Abstract
Anandamide (AEA) and other bioactive N-acylethanolamines (NAEs) are primarily inactivated by the enzyme fatty acid amide hydrolase (FAAH). Recently, FAAH-2 was discovered in humans, suggesting an additional enzyme can mediate NAE inactivation in higher mammals. Here, we performed a biochemical characterization of FAAH-2 and explored its capacity to hydrolyze NAEs in cells. In homogenate activity assays, FAAH-2 hydrolyzed AEA and palmitoylethanolamide (PEA) with activities ∼6 and ∼20% those of FAAH, respectively. In contrast, FAAH-2 hydrolyzed AEA and PEA in intact cells with rates ∼30–40% those of FAAH, highlighting a potentially greater contribution toward NAE catabolism in vivo than previously appreciated. In contrast to endoplasmic reticulum-localized FAAH, immunofluorescence revealed FAAH-2 was localized on lipid droplets. Supporting this distribution pattern, the putative N-terminal hydrophobic region of FAAH-2 was identified as a functional lipid droplet localization sequence. Lipid droplet localization was essential for FAAH-2 activity as chimeras excluded from lipid droplets lacked activity and/or were poorly expressed. Lipid droplets represent novel sites of NAE inactivation. Therefore, we examined substrate delivery to these organelles. AEA was readily trafficked to lipid droplets, confirming that lipid droplets constitute functional sites of NAE inactivation. Collectively, these results establish FAAH-2 as a bone fide NAE-catabolizing enzyme and suggest that NAE inactivation is spatially separated in cells of higher mammals.
Keywords: Eicosanoids/Anandamide, Enzymes/Lipid, Membrane/Trafficking, Metabolism/Lipid, FAAH, FAAH-2, Anandamide, Endocannabinoid
Introduction
N-Acylethanolamines (NAE)2 comprise a diverse family of endogenous signaling lipids (1, 2). The best studied NAEs include the endocannabinoid anandamide (AEA) and palmitoylethanolamide (PEA). AEA and PEA mediate various physiological functions, including nociception and inflammation (2). NAE signaling is terminated primarily through hydrolysis by fatty acid amide hydrolase (FAAH) (3–5). FAAH is an endoplasmic reticulum (ER)-localized enzyme belonging to the amidase signature family of serine hydrolases (6–9). FAAH knock-out mice possess elevated levels of NAEs and exhibit anti-nociceptive and anti-inflammatory phenotypes (5, 10), confirming the central role of FAAH in regulating in vivo NAE signaling in mice.
Recently, FAAH-2 was identified as a second FAAH enzyme capable of hydrolyzing NAEs in vitro (11). FAAH-2 is expressed in primates, humans, and a variety of other species, but is absent from murid genomes. Similar to FAAH, FAAH-2 possesses an amidase signature sequence and hydrolyzes numerous NAEs, including AEA and PEA in vitro (11). FAAH and FAAH-2 are co-expressed in several human tissues, including kidney, liver, lung, and prostate (11). In contrast, FAAH-2 but not FAAH is expressed in heart and ovary, suggesting that these enzymes may regulate overlapping yet distinct NAE pools in vivo. In this study, we examined the contribution of FAAH-2 toward NAE hydrolysis in intact cells and explored how its subcellular localization affects its activity.
EXPERIMENTAL PROCEDURES
Materials
COS-7 and HeLa cells were obtained from the American Type Culture Collection (Manassas, VA). DMEM, streptomycin, penicillin, sodium pyruvate, and fetal bovine serum were from Invitrogen (Carlsbad, CA). AEA and PEA were from Cayman Chemical (Ann Arbor, MI). [14C]AEA (ethanolamine-1-14C, 53 mCi/mmol) and [14C]PEA (ethanolamine-1-14C, 53 mCi/mmol) were from the National Institute on Drug Abuse Drug Supply Program. Fatty acid-free bovine serum albumin (BSA) was from Sigma. BODIPY 493/503 was from molecular probes (Carlsbad, CA).
Molecular Biology
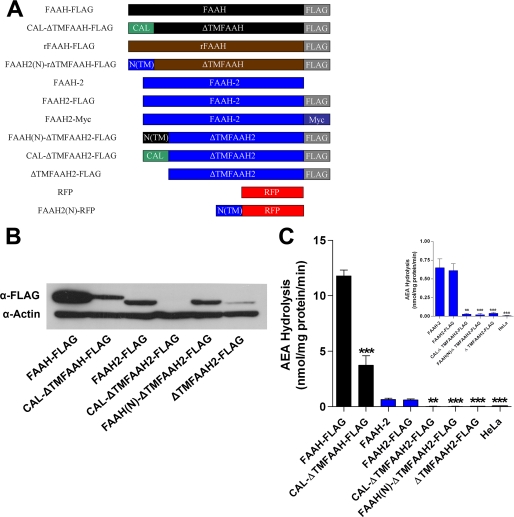
FAAH-2 cDNA was kindly provided by Benjamin F. Cravatt (The Scripps Research Institute). The sequence of the FAAH-2 cDNA provided to us slightly differed from the published mRNA sequence (NM_174912) through the following alterations: T99C, C822T, and G1077A. These changes did not alter the corresponding amino acid sequence. To generate FLAG-tagged FAAH-2, a FLAG epitope was incorporated into the C terminus of FAAH-2 by PCR and was inserted into pcDNA4. To generate Myc-tagged FAAH-2, the FAAH-2 cDNA lacking a stop codon was inserted into the pcDNA4/Myc-His plasmid (Clontech). To generate ER lumen-localized FAAH-2 (CAL-ΔTMFAAH2-FLAG), the putative N-terminal transmembrane region (TM) of FAAH-2 (residues 1–28) was replaced with the signal sequence (1–26) from mouse calreticulin. CAL-ΔTMFAAH-FLAG was generated by fusion of the calreticulin signal sequence to ΔTMFAAH-FLAG (residues 30–579). In CAL-ΔTMFAAH(N/Q)-FLAG, asparagine 334 in the consensus sequence for N-linked glycosylation was mutated to glutamine. FAAH(N)-ΔTMFAAH2-FLAG contains the N terminus of human FAAH (residues 1–32). ΔTMFAAH2-FLAG contains residues 35–532 of FAAH-2 and an N-terminal methionine. FAAH2(N)-RFP and FAAH2(N)-rΔTMFAAH-FLAG contain the N terminus of FAAH-2 (residues 1–35) fused to the N termini of DsRed2 or rat ΔTMFAAH-FLAG (residues 30–579), respectively. See Fig. 4A for a schematic of the above-mentioned constructs. The adipose differentiation-related protein (ADRP) expression plasmid has been previously described (12). All constructs were confirmed by sequencing.
