Abstract
Phosphorylation of α-synuclein (α-syn) at Ser-129 is a hallmark of Parkinson disease and related synucleinopathies. However, the identity of the natural kinases and phosphatases responsible for regulating α-syn phosphorylation remain unknown. Here we demonstrate that three closely related members of the human Polo-like kinase (PLK) family (PLK1, PLK2, and PLK3) phosphorylate α-syn and β-syn specifically at Ser-129 and Ser-118, respectively. Unlike other kinases reported to partially phosphorylate α-syn at Ser-129 in vitro, phosphorylation by PLK2 and PLK3 is quantitative (>95% conversion). Only PLK1 and PLK3 phosphorylate β-syn at Ser-118, whereas no phosphorylation of γ-syn was detected by any of the four PLKs (PLK1 to -4). PLK-mediated phosphorylation was greatly reduced in an isolated C-terminal fragment (residues 103–140) of α-syn, suggesting substrate recognition via the N-terminal repeats and/or the non-amyloid component domain of α-syn. PLKs specifically co-localized with phosphorylated Ser-129 (Ser(P)-129) α-syn in various subcellular compartments (cytoplasm, nucleus, and membranes) of mammalian cell lines and primary neurons as well as in α-syn transgenic mice, especially cortical brain areas involved in synaptic plasticity. Furthermore, we report that the levels of PLK2 are significantly increased in brains of Alzheimer disease and Lewy body disease patients. Taken together, these results provide biochemical and in vivo evidence of α-syn and β-syn phosphorylation by specific PLKs. Our results suggest a need for further studies to elucidate the potential role of PLK-syn interactions in the normal biology of these proteins as well as their involvement in the pathogenesis of Parkinson disease and other synucleinopathies.
Keywords: Cell/Fractionation, Diseases/Amyloid, Neurochemistry, Neurobiology/Neuroscience, Nucleus, Phosphorylation/Kinases/Serine-Threonine, Protein/Structure, RNA/RNAi
Introduction
Increasing evidence suggests that phosphorylation may play an important role in the oligomerization and fibrillogenesis (1), Lewy body formation (1, 2) and neurotoxicity of α-synuclein (α-syn)5 in vivo (3). The majority of α-syn within Lewy bodies (LBs) in diseased human brains and animal models of Parkinson disease (PD) and related synucleinopathies is phosphorylated at Ser-129 (Ser(P)-129) (1, 2, 4–7). Although recent studies support the notion that phosphorylation at Ser-129 is related to pathology and blocks α-syn fibrillization in vitro (8, 9), the exact mechanisms by which phosphorylation at Ser-129 modulates α-syn aggregation and toxicity in vivo remain elusive. Unraveling the role of phosphorylation in modulating the physiological and pathogenic activities of α-syn requires identification of the kinases and phosphatases involved in regulating its phosphorylation in vivo.
Several kinases that phosphorylate α-syn at serine and tyrosine residues, primarily in its C-terminal region, have been identified using in vitro kinase assays and co-transfection studies. Casein kinase I and II, G-protein-coupled receptor kinases (GRK1, GRK2, GRK5, and GRK6), and calmodulin-dependent kinase II (10–12) phosphorylate α-syn at Ser-129. Ser-87 is the only residue outside the C-terminal region reported to undergo phosphorylation by casein kinase I (12) and the dual specificity tyrosine-regulated kinase 1A (13). Tyrosine phosphorylation has also been reported at Tyr-125 by Fyn (14), Syk (15), Lyn (15), c-Frg (15), and Src (16) tyrosine kinases, with Syk (15) also phosphorylating at Tyr-133 and Tyr-136.
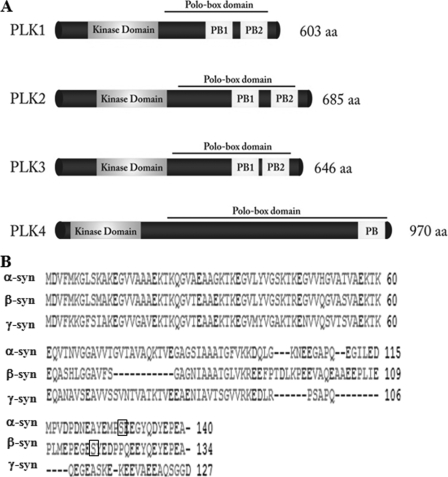
The Polo-like kinases (PLKs) comprise a family of conserved Ser/Thr protein kinases that play key roles in cell cycle regulation, cellular response to stress, and carcinogenesis. In mammalian cells, the PLK family consists of three closely related kinases PLK1, PLK2/Snk (serum-inducible kinase), PLK3/Fnk (fibroblast growth factor-inducible kinase) also designated Prk (proliferation-related kinase), and a distant member PLK4/Sak (Snk akin kinase) (17–21). The four PLKs share a conserved sequence motif characterized by two regions: a highly conserved N-terminal serine/threonine catalytic domain and a C-terminal non-catalytic termed the Polo box domain (PBD) (Fig. 1) (22, 23). The PBD plays important roles in regulating substrate interactions, targeting, subcellular localization, and autoinhibition of the PLKs (24). The PBD of PLK1, PLK2, and PLK3 contain two tandem Polo boxes (∼80 residues in length) that associate to form a phosphopeptide binding site, whereas PLK4 possesses only a single PB, which mediates the dimerization of PLK4, resulting in a structure that resembles the PBD of the other PLKs (24–26).
FIGURE 1.
Schematic depiction of the sequences of synucleins and PLK1s-4. A, schematic depictions illustrating the sequence similarities and differences among the four PLKs and members of the synuclein family of proteins. The four members of the PLK family share a conserved N-terminal kinase domain. The PBD of PLK1, PLK2, and PLK3 contain two tandem Polo boxes (∼80 residues in length) that associate to form a phosphopeptide binding site, whereas PLK4 possesses only a single PB, which mediates the dimerization of PLK4. B, sequence alignment of the human WT α-, β-, and γ-syn generated by the Clustal 2.0.8 multiple sequence alignment program. Ser-129 in α-syn and Ser-118 are highlighted within the square. aa, amino acids.
Recent reports demonstrate that α-syn is phosphorylated by PLK2 (27, 28). In this report, we confirm these findings and demonstrate for the first time, using in vitro kinase assays, co-transfection, and small interference RNA (siRNA)-mediated knockdown of PLKs, that α- and β-, but not γ-syn are phosphorylated by specific members of the PLK family. PLK phosphorylation of synucleins occurs specifically at Ser-129 in α-syn and Ser-118 in β-syn and appears to be mediated by specific interactions between the PLKs and the N-terminal region (residues 1–95) of syn. These findings were validated by colocalization of α-syn and PLKs in different subcellular compartments and co-transfection studies as well as siRNA-mediated knockdown of PLKs in mammalian cells and primary neurons. PLK2 and PLK3 partly co-localized with Ser(P)-129 α-syn in primary hippocampal neurons as well as in cortical brain areas of α-syn transgenic mice. Furthermore, we demonstrate that the level of neuronal PLK2 is elevated in the Alzheimer disease (AD) and Lewy body disease (LBD) brains and correlates with the increased levels of Ser(P)-129 in these brains compared with healthy controls. Together, our findings point to PLK2 and PLK3 as the primary PLKs responsible for α-syn phosphorylation and highlight the importance of further studies to elucidate the potential role of the interactions between the PLKs and α-syn and their implications for α-syn aggregation and toxicity in PD and other synucleinopathies.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Proteins
pCMV6-Entry-PLK1 (PLK1), pCMV6-Entry-PLK2 (PLK2), pCMV6-Entry-PLK3 (PLK3) and pCMV6-Entry-PLK4 (PLK4) open reading frame clones of human PLKs were from OriGENE and provided as Myc/DDK-tagged plasmids. pAAV-CMV-α-syn wild type (WT) (α-syn) was kindly provided by Patrick Aebischer's laboratory (Brain Mind Institute, Ecole Polytechnique Federale de Lausanne). The α-syn deletion mutant (73–83Δ) and the β/α-syn chimera (β-syn-(1–72)/α-syn-(73–83)/β-syn-(73–134)) were kindly provided by Prof. Michel Goedert (Medical Research Council Laboratory of Molecular Biology, Cambridge, UK) (29). All constructs were confirmed by DNA sequencing (Mycrosynth, Switzerland) using the primers provided by the supplier. Recombinant PLK enzymes were purchased from Intvitrogen and Cell Signaling Technologies. All recombinant α-syn proteins and mutants used in this study were expressed as described previously (8).
Unless otherwise specified, the following antibodies were used in this study: anti-human PLK1 (sc-5585), anti-human/rodent PLK2 H-90 (sc-25421), and anti-human α-syn (clone 211) antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Anti-human PRK (PLK3) (BD 556518) were purchased from BD Pharmigen. Anti-Ser(P)-129 α-syn was from Wako, β-actin antibody was from Abcam (Cambridge, MA), and goat anti-mouse Cy3 was from (GE Healthcare).
Preparation of α-Syn C-terminal Fragment
WT α-syn in 50 mm Tris, 150 mm sodium chloride, pH 7.6, was trypsinized at a concentration of 3 mg/ml and a mass ratio of α-syn/trypsin (Promega Corp., Madison, WI) of 100:1. The reaction was incubated at 37 °C for 24 h to generate the C-terminal fragment (residues 103–140). The C-terminal fragment was then purified by size exclusion chromatography and using a Superdex 75 HR 10/30 column (GE Healthcare).
In Vitro Kinase Assay
WT or mutant α-syn was phosphorylated by PLK1 to -4 (Cell Signaling Technologies and Invitrogen) at a concentration of 1.44 mg/ml (100 μm), unless otherwise stated. The phosphorylation reactions were carried out in the presence of 1.09 mm ATP (Sigma), 1× reaction solution (20 mm HEPES, 10 mm MgCl2, 2 mm DDT, pH 7.4), and 1 μg of PLK, 144 μg of α-syn at 30 °C for 15 min to 24 h. The reactions were quenched with EDTA disodium salt (25 mm final concentration) (Axon Lab, AG, Le Mont-Sur-Lausanne, Switzerland). The progress of the reaction was monitored by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (30).
