Abstract
FGF2 transgenic mice were developed in which type I collagen regulatory sequences drive the nuclear high molecular weight FGF2 isoforms in osteoblasts (TgHMW). The phenotype of TgHMW mice included dwarfism, decreased bone mineral density (BMD), osteomalacia, and decreased serum phosphate (Pi). When TgHMW mice were fed a high Pi diet, BMD was increased, and dwarfism was partially reversed. The TgHMW phenotype was similar to mice overexpressing FGF23. Serum FGF23 was increased in TgHMW mice. Fgf23 mRNA in bones and fibroblast growth factor receptors 1c and 3c and Klotho mRNAs in kidneys were increased in TgHMW mice, whereas the renal Na+/Pi co-transporter Npt2a mRNA was decreased. Immunohistochemistry and Western blot analyses of TgHMW kidneys showed increased KLOTHO and decreased NPT2a protein. The results suggest that overexpression of HMW FGF2 increases FGF23/FGFR/KLOTHO signaling to down-regulate NPT2a, causing Pi wasting, osteomalacia, and decreased BMD. We assessed whether HMW FGF2 expression was altered in the Hyp mouse, a mouse homolog of the human disease X-linked hypophosphatemic rickets/osteomalacia. Fgf2 mRNA was increased in bones, and Western blots showed increased FGF2 protein in nuclear fractions from osteoblasts of Hyp mice. In addition, immunohistochemistry demonstrated co-localization of FGF23 and HMW FGF2 protein in osteoblasts and osteocytes from Hyp mice. This study reveals a novel mechanism of regulation of the FGF23-Pi homeostatic axis.
Keywords: Calcium/Bone, Development Differentiation/Organ, Diseases, Growth Factors, FGF23, Hyp mice, Klotho, Phosphate
Introduction
Fibroblast growth factor (FGF)2 ligands and their receptors (FGFRs) are important modulators of bone growth and development in mice (1–4). Mutations in FGFRs cause autosomal dominant human chondrodysplasias (1–3). These mutations result in aberrant or amplified signal transduction from the tyrosine kinase domain of the FGFRs (1). However, only recently have ligands for FGFRs been associated with human diseases (1, 5). There are 22 FGF ligands. Recent studies have shown that FGF23 is the phosphaturic factor (5) responsible for autosomal dominant hypophosphatemic rickets (6) and tumor-induced osteomalacia (7). FGF23 also mediates phosphate-wasting disorders, such as X-linked hypophosphatemic rickets/osteomalacia, the most common cause of vitamin D-resistant rickets (5). Loss of function mutations in PHEX, a phosphate-regulatory gene with homology to endopeptidases on the X-chromosome, have been identified in X-linked hypophosphatemic rickets/osteomalacia (8); however, the mechanism by which this elevates FGF23 levels remains unclear. Autosomal recessive hypophosphatemic rickets/osteomalacia, caused by loss of function mutations in DMP1 (dentin matrix protein), is also associated with increased FGF23 (9). Increased FGF23 may occur in McCune-Albright syndrome due to a somatic gain of function mutation in GNAS1 arising during embryogenesis, characterized by chimeric distribution of hyperpigmented skin lesions, fibrous dysplasia of bone, and, often, hypophosphatemia (5). Murine models of this group of disorders include the FGF23 transgenic mouse, a model of autosomal dominant hypophosphatemic rickets (10); the Hyp mouse, a homologue of X-linked hypophosphatemic rickets/osteomalacia with a Phex deletion (11); and the Dmp1 null mouse, a model of autosomal recessive hypophosphatemic rickets/osteomalacia (12). These murine homologues demonstrate many of the phenotypic changes of the human disorders, including elevated serum FGF23.
Another FGF ligand, FGF2 (fibroblast growth factor-2) is widely expressed and is a mitogen for many cell types, including osteoblasts and chondrocytes (1, 13, 14). The FGF2 gene encodes multiple FGF2 high molecular weight (HMW) protein isoforms expressed from unique CUG alternative translation start sites located 5′ to the classical AUG initiation codon for the 18-kDa low molecular weight (LMW) exported isoform (1, 15). HMW FGF2 isoforms are ordinarily not released from the cells but have nuclear localization sequences and function in an intracrine manner (16). In humans, there are three HMW isoforms of 22, 23, and 24 kDa and a LMW/18-kDa FGF2 protein isoform. In rodents, there are two HMW isoforms of 21 and 22 kDa and a LMW/17.5-kDa FGF2 isoform that is exported from cells (17, 18). The effect of the 18-kDa FGF2 isoform has been studied in bone (1); however, there are limited studies on the biological function(s) of the HMW FGF2 isoforms that have been primarily studied in non-osseous tissue (19–22). Because there is interest in FGF2 as a therapeutic target (1), it is important to understand whether there are differential effects of FGF2 protein isoforms in bone. In support of such studies, global overexpression of all isoforms of human FGF2 protein (TgFGF2) in mice resulted in dwarfism (4), decreased bone mass, and bone formation (23). Global knock-out of all isoforms of FGF2 in mice also resulted in reduced bone mass and bone formation (24). Transgenic mice that express human LMW 18-kDa FGF2 under the control of the Col3.6 promoter had increased bone mass with normal calcium/phosphate homeostasis (25).
The goal of the present work was to analyze the role of the HMW isoforms of FGF2 in bone. Initial characterization of the isoforms of FGF2 were described in detail by Florkiewicz et al. (15), and the development of cDNA constructs for ALLFGF2 isoforms and mutants engineered to express individual or sets of FGF2 isoforms was also described by Florkiewicz et al. (26). Mutation to one or more of the upstream CUG codons to CUU eliminated the expression of the corresponding high molecular weight FGF2. Mutation of the AUG codon resulted in ablation of the low molecular weight isoform expression, and a frameshift mutation eliminated all of the high molecular weight isoforms.
We generated transgenic mice expressing HMW isoforms of human FGF2 under the control of the same Col3.6 promoter utilized in development of the LMW 18-kDa FGF2 isoform Tg mice (25). In contrast to the high bone mass phenotype of LMW 18-kDa FGF2 mice, the TgHMW mouse phenotype included dwarfism, decreased bone mineral density (BMD), osteomalacia, and hypophosphatemia. The decreased bone mass and hypophosphatemia are similar to results reported in mice that overexpress FGF23 with the same promoter (10). In FGF23 transgenic mice, phosphate wasting is associated with down-regulation of the renal sodium/phosphate transporter NPT2a. Therefore, we explored the potential involvement of the bone kidney axis in the regulation of phosphate homeostasis in HMW FGF2 transgenic mice.