FIGURE 4.
Expression and activities of cytoplasmically and luminally oriented FAAH-2 variants. A, constructs used in the study. ΔTMFAAH and ΔTMFAAH2 indicate the respective proteins lacking their putative N-terminal TM regions, N(TM) represents the N-terminal TM region of the indicated protein, CAL is the calreticulin signal sequence, and RFP represents red fluorescent protein. FLAG and Myc are the epitope tags used. B, relative expression of FAAH-FLAG, FAAH2-FLAG, and cytoplasmically and luminally facing FAAH and FAAH-2 variants. Transfected HeLa homogenates (20 μg) were separated by SDS-PAGE and probed with anti-FLAG and anti-β-actin antibodies. C, enzymatic activities of cell lysates containing the aforementioned proteins. Activity of CAL-ΔTMFAAH-FLAG was compared with FAAH-FLAG (black bars). For the rest of the proteins, activities were compared with FAAH-2 (blue bars). Inset: activities of FAAH-2 and its corresponding chimeras. **, p < 0.01 and ***, p < 0.001 (n = 4–6).
[14C]AEA and [14C]PEA Cellular Uptake and Hydrolysis
Twenty-four hours following transfection with the indicated plasmids using Lipofectamine 2000 (Invitrogen), the cells were plated at ∼90% confluency in 35-mm dishes. The cells were washed twice in DMEM and subsequently incubated for 5 min, 1 min, or 3 s with 750 μl of [14C]AEA (100 nm) or [14C]PEA (100 nm or 1 μm) that were respectively pre-equilibrated for 75 or 90 min in medium containing 0.15% defatted BSA as previously described (13). Following the incubation, 750 μl of ice-cold DMEM plus 0.15% BSA was added to the plates, the media separated from cells, which were then washed with DMEM plus 0.15% BSA to remove non-specifically bound AEA or PEA. The cells were scraped three times with 400 μl of ice-cold 2 mm EDTA in PBS and two volumes of chloroform:methanol (1:1) added to the media and cells, and the phases were separated by centrifugation. The resulting aqueous (containing [14C]ethanolamine) and organic (containing intact [14C]AEA/PEA) phases were counted by liquid scintillation counting. [14C]AEA/PEA uptake was determined by summing the production of [14C]ethanolamine in the media and cells with intact cellular [14C]AEA/PEA. Hydrolysis of [14C]AEA/PEA was quantified by production of [14C]ethanolamine in the media and cells. Statistical significance was determined between transfected cells and FAAH-transfected controls.
Immunofluorescence
Immunolocalization experiments were performed essentially as described (14). Briefly, cells were fixed with 4% paraformaldehyde for 15 min, permeabilized on ice with 0.2% Triton X-100 in 5% goat serum for 5 min, and incubated for 1 h with primary antibodies as follows: rabbit anti-calreticulin (1:200, Affinity Bioreagents, Golden, CO), mouse anti-FLAG M2 (1:500, Sigma), or mouse anti-Myc (1:500, from Jen-Chih Hsieh, Stony Brook University). To visualize neutral lipids, COS-7 or HeLa cells were respectively treated with 1 or 2 μg/ml BODIPY 493/503 during secondary antibody incubations. All images were acquired using a Zeiss LSM 510 META NLO two-photon laser scanning microscope.
Western Blotting
Proteins in cell homogenates were resolved by SDS-PAGE as described (13). Following transfer to a nitrocellulose membrane at 100 V for 25 min, the blots were blocked for 1 h in 5% nonfat dry milk in PBS Tween (PBST). The blots were probed with mouse anti-FLAG M2 (1:2,000), mouse anti-β-actin (1:20,000, Abcam, Cambridge, MA), or mouse anti-Myc (1:5,000) antibodies for 1 h with shaking. The blots were rinsed three times with PBST and incubated with goat anti-mouse IgG horseradish peroxidase-conjugated antibodies (Molecular Probes, Eugene, OR) for 1 h. The blots were rinsed three times with PBST, developed using the Immun-star horseradish peroxidase substrate (Bio-Rad), and exposed to film.
Membrane Topology Analysis
COS-7 or HeLa cells transiently transfected with the indicated proteins were pelleted and resuspended in buffer containing 10 mm HEPES-NaOH, pH 7.5, containing 1 mm EDTA, 1.5 mm MgCl2, 10 mm KCl, and 250 mm sucrose, and homogenized by passage through a 26-gauge needle. Unbroken cells and nuclei were pelleted by centrifugation at 1,000 × g for 10 min, and the resulting supernatants were subjected to centrifugation at 100,000 × g for 60 min at 4 °C. The membrane pellets were resuspended in buffer containing 50 mm Tris, pH 8, 3 mm CaCl2, 1.5 mm MgCl2, 10 mm KCl, 100 mm NaCl, 250 mm sucrose and treated with 500 μg/ml proteinase K for 30 min at 37 °C in the presence or absence of 1% Triton X-100, or left untreated. The reactions were quenched by the addition of 20 mm phenylmethylsulfonyl fluoride. The samples were separated by SDS-PAGE and visualized by immunoblotting with anti-FLAG (1:2,000) or anti-calreticulin (1:2,000) antibodies.
Enzyme Assays
FAAH activity assays were performed as previously described with modifications (14). Briefly, cell homogenates were incubated with 100 μm AEA or PEA plus 0.1 μCi of [14C]AEA/PEA in Tris-HCl (pH 9) containing 0.1% BSA. For kinetic analyses (apparent Km and Vmax), time and protein concentrations were determined to maintain substrate conversion at ∼10% while varying substrate concentrations from 0.1 to 100 μm. Reactions were stopped by the addition of two volumes of 1:1 chloroform:methanol, and the phases were separated by centrifugation. The methanol phase (containing [14C]ethanolamine) was sampled and quantified using a Beckman LS 6500 scintillation counter.