Real-time NMR Spectroscopy
NMR samples contained ∼0.1 mm 15N-labeled α-syn in 200 mm HEPES, 10 mm MgCl2, 2 mm dithiothreitol, and 1.09 mm ATP, pH 6.9. The real-time assay was started by the addition of kinase into the NMR sample, with a protein/kinase ratio of 100:0.5 μg. NMR experiments were acquired on a Bruker Avance 600-MHz NMR spectrometer. NMR data were processed by Bruker TOPSPIN version 2.0. To allow for efficient phosphorylation, the temperature was set to 303 K. To reduce the impact of signal broadening due to amide proton exchange, the temperature was lowered to 288 K during the measurement of 1H,15N heteronuclear single quantum coherence (HSQC) spectra. In the case of PLK3, the phosphorylation reaction proceeded very rapidly at 303 K. Therefore, we also followed the kinetics of phosphorylation of α-syn by PLK3 at 293 and 288 K using ∼0.075 mm α-syn (protein/kinase = 100:0.5 μg). Backbone resonance assignments of WT and phosphorylated α-syn were obtained previously. The degree of phosphorylation at Ser-129 was calculated from the intensity of the NMR signals of phosphorylated and unphosphorylated Ser-129. Errors were estimated based on the signal/noise ratio in the NMR spectra.
MALDI-TOF MS
MALDI calibrants were from Sigma, trifluoroacetic acid was from Pierce, and sinapinic acid was from Fluka. 3-μl aliquots of in vitro PLK-phosphorylated α-syn were reserved for the MALDI analysis. The samples were diluted 1:10 in 0.1% trifluoroacetic acid and further mixed with an equal volume of 14 mg/ml sinapinic acid solution in 0.1% trifluoroacetic acid, ACN (1:1). 0.5 μl of this solution was deposited on the mirror-polished target and analyzed with a 4700 MALDI-TOF/TOF instrument (Applied Biosystems Inc., Foster City, CA). The spectra were calibrated on the pseudomolecular peak of cytochrome c, apomyoglobin, ubiquitin, and α-syn prepared with the same matrix and deposited close to the samples.
Cell Culture and Transient Transfection
Human Embryonic Kidney cells (HEK 293T) and immortal cell lines from HeLa were grown in Dulbecco's modified Eagle's medium (Invitrogen AG, Basel, Switzerland) supplemented with 10% fetal bovine serum and 5% penicillin/streptomycin in a humidified incubator, 5% CO2, 37 °C. For HEK 293T cells, transient transfection was performed on 6-well plates at a cell confluence of 70–80%, using the standard calcium phosphate (CaPO4) transfection protocol. For HeLa cells, transient transfection was performed employing LipofectamineTM 2000 (Invitrogen AG, Basel, Switzerland) according to the manufacturer's instructions. The overall amount of plasmid used was 4 μg/well.
PLK2 and PLK3 Small Interfering RNA (siRNA) Silencing
ON-TARGET siRNA sequence targeting human PLK2 5′-GGACATGGCTGTGAATCAG-3′ (57) and ON-TARGET siRNA sequence targeting human PLK3 5′-CGGCCTCATGCGCACATCC-3′ (58) as well as non-targeting (ON- TARGET siControl) were purchased from Dharmacon (Lafayette, CO). HeLa cells were grown at a density of 200,000 cells/well and transfected the day after with increased concentration of each siRNA independently using LipofectamineTM 2000 according to the manufacturer's instructions. Cells were treated with siRNA for 48 h followed by transient transfection with α-syn WT using LipofectamineTM 2000 for an additional 24 h before analysis by Western blot and immunofluorescence.
For silencing in primary neurons, hippocampal neurons were prepared from P0 Sprague-Dawley rats (Charles River/Iffa Credo, L'Arbresle Cedex, France) as described previously by Steiner et al. (31) and were plated at a density of 200,000 cells/35-mm dish (Falcon, BD Biosciences) in growth medium (minimum Eagle's medium, 20 mm glucose, 0.5 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 10% horse serum). Neuron's medium is kept, and the cells were transfected for 2 h at day 12 with a 60 nm concentration of each murine PLK2 and PLK3 siRNA (sc-39153 and sc-39151, respectively, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) using LipofectamineTM 2000 reagent (Invitrogen) following the manufacturer's instructions. Neurons were then reincubated in their own and previous media for 72 h. For biochemical analysis by Western blotting, the cells were lysed with in a buffer containing Hepes (0.02 m), EGTA (0.02 m), EDTA (0.02 m), KCl (0.1 m), dithiothreitol (0.1 μm), and 1% Triton X-100. The lysate was centrifuged at 20,000 × g for 15 min, and the supernatant was recovered and analyzed immediately by Western blotting or stored immediately at −80 °C.
Subcellular Fractionation
HEK 293T cells were transiently transfected with PLK1 to -3 and α-syn WT for 24 h. Subcellular fractionation was carried out employing the subcellular proteome extraction kit (Calbiochem) according to the manufacturer's instructions. The fractions were analyzed by Western blot, and the purity was evaluated using control antibodies provided as subcellular markers of cell compartment, including cytosolic, membrane/particulate, and nucleus (anti-Hsp90, anti-calnexin, and anti-PARP-1, respectively).
SDS-PAGE and Immunoblot
Cells were harvested 24 h post-transfection, and total homogenate was prepared by lysis in a buffer containing 150 mm NaCl, 1 mm EDTA, 20 mm Tris, pH 7.40, 0.5% Nonidet P-40, enriched with a 1:200 protease inhibitor mixture (Sigma), 1 mm phenylmethylsulfonyl fluoride (Sigma), and 1 μm okadaic acid (Acros). Cleared lysates were obtained by centrifugation at 14,000 rpm, 4 °C for 20 min. 10 μg of total protein diluted in loading buffer were separated on 1-mm, 12% SDS-PAGE. The proteins were then transferred to nitrocellulose membrane using the iBlotTM dry blotting system (Invitrogen AG, Basel, Switzerland) for 7 min. The membrane was then probed overnight with the primary antibody after 30 min of blocking in Odyssey blocking buffer (Li-Cor Biosciences GmbH) diluted 1:3 in phosphate-buffered saline (Sigma). After four times washes with PBST (phosphate-buffered saline, 0.01% (v/v) Tween 20 (Sigma)), the membrane was incubated for 1 h with secondary antibody (goat or rabbit Alexa Fluor® 680 IgG) protected from light at room temperature. The immunoblot was finally washed four times with PBST and three times with phosphate-buffered saline and scanned in a Li-COR scanner at a wavelength of 700 nm.
Subcellular Localization and Immunocytochemistry in Primary Neurons
For immunocytochemistry, hippocampal rat neurons were prepared as described (32). Hippocampal neurons (200,000 cells in a 35-mm2 dish) were cultured on coverslips (12 mm; VWR) coated with poly-d-lysine (BD Biosciences) and laminin mouse (Invitrogen) and fixed for 12 min at room temperature in a freshly prepared solution (pH 7.4) containing 4% paraformaldehyde and 4% sucrose in phosphate-buffered saline. Neurons were then washed three times in MTBS buffer (66 mm NaCl, 100 mm Tris-HCl, pH 7.4) and then incubated overnight with the first antibody in buffer A containing 5% normal goat serum, 5% normal horse serum, 3% bovine serum albumin, 0.3% Triton X-100 in MTBS for overnight at 4 °C. The following day, cultures were rinsed three times in MTBS and incubated in the buffer A containing goat anti-mouse Cy3 coupled (1:1000; Jackson ImmunoResearch, West Grove, PA) or goat anti-rabbit Alexa Fluor® 488 coupled as secondary antibody (IF 1:500; Molecular Probes Inc.) for 2 h at room temperature. After three washes in MTBS, coverslips were mounted on glass slides with 4′,6-diamidino-2-phenylindole-Vectashield medium (Vector) and imaged with a Zeiss LSM510 confocal microscope.
Immunostaining of Mouse Brain Sections
(Thy1)-[A30P]α-syn mice were sacrificed by cervical dislocation. Brains were dissected and fixed in 4% paraformaldehyde in phosphate buffer (pH 7.4). The fixed brains were embedded in paraffin, and 4-μm-thick sections were cut on a microtome. Serial sections were directly transferred onto SuperFrost Plus coverslips, incubated overnight at 60 °C, and paraffin-decorated with xylene. Rehydration was performed over a descending ethanol series: 100, 95, and 75% Tris-buffered saline, pH 7.4 (TBS). Peroxidase activity was eliminated by treatment with 1% H2O2 in TBS for 30 min, followed by three washes in TBS. Antigen retrieval was carried out by heating the sections up to 90 °C in citrate buffer (pH 6.0) for 30 min, followed by a cooling step of 15 min on ice. Blocking was done in 5% goat antiserum in TBS for 30 min at room temperature. Incubation with rabbit monoclonal antibody against human Ser(P)-129 α-syn (1:500; Abcam) and anti-PLK2 or anti-PLK3 (1:50; both from Santa Cruz Biotechnology, Inc.) was performed in TBS containing 2% goat antiserum at 4 °C overnight. Subsequently, the sections were washed and incubated at room temperature with biotinylated goat anti-rabbit antibody followed by the avidin-biotin complex (VECTASTAIN ABC kit). For visualization, Vector SG-Blue was used followed by nuclear staining with Nuclear Fast Red for 5 min and short rinsing under tap water. Sections were then dehydrated (ethanol 75%, 95%, 100%, and xylene) and mounted with Pertex. Photomicrographs were taken with an Axioplan 2 imaging microscope (Carl Zeiss) and processed with AxioVision version 4.3 imaging software.
Human Samples and Tissue Processing
Analysis of α-syn phosphorylation was performed with post-mortem human temporal cortex samples. The human samples were obtained from the Alzheimer's Disease Research Center at the University of California San Diego. A total of 18 cases were included, of which six were non-demented controls, six were diagnosed as AD, and six were diagnosed as LBD. Cases of LBD were defined based on the clinical presentation of dementia and the pathological findings of LBs in the locus coeruleus, substantia nigra, or nucleus basalis of Meynert as well as in cortical and subcortical regions. LBs were detected using hematoxylin/eosin anti-ubiquitin and anti-α-syn antibodies as recommended by the Consortium on LBD. In all cases, the brains were processed within 8 h after death. Brains were divided sagitally, and the right hemibrain was serially sectioned and preserved at −70 °C for Western blot analysis. To analyze the distribution and levels of α-syn, PLK2, and PLK3 in the brains of patients with AD and LBD, samples from the temporal cortex were homogenized and fractioned into cytosolic and membrane fractions by ultracentrifugation. Approximately 20 μg of protein/well were loaded into 4–12% BisTris gels with MOPS/SDS buffer and blotted onto polyvinylidene difluoride membranes. Blots were incubated overnight with antibodies against PLK2 and PLK3 (1:1000; Invitrogen), Ser(P)-129 α-syn (WAKO), and α-syn (1:1000; Chemicon, Temecula, CA), followed by secondary antibodies tagged with horseradish peroxidase (1:5000; Santa Cruz Biotechnology, Inc.). Immunoreactivity was visualized by enhanced chemiluminescence and analyzed with a Versadoc XL imaging apparatus (Bio-Rad). Analysis of actin levels was used as a loading control.