EXPERIMENTAL PROCEDURES
Generation and Identification of Mice Expressing HMW Isoforms of hFGF2
To elucidate the role of endogenous HMW FGF2 isoforms in a bone-specific manner, we generated an expression vector, called Col3.6-Fgf2 24-, 23-, and 22-kDa isoforms-IRES/GFP (Col3.6-HMW Fgf2 isoforms-IRES-GFPsaph or HMW). Col3.6-HMW Fgf2 isoforms-IRES-GFPsaph (Fig. 1A) was built by replacing a chloramphenicol acetyltransferase fragment in previously made Col3.6-CAT-IRES-GFPsaph with HMW isoforms of human Fgf2 cDNA between AfeI and ScaI sites. This expression vector (27) concurrently overexpresses HMW FGF2 isoforms and GFPsaph from a single bicistronic mRNA. Col3.6-IRES/GFP (Vector) construct was also prepared as control to Col3.6-HMW Fgf2 isoforms-IRES-GFPsaph. The construct inserts were released from Col3.6-IRES/GFP (Vector) or Col3.6-HMW Fgf2 isoforms-IRES-GFPsaph by digestion with AseI and AflII and purified according to standard techniques. Microinjections into the pronucleus of fertilized oocytes were performed at the Gene Targeting and Transgenic Facility at the University of Connecticut Health Center. Founder mice of the F2 (FVBN) strain were mated with wild type mice to establish individual transgenic lines. Mice were maintained in a virus- and parasite-free barrier facility under a 12-h light/12-h dark cycle at the Gene Targeting and Transgenic Facility and weaned at 21 days of age. Mice were sacrificed by CO2 narcosis and cervical dislocation. The University of Connecticut Health Center Institutional Animal Care and Use Committee approved all animal protocols. Homozygote male mice were utilized at 2 months of age except where noted.
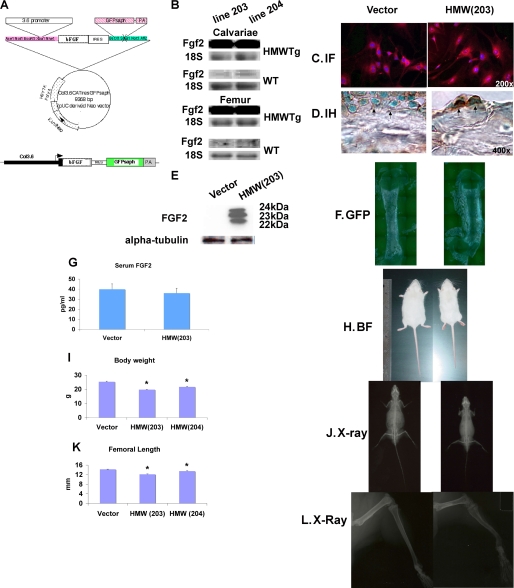
FIGURE 1.
Generation and identification of mice overexpressing hFGF2 HMW isoforms. A, schematic of Col3.6-HMW Fgf2 isoforms-IRES-GFPsaph expression vector. B, Northern blots show the overexpression of Fgf2 mRNA in calvariae and femurs from two lines (line 203 and line 204) of TgHMW compared with their non-Tg WT littermates. Immunofluorescent (IF) staining of cultured calvarial osteoblasts (C) and anti-FGF2 immunostaining (IH) of femurs (D) shows intense FGF2 labeling in nuclei of osteoblast from TgHMW mice compared with TgVector. E, analysis of FGF2 protein isoforms in BMSCs from TgVector and HMW mice. Starting on day 3 of culture, BMSC were fed with osteogenic differentiation media, and cells were cultured for 2 weeks. Western analysis of nuclear proteins revealed increased 22-, 23-, and 24-kDa FGF2 protein in cultures from TgHMW mice. F, scanning fluorescent microscopy shows the GFP expression in femurs of 7-day-old TgVector and TgHMW mice. G, serum FGF2 levels of 2-month-old TgVector and TgHMW mice (n = 8). H, gross appearance (bright field (BF)) of 2-month-old TgVector and TgHMW mice. I, body weight of 2-month-old TgVector and TgHMW mice (n = 13–36). J, x-ray image of 2-month-old TgVector and TgHMW mice. K, femoral length of 2-month-old TgVector and TgHMW mice (n = 6–16). L, x-ray image of lower limbs from 2-month-old TgVector and TgHMW mice. Values are mean ± S.E. *, significantly different from TgVector group, p < 0.05.
Diet
Mice were fed a regular diet of autoclaved Purina rodent chow (catalog number 5010, Ralston Purina) containing 1% calcium, 0.67% phosphorus, and 4.4 IU of vitamin D/g, except in the high phosphate diet rescue experiments. For the high phosphate diet rescue experiments, 21-day-old mice were fed either a regular diet or a high phosphate chow (catalog number TD.87133, Harlan Teklad) consisting of 2% phosphorus, 1.1% calcium, and 2.2 IU/g vitamin D, and 4 weeks later, the mice were sacrificed according to a standard protocol.
Faxitron X-ray
X-ray pictures of the whole mouse and selected areas of interest were taken using a Faxitron x-ray MX20/DX50 (Faxitron X-Ray Corp., Wheeling, IL). X-ray images were taken under constant conditions (25 kV, 20-s exposure at ×4.5 magnification) using Eastman Kodak Co. BIOMAX XAR film.
Dual Beam X-ray Absorptiometry
Third lumbar vertebrae (L3) were harvested and stored in 70% ethanol at 4 °C. BMD and bone mineral content (BMC) were measured using a Piximus Mouse 11 densitometer (GE Medical Systems, Madison, WI).
Microcomputed Tomography (Micro-CT) Scanning of L3
After measurement of the BMD and BMC, L3 vertebrae were analyzed by micro-CT system (μCT-20, Scanco Medical, Zurich), as previously reported (16). Using two-dimensional data from scanned slices, three-dimensional analysis was performed to calculate morphometric indices, including bone volume density (bone volume (BV)/total volume (TV)), trabecular thickness (equal to 2 × BV/bone surface (BS)), trabecular number (equal to (BV/TV)/trabecular thickness), and trabecular separation (equal to (1/trabecular number) − trabecular thickness).