Lipid Droplet Isolation and Subcellular Fractionation
HeLa cells grown in 10-cm dishes were co-transfected with ADRP and the indicated plasmids using Lipofectamine 2000. Five hours later, the media were replaced with DMEM supplemented with 10% fetal bovine serum and 400 μm oleic acid complexed to defatted BSA (oleic acid to BSA ratio of 6.6:1) and incubated overnight as described (12). Cells fed overnight with oleic acid accumulate neutral lipids and possess enlarged lipid droplets. Three plates of FAAH2-FLAG-, one plate of FAAH-FLAG-, and three plates of FAAH2(N)-rΔTMFAAH-FLAG-transfected cells were homogenized in 10 mm Hepes/5 mm EDTA containing 10% sucrose and protease inhibitor mixture (Roche Applied Science), and then centrifuged at 1,000 × g to pellet unbroken cells. The resulting supernatant (700 μl) was overlaid with ∼ 4.3 ml of 10 mm Hepes/5 mm EDTA and spun in a Beckman L8–55 centrifuge at 280,000 × g for 3 h at 4 °C. Fractions (∼700 μl) were collected from the top, and the pellets were resuspended in Hepes buffer. Following acetone precipitation, the samples were separated by SDS-PAGE and immunoblotted with rabbit anti-calnexin (1:5,000, Novus Biologicals, Littleton, CO), mouse anti-FLAG (1:2,000), or guinea pig anti-ADRP antibodies (1:400, RDI Division of Fitzgerald Industries, Concord, MA). The blots were subsequently incubated with goat anti-mouse, goat anti-rabbit, or rabbit anti-guinea pig IgG horseradish peroxidase-conjugated antibodies (1:8,000, Molecular Probes, Eugene, OR) and visualized on film.
Deglycosylation of Proteins by PNGase F
HeLa cells transfected with FAAH-FLAG, FAAH2(N)-rΔTMFAAH-FLAG, or CAL-ΔTMFAAH-FLAG were homogenized, and membrane fractions were generated as described above. A small pool (∼5 μg) of the cytosolic fraction (expressing β-actin) was added to the membranes (40 μg) prior to PNGase F treatment. The membranes were subsequently boiled for 10 min at 100 °C and incubated in the presence or absence of PNGase F (New England Biolabs, Ipswich, MA) for 60 min at 37 °C. The samples were then separated by SDS-PAGE and visualized by immunoblotting with anti-FLAG (1:2,000) or anti-β-actin (1:20,000) antibodies.
Statistical Analysis
All results represent means ± S.E. of at least three independent experiments. Statistical analyses were performed using two-tailed unpaired Student's t tests with GraphPad Prism.
RESULTS
Enzymatic Activity and Kinetics of FAAH-2
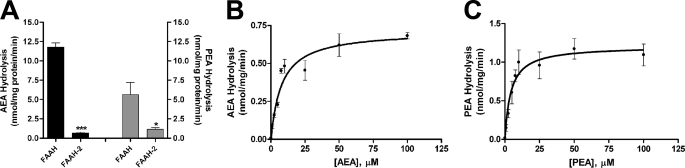
FAAH-2 displayed optimal in vitro activity at pH 9 (data not shown), confirming previous results (11). Enzyme kinetics of FAAH-2 were analyzed in homogenates of FAAH-2-transfected HeLa cells using AEA and PEA as substrates. FAAH-2 hydrolyzed AEA with rates that were ∼6% those of FAAH (Fig. 1A). PEA hydrolysis by FAAH-2 was ∼20% that of FAAH. The apparent Vmax and Km values of FAAH-2 for AEA were 0.71 ± 0.04 nmol/mg/min and 7.9 ± 1.5 μm, respectively (Fig. 1B). For PEA, the apparent Vmax and Km values were 1.21 ± 0.1 nmol/mg/min and 4.3 ± 1.4 μm, respectively (Fig. 1C).
FIGURE 1.
Enzymatic activity and kinetics of FAAH-2. A, hydrolysis of [14C]AEA (black bars) or [14C]PEA (gray bars) by HeLa homogenates expressing FAAH or FAAH-2. AEA/PEA hydrolysis by vector-transfected cells was subtracted from all samples. *, p < 0.05; ***, p < 0.001 (n = 3–4). B, [14C]AEA (0.1–100 μm) hydrolysis by HeLa homogenates expressing FAAH-2. [14C]AEA was hydrolyzed with an apparent Vmax and Km of 0.71 ± 0.04 nmol/mg/min and 7.9 ± 1.5 μm, respectively (n = 7). C, [14C]PEA (0.1–100 μm) was hydrolyzed with an apparent Vmax and Km of 1.21 ± 0.1 nmol/mg/min and 4.3 ± 1.4 μm, respectively (n = 7).
Hydrolysis of AEA and PEA by FAAH-2 in Cells
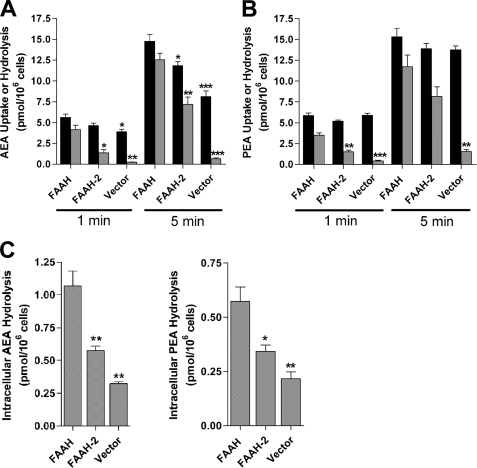
The contributions of FAAH and FAAH-2 toward NAE hydrolysis were examined in transiently transfected HeLa cells using an established protocol and physiological (100 nm to 1 μm) substrate concentrations (13, 15). A time course of [14C]AEA (100 nm) uptake and hydrolysis revealed that HeLa cells expressing FAAH hydrolyzed ∼74% of the [14C]AEA taken up by cells at 1 min and ∼85% at 5 min (Fig. 2A). FAAH-2 expressing cells hydrolyzed ∼29% of the [14C]AEA taken up by cells at 1 min and ∼61% at 5 min. Therefore, FAAH-2-transfected cells hydrolyzed [14C]AEA with moderately lower efficiency (p < 0.05) compared with cells expressing FAAH (Fig. 2A), but at a rate ∼5-fold higher than in the homogenate activity assay. Because AEA uptake is coupled to its hydrolysis (16, 17), FAAH-2-transfected cells internalized less [14C]AEA compared with FAAH-transfected controls at 5 min due to reduced AEA hydrolysis. Mock transfected cells hydrolyzed ∼7 and ∼8% of the AEA taken up by cells at 1 and 5 min, respectively. AEA hydrolysis was also examined using the physiologically more relevant time point of 3 s. Similar to the results above, intracellular [14C]AEA hydrolysis in FAAH-2-expressing cells was moderately lower (p < 0.05) compared with FAAH-transfected cells (Fig. 2C). Following subtraction of mock transfected controls, the data revealed that FAAH-2 hydrolyzed AEA with a rate ∼30% that of FAAH. Similar results were observed in COS-7 cells (data not shown).
FIGURE 2.
