced Act expression, increased histone methylation at the promoter regions of Crem target genes, and suppressed Crem recruitment to its target promoters. This led to significant decreases in the expression of all examined target genes, including Tnp1, Tnp2, Prm1, Prm2, Odf1, and Gsg3. The failure of Crem-regulated gene expression is likely responsible, at least in part, for the abnormal morphology of elongating spermatids and blockade of spermiogenesis at spermatid-elongating steps in Jmjd1a−/− mice (Fig. 7). This conclusion is consistent with previous findings showing defective spermatogenesis in Tnp1, Tnp2, Prm1, and Prm2 mutant mice (4–7).
We also found that disruption of the Jmjd1a gene caused extensive apoptosis of pachytene, diplotene, and secondary spermatocytes as well as spermatids. Our findings indicate that Jmjd1a is required not only for supporting the survival of Jmjd1a-positive pachytene, diplotene, secondary spermatocytes, and step 1 round spermatids but also for maintaining the viability of other Jmjd1a-negative spermatids (Fig. 7). These results suggest that Jmjd1a-regulated epigenetic chromatin modifications and gene expression in Jmjd1a-positive germ cells continue to play a role in maintaining the viability and development of late step Jmjd1a-negative spermatids. Extensive germ cell apoptosis is likely responsible for the phenotype of small testes observed in Jmjd1a−/− mice, whereas both the apoptosis and the blockade of spermiogenesis at elongating steps are likely responsible for the phenotype of severe oligozoospermia and male infertility in Jmjd1a−/− mice.
While this work was in progress, another Jmjd1a mutant mouse model generated by a gene-trap strategy was recently reported (57). Although the overall phenotype of defective spermiogenesis and male infertility in this model is similar to that observed in our Jmjd1a knock-out mice, major differences exist between these two studies. First, the previous study reported Jmjd1a protein in stages IX and X pachytene, diplotene, and secondary spermatocytes, steps 1–13 spermatids, and Sertoli cells. However, our analyses revealed that Jmjd1a protein is present in stages I–X pachytene, diplotene, and secondary spermatocytes and step 1 round spermatids but not in steps 2–13 spermatids and Sertoli cells. Second, the residual full-length Jmjd1a protein was still detectable in the testes of the former Jmjd1a gene-trap mice, suggesting it is a Jmjd1a hypomorphic mouse model, whereas the Jmjd1a−/− mouse line in this study is a Jmjd1a null model. Third, the previous study did not examine H3K9 methylation profiles in a germ cell stage-specific manner and found no global alterations of H3K9me1 and H3K9me2 levels in the gene-trap model by Western blotting. Our Western blot analyses revealed that H3K9me1 and H3K9me2 levels significantly increased in the testes of Jmjd1a−/− mice. Fourth, the previous study showed direct association of Jmjd1a with the Tnp1 and Prm1 promoters and suggested that Jmjd1a directly regulates transcriptional activation of the Tnp1 and Prm1 genes. However, our study demonstrated that Jmjd1a regulates Tnp1, Tnp2, Prm1, and Prm2 expression through Crem/Act-mediated gene expression, an indirect mechanism (see below and Fig. 8). Finally, Jmjd1a−/− mice in this study showed more severe phenotype than the previous mutant model. For example, morphological defects of spermatids were observed at step 9, which was earlier than the defects found in the previous model at step 11; the sperm number decreased to 1:1000 in Jmjd1a−/− mice versus WT mice in this study, whereas it only decreased to 1:500 versus WT mice in the previous model.
FIGURE 8.
A working model of Jmjd1a function in spermatogenesis.
In summary, our findings support a simplified working model sketched in Fig. 8 for the physiological function of Jmjd1a in the process of spermatogenesis. The levels of H3K9me1 and H3K9me2 are high in pre-pachytene and early stage pachytene spermatocytes. Jmjd1a is first expressed in early stage pachytene spermatocytes and functions to decrease H3K9me1 and H3K9me2 levels in later stage pachytene spermatocytes, which allows an increase in histone acetylation and Act expression. Crem is also simultaneously expressed with Act starting in stage V pachytene spermatocytes (Fig. 7). The hypomethylation of H3K9 continues in Jmjd1a-negative spermatids, which creates a chromatin epigenetic modification environment suitable for recruitment of Crem and for transcriptional activation of Crem and Act target genes, including Tnp1, Tnp2, Prm1, and Prm2 in step 7 round spermatids and elongating spermatids.
Acknowledgments
We thank Dr. Ming Zhao for experimental assistance; Eric Dale Buras and Jean Ching-Yi Tien for critical reading; Dr. James Martin for the pFrt-LoxP plasmid, and Dr. Stephen Kistler for Tnp1 antibody. We thank Dr. F. DeMayo, the leader of the Transgenic Mouse Core, for microinjection.
This work was supported, in whole or in part, by National Institutes of Health Grants DK058242, CA112403, CA119689, and DK059820.