Bone Histomorphometry
For static bone histomorphometry, L3 from 2-month-old Vector or HMW (line 203) mice were fixed in 4% paraformaldehyde at 4 °C, decalcified in 15% EDTA, dehydrated in progressive concentrations of ethanol, cleared in xylene, and embedded in paraffin. 5-μm sections were cut and stained for tartrate-resistant acid phosphatase to visualize osteoclasts and counterstained with hematoxylin. Histomorphometric analysis was performed in a blinded, nonbiased manner using a computerized image analysis system. BV/TV, osteoblast surface, osteoclast surface, and osteoclast number/bone surface were measured. For dynamic histomorphometric measurements, Vector and HMW (line 203) mice were weighed and injected on day 1 with calcein at 1 mg/100 g body weight, a second injection was administered on day 8, and mice were sacrificed on day 10. L3 vertebrae were dissected free of tissue and fixed in 70% ethanol at the time of sacrifice. The L3 were dehydrated in increasing concentrations of ethanol, cleared in xylene, and embedded in methyl methacrylate. 5-μm-thick longitudinal serial sections were cut on a Reichert-Jung Polycut S microtome (Reichert-Jung) with a D profile knife (Delaware Diamond Knives Corp., Wilmington, DE). Sections were taken from the middle of the L3. Unstained sections were evaluated for dynamic parameters. Additional sections were stained with modified Masson trichrome stain. Osteoblasts were identified as cuboidal cells lining the trabecular bone. Osteoclasts were identified as multinucleated cells on the trabecular bone surface. Histomorphometric measurements were made in a blinded, nonbiased manner using the Bio-Quant computerized image analysis system (R & M Biometrics, Nashville, TN) interfaced with a Nikon E400 microscope (Nikon Inc., Melville, NY). The terminology and units used are those recommended by the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research (28). Mineralizing surface/BS, the bone formation rate/BS, interlabel thickness, and mineral apposition rate (μm/day) were measured.
Biochemistry
Mice were fasted overnight, and then blood was collected from euthanized animals by cardiac puncture. After clotting, the blood was spun, and serum was collected. Serum phosphate and calcium were measured using the FAST 340 phosphorous reagent SET/calcium reagent SET (Eagle Diagnostics, Cedar Hill, TX). Creatinine was measured using the QuantiChromTM creatinine assay kit (DICT-500) (BioAssay Systems, Haywood, CA). Serum FGF23, 1,25-(OH)2-vitamin D3, and parathyroid hormone (PTH) were measured by a full-length FGF23 enzyme-linked immunosorbent assay kit (Kainos Laboratories, Inc., Tokyo, Japan), a 1,25-dihydroxyvitamin D enzyme immunoassay kit (ALPCO Diagnostic, Windham, NH), and a mouse intact PTH enzyme-linked immunosorbent assay kit (Immutopics, Carlsbad, CA), respectively. Serum human FGF2 was measured using the Quantikine® HS kit (R&D Systems, Minneapolis, MN). For the urine phosphate assay, mice were kept in metabolic cages, and 24-h urine was collected. Urine phosphate was measured using the FAST 340 phosphorous reagent SET/calcium reagent SET (Eagle Diagnostics). For measurement of the serum levels of the bone turnover marker C-terminal telopeptide of type 1 collagen (CTX), mice were fasted for ∼14 h with free access to water before being sacrificed. At sacrifice, blood was collected for serum, which was assayed for CTX using a mouse sandwich enzyme-linked immunoassay kit according to the manufacturer's instructions (RatLaps Elisa kit, Nordic Bioscience (Chesapeake, VA)).
RNA Isolation, Northern Blot Analysis, and Real-time PCR
Total RNA was extracted from bones or kidneys by TRI REAGENT according to the manufacturer's protocol. For Northern analysis, 20 μg of total RNA was denatured and fractionated on a 0.8% agarose, 1.1 m formaldehyde gel, transferred to filters by capillary blotting, and fixed to the filter by UV irradiation. After a 4-h prehybridization, filters were hybridized overnight with a [32P]cDNA probe for the mRNAs of Fgf2. For real-time quantitative reverse transcription (RT)-PCR analysis, RNA was reverse-transcribed by the SuperScriptTM first strand synthesis system for RT-PCR (Invitrogen). Quantitative PCR was carried out using the iQTM SYBR Green on a MyiQTM instrument (Bio-Rad). β-Actin was used as an internal reference for each sample.
Primary Calvarial Osteoblast (OB) Cultures for Immunofluorescent Staining
Calvarial OBs were obtained from newborn mice by sequential digestion with 0.1% collagenase (Roche Applied Science), as previously described (24). OBs were pooled and then cultured on a coverslide in 6-well culture dishes in Dulbecco's modified Eagle's medium (Invitrogen) with 10% heat-inactivated fetal bovine serum, penicillin, and streptomycin. Cells were grown for 7 days to ∼80% confluence. Then cells were briefly rinsed with 0.1 m PBS, pH 7.4, and fixed in 4% paraformaldehyde for 20 min at room temperature. Cells were washed three times in PBS, permeabilized with 0.3% Triton X-100 for 30 min, and incubated with 0.5% bovine serum albumin diluted in PBS for 20 min at room temperature. Finally, cells were incubated with mouse anti-FGF2 antibody (BD Biosciences) diluted 1:50 in PBS for 2 h at room temperature. After rinsing, cells were incubated with rhodamine (TRITC)-conjugated donkey anti-mouse IgG. After washing, coverslides were mounted on slides with UltraCruz mounting medium (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), on which nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Slides were imaged using a fluorescent microscope, and photographs were stored as electronic .tif files.
Western Blot Analysis of FGF2 Nuclear Proteins
Bone marrow stromal cells (BMSCs) were isolated using a modification of previously published methods (25). Tibiae and femurs from TgVector and HMW mice were dissected free of adhering tissue. Bone ends were removed, and the marrow cavity was flushed with α-minimum Eagle's medium by slowly injecting medium into one end of the bone using a sterile 25-gauge needle. Marrow cells were collected into tubes washed twice with serum-free α-minimum Eagle's medium. BMSCs were plated in 6-well plates (2 × 106 cells/well) in α-minimum Eagle's medium containing 10% heat-inactivated FCS and on day 3 were changed to osteogenic differentiation medium, and cultures were fed every 3 days by replacing 100% of the medium with fresh medium. On day 14, nuclear protein of BMSCs was extracted using a nuclear extraction kit according to the manufacturer's instructions (Panomics, Fremont, CA). Total protein of kidneys was extracted using 1× radioimmune precipitation buffer (Cell Signaling Technology, Inc., Beverly, MA), and protein concentration was assayed with BCA protein assay reagent (Pierce).