Uptake and hydrolysis of AEA and PEA in transfected HeLa cells. A, HeLa cells transfected with FAAH, FAAH-2, or vector controls were incubated with 100 nm [14C]AEA for 1 or 5 min and uptake (black bars) and hydrolysis (gray bars) determined. Significance was determined between FAAH-transfected controls and FAAH-2/vector-transfected cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (n = 3). B, uptake and hydrolysis of 100 nm [14C]PEA by HeLa cells transfected with FAAH, FAAH-2, or vector controls. **, p < 0.01; ***, p < 0.001 (n = 3). C, intracellular hydrolysis of [14C]AEA and [14C]PEA (both at 100 nm) following uptake at 3 s in HeLa cells transfected with FAAH, FAAH-2 or vector controls. *, p < 0.05, **, p < 0.01 (n = 3).
PEA inactivation followed a similar trend. FAAH hydrolyzed ∼60 and ∼77% of the [14C]PEA (100 nm) taken up by cells at 1 and 5 min, respectively (Fig. 2B). [14C]PEA metabolism was lower in cells transfected with FAAH-2, with ∼29 and ∼59% hydrolysis at 1 and 5 min, respectively. [14C]PEA hydrolysis in mock transfected cells reached ∼7 and 11% at 1 and 5 min, respectively. Similar results were obtained with 1 μm PEA (data not shown). It is noteworthy that, in contrast to AEA, PEA uptake does not appear to be coupled to its hydrolysis by FAAH (18). Similar results were obtained in this study (Fig. 2B). This may result from intracellularly sequestered PEA that is not in equilibrium with extracellular PEA. Similar to AEA, [14C]PEA hydrolysis at 3 s was lower (p < 0.05) in cells expressing FAAH-2 compared with FAAH (Fig. 2C). Following subtraction of [14C]PEA metabolism in mock transfected controls, FAAH-2 hydrolyzed [14C]PEA with a rate ∼40% that of FAAH. Collectively, these data provide evidence that FAAH-2 is an efficient NAE-hydrolyzing enzyme in intact cells.
Subcellular Localization of FAAH-2
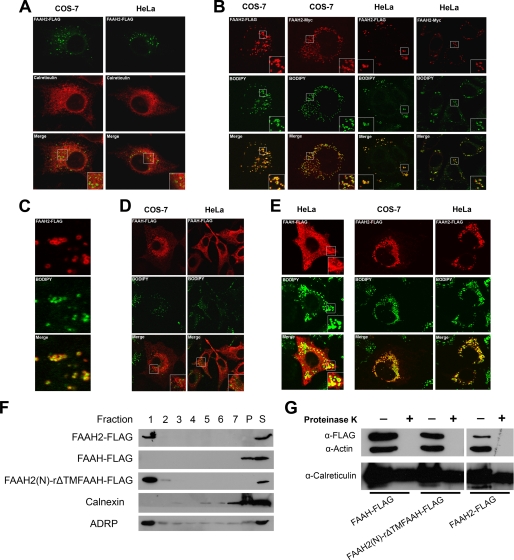
Because the efficiency of enzyme activity is related to substrate delivery, the subcellular localization of FAAH-2 was compared with FAAH. FAAH-2 constructs containing FLAG or Myc epitope tags were employed for the immunolocalization studies. Immunofluorescence revealed that, in transfected COS-7 and HeLa cells, FAAH2-FLAG localized to ring-like, hollow cytoplasmic structures that did not overlap with the ER marker calreticulin (Fig. 3A). In addition to these structures, a minor pool of ER-localized FAAH-2 was typically observed in COS-7 cells (in ∼60% of cells) and less frequently in HeLa cells (<10% of cells).
FIGURE 3.
Subcellular localization of FAAH-2 in COS-7 and HeLa cells. A, COS-7 and HeLa cells were transiently transfected with FAAH2-FLAG, fixed, permeabilized, and stained with anti-FLAG or anti-calreticulin antibodies. The top panel shows FAAH-2, the middle panel represents calreticulin, and the bottom panel shows the merged images. B, COS-7 and HeLa cells expressing FAAH2-FLAG or FAAH2-Myc were incubated with anti-FLAG or anti-Myc antibodies and stained with BODIPY493/503. The top panel shows FAAH-2, the middle panel represents BODIPY493/503, and the bottom panel sows the merged images. C, close-up image of a FAAH2-FLAG expressing HeLa cell stained with BODIPY493/503. D, localization of FAAH-FLAG in COS-7 and HeLa cells treated with BODIPY493/503. E, subcellular localization of FAAH-FLAG and FAAH2-FLAG following an overnight incubation with 400 μm oleic acid. F, distribution of FAAH-FLAG, FAAH2-FLAG, and FAAH2(N)-rΔTMFAAH-FLAG after subcellular fractionation. HeLa cells were co-transfected with ADRP and the indicated plasmids and were grown overnight in media supplemented with 400 μm oleic acid. The post-nuclear supernatants were subjected to ultracentrifugation. Fractions were collected from the top with fraction 1 corresponding to the floating lipid droplet layer, P representing the pellet, and S the unfractionated post-nuclear supernatant. The fractions were resolved by SDS-PAGE and probed with anti-FLAG, anti-calnexin, or anti-ADRP antibodies. G, proteinase K protection analysis of FAAH-FLAG, FAAH2(N)-rΔTMFAAH-FLAG, and FAAH2-FLAG in post-nuclear supernatants of COS-7 cells. Supernatants (60 μg) were either left untreated or were incubated with 500 μg/ml proteinase K for 30 min at 37 °C. The samples were subsequently resolved by SDS-PAGE and probed with anti-FLAG, anti-β-actin, and anti-calreticulin antibodies.
These cytoplasmic structures are morphologically similar to lipid droplets/adiposomes (19). Lipid droplets are composed of a neutral lipid core surrounded by a phospholipid monolayer and serve as sites of triglyceride and cholesteryl ester storage and mobilization (20). The neutral lipid/lipid droplet-specific dye BODIPY 493/503 was employed to determine whether FAAH-2 localized to lipid droplets (12). In COS-7 and HeLa cells, FLAG- and Myc-tagged FAAH-2 co-localized with BODIPY 493/503 (Fig. 3B). These results confirmed that FAAH-2 is present on lipid droplets and that its subcellular distribution is independent of the epitope tag and cell-type used. FAAH-2 was present solely on the phospholipid surface of lipid droplets and was excluded from BODIPY493/503-positive neutral lipid cores (Fig. 3C) (21). In contrast to FAAH-2, FAAH did not localize to lipid droplets (Fig. 3D).