RESULTS
To determine whether α-syn is a substrate for members of the PLK family of kinases (PLK1, PLK2, PLK3, and PLK4), we performed in vitro kinase assays using purified recombinant α-syn together with each of the four PLKs, and co-transfection experiments in mammalian cells (human embryonic kidney HEK 293T, HeLa and SH-SY5Y neuroblastoma cells).
PLK1 to -3 Phosphorylate α-Syn Specifically at Ser-129 in Vitro
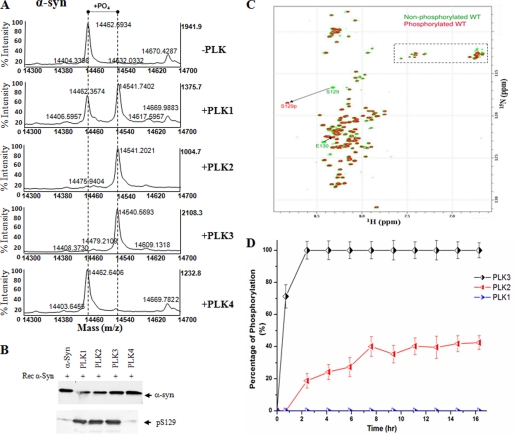
To assess the phosphorylation of α-syn by the PLKs, α-syn (100 μm) was incubated with either of the four kinases at 1 μg of PLK, 144 μg of α-syn. The reactions were incubated at 30 °C, and the extent of phosphorylation was monitored by MALDI-TOF mass spectrometry. Upon incubation with PLK1, PLK2, and PLK3, we observed a shift in the molecular mass of α-syn by 80 Da (from 14,462 to 14,542 Da) for PLK1, -2, and -3, which corresponds to the addition of one phosphate group (Fig. 2). A closer examination of Fig. 2A demonstrates that PLK2, and PLK3 phosphorylate α-syn quantitatively, whereas ∼60–70% conversion was observed for PLK1 (Fig. 2A). No other kinase has been observed (data not shown) or reported to phosphorylate α-syn with the same efficiency. The MALDI-TOF results suggest that PLK1 to -3 phosphorylate α-syn at a single site. To determine if PLK1 to -3-mediated phosphorylation occurs at Ser-129, the phosphorylation reactions were analyzed by Western blotting using an antibody against Ser(P)-129 α-syn and subjected to trypsin digestion and peptide mapping by MALDI-TOF and liquid chromatography-electrospray ionization MS/MS. α-Syn samples incubated with the PLK1 to -3 showed a band at 14 kDa that was detectable with both anti-α-syn and anti-Ser(P)-129 antibodies (Fig. 2B). Comparative analysis of the tryptic fragments of these bands revealed that phosphorylation of α-syn occurs only within peptide fragments that encompass Ser-129. Trypsin digestion resulted in a peptide with a molecular mass of 4353.0 Da (M + H)+ corresponding to the monophosphorylated C-terminal fragment 103–140 containing Ser-129 (calculated mass 4272.4 Da (M) (NEEGAPQ EGILEDMPVDPDNEAYEMPpSEEGYQDYEPEA, where pS represents phosphoserine) (data not shown). This peptide fragment does not contain other serine or threonine residues, suggesting that phosphorylation indeed occurs at Ser-129. To further confirm these results, in vitro phosphorylation was also performed with a mutant that cannot be phosphorylated at position 129, S129A. As predicted, none of the PLKs could phosphorylate S129A mutant α-syn (data not shown).
FIGURE 2.
In vitro phosphorylation of synucleins by PLK1 to -4. A, MALDI-TOF analysis of the WT α-syn after phosphorylation by PLK1 to -4. For PLK1 to -3, there is an 80-Da increase in the molecular mass of WT α-syn (14,461 + 80 = 14,541), corresponding to one phosphorylation. B, Western blot analysis of the same samples in A. The anti-Ser(P)-129 antibody detected a band, suggesting that the phosphorylation detected by mass spectrometry is at position 129. C, comparison of two-dimensional 1H,15N HSQC spectra of unphosphorylated WT (green) and α-syn phosphorylated by PLK3 (red). A dashed rectangle marks glutamine (Q) and asparagine (N) side chain resonances. D, kinetics of in vitro phosphorylation of Ser-129 in α-syn by PLK3 (black), PLK2 (red), and PLK1 (blue) as monitored by real-time NMR spectroscopy. NMR samples contained ∼0.1 mm 15N-labeled α-syn in 200 mm HEPES, 10 mm MgCl2, 2 mm dithiothreitol, and 1.09 mm ATP, pH 6.9. The real-time assay was started by the addition of kinase into the NMR sample using a protein/kinase ratio of 100:0.5 mg. The error bars were determined based on the signal/noise ratio observed in the NMR spectra.
Real-time Spectroscopy of the Phosphorylation Reaction
Hetereonuclear NMR spectroscopy on 15N-labeled protein modified by PLK allows identification of all phosphorylation sites, measures the level of integration, and yields kinetic data for the enzymatic modification of the individual sites. Although the reaction mixture is complex with enzyme, ATP and α-syn, filtering through the 15N label allows the monitoring of the kinase activity in the NMR tube without any further sample purification. To obtain single-residue resolution and identify all potential phosphorylation sites, the enzymatic reaction was followed by two-dimensional 1H-15N heteronuclear correlation spectra. Phosphorylated serine and threonine residues are readily detected because phosphorylation shifts their amide proton resonance downfield 8.8 ppm, to an empty region of the 15N-α-syn HSQC spectrum (Fig. 2C). The real-time NMR assay was applied to PLK1, PLK2, and PLK3. In the case of PLK3, the peak of phosphorylated Ser-129 was clearly observed already after 45 min (Fig. 2D). With increasing incubation time, the intensity of the NMR signal of phosphorylated Ser-129 increased. Quantitative analysis of the increase of the NMR signal of phosphorylated Ser-129 indicated that PLK3 fully phosphorylates α-syn within 2.5 h under the conditions of the assay (Fig. 2D). Importantly, no additional NMR signals of phosphorylated residues appeared, indicating that PLK3 exclusively phosphorylated α-syn at Ser-129. In the case of PLK1, no phosphorylation of α-syn could be detected during the time course of the experiment. The significantly reduced PLK1 phosphorylation of α-syn in the NMR experiments, compared with the in vitro kinase assay, is most likely due to the fact that the NMR phosphorylation experiments were performed at lower temperature (15 °C versus 30 °C for in vitro kinase assays) to reduce the impact of signal broadening due to amide proton exchange. Real-time spectroscopy of α-syn phosphorylation by PLK2 showed that PLK2 phosphorylates α-syn only at Ser-129. However, the kinetics of the enzymatic reaction is significantly slower for PLK2 than for PLK3, and α-syn was not fully phosphorylated at the end of the assay.
α- and β-Syn but Not γ-Syn Are Phosphorylated by PLK1 to -3
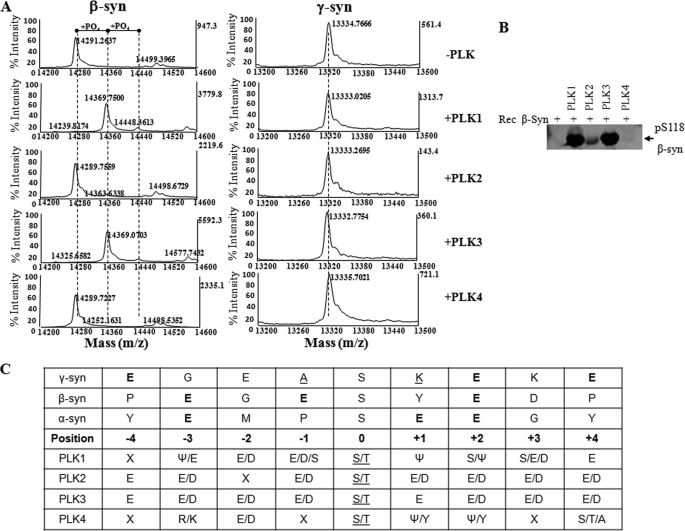
To evaluate the specificity of PLKs for α-syn, we also performed in vitro phosphorylation employing purified β- and γ-syn. β-Syn is a 134-amino acid-long protein sharing 61% sequence homology with α-syn. The sequence homology of the two synucleins is more prominent in their N-terminal regions (∼90% sequence similarity) (33). γ-Syn is 127 amino acids long and shares 55.9 and 54.3% sequence homology with α-syn and β-syn, respectively (34) (Fig. 1B). Unlike α-syn, both β- and γ-syn are not found in LBs (35). As illustrated in Fig. 3, incubation with PLK1 and PLK3 resulted in quantitative conversion of β-syn into its monophosphorylated form, as evidenced by the shift of the β-syn molecular mass, m/z, from 14,291 to 14,369.7 Da. A minor (<5%) second peak corresponding to diphosphorylated β-syn (m/z 14,448) was also detected under these conditions. Using an anti-Ser(P)-118 antibody generated in our laboratory and trypsin digestion and peptide mapping (data not shown), we showed that PLK1- and PLK3-mediated phosphorylation of β-syn occurs at Ser-118 (Fig. 3B). We did not observe any detectable phosphorylation of β-syn by PLK2. Interestingly, all four PLKs failed to phosphorylate γ-syn in vitro. These differences may reflect differences in substrate specificity among the four PLKs (Fig. 3C) and/or suggest that PLK-mediated phosphorylation of synucleins involves specific interactions between the various kinases and specific domains in synucleins.
FIGURE 3.
Recombinant β-syn is phosphorylated by PLK1 and PLK3, whereas γ-syn does not undergo phosphorylation by PLK1 to -4. A, MALDI-TOF analysis of the in vitro phosphorylation reaction of WT β- and γ-syn by PLK1 to -4. WT β-syn undergoes phosphorylation by PLK1 and PLK3 but not PLK2 and PLK4. None of the recombinant PLKs were observed to phosphorylate γ-syn in vitro. B, Western blot analysis of β-syn samples phosphorylated by PLK1 to -4, using our anti-phosphoserine 118 (pS118) antibody. The table in C shows the consensus substrate specificity of PLK1 to -4 (ψ, hydrophobic amino acid) and the amino acids adjacent to the Ser-129 in α-syn and its equivalent residues in β- and γ-syn.