400 μg of nuclear protein from BMSC or 300 μg of total protein from kidney were diluted with 1× radioimmune precipitation buffer and incubated at 4 °C overnight with Protein A/G PLUS-agarose (20 μl) (Santa Cruz Biotechnology) as well as FGF2 (Millipore), KLOTHO (R&D Systems), Pp44/42 (Cell Signaling Technology, Beverly, MA), or NPT2a (Alpha Diagnostic International Inc. San Antonio, TX) antibody. Beads were washed three times, and precipitated proteins were followed by Western blot as described previously (25), using FGF2, KLOTHO, Pp44/42, and NPT2a antibodies. Equivalent amounts of total proteins were run on a separate gel and probed for α-tubulin or total p44/42 as a loading control.
Immunohistochemistry
Paraffin-embedded sections of kidneys from 2-month-old mice or femurs from 7-day-old mice were used. The primary antibodies were a monoclonal anti-phospho-p44/42 MAPK antibody (Cell Signaling Technology, Beverly, MA) utilized at a 1:100 dilution, anti-NPT2a antibody (Alpha Diagnostic International Inc.) used at a 1:200 dilution, and a monoclonal anti-KLOTHO antibody (Santa Cruz Biotechnology, Inc.) used at 1:100 dilutions. Anti-FGF2 antibody (BD Biosciences) was utilized at a concentration of 5 μg/ml. Briefly, the sections were blocked (0.3% H2O2/methanol) for 30 min, followed by blocking in 1:200 normal serum for 30 min, and then incubated with primary antibodies at 4 °C overnight. The sections were washed three times for 5 min in PBS, and then a horseradish peroxidase-conjugated secondary antibody was applied for 30 min. Following three 5-min PBS washes, color was developed with ABC reagent for 30 min. Slides were counterstained with methyl green or hematoxylin. Slides were mounted, examined, and photographed using bright field microscopy (Nikon Corp., Shinagawa-ku, Tokyo, Japan).
Construction and Production of CMV/Fgf2/IRES/eGFP Retroviral Vectors
CMV/IRES/eGFP (pMg1cla) vector containing the enhanced GFP (eGFP) gene driven from an internal cytomegalovirus (CMV) promoter was obtained from Dr A. Lichtler (29). The coding sequence of the HMW or LMW Fgf2 was cloned into the EcoRI site of the CMV/IRES/eGFP retroviral vector. eGFP was used to visualize expression of protein mediated by CMV vectors in cells. Control virus was CMV/IRES/eGFP vector without inserted HMW or LMW Fgf2 cDNA.
293GPG packaging cell lines were infected by CMV/IRES/eGFP, CMV/HMW Fgf2/IRES/eGFP, or CMV/LMW Fgf2/IRES/eGFP vector using the LipofectamineTM 2000 reagent (Invitrogen), following the product protocol. Infected 293GPG packaging cell lines were grown in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum, penicillin, and streptomycin, 2 μg/ml puromycin, 0.1 μg/ml tetracycline, and 0.4 mg/ml G418. To produce vector virus, tetracycline and G418 were removed from the media, and then condition media were collected after 24, 48, 72, and 96 h and passed through a 0.2-μm Nalgene filter. Condition medium was measured for reverse transcript activity then frozen at −80 °C.
Transduction of Mouse Primary Calvarial Osteoblasts
Primary calvarial osteoblasts from Fgf2 KO mice were plated in 6-well dishes and grown in Dulbecco's modified Eagle's medium (Sigma) containing 10% heat-inactivated fetal bovine serum plus 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were transduced with virus when 50% confluent. Medium, including 1 ml of virus-containing medium and 1 ml of Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum and 8 mg/ml protamine sulfate, was added to the cell cultures for ∼16 h, followed by fresh medium replacement for 8 h. The cells were exposed to three cycles of virus infection. Forty-eight hours after infection, total RNA was extracted.
Analysis of Phex, Fgf23, and Fgf2 Expression in Hyp Mouse
Two-month-old WT and Phex mutant mice were purchased from Jackson Laboratory (Bar Harbor, ME). Bones were harvested from WT and Hyp mice as described above. For mRNA expression of Phex, Fgf23, and Fgf2, total RNA was extracted from the shaft of bones of WT and Hyp mice, and quantitative RT-PCR analysis for Phex, Fgf2, and Fgf23 mRNA was performed as described above.
For Western blot analysis, bone marrow was flushed, and cells were cultured in osteogenic media as described above. Cultures were terminated at 14 and 21 days, and cytosolic and nuclear protein fractions were obtained for Western analysis for FGF23 and FGF2.
Immunofluorescent microscopy for FGF2 and FGF23 (monoclonal FGF23 antibody was purchased from R&D Systems) were also performed on BMSCs cultured in differentiation medium for 1 week and on frozen sections of tibiae from WT and Hyp mice.
Statistical Analyses
All results were expressed as means ± S.E. Differences between groups were analyzed using Student's t test, and differences were considered significant at p values of less than 0.05.
RESULTS
TgHMW FGF2 Transgenic Mouse Phenotypes
Several founder mice were identified, and two viable FGF2 transgenic lines carrying HMW (line 203 and line 204) isoforms of the hFGF2 transgene were established as well as a vector-only control transgenic mouse (TgVector). Northern blots (Fig. 1B) show overexpression of Fgf2 mRNA in calvariae and femurs from both lines. Homozygous breeding pairs were maintained to generate sufficient mice necessary for experimentation. Two-month-old mice were studied unless stated otherwise. Localized expression of HMW FGF2 isoforms from the transgene was confirmed by immunofluorescent staining (Fig. 1, C and D). There was intense FGF2 labeling in nuclei of cultured calvarial OB from TgHMW(203) mice (Fig. 1C) and intracellular localization in OBs of femurs (Fig. 1D). As shown in Fig. 1E, there was overexpression of the nuclear isoforms of human FGF2 protein in BMSCs from TgHMW mice compared with the endogenous HMW isoform of the vector mice that did not express the transgene. Fig. 1F shows GFP fluorescence in cortical and trabecular bone of TgVector and TgHMW mice but no expression in the growth plate or bone collar. There was no difference in serum FGF2 levels between TgVector and TgHMW mice, suggesting that HMW FGF2 expression was not secreted (Fig. 1G). Phenotypic differences in body size (Fig. 1H) were observed between TgVector and TgHMW(203) mice. The mean body weights (Fig. 1I) of TgHMW (line 203; 19.8 ± 0.2 g) and TgHMW (line 204, 21.7 ± 0.4 g) mice were both significantly different (p < 0.05) from TgVector mice (25.3 ± 0.5 g). X-rays (Fig. 1, J and L) showed shortened misshaped hypolucent bones with widened growth plates in TgHMW mice with significantly shorter femurs (p < 0.05) (Fig. 1K).