Our next goal was to confirm the localization of FAAH-2 to lipid droplets by subcellular fractionation. To facilitate isolation of lipid droplets by gradient centrifugation, we needed to enlarge lipid droplets by overnight incubation with oleic acid. For this reason, we first verified that oleic acid incubation did not affect the localization of FAAH-2 as judged by fluorescence microscopy. Indeed, incubation of cells in oleic acid did not alter the localization of FAAH-2, although a minor pool of FAAH appeared to have redistributed to lipid droplets (Fig. 3E). HeLa cells were co-transfected with the lipid droplet-resident protein ADRP and either FAAH-FLAG or FAAH2-FLAG, cultured overnight in media supplemented with oleic acid, and subsequently fractionated by ultracentrifugation as previously described (12). Following ultracentrifugation, fractions were collected from the top to isolate lipid droplets. The separation of lipid droplets from ER membranes was confirmed by the distribution of ADRP to fraction 1 and the ER marker calnexin to fraction 7 and the pellet (Fig. 3F). FAAH2-FLAG was present solely in the ADRP-positive fraction 1 and was excluded from calnexin-positive fractions. As expected, FAAH-FLAG distributed solely to the calnexin-positive fraction and was excluded from lipid droplets.
Lastly, we used proteinase K protection analysis of post-nuclear supernatants of FAAH-2-expressing cells to verify that FAAH-2 is a cytoplasmically oriented enzyme, as predicted by its lipid droplet localization. This was necessary, because FAAH-2 was proposed to reside in luminal membrane compartments (11). FAAH-2 was efficiently degraded by proteinase K, showing that little or none of the protein was present in the lumen of the ER or other organelles (Fig. 3G). As expected, the cytoplasmically oriented FAAH (11, 13) was also completely degraded by proteinase K. Taken together, the fractionation, immunofluorescence, and protease protection results confirm the lipid droplet localization of FAAH-2.
FAAH-2 Activity Is Dependent upon Lipid Droplet Localization
We next sought to determine the functional significance of lipid droplet localization upon FAAH-2 activity. Toward this end, we attempted to generate FAAH-2 variants that were excluded from lipid droplets (Fig. 4A). We first decided to target FAAH-2 to the cytoplasmic face of the ER by replacing its putative N-terminal TM region (residues 1–34) with the analogous domain of FAAH (residues 1–32). The resulting construct, FAAH(N)-ΔTMFAAH2-FLAG, was expressed at a comparable level to FAAH-2 (Fig. 4B). Protease protection analysis revealed that FAAH(N)-ΔTMFAAH2-FLAG was localized to the cytoplasmic face of membranes (Fig. 5A). Immunofluorescence analysis showed that the protein did not co-localize with BODIPY 493/503, confirming its exclusion from lipid droplets (Fig. 5B). Unexpectedly, enzymatic analysis revealed that FAAH(N)-ΔTMFAAH2-FLAG was catalytically inactive (Fig. 4C).
FIGURE 5.
Membrane orientation and localization of cytoplasmically and luminally oriented ER FAAH-2 chimeras. A, membrane orientation of FAAH-2 variants. Membrane fractions of COS-7 cells transfected with the indicated proteins were treated with 500 μg/ml proteinase K in the presence or absence of 1% Triton X-100 for 30 min at 37 °C. Control samples were left untreated. The samples were resolved by SDS-PAGE and probed with FLAG or calreticulin antibodies. B, immunolocalization of cytoplasmically and luminally oriented FAAH and FAAH-2 chimeras. COS-7 cells expressing CAL-ΔTMFAAH2-FLAG, CAL-ΔTMFAAH-FLAG, or FAAH(N)-ΔTMFAAH2-FLAG were probed with anti-FLAG antibodies and stained with BODIPY 493/503. The top panel represents the FAAH/FAAH-2 variants, the middle panel shows BODIPY 493/503, and the bottom panel shows the merged image. C, PNGase F sensitivity of FAAH chimeras. Membrane fractions of HeLa cells expressing FAAH-FLAG, FAAH2(N)-rΔTMFAAH-FLAG, CAL-ΔTMFAAH-FLAG, or CAL-ΔTMFAAH(N/Q)-FLAG were incubated in the presence or absence of PNGase F as described under “Experimental Procedures.” The proteins were subsequently separated by SDS-PAGE and probed with anti-FLAG and anti-β-actin antibodies. Note that CAL-ΔTMFAAH(N/Q)-FLAG was poorly expressed.
We next made a second FAAH-2 variant, ΔTMFAAH2-FLAG, by deleting the N-terminal hydrophobic domain from FAAH-2 (Fig. 4A). We predicted that, similar to ΔTM-FAAH (7, 22), this protein would localize to the cytoplasmic face of the ER. Unexpectedly, the expression of this construct was significantly reduced compared with FAAH-2 (Fig. 4B). This suggested that FAAH-2, unlike FAAH (7, 22), requires an N-terminal hydrophobic region for stable expression. Nevertheless, ΔTMFAAH2-FLAG behaved in a similar manner to FAAH(N)-ΔTMFAAH2-FLAG: it localized to the cytoplasmic face of membranes (Fig. 5A) and was enzymatically inactive (Fig. 4C).
FAAH-2 was previously suggested to reside in the ER lumen (11). Therefore, we sought to determine whether a FAAH-2 variant that was re-localized to the ER lumen would retain its activity. A luminally oriented FAAH-2 chimera was generated by replacing the hydrophobic N terminus of FAAH2 with the signal sequence of mouse calreticulin (residues 1–26) (Fig. 4A). CAL-ΔTMFAAH2-FLAG was protected from protease digestion unless membranes were dissolved with Triton X-100, consistent with translocation into the ER lumen (Fig. 5A). Unexpectedly, though, its expression was extremely low (Fig. 4B). The reduced expression of CAL-ΔTMFAAH2-FLAG may have resulted from instability of the protein following cleavage of the calreticulin signal sequence within the ER lumen. Therefore, we engineered a luminally oriented fusion protein that would retain the N terminus of FAAH-2 following signal sequence cleavage. However, similar to CAL-ΔTMFAAH2-FLAG, this chimera was also poorly expressed (data not shown). These proteins were not detected in the media, ruling out secretion as a cause for the apparent low protein expression (data not shown).
Similar to the cytoplasmically oriented FAAH-2 variants, the luminally oriented CAL-ΔTMFAAH2-FLAG was also catalytically inactive (Fig. 4C). To determine whether the lack of FAAH-2 expression in the ER lumen was intrinsic to this enzyme, a luminally oriented FAAH construct was prepared (Fig. 4A). Similar to its FAAH-2 counterpart, CAL-ΔTMFAAH-FLAG localized to the luminal face of ER membranes (Fig. 5, A and B). However, despite a reduction in expression, CAL-ΔTMFAAH-FLAG retained its enzymatic activity (Fig. 4, B and C). These results suggest that FAAH is a versatile enzyme capable of functioning in both reducing (cytosol) and oxidizing (ER) environments. In contrast, FAAH-2 is an enzyme adapted to enzymatic activity on the lipid droplet surface.