Specificity of α-Syn Phosphorylation by PLK1 to -3
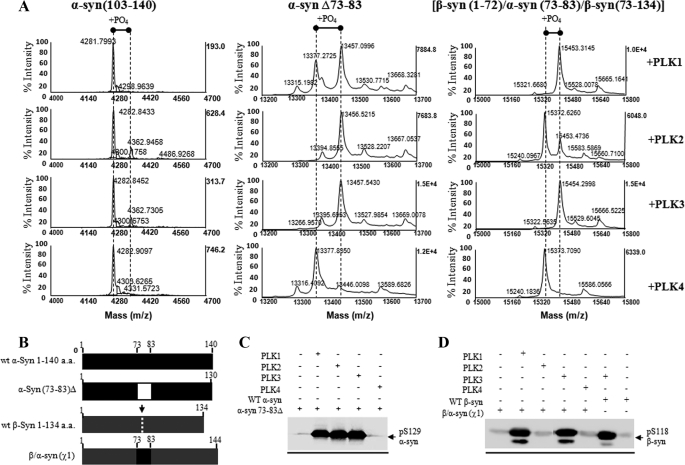
Intrigued by the lack of phosphorylation of β-syn by PLK2, we sought to determine the sequence determinants of α- and β-syn by the PLKs. More specifically, we tried to assess if the differences in phosphorylation efficiency are due to 1) the sequence divergence between α and β-syn; 2) differences in the substrate specificity of the PLKs as determined by the amino acid residues flanking the phosphorylated serine residue; 3) specific protein-protein interaction; or 4) differences in the conformational properties of the two proteins. α-Syn and β-syn share high sequence homology at the N- and C-terminal region and differ mainly in the fact that β-syn lacks residues 73–83 in the non-amyloid component (NAC) region of α-syn, suggesting that NAC region is important for synuclein interactions and phosphorylation by PLKs. To determine if phosphorylation by the PLKs is mediated by protein-protein interactions involving the N-terminal region of α-syn (residues 1–102), we first generated a C-terminal fragment of α-syn comprising residues 103–140, which contains the PLK phosphorylation site, Ser-129. All four PLKs failed to efficiently phosphorylate 103–140 α-syn. Only minor phosphorylation was observed in the case of PLK2 and PLK3 (Fig. 4A). This finding further suggested that the N-terminal and NAC regions of α-syn might influence the efficiency of the PLK1 to -3 phosphorylation. It is also plausible that α-syn interaction with the PLKs is mediated by specific conformations of the C terminus that only exist in the full-length protein.
FIGURE 4.
Removal of the entire N-terminal residues (positions 1–103) abolishes Ser-129 phosphorylation, whereas deletion of the NAC residues 73–83 has no effect on Ser-129 phosphorylation by PLK1 to -3. A, MALDI-TOF analysis of the PLK-phosphorylated Δ1–103 α-syn, Δ73–83 α-syn, and chimeric β-syn(χ1) proteins. B, schematic representation of the WT α- and β-syn, deletion α-syn Δ73–83 and chimeric β-syn(χ1) proteins. C, Western blotting analysis of the in vitro phosphorylation of these constructs at Ser-129 by PLK1 to -4 using anti Ser(P)-129 antibody (1:5000; Wako). D, Western blotting analysis of the PLK-phosphorylated WT and chimeric β-syn(χ1) probed with Ser(P)-118 antibody (1:250) (1000 ng/lane). a.a., amino acids.
To test these hypotheses, we investigated the in vitro phosphorylation of an α-syn mutant lacking residues 73–83 within the NAC region (α-syn-(73–83)Δ), which are absent in β-syn (Fig. 4B). Removal of these residues did not have an effect on Ser-129 phosphorylation by PLK1, PLK2, and PLK3 as discerned by MALDI-TOF and Western blotting analysis using an anti-Ser(P)-129 antibody (Fig. 4, A and C). Similarly, the incorporation of these NAC residues into corresponding sites within β-syn (β-syn-(1–72)/α-syn-(73–83)/β-syn-(73–134)), called β/α-syn(χ1) in this study, did not significantly influence the levels of phosphorylation at Ser-118 by all three PLKs (Fig. 4, A and D). These findings suggest that the absence of 73–83 in β-syn is not sufficient to explain the lack of phosphorylation by PLK2 and that the NAC region may not play an important role in modulating α-syn phosphorylation by PLK1 to -3.
To further test this hypothesis and probe the effect of other regions within α-syn, we evaluated the effect of various mutations in the N-terminal, NAC, and C-terminal regions of α-syn on the PLK1 to -3 phosphorylation efficiency. Mutations at Ser-87 (S87A and S87E) abolished phosphorylation by PLK1, whereas phosphorylation by PLK2 and PLK3 was not affected (supplemental Figs. 1 and 2). To confirm these findings, we investigated the phosphorylation of mouse α-syn by PLK1 to -4. Mouse α-syn shares high (>94%) sequence homology with human WT α-syn, and their sequence differences are mostly located in the NAC and C-terminal regions (residues 83, 87, 100, 103, 107, 121, and 122), including a serine → asparagine mutation at position 87 (S87N). These mutations appear to reduce PLK1-mediated phosphorylation of α-syn without influencing phosphorylation by PLK2 and PLK3 (supplemental Fig. 3). The effect of mutations at the C terminus of α-syn was also assessed. Mutating Ser-129 to alanine blocked the phosphorylation by PLK1 to -3. In addition, mutations near Ser-129 alter the specificity of the three kinases, because only PLK2 was observed to phosphorylate E126A (supplemental Fig. 3C).
In Vitro Phosphorylation of Fibrillar α-Syn by PLK1 to -4
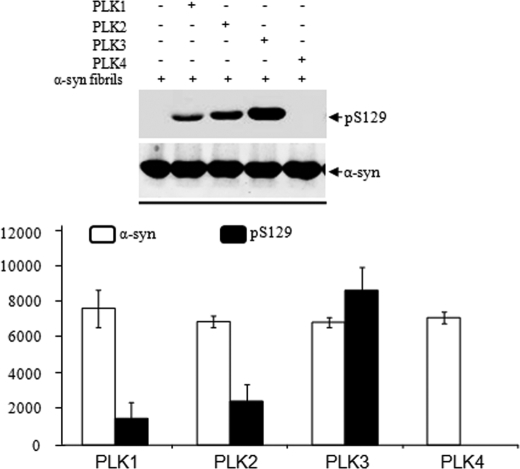
Whether phosphorylation at Ser-129 occurs prior to or after α-syn fibrillization and LB formation in vivo remains unclear. Recent studies by Waxman and Giasson (36) and our laboratory demonstrated that fibrillar α-syn is a better substrate for casein kinase I phosphorylation. To determine whether α-syn fibrils are good substrates for phosphorylation by PLKs, we generated fibrils of WT α-syn and subjected them to in vitro phosphorylation with PLK1 to -4. Briefly, WT α-syn was aggregated at a concentration of 200–400 μm at 37 °C with continuous shaking for 24–72 h. The fibrils formed were harvested by centrifugation, sonicated, and phosphorylated at a concentration of 100 μm as described above. As shown in Fig. 5, the efficiency of in vitro phosphorylation of α-syn fibril was as follows: PLK3 > PLK2 > PLK1. As is the case with monomeric α-syn, PLK1 exhibited decreased phosphorylation activity toward fibrillar α-syn relative to PLK2 and PLK3.
FIGURE 5.
In vitro phosphorylation of α-syn fibrils by PLK1 to -4. Top, preformed α- fibrils were incubated with recombinant PLKs in the appropriate reaction buffers, and phosphorylation at Ser-129 was assessed by anti-Ser(P)-129 antibodies. Bottom, quantification of the level of Ser(P)-129 signal normalized against α-syn and β-actin (pS118/(α-syn + β actin)), n = 3.
PLK Phosphorylation of α-Syn in Mammalian Cell Lines and Primary Neurons
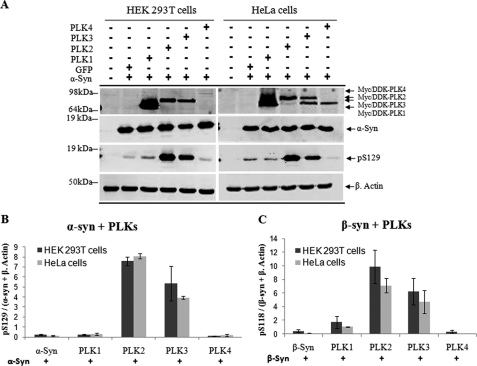
Having established that PLK1 to -3 phosphorylate α-syn at Ser-129 in vitro, we then sought to determine whether α-syn phosphorylation by PLKs occurs in mammalian cells. HEK 293T and HeLa were co-transfected with expression vectors encoding α-syn and PLK1, -2, -3, or -4. Western blot analysis of cell lysates demonstrated that PLK2 and PLK3, but not PLK4, phosphorylate α-syn at Ser-129 (Fig. 6A). Only minor phosphorylation by PLK1 was detected in both cell lines. Quantification of the Ser(P)-129 bands in Fig. 6A demonstrates that PLK2 is more efficient in phosphorylating α-syn in these cell lines (Fig. 6B). Similar results were obtained in SH-SY5Y cells (data not shown). Interestingly, co-transfection of β-syn with PLK1, -2, -3, or -4 in HEK 293T cells and HeLa resulted in significant phosphorylation at Ser-118 by PLK1, PLK2, and PLK3, with PLK2 showing the highest efficiency in phosphorylating β-syn in vivo (Fig. 6C), in contrast to the in vitro results (Fig. 3). The lack of phosphorylation of β-syn by PLK2 in vitro suggests that other factor(s) within the cell may influence the conformation of β-syn and/or its interactions with PLK2 in a manner that results in significant enhancement in Ser-118 phosphorylation by PLK2 in the cell.
FIGURE 6.
Phosphorylation of α-syn and β-syn by PLKs in mammalian cells (HEK-293T and HeLa). A, the cells were co-transfected with pAAV-CMV-α-syn-WT and different pCMV6-Èntry-PLKs and lysed 24 h post-transfection. The expression level of PLKs and α-syn phosphorylation at Ser-129 were detected by immunoblot using antibodies against FLAG tag and anti-Ser(P)-129. B, quantification of the level of Ser(P)-129 signal intensity was normalized against total fluorescence signal of α-syn WT and β-actin (Ser(P)-129/(α-syn + β-actin), n = 3. C, expression of β-syn-WT and Ser(P)-118 in HEK 293T and HeLa cells after co-transfection as previously described was detected by Western blot (data not shown), and quantification of the level of β-syn Ser(P)-118 signal intensity was normalized against the total fluorescence signal of β-syn-WT and β-actin (pS118/(β-syn + β-actin), n = 3.