Dual Beam X-ray Absorptiometry and Micro-CT
BMD of the L3 vertebra (Fig. 2A) was decreased (p < 0.05) in TgHMW(203) by 13% and in TgHMW(204) by 8%. BMC (Fig. 2B) was also significantly decreased in comparison with TgVector mice (p < 0.05), by 26% in TgHMW(203) mice and by 10% in TgHMW(204) mice.
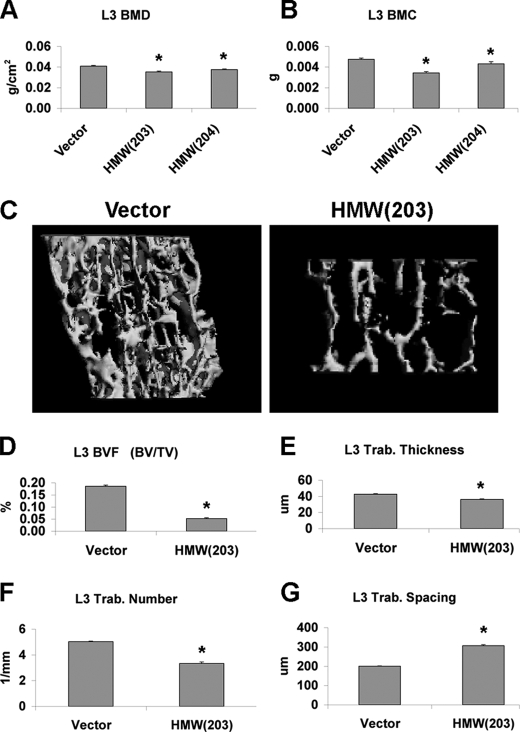
FIGURE 2.
Dual beam x-ray absorptiometry and micro-CT analysis of L3 vertebrae from TgVector and TgHMW mice. A, L3 BMD (n = 13–36); B, L3 BMC (n = 13–36); C, three-dimensional trabecular structure of L3 of 2-month-old male homozygous TgVector and TgHMW mice (n = 5–11). D–G, three-dimensional microstructural parameters were calculated using two-dimensional data obtained from micro-CT of vertebral bone. Calculated morphometric indices included bone volume density (BV/TV) (D), trabecular thickness (Trab. thickness) (DT-Tb-Th) (E), trabecular number (Trab. number) (F), and trabecular spacing (Trab. spacing) (G). *, significantly different from TgVector group (p < 0.05).
Micro-CT was used to compare bone microarchitecture between TgVector and TgHMW(203). The platelike structure of the trabecular bone was markedly decreased in TgHMW mice compared with TgVector mice (Fig. 2C). BV/TV (Fig. 2D) was decreased by 72% in TgHMW (p < 0.05). Trabecular thickness (Fig. 2E) was decreased by 15% (p < 0.05). Trabecular number (Fig. 2F) was decreased, and trabecular spacing (Fig. 2G) was (p < 0.001) increased.
Bone Histomorphometry
To determine whether decreased bone mass was caused by a decrease in bone formation and/or an increase in bone resorption, histomorphometric analysis was performed on the L3 vertebrae. There was a 60% reduction in BV/TV (Fig. 3A) in TgHMW mice. Osteoblast surface/BS (%ObS) (Fig. 3B) was reduced by 54% in TgHMW mice compared with TgVector mice. Osteoclast surface (%OcS) (Fig. 3C) and osteoclast number (OcNo)/BS (Fig. 3D) were increased by 179 and 190%, respectively, in TgHMW. Mineralizing surface (MS)/BS (Fig. 3E), bone formation rate (BFR)/BS (Fig. 3F), and interlabel thickness (Fig. 3, G and J) were decreased by 23, 27, and 14%, respectively, in TgHMW mice. The mineral apposition rate (Fig. 3H) was also decreased by 14% in TgHMW mice. von Kossa staining of non-decalcified sections from L3 vertebrae showed decreased mineralized trabeculae in TgHMW mice (Fig. 3I). These data suggest that decreased bone mass in TgHMW mice is due to both decreased bone formation and increased bone resorption.
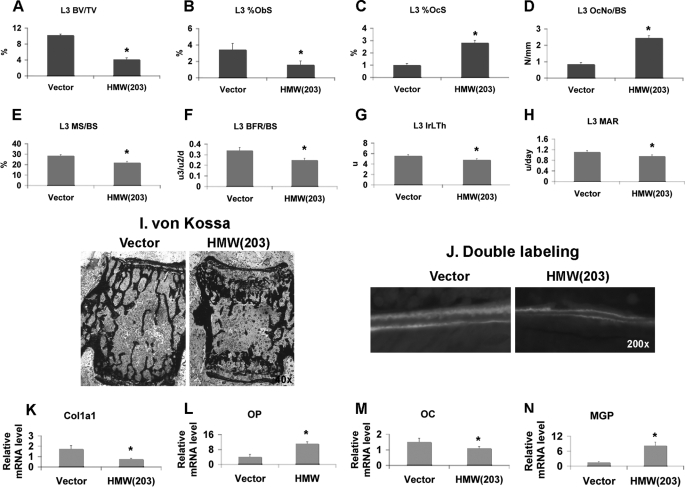
FIGURE 3.
Bone histomorphometry analysis of L3 vertebrae and bone marker gene expression in tibiae from 2-month-old TgVector and TgHMW mice. A–D, static bone histomorphometry analysis of L3 vertebrae from TgVector and TgHMW mice (n = 6). A, BV/TV. B, osteoblast surface (ObS)/BS. C, osteoclast surface (OcS). D, osteoclast number (OcNo)/BS. E–H, dynamic bone histomorphometry analysis of L3 vertebrae from TgVector and TgHMW mice (n = 7). E, mineralizing surface (MS)/BS. F, bone formation rate (BFR)/BS. G, interlabel thickness (IrLTh). H, mineral apposition rate (MAR). Values are mean ± S.E. *, significantly different from TgVector group, p < 0.05. I, von Kossa staining of non-decalcified sections from L3 vertebrae showed decreased mineralized trabuculae in TgHMW mice. J, calcein double labeling showed decreased interlabel thickness in TgHMW mice compared with TgVector mice. K–N, real-time RT-PCR analysis of Col1a1, osteopontin (OP), osteocalcin (OC), and matrix γ-carboxyglutamic acid protein (MGP) mRNA expression in tibiae from both genotypes. Data are expressed as mean ± S.E. from three independent experiments. *, significantly different from TgVector, p < 0.05.