The N Terminus of FAAH-2 Is a Lipid Droplet Localization Sequence
We next sought to identify the lipid droplet-targeting sequence of FAAH-2. Our efforts focused upon the N-terminal hydrophobic region of FAAH-2, because its removal abrogated the activity and lipid droplet localization of the enzyme (Figs. 4C and 5B). To determine whether this region mediates lipid droplet targeting, we fused this sequence to red fluorescent protein (RFP), generating FAAH2(N)-RFP. In transfected COS-7 cells, RFP localized to the cytoplasm and nucleus and did not co-localize with BODIPY 493/503 (Fig. 6A). In contrast, FAAH2(N)-RFP co-localized with BODIPY 493/503, confirming that the N terminus of FAAH-2 is sufficient to target RFP to lipid droplets (Fig. 6A). We next determined whether this domain could target FAAH to lipid droplets. The N-terminal hydrophobic domain (residues 1–29) of rat FAAH was replaced with that of human FAAH-2 (residues 1–35) to generate FAAH2(N)-rΔTMFAAH-FLAG (Fig. 4A). Note that rat, rather than human FAAH, was used in these studies due to poor expression of this human FAAH variant. In contrast to ER-localized rat FAAH (13), immunofluorescence confirmed that FAAH2(N)-rΔTMFAAH-FLAG localized to lipid droplets in COS-7 and HeLa cells (Fig. 6B). The intracellular distribution of this protein to lipid droplets was confirmed by subcellular fractionation (Fig. 3F). These results establish the N terminus of FAAH-2 as a lipid droplet-targeting sequence.
FIGURE 6.
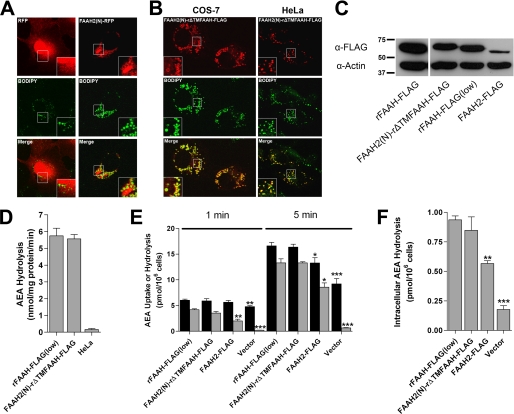
The N terminus of FAAH-2 is a lipid droplet localization sequence. A, COS-7 cells transfected with RFP or FAAH2(N)-RFP were fixed and stained with BODIPY493/503. The top panel shows RFP, the middle panel depicts BODIPY493/503, and the bottom panel shows the merged images. Note that RFP aggregates were present in every FAAH2(N)-RFP-expressing cell examined. B, co-localization of FAAH2(N)-rΔTMFAAH-FLAG with BODIPY493/503 in COS-7 and HeLa cells. C, expression of rFAAH-FLAG, rFAAH-FLAG(low), FAAH2(N)-rΔTMFAAH-FLAG, and FAAH2-FLAG in HeLa cells. rFAAH-FLAG(low) represents rat FAAH-FLAG expressed at levels comparable to FAAH2(N)-rΔTMFAAH-FLAG. The proteins were resolved by SDS-PAGE and probed with anti-FLAG or anti-β-actin antibodies. D, similar (p > 0.05) enzymatic activities of rFAAH-FLAG(low) and FAAH2(N)-rΔTMFAAH-FLAG in homogenates of HeLa cells (n = 3). E, uptake (black bar) and hydrolysis (gray bar) of 100 nm [14C]AEA by HeLa cells transiently transfected with rFAAH-FLAG(low), FAAH2(N)-rΔTMFAAH-FLAG, FAAH2-FLAG, or vector controls following 1- or 5-min incubations. Statistical significance was determined between transfected cells and rFAAH-FLAG(low)-transfected controls. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (n = 3). F, intracellular hydrolysis of [14C]AEA in transfected HeLa cells following uptake at 3 s. **, p < 0.01; ***, p < 0.001 (n = 3).
FAAH-2 Resides Solely in Cytoplasmic Membrane Compartments
A prior report concluded that FAAH-2 resides in the ER lumen and does not undergo N-glycosylation, because it was insensitive to PNGase F (11). In sharp contrast, the data presented here support the localization of FAAH-2 on cytoplasmic lipid droplets. In addition, we did not detect any protease-protected FAAH-2 in post-nuclear supernatants (Fig. 3G). To examine luminal orientation more thoroughly, we used PNGase F sensitivity as an indicator of protein translocation into the ER lumen, because a consensus site for N-glycosylation is present in both FAAH and FAAH-2. In validation of this approach, gel mobility of the luminally oriented FAAH variant CAL-ΔTMFAAH-FLAG was altered by PNGase F (Fig. 5C). The specificity of the deglycosylation reaction was confirmed in cells expressing CAL-ΔTMFAAH(N/Q)-FLAG, a luminally oriented FAAH variant in which the asparagine that undergoes N-glycosylation was mutated to glutamine. As expected, this protein was insensitive to PNGase F (Fig. 5C).
We next applied the PNGase F sensitivity assay toward analyzing the membrane orientation of FAAH-2. We employed the lipid droplet-targeted FAAH variant, FAAH2(N)-rΔTMFAAH-FLAG, which mimics the localization pattern of FAAH-2, to determine if FAAH-2 is luminally oriented. Similar to FAAH-2, FAAH2(N)-rΔTMFAAH-FLAG was insensitive to PNGase F (Fig. 5C), confirming its localization to the cytoplasmic face of lipid droplets. As expected, wild-type FAAH was insensitive to PNGase F. These data further support the notion that FAAH-2 is not translocated into the ER lumen.
AEA Is Efficiently Delivered to Lipid Droplets for Hydrolysis
Lipid droplets represent novel intracellular sites for NAE catabolism. Therefore, we sought to determine whether NAEs were efficiently delivered to lipid droplets for hydrolysis. Our approach was to compare AEA uptake and hydrolysis in HeLa cells expressing either ER- or lipid droplet-targeted FAAH. We first needed to ensure that the two proteins were expressed at the same level, and had similar intrinsic enzymatic activities. Rat FAAH (rFAAH-FLAG) was expressed at a higher level than lipid droplet-localized FAAH2(N)-rΔTMFAAH-FLAG (Fig. 6C). Therefore, its expression was reduced by transfecting cells with less plasmid (0.5 μg versus 1.6 μg/35-mm dish), yielding rFAAH-FLAG(low). Western blotting and enzymatic assays confirmed similar expression and intrinsic activities of both proteins (Fig. 6, C and D).