- Crem
- cAMP-response element modulator
- PBS
- phosphate-buffered saline
- WT
- wild type
- KO
- knock-out
- IHC
- immunohistochemistry
- ES
- embryonic stem
- ChIP
- chromatin immunoprecipitation
- AR
- androgen receptor.
REFERENCES
- 1.Russell L. D., Ettlin R. A., Sinha-Hikim A. P., Clegg E. D. (1990) Histological and Histopathological Evaluation of the Testis, pp. 1–161, Cache River Press, Clearwater, FL [Google Scholar]
- 2.Mali P., Kaipia A., Kangasniemi M., Toppari J., Sandberg M., Hecht N. B., Parvinen M. (1989) Reprod. Fertil. Dev. 1, 369–382 [DOI] [PubMed] [Google Scholar]
- 3.Nantel F., Monaco L., Foulkes N. S., Masquilier D., LeMeur M., Henriksén K., Dierich A., Parvinen M., Sassone-Corsi P. (1996) Nature 380, 159–162 [DOI] [PubMed] [Google Scholar]
- 4.Cho C., Willis W. D., Goulding E. H., Jung-Ha H., Choi Y. C., Hecht N. B., Eddy E. M. (2001) Nat. Genet. 28, 82–86 [DOI] [PubMed] [Google Scholar]
- 5.Yu Y. E., Zhang Y., Unni E., Shirley C. R., Deng J. M., Russell L. D., Weil M. M., Behringer R. R., Meistrich M. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4683–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao M., Shirley C. R., Yu Y. E., Mohapatra B., Zhang Y., Unni E., Deng J. M., Arango N. A., Terry N. H., Weil M. M., Russell L. D., Behringer R. R., Meistrich M. L. (2001) Mol. Cell. Biol. 21, 7243–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao M., Shirley C. R., Mounsey S., Meistrich M. L. (2004) Biol. Reprod. 71, 1016–1025 [DOI] [PubMed] [Google Scholar]
- 8.Fimia G. M., De Cesare D., Sassone-Corsi P. (1999) Nature 398, 165–169 [DOI] [PubMed] [Google Scholar]
- 9.Deng W., Lin H. (2002) Dev. Cell 2, 819–830 [DOI] [PubMed] [Google Scholar]
- 10.Giorgini F., Davies H. G., Braun R. E. (2002) Development 129, 3669–3679 [DOI] [PubMed] [Google Scholar]
- 11.Kotaja N., De Cesare D., Macho B., Monaco L., Brancorsini S., Goossens E., Tournaye H., Gansmuller A., Sassone-Corsi P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govin J., Caron C., Lestrat C., Rousseaux S., Khochbin S. (2004) Eur. J. Biochem. 271, 3459–3469 [DOI] [PubMed] [Google Scholar]
- 13.Martianov I., Brancorsini S., Catena R., Gansmuller A., Kotaja N., Parvinen M., Sassone-Corsi P., Davidson I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2808–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka H., Iguchi N., Isotani A., Kitamura K., Toyama Y., Matsuoka Y., Onishi M., Masai K., Maekawa M., Toshimori K., Okabe M., Nishimune Y. (2005) Mol. Cell. Biol. 25, 7107–7119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couldrey C., Carlton M. B., Nolan P. M., Colledge W. H., Evans M. J. (1999) Hum. Mol. Genet. 8, 2489–2495 [DOI] [PubMed] [Google Scholar]
- 16.Meistrich M. L., Trostle-Weige P. K., Lin R., Bhatnagar Y. M., Allis C. D. (1992) Mol. Reprod. Dev. 31, 170–181 [DOI] [PubMed] [Google Scholar]
- 17.Govin J., Escoffier E., Rousseaux S., Kuhn L., Ferro M., Thévenon J., Catena R., Davidson I., Garin J., Khochbin S., Caron C. (2007) J. Cell Biol. 176, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazzouri M., Pivot-Pajot C., Faure A. K., Usson Y., Pelletier R., Sèle B., Khochbin S., Rousseaux S. (2000) Eur. J. Cell Biol. 79, 950–960 [DOI] [PubMed] [Google Scholar]
- 19.Tachibana M., Nozaki M., Takeda N., Shinkai Y. (2007) EMBO J. 26, 3346–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godmann M., Auger V., Ferraroni-Aguiar V., Di Sauro A., Sette C., Behr R., Kimmins S. (2007) Biol. Reprod. 77, 754–764 [DOI] [PubMed] [Google Scholar]
- 21.Payne C., Braun R. E. (2006) Dev. Biol. 293, 461–472 [DOI] [PubMed] [Google Scholar]
- 22.Lahn B. T., Tang Z. L., Zhou J., Barndt R. J., Parvinen M., Allis C. D., Page D. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8707–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi K., Yoshida K., Matsui Y. (2005) Nature 438, 374–378 [DOI] [PubMed] [Google Scholar]
- 24.O'Carroll D., Scherthan H., Peters A. H., Opravil S., Haynes A. R., Laible G., Rea S., Schmid M., Lebersorger A., Jerratsch M., Sattler L., Mattei M. G., Denny P., Brown S. D., Schweizer D., Jenuwein T. (2000) Mol. Cell. Biol. 20, 9423–9433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akimoto C., Kitagawa H., Matsumoto T., Kato S. (2008) Genes Cells 13, 623–633 [DOI] [PubMed] [Google Scholar]