Differential Bone Marker Gene Expression
To further assess the reduced bone formation, expression of mRNA for genes important in osteoblast precursor differentiation and mineralization was measured. Whole tibiae were analyzed by real-time PCR (Fig. 3, K–N). Type 1 collagen (Col1a1) (Fig. 3K) was decreased by 56% in bones from TgHMW compared with TgVector mice. Previous reports showed that osteopontin was regulated by FGF2 in periodontal cells (30). A 174% increase in osteopontin mRNA was observed in TgHMW bone. Osteocalcin expression that occurs later in osteoblast differentiation was decreased by 27% in TgHMW bone. Matrix γ-carboxyglutamic acid protein that inhibits matrix mineralization (31) was increased by 446% in TgHMW bone.
Hematology and Biochemistry
Anemia and kidney abnormalities have been reported in rats following systemic administration of FGF2 (32). To determine whether these effects developed in TgHMW mice, hematocrit and serum creatinine were measured in 2-month old mice. As shown in Table 1, hematocrit and creatinine were similar between TgVector and TgHMW mice. The TgHMW mice had low serum phosphate (Pi) but normal serum calcium compared with TgVector controls. To determine whether hypophosphatemia is due to renal Pi wasting, urinary Pi/g creatinine was determined. Urinary Pi/g creatinine was increased (p < 0.05) by 71% in TgHMW mice. Because PTH and vitamin D are important hormones involved in kidney Pi physiology, serum PTH and 1,25-(OH2)-vitamin D were also measured. There was an 86% increase (p < 0.05) in serum PTH in TgHMW mice compared with TgVector mice. There was no difference in serum 1,25-(OH2)-vitamin D. To further assess bone resorption, serum levels of CTX (33) were measured and were significantly higher in TgHMW mice.
TABLE 1.
Hematology and urine biochemistry
Blood, serum and urine were collected from 2-month-old male TgVector and TgHMW mice. Values are mean ± SE.
| Vector | HMW | n | |
|---|---|---|---|
| Hematocrit (%) | 45.79 ± 0.28 | 46.47 ± 0.22 | 4–17 |
| Serum creatinine (mg/dl) | 0.98 ± 0.06 | 1.07 ± 0.08 | 13–14 |
| Serum calcein (mg/dl) | 7.17 ± 0.18 | 7.74 ± 0.14 | 13–16 |
| Serum PTH (pg/ml) | 132.20 ± 29.41 | 245.60 ± 42.32a | 8 |
| Serum 1,25-(OH)2-vitamin D3 (pg/ml) | 54.11 ± 5.41 | 62.05 ± 6.52 | 9–10 |
| Serum CTX (ng/ml) | 17.76 ± 3.73 | 63.15 ± 7.52a | 8 |
| Serum Pi (mg/dl) | 9.52 ± 0.38 | 8.41 ± 0.16a | 13–16 |
| Urinary Pi (mol)/creatinine (g) | 0.011 ± 0.001 | 0.019 ± 0.002a | 3–4 |
a p < 0.05.
The comparatively normal levels for serum 1,25-(OH2)-vitamin D in TgHMW mice could be considered inappropriately low in the presence of hypophosphatemia and increased PTH. In addition, there were no significant differences in mRNA levels for 25-hydroxyvitamin D-1α-hydroxylase (Cyp27b1) or 24-hydroxylase (Cyp24) (supplemental Fig. 1).
Effects of High Phosphate in TgHMW Mice
The phenotype of TgHMW mice includes osteomalacia, decreased BMD, and dwarfism, which could be due to hypophosphatemia. To determine whether an increase in dietary phosphate could rescue TgHMW mice, 21-day-old TgVector mice and TgHMW mice were fed a normal or high phosphate diet for 4 weeks. The high phosphate diet increased the body weight (Fig. 4A), but it was still different from the TgVector normal diet group. Serum Pi was increased to the level of TgVector mice on a normal diet (Fig. 4B). BMD (Fig. 4D) and BMC (Fig. 4E) in TgHMW normal diet mice were increased to the level of TgVector mice on a normal diet. Reversal of the body weights, BMD, and BMC by the high phosphate diet also led to partial reversal of the TgHMW dwarfism phenotype in mice maintained on the high Pi diet for 4 weeks (Fig. 4, F–H). Because the TgHMW phenotype was similar to that reported for FGF23 transgenic mice (10), serum FGF23 was measured in TgVector normal diet, TgHMW normal diet, and TgHMW high Pi diet mice (Fig. 4C). Serum FGF23 was significantly increased in TgHMW normal diet mice compared with TgVector normal diet mice. Serum FGF23 levels were further increased after TgHMW mice were fed the high Pi diet.
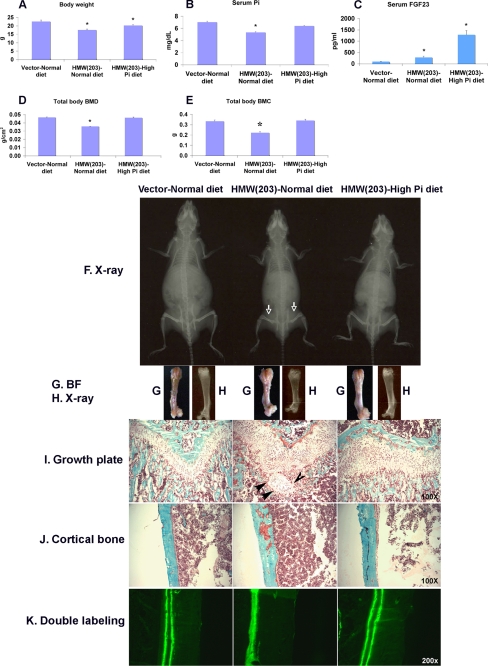
FIGURE 4.
High phosphate diet partially rescues the bone phenotype of TgHMW mice. TgVector and TgHMW mice were fed either a normal or high phosphate diet for 1 month immediately after weaning. They were sacrificed at 51 days old, and the following parameters were determined. A, body weight of TgVector and TgHMW mice (n = 4–7). B, serum phosphate level of TgVector and TgHMW mice (n = 4–7). C, serum FGF23 level of TgVector and TgHMW mice (n = 5–7). D, total body BMD of TgVector and TgHMW mice (n = 4–7). E, total body BMC of TgVector and TgHMW mice (n = 4–7). F, x-ray of whole body of TgVector and TgHMW mice. G, bright field (BF) of femurs from TgVector and TgHMW mice. H, x-ray of femurs from TgVector and TgHMW mice. Shown is Masson trichrome stain of growth plate (I) (note the arrows pointing to islands of cartilage surrounded by chondrocytes and mineralized bone) and cortical bone (magnification, ×100) of femurs from TgVector and TgHMW mice (J). K, calcein double labeling (magnification, ×200) of femurs from TgVector and TgHMW male mice. *, p < 0.05.