We next examined the capacity of lipid droplet-localized FAAH to hydrolyze AEA. AEA uptake and hydrolysis were similar between cells expressing rFAAH-FLAG(low) or FAAH2(N)-rΔTMFAAH-FLAG (Fig. 6, E and F). In contrast, AEA hydrolysis was significantly lower in cells expressing FAAH-2, confirming an intrinsically lower activity of FAAH-2 toward NAEs. Taken together, these data confirm that AEA is readily delivered to lipid droplets, thereby establishing lipid droplets as functional sites of NAE catabolism.
DISCUSSION
Regulation of cellular NAE pools requires a coordinated set of synthetic and catabolic enzymes. FAAH is the principal enzyme that degrades NAEs in mice (5, 10). In addition to FAAH, NAE-hydrolyzing acid amidase hydrolyzes PEA, although its role in PEA degradation is restricted to macrophages (23, 24). The recent discovery of FAAH-2 in humans suggests an additional enzyme capable of hydrolyzing NAEs in higher mammals (11).
The in vitro activity profile of FAAH-2 in the current study is in general agreement with the findings of Wei et al. (11), confirming that FAAH-2 homogenates hydrolyze NAEs with rates 5- to 30-fold lower than FAAH. In contrast, FAAH-2 hydrolyzed AEA and PEA with rates ∼30–40% those of FAAH in intact cells. This suggests that in vitro activities, employing high substrate concentrations, do not reveal the true catabolic capacities of NAE-inactivating enzymes in cells. The elevated activity of FAAH-2 in intact cells is supported by its low Km values for AEA and PEA, which are severalfold lower than those reported for FAAH (25, 26). This suggests higher enzymatic activity at physiological substrate concentrations (i.e. ≤1 μm), such as those used in our intact cell system. Therefore, FAAH and FAAH-2 may coordinate NAE inactivation in vivo, especially in tissues with similar expression of both enzymes such as the kidney, liver, lung, and prostate (11). In heart and ovary, which lack significant FAAH expression, FAAH-2 may constitute the sole NAE-catabolizing enzyme.
Immunofluorescence and biochemical analyses revealed that FAAH-2 localized to cytoplasmic lipid droplets. Lipid droplets function as dynamic sites of triacylglyceride and cholesteryl ester storage and mobilization (20, 27–31). Three classes of proteins localize to lipid droplets: lipid-anchored proteins such as RAB18, structural lipid droplet proteins such as the perilipin, adipophilin, and Tip47 family of proteins, and monotopic membrane proteins (20, 31). FAAH-2 belongs to the last category of monotopic membrane proteins.
In contrast to FAAH-2, FAAH did not localize to lipid droplets. Instead, FAAH localization was restricted to ER membranes, in agreement with previous studies (7, 14, 22, 32, 33). However, it was recently proposed that a small pool of FAAH partitioned to lipid droplets in HaCaT keratinocytes (34). We observed a minor fraction of lipid droplet-localized FAAH in oleic acid fed cells by immunofluorescence but not subcellular fractionation. This may suggest redistribution of FAAH to lipid droplets following oleic acid supplementation. Alternatively, the apparent localization of FAAH on lipid droplets may reflect close apposition of lipid droplets to ER membranes containing FAAH.
The N terminus of FAAH-2 was necessary and sufficient to mediate lipid droplet localization. This hydrophobic domain bears striking similarity to the N-terminal lipid droplet localization sequence of methyltransferase-like 7B (35). Both peptides are hydrophobic with the sequence of FAAH-2 possessing interspersed basic residues. Curiously, the N terminus of FAAH-2 contains several α-helix disrupting glycines and prolines (11), a common feature in lipid droplet-targeting sequences (20).
We hypothesize that the N terminus of FAAH-2 is required for proper enzyme folding, as removal of this region greatly reduces expression and renders the enzyme inactive, likely by destabilizing its structure. We showed that replacement of the N terminus with a complementary transmembrane segment from FAAH restored FAAH-2 expression but not its enzymatic activity. These data suggest that the N terminus of FAAH-2 may adopt a distinct conformation (e.g. amphipathic helix) from that of the FAAH N terminus. This is consistent with the exclusion of proteins bearing transmembrane helices from lipid droplets (20). It is likely that this conformational difference is required for proper positioning of the enzyme upon the lipid droplet phospholipid monolayer in a catalytically competent state. Altogether, these data suggest that FAAH-2 is adapted for enzymatic catalysis on the lipid droplet surface.
Lipid droplets represent novel intracellular sites of NAE catabolism. Therefore, it was essential to establish that intracellular NAE delivery to lipid droplets was efficient. We found that AEA readily reached lipid droplets and ER membranes, consistent with intracellular trafficking mediated by fatty acid binding proteins, Hsp70.2, or as yet unidentified intracellular carriers (13, 36). These data are in agreement with the recent finding that AEA is capable of partitioning to lipid droplets in cells following inhibition of FAAH (34).
Why have two FAAH enzymes been retained in the genomes of higher mammals? Both enzymes are capable of hydrolyzing NAEs with nearly comparable efficiencies in cells. The differential tissue distribution of the two enzymes (11) may in part account for the retention of FAAH-2. FAAH and FAAH-2 may coordinate NAE inactivation in some tissues, with FAAH-2 comprising the sole NAE-hydrolyzing enzyme in the heart and ovary. Additionally, lipid droplet localization may provide FAAH-2 with access to unique lipid substrates present in the lipid droplet lipidome (30).
Collectively, this study establishes FAAH-2 as a bone fide endocannabinoid (and NAE)-inactivating enzyme. FAAH-2 efficiently hydrolyzed AEA and PEA in intact cells while attaining a unique subcellular localization among NAE-hydrolyzing enzymes. From a therapeutic perspective, it is noteworthy that ablation of FAAH promotes analgesic, anti-inflammatory, and cardioprotective effects (37, 38), suggesting that targeting FAAH and possibly FAAH-2 may be therapeutically useful.
Acknowledgments
We thank Dr. Benjamin F. Cravatt for providing the FAAH-2 cDNA and Dr. Guo-Wei Tian for help with confocal microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants DA9374 and DA16419.