Subcellular Localization of α-Syn and PLK1 to -3
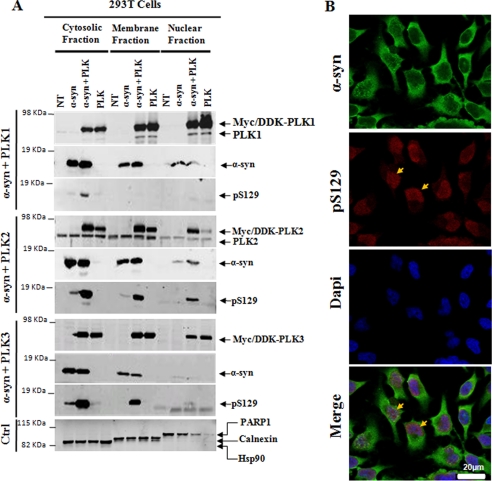
To determine whether the level of phosphorylation correlates with the degree of colocalization of α-syn and the PLKs, the expression and subcellular localization of α-syn, PLKs, or α-syn co-transfected with PLK1, PLK2, or PLK3 was assessed by subcellular fractionation, Western blotting, and immunocytochemistry on non-transfected HeLa cells (Fig. 7). Western blot analysis revealed that PLK1, PLK2, PLK3, and α-syn were strongly enriched in the cytosolic and membrane fractions. PLK1 and PLK3 were also highly enriched in the nuclear fraction relative to PLK2, although the level of nuclear PLK2 seemed to increase in cells co-transfected with α-syn (Fig. 7A). The colocalization of α-syn with PLK2 or PLK3 correlated well with an increase in Ser(P)-129. Despite the increased expression and colocalization of α-syn and PLK1 in the cytosolic, membrane, and nuclear fractions, the level of Ser(P)-129 in these fractions was minimal. These findings are consistent with the results from the in vitro kinase assays (Fig. 2) and co-transfection experiments (Fig. 6) and suggest that PLK2 might phosphorylate α-syn throughout the cell, whereas PLK3 phosphorylates cytosolic and nuclear α-syn. PLK1 does not contribute significantly to Ser-129 phosphorylation, at least in HEK 293T, HeLa, and SH SY5Y cells. Interestingly, we have consistently observed that constitutively phosphorylated α-syn (Ser(P)-129) is localized primarily in the nucleus (Fig. 7B). These findings are consistent with the nuclear distribution of α-syn (37) and with recent studies demonstrating nuclear enrichment of Ser(P)-129 immunostaining in HEK 293T cell as well as in aged and young presymptomatic mice overexpressing α-syn (38). In these studies, the specificity of nuclear localization of Ser(P)-129 was further verified using different anti-Ser(P)-129 antibodies and treatments with phosphatases (38).
FIGURE 7.
Subcellular localization of α-syn and PLKs in mammalian cell. A, HEK 293T cells were co-transfected with pAAV-CMV-α-syn-WT and different pCMV6-Èntry-PLKs and then subjected to subcellular fractionation 24–48 h post-transfection with the ProteoExtract® subcellular proteome extraction kit from Calbiochem. Purified fractions were then immunoblotted with corresponding antibodies. PARP1, Hsp90, and calnexin proteins were used as control for the nuclear, cytosolic, and membrane particulate fractions, respectively. B, confocal microscopy of HeLa cells co-stained with anti-Ser(P)-129 and anti-α-syn antibodies. Endogenous Ser(P)-129 (red) accumulate exclusively in the nucleus stained with 4′-6′-diamidino-2-phenylindole (Dapi), whereas α-syn WT (green) is mostly membrane and cytosolic in non-transfected cells (NT).
siRNA-mediated Gene Silencing of PLK2 and/or PLK3 Results in Significant Reduction (∼50–60%) in Ser-129 Phosphorylation in Cell Lines and Primary Neurons
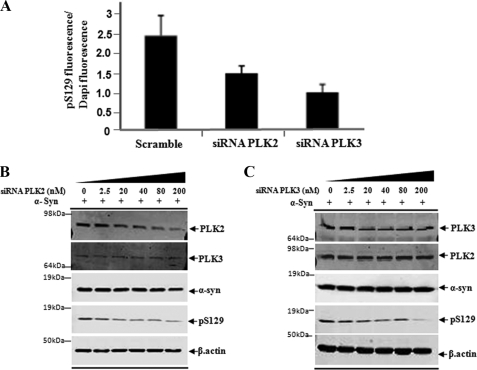
In order to determine the relative contribution of PLK2 and PLK3 to the phosphorylation of endogenous α-syn at Ser-129, we silenced each kinase in HeLa cells and primary neurons using siRNA against human and mouse PLKs, respectively. Given the low levels of endogenously expressed cytosolic α-syn in HEK 293T and HeLa cells, the levels of Ser(P)-129 were measured and quantified by immunofluorescence using anti-Ser(P)-129-specific antibodies. These results provide a measure of only nuclear Ser(P)-129. To determine the consequences of siRNA-mediated gene silencing of PLK2 and/or PLK3 on total Ser(P)-129 levels, the siRNAs were introduced 48 h prior-α-syn transfection in HeLa cells.
Upon silencing of PLK2 or PLK3 in HeLa cells, we observed ∼45–50% reduction in the levels of endogenous Ser(P)-129 in the nucleus as quantified by immunofluorescence using an anti-Ser(P)-129 antibody (Fig. 8A). Given the efficiency of both kinases in phosphorylating α-syn, we thought that silencing both kinases might be necessary to abolish phosphorylation at Ser-129. Interestingly, silencing of both kinases did not result in further reduction in nuclear Ser(P)-129 signal. These findings are consistent with our subcellular fractionation results and previous studies (39) demonstrating significant nuclear localization of PLK3 and PLK1 but not PLK2. Transient overexpression of α-syn results in increased levels of nuclear and cytoplasmic Ser(P)-129 in cell lines, including HeLa and HEK 293T, thus facilitating the detection of Ser(P)-129 by Western blot analysis. Interestingly, when we examined the effect of silencing either kinases on transiently expressed α-syn by Western blotting, we observed greater than 60% reduction in Ser(P)-129 levels (Fig. 8, B and C). Together, these findings demonstrate that PLKs contribute significantly to nuclear and cytoplasmic Ser-129 α-syn phosphorylation. The fact that we were not able to abolish Ser(P)-129 levels upon siRNA-mediated knockdown of each kinase or both kinases suggests that other kinases are involved in modulating the phosphorylation of nuclear and cytoplasmic α-syn at this residue and/or incomplete knockdown efficiency.
FIGURE 8.
Silencing of PLK2 and PLK3 results in significant reduction of Ser(P)-129 levels in HeLa cell. A, detection of nuclear Ser(P)-129 in non-transfected cells and cells transfected with 100 nm ON-TARGET siRNA PLK2 and 100 nm ON-TARGET siRNA PLK3. Quantitative analysis of total Ser(P)-129 fluorescence of individual cells was normalized against 4′,6-diamidino-2-phenylindole (Dapi) intensity, n = 3. B and C, immunoblot analysis of crude lysate from HeLa cells sequentially transfected with α-syn and siRNA PLK2 and siRNA PLK3 at the indicated concentration. Anti-β-actin was used as a control of the total amount of proteins loaded.
PLK1 to -3 Phosphorylate α-Syn Specifically at Ser-129 in Primary Neurons
To further evaluate the relevance of these findings in neurons, we investigated the expression of PLK2 and PLK3 and their subcellular localization with respect to α-syn and Ser(P)-129 α-syn. In addition to their prominent role in cell cycle regulation, the PLKs are also expressed to different levels in postmitotic cells, including neurons. Interestingly, we observed that constitutively phosphorylated α-syn is localized primarily in the nucleus of primary hippocampal neurons (Fig. 9A), consistent with the observations made in cell lines (Fig. 7B). Among the two PLKs, PLK2 yielded strong signals and colocalization to the nucleus of primary rat neurons with Ser(P)-129. PLK3 was distributed homogeneously in hippocampal neurons with low expression levels in the nucleus. More strikingly, we found that Ser(P)-129 colocalized at a lower level with PLK3 than with PLK2 in the nucleus (Fig. 9A). Similarly, PLK2 co-localized with Ser(P)-129 in neuronal nuclei and cell bodies in hippocampal and cortical brain areas, whereas PLK3 showed a distinct neuropil staining (Fig. 9, A and B).
FIGURE 9.
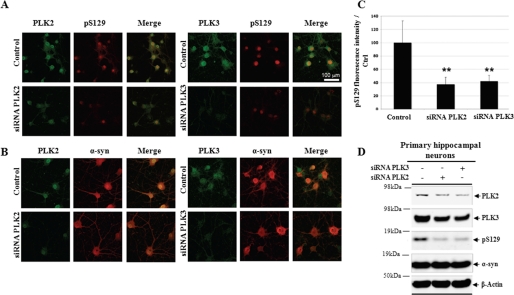
Silencing of PLK2 and PLK3 significantly inhibit α-syn phosphorylation at Ser-129 in primary neurons. Shown is colocalization (first panel) of Ser(P)-129 and PLKs (A) or WT α-syn versus PLKs (B). C, Western blot analysis of total cell homogenate from 72-h silencing of PLKs in hippocampal neurons. D, quantification of Ser(P)-129 fluorescence signal relative to control (from non-transfected cells). Data are expressed as the mean ± S.D; bar, 100 μm; n = 3.
To assess phosphorylation of Ser-129 α-syn by PLK1 to -3, hippocampal primary neurons were transfected with different siRNA targeting RNA coding for PLK1, PLK2, and PLK3. We observed that PLK2 and PLK3 are the major PLKs contributing to Ser-129 phosphorylation. We quantified the part of each PLK in the phosphorylation at Ser-129. Compared with the control (Fig. 9C), we found that PLK2 and PLK3 knockdown decrease by 63 and 58%, respectively, the total amount of Ser(P)-129, indicating that they are the major kinase involved in phosphorylation of endogenous α-syn. Moreover, we observed that the decrease of the protein level of each PLK modifies neither the levels of expressions of the other PLKs nor the total amount of α-syn as shown by the confocal imaging and by Western blot analysis (Fig. 9D). These results suggest that PLK-mediated phosphorylation of α-syn at position Ser-129 strongly involves PLK2 and PLK3.