Bones (Fig. 4I) showed an organized epiphyseal growth plate in TgVector normal diet, a widened and largely disorganized growth plate was observed in TgHMW normal diet mice that was partially rescued in the TgHMW high Pi diet group. Mineralized bony trabeculae in the primary and secondary spongiosa were decreased in TgHMW normal diet mice, an effect that was partially rescued by the high Pi diet. Increased osteoid around the growth plate, in cortical bone and trabecular bone (Figs. 4, I and J), was observed in TgHMW normal diet mice. This mineralization defect was corrected in TgHMW mice fed a high Pi diet. The indistinct fluorescent double labeling in TgHMW normal diet also improved after high Pi diet (Fig. 4K). Finally, similar to FGF23 Tg mice (10), islands of cartilage surrounded by chondrocytes and mineralized bone were observed in the metaphysis of mice overexpressing TgHMW hFGF2 (Fig. 4I). These islands disappeared on the high Pi diet.
TgHMW FGF2 Affects the FGF23 Regulatory Axis
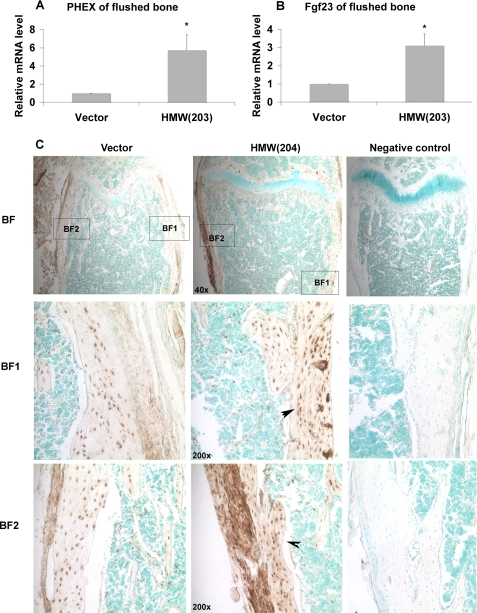
To determine whether overexpression of HMW FGF2 was associated with increased FGF23 expression in bone, mRNA and protein levels were determined by real-time PCR. There was a 3.1-fold increase in Fgf23 mRNA in flushed tibiae from TgHMW mice (Fig. 5B). Also, the mRNA for Phex, an osteoblast/osteocyte enzyme, which when disrupted leads to increased circulating FGF23 levels (34), was increased 5.7-fold (Fig. 5A). There was increased staining for FGF23 protein in osteoblasts and osteocytes (arrows) of femurs from TgHMW mice compared with TgVector mice (Fig. 5C).
FIGURE 5.
Effect of hFGF2 HMW isoform overexpression on Phex mRNA and Fgf23 mRNA and protein in bone. Real-time RT-PCR analysis of PHEX (A) and Fgf23 (B) expression in flushed bone. Data are expressed as mean ± S.E. from three experiments. *, significantly different from TgVector, p < 0.05. C, detection of FGF23 protein expression in femurs from TgVector and TgHMW by immunohistochemistry staining. Increased FGF23 staining in osteocytes (arrows) and osteoblasts in femurs of TgHMW mice compared with TgVector mice. BF, bright field.
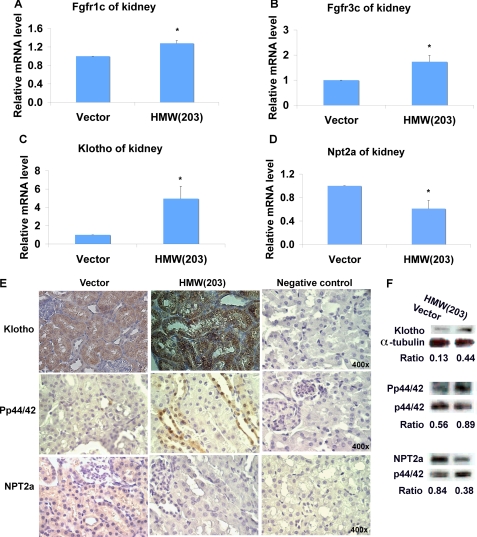
FGF23 activates FGF receptors, which require the presence of the co-receptor, KLOTHO, forming a FGF23-KLOTHO-FGFR1c complex, necessary for the transduction of the FGF23 signal, which results in phosphorylation of ERK (Pp44/42) and down-regulation of NPT2a (5, 34). To evaluate whether the FGF23-KLOTHO axis is involved in renal regulation of phosphate homeostasis in TgHMW mice, mRNA levels of whole kidneys were determined by real-time PCR. We found significant increases in mRNA for Fgfr1c, Fgfr3c, and Klotho in TgHMW (Fig. 6, A–C). By contrast, mRNA of the renal Na+/Pi co-transporter NPT2a was significantly decreased (Fig. 6D). Immunohistochemistry for KLOTHO and Pp44/42 (Fig. 6E) showed increased expression in kidneys of TgHMW mice, whereas NPT2a protein was decreased. As shown in Fig. 6F, Western blot confirmed the increase in KLOTHO and Pp44/42 and decreased NPT2a protein in kidneys of TgHMW mice.
FIGURE 6.
Effect of hFGF2 HMW isoform overexpression on FGF23-KLOTHO axis in kidney. Shown is real-time RT-PCR analysis of Fgfr1c (A), Fgfr3c (B), Klotho (C), and Npt2a (D) mRNA expression in kidney from 2-month-old male mice. Data are expressed as mean ± S.E. from three experiments. *, significantly different from TgVector, p < 0.05. E, immunohistochemistry for detection of KLOTHO, Pp44/42, and NPT2a protein expression in mouse kidneys from 2-month-old male TgVector and TgHMW mice. Note increased KLOTHO protein expression in both proximal and distal tubules of kidney from TgHMW mice. Negative control had no primary antibody during immunostaining. F, Western blot analysis of KLOTHO, Pp44/42, and NPT2a protein. Protein extracted from kidneys showed increased expression of KLOTHO and Pp44/42 but decreased expression of NPT2a from TgHMW mice compared with TgVector mice.