- NAE
- N-acylethanolamine
- AEA
- anandamide
- PEA
- palmitoylethanolamide
- FAAH
- fatty acid amide hydrolase
- DMEM
- Dulbecco's modified Eagle's medium
- BSA
- bovine serum albumin
- TM
- transmembrane
- ADRP
- adipose differentiation-related protein
- PBS
- phosphate-buffered saline
- PNGase F
- peptide N-glycosidase F
- RFP
- red fluorescent protein
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Lambert D. M., Fowler C. J. (2005) J. Med. Chem. 48, 5059–5087 [DOI] [PubMed] [Google Scholar]
- 2.Walker J. M., Krey J. F., Chu C. J., Huang S. M. (2002) Chem. Phys. Lipids 121, 159–172 [DOI] [PubMed] [Google Scholar]
- 3.Deutsch D. G., Chin S. A. (1993) Biochem. Pharmacol. 46, 791–796 [DOI] [PubMed] [Google Scholar]
- 4.Cravatt B. F., Giang D. K., Mayfield S. P., Boger D. L., Lerner R. A., Gilula N. B. (1996) Nature 384, 83–87 [DOI] [PubMed] [Google Scholar]
- 5.Cravatt B. F., Demarest K., Patricelli M. P., Bracey M. H., Giang D. K., Martin B. R., Lichtman A. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9371–9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giang D. K., Cravatt B. F. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2238–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arreaza G., Deutsch D. G. (1999) FEBS Lett. 454, 57–60 [DOI] [PubMed] [Google Scholar]
- 8.Chebrou H., Bigey F., Arnaud A., Galzy P. (1996) Biochim. Biophys. Acta 1298, 285–293 [DOI] [PubMed] [Google Scholar]
- 9.McKinney M. K., Cravatt B. F. (2005) Annu. Rev. Biochem. 74, 411–432 [DOI] [PubMed] [Google Scholar]
- 10.Cravatt B. F., Saghatelian A., Hawkins E. G., Clement A. B., Bracey M. H., Lichtman A. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10821–10826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei B. Q., Mikkelsen T. S., McKinney M. K., Lander E. S., Cravatt B. F. (2006) J. Biol. Chem. 281, 36569–36578 [DOI] [PubMed] [Google Scholar]
- 12.Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., Brown D. A. (2007) J. Lipid Res. 48, 2751–2761 [DOI] [PubMed] [Google Scholar]
- 13.Kaczocha M., Glaser S. T., Deutsch D. G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6375–6380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser S. T., Abumrad N. A., Fatade F., Kaczocha M., Studholme K. M., Deutsch D. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4269–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaczocha M., Hermann A., Glaser S. T., Bojesen I. N., Deutsch D. G. (2006) J. Biol. Chem. 281, 9066–9075 [DOI] [PubMed] [Google Scholar]
- 16.Day T. A., Rakhshan F., Deutsch D. G., Barker E. L. (2001) Mol. Pharmacol. 59, 1369–1375 [DOI] [PubMed] [Google Scholar]
- 17.Deutsch D. G., Glaser S. T., Howell J. M., Kunz J. S., Puffenbarger R. A., Hillard C. J., Abumrad N. (2001) J. Biol. Chem. 276, 6967–6973 [DOI] [PubMed] [Google Scholar]
- 18.Jacobsson S. O., Fowler C. J. (2001) Br. J. Pharmacol. 132, 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smirnova E., Goldberg E. B., Makarova K. S., Lin L., Brown W. J., Jackson C. L. (2006) EMBO Rep. 7, 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiele C., Spandl J. (2008) Curr. Opin. Cell Biol. 20, 378–385 [DOI] [PubMed] [Google Scholar]
- 21.Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. (2002) J. Biol. Chem. 277, 44507–44512 [DOI] [PubMed] [Google Scholar]
- 22.Patricelli M. P., Lashuel H. A., Giang D. K., Kelly J. W., Cravatt B. F. (1998) Biochemistry 37, 15177–15187 [DOI] [PubMed] [Google Scholar]
- 23.Sun Y. X., Tsuboi K., Zhao L. Y., Okamoto Y., Lambert D. M., Ueda N. (2005) Biochim. Biophys. Acta 1736, 211–220 [DOI] [PubMed] [Google Scholar]
- 24.Tsuboi K., Sun Y. X., Okamoto Y., Araki N., Tonai T., Ueda N. (2005) J. Biol. Chem. 280, 11082–11092 [DOI] [PubMed] [Google Scholar]
- 25.Omeir R. L., Chin S., Hong Y., Ahern D. G., Deutsch D. G. (1995) Life Sci. 56, 1999–2005 [DOI] [PubMed] [Google Scholar]
- 26.Bisogno T., Maurelli S., Melck D., De Petrocellis L., Di Marzo V. (1997) J. Biol. Chem. 272, 3315–3323 [DOI] [PubMed] [Google Scholar]
- 27.Olofsson S. O., Boström P., Andersson L., Rutberg M., Levin M., Perman J., Borén J. (2008) Curr. Opin. Lipidol. 19, 441–447 [DOI] [PubMed] [Google Scholar]
- 28.Olofsson S. O., Boström P., Andersson L., Rutberg M., Perman J., Borén J. (2009) Biochim. Biophys. Acta 1791, 448–458 [DOI] [PubMed] [Google Scholar]
- 29.Brown D. A. (2001) Curr. Biol. 11, R446–R449 [DOI] [PubMed] [Google Scholar]
- 30.Bartz R., Li W. H., Venables B., Zehmer J. K., Roth M. R., Welti R., Anderson R. G., Liu P., Chapman K. D. (2007) J. Lipid Res. 48, 837–847 [DOI] [PubMed] [Google Scholar]
- 31.Zehmer J. K., Huang Y., Peng G., Pu J., Anderson R. G., Liu P. (2009) Proteomics 9, 914–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gulyas A. I., Cravatt B. F., Bracey M. H., Dinh T. P., Piomelli D., Boscia F., Freund T. F. (2004) Eur. J. Neurosci. 20, 441–458 [DOI] [PubMed] [Google Scholar]
- 33.Oddi S., Bari M., Battista N., Barsacchi D., Cozzani I., Maccarrone M. (2005) Cell. Mol. Life Sci. 62, 386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oddi S., Fezza F., Pasquariello N., De Simone C., Rapino C., Dainese E., Finazzi-Agrò A., Maccarrone M. (2008) Cell. Mol. Life Sci. 65, 840–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zehmer J. K., Bartz R., Liu P., Anderson R. G. (2008) J. Cell Sci. 121, 1852–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oddi S., Fezza F., Pasquariello N., D'Agostino A., Catanzaro G., De Simone C., Rapino C., Finazzi-Agrò A., Maccarrone M. (2009) Chem. Biol. 16, 624–632 [DOI] [PubMed] [Google Scholar]
- 37.Ahn K., McKinney M. K., Cravatt B. F. (2008) Chem. Rev. 108, 1687–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bátkai S., Rajesh M., Mukhopadhyay P., Haskó G., Liaudet L., Cravatt B. F., Csiszár A., Ungvári Z., Pacher P. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H909–H918 [DOI] [PMC free article] [PubMed] [Google Scholar]