PLK2 and PLK3 Expression in Mouse Model of Synucleinopathy
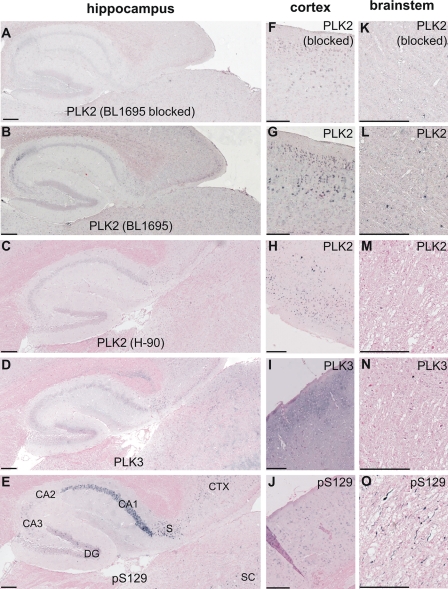
To investigate the relationship of α-syn phosphorylation with PLKs in vivo, we performed immunostaining on brain sections derived from α-syn transgenic (Thy1)-[A30P]α-syn, which develop age-dependent α-synucleinopathies that can be labeled with anti-Ser(P)-129-specific antibodies (5, 38, 40, 41). In these mice, PLK2 and PLK3 were widely distributed throughout the brain. PLK2 and PLK3 were generally localized diffusely in the neuropil. In addition, some neuronal subpopulations showed enriched somal and nuclear immunostainings for PLK2 and PLK3, respectively. In the hippocampus, PLK2 was weakly immunostained in scattered neuronal cell bodies throughout the cornu ammonis, whereas PLK3 showed a more diffuse staining pattern in the neuropil (Fig. 10, A–D). By comparison, Ser(P)-129 immunoreactivity in old mice was very strong in CA1, weak in CA3, and barely detectable in CA2 (Fig. 10E). Interestingly, we noted the presence of PLK3 and Ser(P)-129 immunoreactivity in neuronal cell bodies in the subiculum (Fig. 10, D and E). In the neocortex, there was a very similar staining pattern for non-classical somal/nuclear Ser(P)-129 immunostaining (38) and both PLK2 antibodies, whereas PLK3 showed almost exclusive neuropil staining in superficial cortical layers (Fig. 10, F–J). The tendency of PLK2 to co-localize with Ser(P)-129 in neuronal nuclei and a more cytosolic/neuritic distribution of PLK3 is in accordance with the hippocampal neuron culture staining (Fig. 9). None of the PLK antibodies used here specifically immunostained the Lewy-like Ser(P)-129 neuropathology in the brain stem of old (Thy1)-[A30P]α-syn mice (Fig. 10, K–O). Two different anti-PLK2 showed the cortical somal/nuclear localization (Fig. 10, G and H) and its absence in brain stem neuropathologies (Fig. 10, L and M), and antibody neutralization reduced the anti-PLK2 staining in the neuropil completely and in cell bodies to a good extent (Fig. 10, A, F, and K), indicating specificity of the immunostaining.
FIGURE 10.
Paraffin-embedded brain serial sections from 1-year-old (Thy1)-[A30P]-αSYN transgenic mice were immunostained with BL1695 anti-PLK2 with (A, F, and K) or without (B, G, and L) preincubation with neutralizing PLK2 protein, H-90 anti-SNK/PLK2 (C, H, and M) anti-PLK3 (D, I, and N), and anti-Ser(P)-129 (E, J, and O). Counterstaining was performed with Nuclear Fast Red. Pictures were taken from the hippocampal region (A–E), neocortex (F–J), and brain stem (K–O). Size bars, 200 μm.
Collectively, these results suggest that PLK2 may contribute to some physiological α-syn phosphorylation in cortical brain areas. Up-regulation of CA3 Ser(P)-129 in old transgenic mice might involve further, post-translational activations of PLKs and/or additional kinases (priming or additive). As for the pathological α-syn kinase, the lack of co-localization in Lewy neurites in this mouse model raises the speculation that neuropil PLK3 might phosphorylate α-syn in the synapse, and after that retrogradely transported Ser(P)-129 α-syn fibrilizes within axon (Lewy neurites) and neuronal cell bodies (Lewy bodies). Cross-breedings with PLK knock-out mice and long term treatments with specific PLK inhibitors are necessary to answer these questions and determine to what extent these kinases, and more specifically Ser-129 phosphorylation, contribute to the neuropathological processes in synucleinopathies. Further studies are required to determine whether Ser-129 phosphorylation in the nucleus is mediated primarily by members of the PLK family or selectively translocated from the cytoplasm upon phosphorylation by PLKs and other kinases.
PLK Levels Are Elevated in AD and LBD Brains
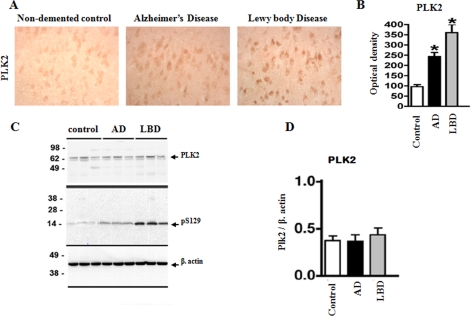
In order to determine the physiological and pathological relevance of Ser-129 phosphorylation by PLKs, we sought to assess the levels of PLK2 and PLK3 in human brain (temporal cortex) homogenates from non-demented controls and AD and LBD cases. Compared with controls, in the LBD, there was an increase in the levels of PLK2 immunoreactivity in the neurons (Fig. 11A). In the AD cases, there was also an increase in PLK2 immunoreactivity but to a lesser extent compared with the LBD cases (Fig. 11, A and B). By immunoblot analysis we confirmed the presence of PLK2 and the specificity of the antibody at detecting a doublet band at an estimated molecular mass of 62 kDa in the membrane fractions (Fig. 11C). However, immunoblot analysis did not reveal any significant differences in the levels of PLK2 in control versus AD and LBD cases (Fig. 11C). In addition to its localization in neurons, PLK2 appears to be highly enriched in the processes in the neuropil, where it is highly enriched. When performing the image analysis, it is possible to differentiate the levels of PLK2 in the different diseased tissues by immunocytochemistry, but by immunoblotting this is more difficult because of all the PLK2 in the neuropil, which might have a dilutional effect, thus masking the differences observed by immunocytochemistry (Fig. 11D). In the case of PLK3, we tested several commercial antibodies but failed to identify an antibody that allows for specific and reliable detection of PLK3 in human brain tissues by immunohistochemistry and immunoblotting.
FIGURE 11.
PLK2 expression in diseased brains (AD and LBD). A and B, immunohistochemical analysis of levels of PLK2 in control, AD, and LBD cases. C and D, immunoblot analyses with antibodies against cytosolic α-syn, PLK2, Ser(P)-129, and actin was performed using temporal cortex homogenates from non-demented controls, AD, and LBD cases. Total cellular protein concentrations in each sample were confirmed by comparison with levels of actin (bottom). Bar graph, quantitative image analysis of both immunocytochemical and immunoblotting results are shown.
DISCUSSION
Several post-translational modifications of α-syn have been described, including phosphorylation, nitration, ubiquitination, and truncations, but the role of these modifications in modulating the physiologic and/or pathological properties of α-syn remains poorly understood. One can speculate that many α-syn properties and functions are regulated by its interactions with other proteins and its subcellular localization/targeting, which in turn might be regulated by post-translational modifications. Therefore, identifying the key molecules responsible for regulating α-syn post-translational modifications and elucidating their mechanism of action are essential for unraveling the molecular basis of α-syn function(s) in health and disease.
Here we showed using in vitro kinase assays that α- and β-syn but not γ-syn are phosphorylated by members of the PLK family of Ser/Thr protein kinases. Phosphorylation of α- and β-syn occurs primarily on Ser-129 and Ser-118, respectively. Interestingly, neither α-, β- or γ-syn appear to be a substrate for PLK4. PLK4 is significantly larger (970 amino acids) than PLK1 to -3 (Fig. 1A) and contains only one PBD, which is essential for PLK4 dimerization (24, 26) (Fig. 1A). Given the reported roles of Polo box domains in modulating the subcellular localization of PLKs (42) and the interactions of PLKs with their substrates, it is tempting to propose that the PBD in PLK1 to -3 plays a critical role in modulating the interactions α-syn phosphorylation by PLK1 to -3. However, our results suggest that the propensity of PLKs to phosphorylate α-syn is determined by their substrate specificity and sequence complementarities between the catalytic site of each kinase and the amino acid sequence flanking the phosphorylated serine residue as well as the conformation of the protein. Sequence analysis and three-dimensional homology modeling studies revealed that PLK2 and PLK3 show the highest pairwise identity of the catalytic domain and ATP binding site. The sequence identity between PLK2 and PLK3 at the catalytic domain and ATP binding site is 68 and 97%, and that of PLK2 and PLK3 to PLK1 is ∼53 and 86–90%, respectively (43). Furthermore, the consensus sequences of PLK2 and PLK3 are very similar and characterized by the presence of acidic residues within the 3–4 residues flanking the phosphorylation site ((D/E)X(S/T)ψ(D/E), where ψ denotes a hydrophobic amino acid). PLK1 to -3 have a preference for substrates in which at least one of the three residues flanking the serine residue is negatively charged. Both α- and β-syn fulfill this criterion (Fig. 3C). This charge distribution on the substrates complements the electropositive distribution on the substrate binding sites of PLK1 to -3. PLK4, which lacks the preference for acidic residues, has much reduced electropositive environment in its substrate biding site. The lack of phosphorylation of γ-syn by the PLKs can also be explained by its sequence divergence from α-syn and β-syn. The amino acids flanking the equivalent serine residue in γ-syn are alanine (−1-position) and lysine (+1-position), thus explaining the lack of phosphorylation by PLK1 to -4.
Although α-syn is phosphorylated by PLK1, PLK2, and PLK3, only PLK1 and PLK3 were observed to phosphorylate β-syn in vitro. α-Syn and β-syn share high sequence homology at the N- and C-terminal region and differ mainly in the fact that β-syn lacks residues 73–83 in the NAC region of α-syn, suggesting that these residues might be important for synuclein interaction and phosphorylation by PLKs. To test this hypothesis, we investigated the effect of removal of the entire N-terminal region 1–102 or the NAC region 73–83 on PLK-mediated phosphorylation of α-syn. In addition, we examined the ability of the PLKs to phosphorylate a chimeric construct, in which the NAC domain comprising residues 73–83 has been reintroduced to β-syn, β/α-syn(χ1). Interestingly, removal of the entire N-terminal domain 1–102 abolished Ser-129 phosphorylation by all three PLKs 1–3, whereas removal of the NAC domain 73–83 did not have a significant effect on Ser-129 phosphorylation by the PLKs. Similarly, the introduction of the NAC domain 73–83 derived from α-syn into β-syn did not change the level of phosphorylation and/or specificity of PLKs toward β-syn. Both WT and chimeric β-syn undergo phosphorylation by PLK1 and PLK3 but not PLK2. Our observations that PLK2 phosphorylates β-syn in HEK 293T and HeLa cells suggest that other factor within the cell may influence the conformation of β-syn and/or its interactions with PLKs. The potential role of the conformational state of α-syn and β-syn in modulating their phosphorylation by the PLKs is supported by our findings that 1) mutations within the NAC region (S87A, S87E, and S87N in mouse α-syn), which are distant from Ser-129, result in significant reduction in PLK1-mediated Ser- 129 phosphorylation; 2) none of the PLKs phosphorylate the C-terminal fragment 102–140. These findings strongly suggest that phosphorylation at Ser-87 and/or tyrosine residues within the C terminus (Tyr-125, Tyr-133, and Tyr-136) would significantly influence the efficiency and specificity of Ser-129 phosphorylation by PLKs and possibly other kinases (44).