- 26.Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T. (2001) Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 27.Höög C., Schalling M., Grunder-Brundell E., Daneholt B. (1991) Mol. Reprod. Dev. 30, 173–181 [DOI] [PubMed] [Google Scholar]
- 28.Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) Cell 125, 483–495 [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Bassets I., Kwon Y. S., Telese F., Prefontaine G. G., Hutt K. R., Cheng C. S., Ju B. G., Ohgi K. A., Wang J., Escoubet-Lozach L., Rose D. W., Glass C. K., Fu X. D., Rosenfeld M. G. (2007) Cell 128, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellmann S., Bettkober M., Zelmer A., Seeger K., Faigle M., Eltzschig H. K., Bührer C. (2008) Biochem. Biophys. Res. Commun. 372, 892–897 [DOI] [PubMed] [Google Scholar]
- 31.Pollard P. J., Loenarz C., Mole D. R., McDonough M. A., Gleadle J. M., Schofield C. J., Ratcliffe P. J. (2008) Biochem. J. 416, 387–394 [DOI] [PubMed] [Google Scholar]
- 32.Lockman K., Taylor J. M., Mack C. P. (2007) Circ. Res. 101, e115–e123 [DOI] [PubMed] [Google Scholar]
- 33.Ko S. Y., Kang H. Y., Lee H. S., Han S. Y., Hong S. H. (2006) Cell Struct. Funct. 31, 53–62 [DOI] [PubMed] [Google Scholar]
- 34.Loh Y. H., Zhang W., Chen X., George J., Ng H. H. (2007) Genes Dev. 21, 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma D. K., Chiang C. H., Ponnusamy K., Ming G. L., Song H. (2008) Stem Cells 26, 2131–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tateishi K., Okada Y., Kallin E. M., Zhang Y. (2009) Nature 458, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng C., Wynshaw-Boris A., Zhou F., Kuo A., Leder P. (1996) Cell 84, 911–921 [DOI] [PubMed] [Google Scholar]
- 38.Liu W., Selever J., Wang D., Lu M. F., Moses K. A., Schwartz R. J., Martin J. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4489–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., pp. 15.44–15.48, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40.Wu J. Y., Ribar T. J., Cummings D. E., Burton K. A., McKnight G. S., Means A. R. (2000) Nat. Genet. 25, 448–452 [DOI] [PubMed] [Google Scholar]
- 41.Mark M., Yoshida-Komiya H., Gehin M., Liao L., Tsai M. J., O'Malley B. W., Chambon P., Xu J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeyaraj D. A., Grossman G., Petrusz P. (2005) Steroids 70, 704–714 [DOI] [PubMed] [Google Scholar]
- 43.Latendresse J. R., Warbrittion A. R., Jonassen H., Creasy D. M. (2002) Toxicol. Pathol. 30, 524–533 [DOI] [PubMed] [Google Scholar]
- 44.Boulanger C. A., Mack D. L., Booth B. W., Smith G. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin L., Liu Z., Chen H., Xu J. (2009) Cancer Res. 69, 3819–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A. E., Helin K. (2007) Nature 449, 731–734 [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., Reinberg D. (2001) Genes Dev. 15, 2343–2360 [DOI] [PubMed] [Google Scholar]
- 48.Thomas T., Loveland K. L., Voss A. K. (2007) Gene Expr. Patterns 7, 657–665 [DOI] [PubMed] [Google Scholar]
- 49.Delmas V., van der Hoorn F., Mellström B., Jégou B., Sassone-Corsi P. (1993) Mol. Endocrinol. 7, 1502–1514 [DOI] [PubMed] [Google Scholar]
- 50.Martin C., Zhang Y. (2005) Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 51.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. (2004) Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 52.Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. (2006) Nature 439, 811–816 [DOI] [PubMed] [Google Scholar]
- 53.Chang B., Chen Y., Zhao Y., Bruick R. K. (2007) Science 318, 444–447 [DOI] [PubMed] [Google Scholar]
- 54.Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) Nature 442, 312–316 [DOI] [PubMed] [Google Scholar]
- 55.De Gendt K., Swinnen J. V., Saunders P. T., Schoonjans L., Dewerchin M., Devos A., Tan K., Atanassova N., Claessens F., Lécureuil C., Heyns W., Carmeliet P., Guillou F., Sharpe R. M., Verhoeven G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Cesare D., Fimia G. M., Brancorsini S., Parvinen M., Sassone-Corsi P. (2003) Mol. Endocrinol. 17, 2554–2565 [DOI] [PubMed] [Google Scholar]
- 57.Okada Y., Scott G., Ray M. K., Mishina Y., Zhang Y. (2007) Nature 450, 119–123 [DOI] [PubMed] [Google Scholar]