Ser-129 Phosphorylation by the PLKs Occurs in Mammalian Primary Neurons and Human Brain
To determine the relevance of the in vitro kinase assays, we probed the possibility that α-syn phosphorylation by the PLKs occurs in mammalian cells and investigated the expression levels and subcellular colocalization of α-syn and PLKs in primary rat neurons and brain tissues from symptomatic α-syn transgenic mice and human brain (temporal cortex) homogenates from AD and LBD cases and non-demented controls.
By co-transfection of α-syn and PLKs or siRNA-mediated silencing of PLKs, we demonstrated that PLK2 and PLK3 colocalize with α-syn in different cellular compartments and are major contributors to Ser-129 phosphorylation of endogenous and transiently expressed α-syn in mammalian cells (HEK 293T and HeLa) and primary neurons. These findings are consistent with the results we obtained from the in vitro kinase assays using purified recombinant α-syn and PLKs. Despite the high degree of colocalization of PLK1 and α-syn in nuclear, cytosolic, and membrane fractions, very minimal levels of Ser-129 phosphorylation were detected upon co-transfection of PLK1 and α-syn, consistent with observed low efficiency of PLK1 phosphorylation of α-syn in vitro (Fig. 2).
In addition to their prominent role in cell cycle regulation, the PLKs are also expressed to different levels in postmitotic cells, including neurons. PLK2 and PLK3 are expressed in response to synaptic activation and appear to be involved in synaptic plasticity, remodeling, and homeostasis (45–47) and have been implicated in the regulation of dendritic spin morphology. Moreover, a recent study using whole-genome microarray hybridization analysis identified PLK2 as indispensable for nerve growth factor-driven neuronal differentiation because its silencing inhibits neuronal differentiation (48). However, little is known about their substrates and function in neurons. In neurons, PLK2 and PLK3 were shown to interact with Cib, a Ca2+- and integrin-binding protein (49), SPAR (spine-associated Rap guanosine triphosphatase-activating protein), and the postsynaptic scaffolding protein PSD-95 (50). Seeburg et al. (47) demonstrated that induction of PLK2 in hippocampal neurons results in phosphorylation and degradation of PSD95 and SPAR by the ubiquitin-proteasome system, resulting in the loss of mature dendrite spines and synapse in cell lines and primary neuron cultures. The work presented here establishes the presynaptic proteins α- and β-syn as neuronal substrates for PLK2 and -3 and suggest a potential role for these kinases in modulating the normal physiology of α-syn.
The physiological relevance of α-syn phosphorylation by members of the PLK family of kinases is supported by the following: 1) PLK2 and PLK3 partially colocalize with Ser(P)-129 α-syn in mammalian cells and primary hippocampal neurons and in cortical brain areas of mice overexpressing α-syn; 2) siRNA-mediated silencing of PLK2 or PLK3 results in significant reduction of nuclear and cytoplasmic Ser(P)-129 levels in mammalian cells and primary neurons; 3) PLK2 is expressed in normal brain, and neuronal PLK2 is elevated in AD and LBD brains, consistent with previous reports demonstrating increased levels of PLK in the cytosol of vulnerable hippocampal and cortical neurons and homogenates of AD brain compared with control brains (51); 4) Inglis et al. (27) identified PLK2 as a major contributor to α-syn phosphorylation in vivo and showed that the level of Ser(P)-129 in the PLK2 knock-out mice is reduced by ∼70%. Together, these findings point to PLK2 and PLK3 as the primary PLKs and major kinases responsible for α-syn phosphorylation at Ser-129.
Implications for Elucidating the Role of Ser(P)-129 in PD Pathogenesis
The identification of α-syn as a substrate of PLK2 and PLK3 has significant implications for understanding the role of Ser-129 phosphorylation in modulating α-syn aggregation and toxicity and development of therapeutic strategies to treat PD and related synucleinopathies. Previous in vivo studies attempting to elucidate the role of Ser-129 phosphorylation have relied on the overexpression of phosphomimicking mutations (S129E/D) and blocking phosphorylation (S129A) have yielded inconclusive and/or conflicting results (52–55). Furthermore, in vitro biophysical comparison of the phosphomimicking (S129E/D) mutants to Ser(P)-129 α-syn revealed that these mutations do not reproduce all aspects of phosphorylation (8, 9).6 Together, these findings suggest that modeling α-syn phosphorylation in vivo can only be achieved by modulating the expression and/or activity of the kinases and phosphatases involved. PLK2 and PLK3 are the only kinases we have identified thus far to yield selective and quantitative phosphorylation of α-syn (PLK2 and -3) and β-syn (PLK1 and -3) in vitro. The specificity and efficiency with which these kinases phosphorylate α-syn in vitro and in vivo provide unique opportunities to elucidate the molecular mechanisms and functional consequences of α-syn phosphorylation in vivo and validate the potential of PLKs as therapeutic targets for treating PD and related synucleinopathies.
Therapeutic Implications
Whether inhibition of PLK2 and PLK3 kinase activity will increase or diminish α-syn aggregation and toxicity and PD still awaits the results from in vivo studies aimed at elucidating the consequences of selective overexpression and/or silencing of these PLKs in the affected brain regions on animal models of synucleinopathies. If it turns out that phosphorylation of Ser-129 is a causative event in the pathogenesis of PD, then inhibiting PLK2 and PLK3 would be an attractive strategy for the treatment of PD. The PLKs, mainly PLK1 and PLK3, have been the focus of drug discovery efforts for the treatment of cancer. However, the use of kinase inhibitors as potential drugs in PD would require the development of potent and selective inhibitors that can cross the blood brain barrier and target each member of the PLK family. The four PLKs share high sequence identity (25–34%) at the kinase domain and ∼80% identity of the ATP binding sites. Studies by Kothe et al. (56) suggested that subtle differences at the catalytic site may offer opportunities for potent selective targeting of PLK2. For example, the presence of Leu-132 in PLK1 and PLK3, creates a pocket in the ATP binding site that does not exist in PLK2 or other kinases in which this residue is conserved as tyrosine. As of today, the only structural information available is derived from the x-ray structural data of the catalytic domain of PLK1. Therefore, solving the x-ray structure of PLK2 and PLK3 may shed new light onto sequence and structural features that could be exploited to develop selective inhibitors targeting each of the PLKs. An alternative approach that could allow specific modulation of PLKs activity would be to target interactions involving the Polo box domains that are specific to this family of kinases.
Supplementary Material
Acknowledgments
We thank Dr. David Eliezer for reviewing the manuscript and Dr. Marc Moniatte, Diego Chiappe, and Jerome Vialaret from the Ecole Polytechnique Federale de Lausanne proteomic core facility for assistance with mass spectrometry data collection, Liliane Glauser for the assistance with primary neuronal cell culture, Maria Laura Orcellet for technical assistance, and Dr. Romain Zufferey for thoughtful discussions. We are also grateful to Prof. Michel Geodert for providing the plasmids of the α-syn deletion mutant (73–83Δ) and the β/α-syn chimera (β-syn-(1–72)/α-syn-(73–83)/β-syn-(73–134)).
This work was supported by the Swiss Federal Institute of Technology Lausanne and grants to the Laboratory of Molecular Neurobiology and Neuroproteomics from the Swiss National Science Foundation (Grant 310000-110027) and the Michael J. Fox Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
K. E. Paleologou, A. Oueslati, S. Gideo, C. C. Rospigliosi, H. Y. Kim, G. R. Lamberto, C. O. Fernandez, A. Schmid, F. Chegini, P. W. Gai, D. Chiappe, M. Moniatte, B. L. Schneider, P. Aebischer, M. Zweckstetter, D. Eliezer, and H. A. Lashuel, submitted for publication.
- α-
- β- and γ-syn, α-, β-, and γ-synuclein, respectively
- LB
- Lewy body
- PD
- Parkinson disease
- PLK
- Polo-like kinase
- PBD
- Polo box domain
- AD
- Alzheimer disease
- LBD
- Lewy body disease
- WT
- wild type
- MALDI
- matrix-assisted laser desorption ionization
- TOF
- time-of-flight
- HSQC
- heteronuclear single quantum coherence
- siRNA
- small interference RNA
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- TBS
- Tris-buffered saline
- NAC
- non-amyloid component.
REFERENCES
- 1.Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M. S., Shen J., Takio K., Iwatsubo T. (2002) Nat. Cell Biol. 4, 160–164 [DOI] [PubMed] [Google Scholar]
- 2.Anderson J. P., Walker D. E., Goldstein J. M., de Laat R., Banducci K., Caccavello R. J., Barbour R., Huang J., Kling K., Lee M., Diep L., Keim P. S., Shen X., Chataway T., Schlossmacher M. G., Seubert P., Schenk D., Sinha S., Gai W. P., Chilcote T. J. (2006) J. Biol. Chem. 281, 29739–29752 [DOI] [PubMed] [Google Scholar]
- 3.Chen L., Feany M. B. (2005) Nat. Neurosci. 8, 657–663 [DOI] [PubMed] [Google Scholar]
- 4.Kahle P. J., Neumann M., Ozmen L., Haass C. (2000) Ann. N.Y. Acad. Sci. 920, 33–41 [DOI] [PubMed] [Google Scholar]
- 5.Neumann M., Kahle P. J., Giasson B. I., Ozmen L., Borroni E., Spooren W., Müller V., Odoy S., Fujiwara H., Hasegawa M., Iwatsubo T., Trojanowski J. Q., Kretzschmar H. A., Haass C. (2002) J. Clin. Invest. 110, 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi M., Kanuka H., Fujiwara H., Koyama A., Hasegawa M., Miura M., Iwatsubo T. (2003) Neurosci. Lett. 336, 155–158 [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa M., Fujiwara H., Nonaka T., Wakabayashi K., Takahashi H., Lee V. M., Trojanowski J. Q., Mann D., Iwatsubo T. (2002) J. Biol. Chem. 277, 49071–49076 [DOI] [PubMed] [Google Scholar]
- 8.Paleologou K. E., Schmid A. W., Rospigliosi C. C., Kim H. Y., Lamberto G. R., Fredenburg R. A., Lansbury P. T., Jr., Fernandez C. O., Eliezer D., Zweckstetter M., Lashuel H. A. (2008) J. Biol. Chem. 283, 16895–16905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waxman E. A., Giasson B. I. (2008) J. Neuropathol. Exp. Neurol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pronin A. N., Morris A. J., Surguchov A., Benovic J. L. (2000) J. Biol. Chem. 275, 26515–26522 [DOI] [PubMed] [Google Scholar]
- 11.Arawaka S., Wada M., Goto S., Karube H., Sakamoto M., Ren C. H., Koyama S., Nagasawa H., Kimura H., Kawanami T., Kurita K., Tajima K., Daimon M., Baba M., Kido T., Saino S., Goto K., Asao H., Kitanaka C., Takashita E., Hongo S., Nakamura T., Kayama T., Suzuki Y., Kobayashi K., Katagiri T., Kurokawa K., Kurimura M., Toyoshima I., Niizato K., Tsuchiya K., Iwatsubo T., Muramatsu M., Matsumine H., Kato T. (2006) J. Neurosci. 26, 9227–9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okochi M., Walter J., Koyama A., Nakajo S., Baba M., Iwatsubo T., Meijer L., Kahle P. J., Haass C. (2000) J. Biol. Chem. 275, 390–397 [DOI] [PubMed] [Google Scholar]
- 13.Kim E. J., Sung J. Y., Lee H. J., Rhim H., Hasegawa M., Iwatsubo T., Min do S., Kim J., Paik S. R., Chung K. C. (2006) J. Biol. Chem. 281, 33250–33257 [DOI] [PubMed] [Google Scholar]
- 14.Nakamura T., Yamashita H., Takahashi T., Nakamura S. (2001) Biochem. Biophys. Res. Commun. 280, 1085–1092 [DOI] [PubMed] [Google Scholar]
- 15.Negro A., Brunati A. M., Donella-Deana A., Massimino M. L., Pinna L. A. (2002) FASEB J. 16, 210–212 [DOI] [PubMed] [Google Scholar]
- 16.Ellis C. E., Schwartzberg P. L., Grider T. L., Fink D. W., Nussbaum R. L. (2001) J. Biol. Chem. 276, 3879–3884 [DOI] [PubMed] [Google Scholar]
- 17.Clay F. J., McEwen S. J., Bertoncello I., Wilks A. F., Dunn A. R. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4882–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamanaka R., Maloid S., Smith M. R., O'Connell C. D., Longo D. L., Ferris D. K. (1994) Cell Growth Differ. 5, 249–257 [PubMed] [Google Scholar]
- 19.Golsteyn R. M., Schultz S. J., Bartek J., Ziemiecki A., Ried T., Nigg E. A. (1994) J. Cell Sci. 107, 1509–1517 [DOI] [PubMed] [Google Scholar]
- 20.Fode C., Motro B., Yousefi S., Heffernan M., Dennis J. W. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6388–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B., Ouyang B., Pan H., Reissmann P. T., Slamon D. J., Arceci R., Lu L., Dai W. (1996) J. Biol. Chem. 271, 19402–19408 [DOI] [PubMed] [Google Scholar]
- 22.Barr F. A., Silljé H. H., Nigg E. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 23.Xie S., Xie B., Lee M. Y., Dai W. (2005) Oncogene 24, 277–286 [DOI] [PubMed] [Google Scholar]
- 24.Cheng K. Y., Lowe E. D., Sinclair J., Nigg E. A., Johnson L. N. (2003) EMBO J. 22, 5757–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai W. (2005) Oncogene 24, 214–216 [DOI] [PubMed] [Google Scholar]
- 26.Leung G. C., Hudson J. W., Kozarova A., Davidson A., Dennis J. W., Sicheri F. (2002) Nat. Struct. Biol. 9, 719–724 [DOI] [PubMed] [Google Scholar]
- 27.Inglis K. J., Chereau D., Brigham E. F., Chiou S. S., Schöbel S., Frigon N. L., Yu M., Caccavello R. J., Nelson S., Motter R., Wright S., Chian D., Santiago P., Soriano F., Ramos C., Powell K., Goldstein J. M., Babcock M., Yednock T., Bard F., Basi G. S., Sham H., Chilcote T. J., McConlogue L., Griswold-Prenner I., Anderson J. P. (2009) J. Biol. Chem. 284, 2598–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mbefo M. K., Paleologou K. E., Boucharaba A., Oueslati A., Olschewski D., Hirling H., Lashuel H. (2008) Synuclein in Health and Disease, Lausanne, Switzerland, September 24–26, p. 21 (Abstr) [Google Scholar]
- 29.Zibaee S., Jakes R., Fraser G., Serpell L. C., Crowther R. A., Goedert M. (2007) J. Mol. Biol. 374, 454–464 [DOI] [PubMed] [Google Scholar]
- 30.Wilkins D. K., Grimshaw S. B., Receveur V., Dobson C. M., Jones J. A., Smith L. J. (1999) Biochemistry 38, 16424–16431 [DOI] [PubMed] [Google Scholar]
- 31.Steiner P., Sarria J. C., Glauser L., Magnin S., Catsicas S., Hirling H. (2002) J. Cell Biol. 157, 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirling H., Steiner P., Chaperon C., Marsault R., Regazzi R., Catsicas S. (2000) Eur. J. Neurosci. 12, 1913–1923 [DOI] [PubMed] [Google Scholar]
- 33.Jakes R., Spillantini M. G., Goedert M. (1994) FEBS Lett. 345, 27–32 [DOI] [PubMed] [Google Scholar]
- 34.Lavedan C. (1998) Genome Res. 8, 871–880 [DOI] [PubMed] [Google Scholar]
- 35.Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6469–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waxman E. A., Giasson B. I. (2008) J. Neuropathol. Exp. Neurol. 67, 402–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maroteaux L., Campanelli J. T., Scheller R. H. (1988) J. Neurosci. 8, 2804–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schell H., Hasegawa T., Neumann M., Kahle P. J. (2009) Neuroscience 160, 796–804 [DOI] [PubMed] [Google Scholar]
- 39.Zimmerman W. C., Erikson R. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1847–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freichel C., Neumann M., Ballard T., Müller V., Woolley M., Ozmen L., Borroni E., Kretzschmar H. A., Haass C., Spooren W., Kahle P. J. (2007) Neurobiol. Aging 28, 1421–1435 [DOI] [PubMed] [Google Scholar]
- 41.Fournier M., Vitte J., Garrigue J., Langui D., Dullin J. P., Saurini F., Hanoun N., Perez-Diaz F., Cornilleau F., Joubert C., Ardila-Osorio H., Traver S., Duchateau R., Goujet-Zalc C., Paleologou K., Lashuel H. A., Haass C., Duyckaerts C., Cohen-Salmon C., Kahle P. J., Hamon M., Brice A., Corti O. (2009) PLoS ONE 4, e6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee K. S., Grenfell T. Z., Yarm F. R., Erikson R. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9301–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson E. F., Stewart K. D., Woods K. W., Giranda V. L., Luo Y. (2007) Biochemistry 46, 9551–9563 [DOI] [PubMed] [Google Scholar]
- 44.Chen L., Periquet M., Wang X., Negro A., McLean P. J., Hyman B. T., Feany M. B. (2009) J. Clin. Invest., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kauselmann G., Weiler M., Wulff P., Jessberger S., Konietzko U., Scafidi J., Staubli U., Bereiter-Hahn J., Strebhardt K., Kuhl D. (1999) EMBO J. 18, 5528–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeburg D. P., Pak D., Sheng M. (2005) Oncogene 24, 292–298 [DOI] [PubMed] [Google Scholar]
- 47.Seeburg D. P., Feliu-Mojer M., Gaiottino J., Pak D. T., Sheng M. (2008) Neuron 58, 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Draghetti C., Salvat C., Zanoguera F., Curchod M. L., Vignaud C., Peixoto H., Di Cara A., Fischer D., Dhanabal M., Andreas G., Abderrahim H., Rommel C., Camps M. (2009) J. Biol. Chem. 284, 32053–32065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma S., Charron J., Erikson R. L. (2003) Mol. Cell. Biol. 23, 6936–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pak D. T., Sheng M. (2003) Science 302, 1368–1373 [DOI] [PubMed] [Google Scholar]
- 51.Harris P. L., Zhu X., Pamies C., Rottkamp C. A., Ghanbari H. A., McShea A., Feng Y., Ferris D. K., Smith M. A. (2000) Neurobiol. Aging 21, 837–841 [DOI] [PubMed] [Google Scholar]
- 52.Smith W. W., Margolis R. L., Li X., Troncoso J. C., Lee M. K., Dawson V. L., Dawson T. M., Iwatsubo T., Ross C. A. (2005) J. Neurosci. 25, 5544–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorbatyuk O. S., Li S., Sullivan L. F., Chen W., Kondrikova G., Manfredsson F. P., Mandel R. J., Muzyczka N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azeredo da Silveira S., Schneider B. L., Cifuentes-Diaz C., Sage D., Abbas-Terki T., Iwatsubo T., Unser M., Aebischer P. (2009) Hum Mol. Genet. 18, 872–887 [DOI] [PubMed] [Google Scholar]
- 55.McFarland N. R., Fan Z., Xu K., Schwarzschild M. A., Feany M. B., Hyman B. T., McLean P. J. (2009) J. Neuropathol. Exp. Neurol. 68, 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kothe M., Kohls D., Low S., Coli R., Cheng A. C., Jacques S. L., Johnson T. L., Lewis C., Loh C., Nonomiya J., Sheils A. L., Verdries K. A., Wynn T. A., Kuhn C., Ding Y. H. (2007) Biochemistry 46, 5960–5971 [DOI] [PubMed] [Google Scholar]
- 57.Warnke S., Kemmler S., Hames R. S., Tsai H. L., Hoffmann-Rohrer U., Fry A. M., Hoffmann I. (2004) Curr. Biol. 14, 1200–1207 [DOI] [PubMed] [Google Scholar]
- 58.Bahassi el M., Hennigan R. F., Myer D. L., Stambrook P. J. (2004) Oncogene 23, 2658–2663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.