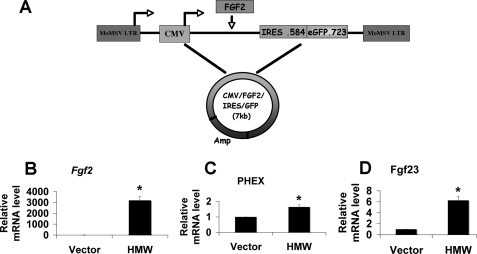
In vitro experiments were performed to evaluate whether increased production of FGF23 and PHEX was a direct effect from HMW FGF2 isoform overexpression in osteoblasts. To control for endogenous FGF2 expression, Fgf2 KO osteoblasts (15) were transduced with CMV/IRES/eGFP (Fig. 7A) as a control or CMV/HMW Fgf2/IRES/eGFP retrovirus for 48 h. Overexpression of the HMW FGF2 isoforms in Fgf2 KO osteoblasts was confirmed by real-time PCR (Fig. 7B). There was no Fgf2 mRNA in Fgf2 KO osteoblasts transduced with the CMV/IRES/eGFP control vector. Transduction of Fgf2 KO osteoblasts with CMV/HMW Fgf2/IRES/eGFP retrovirus for 48 h increased Fgf23 (Fig. 7D) and Phex (Fig. 7C) mRNA levels 6.3- and 1.6-fold, respectively.
FIGURE 7.
Effect of hFGF2 HMW isoform overexpression on Fgf23 and Phex transcripts in cultured Fgf2 KO calvarial osteoblasts. A, schematic of CMV-HMW Fgf2 isoforms-IRES-eGFP expression vector. Shown is real-time RT-PCR analysis of Fgf2 (B), Phex (C), and Fgf23 (D) mRNA expression in Fgf2 KO calvarial osteoblasts. Data are expressed as mean ± S.E. from three experiments. *, significantly different from vector transduction, p < 0.05.
Analysis of FGF2 mRNA and Protein in Bones and Osteoblasts from Hyp Mouse
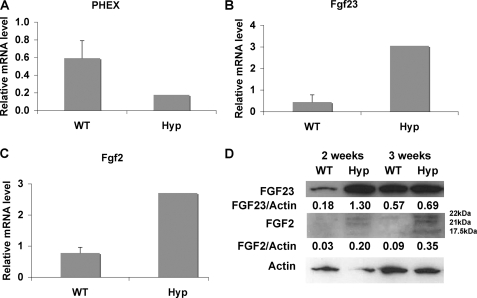
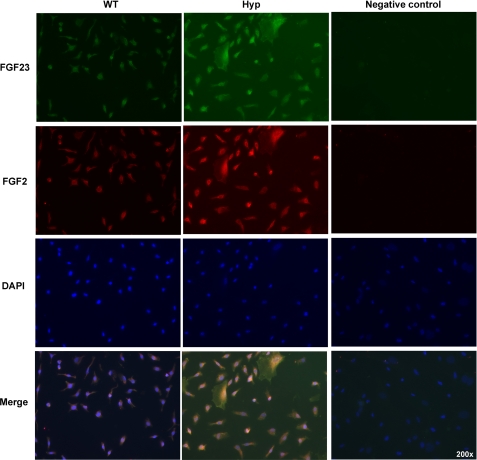
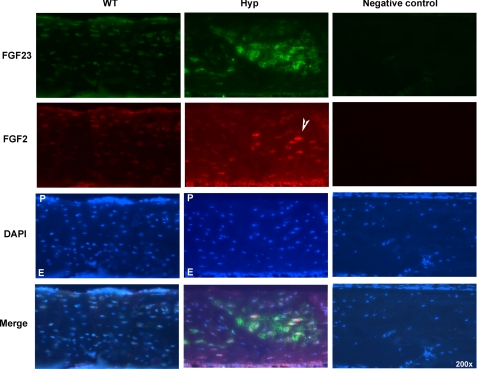
We determined whether HMW FGF2 expression was altered in the Hyp mouse, a model of human X-linked hypophosphatemic rickets/osteomalacia. As shown in Fig. 8, A–C, Phex mRNA was decreased (Fig. 8A), and Fgf23 mRNA was increased (Fig. 8B), as was the mRNA for FGF2 (Fig. 8C). Western blot (Fig. 8D) shows increased nuclear accumulation of FGF2 compared with WT cultures. As shown in Figs. 9 and 10, immunofluorescent microscopy showed increased FGF23 and FGF2 protein in cultured BMSCs and in osteocyte in tibiae from Hyp mice, and merged images show co-localization of both proteins.
FIGURE 8.
Analysis of Phex, Fgf23, and Fgf2 mRNA and FGF2 protein in bones from Hyp mice. Shown is real-time RT-PCR analysis of Phex (A), Fgf23 (B), and Fgf2 (C) mRNA expression in flushed bones. D, Western blot detection of FGF2 and FGF23 protein expression in nuclear fractions of BMSCs from WT and Hyp cultured for 2 and 3 weeks in osteogenic media. Increased HMW FGF2 and FGF23 protein is observed in nuclear fractions from Hyp mice compared with WT mice.
FIGURE 9.
Immunohistochemistry for detection of FGF23 and FGF2 protein in osteoblasts from Hyp mice. BMSCs from WT and Hyp mice were grown to confluence for 7 days and then cultured in osteogenic medium for an additional 7 days. Immunohistochemistry for detection of FGF23 (green), FGF2 (red), and 4′,6-diamidino-2-phenylindole (DAPI; blue) labeling of nuclei was performed. Note increased FGF23 and FGF2 protein expression in Hyp cultures. Merge images show co-localization of FGF23 and FGF2 in many cells from Hyp mice.
FIGURE 10.
Immunohistochemistry for detection of FGF23 and FGF2 in cortical bone from WT and Hyp mice. Tibias were collected from WT and Hyp mice, and immunohistochemistry was performed on frozen sections for detection of FGF23 (green) and FGF2 (red). The periosteal (P) and endosteal (E) regions of the cortical bones are shown. Nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI; blue). Increased FGF23 and FGF2 staining was observed in osteocytes (note the arrowheads) in tibiae of Hyp mice compared with WT mice. Merge images show co-localization (yellow) of FGF23 and FGF2 in osteocytes from Hyp mice.
DISCUSSION
These studies demonstrate that mice overexpressing the nuclear HMW human FGF2 isoforms in bone manifest an abnormal skeletal phenotype characterized by dwarfism, decreased BMD, rickets/osteomalacia, and a disorganized growth plate. This phenotype is accompanied by local and systemic increases in the phosphatonin FGF23 (22), resulting in hypophosphatemia due to increased renal Pi wasting. Increased renal Pi clearance is mediated by FGF receptors and the co-receptor protein, KLOTHO, to activate downstream signaling via the MAPK/ERK pathway, which in turn results in reduced expression of the Na+/Pi co-transporter, NPT2a.
The HMW FGF2 isoforms are ordinarily not released from the cells but function in an intracrine manner (16). Consistent with this, we observed increased nuclear labeling but no increased FGF2 either in osteoblast-conditioned medium or sera from HMW mice. Thus, the HMW FGF2 isoforms may mediate their effects on bone and kidney by modulating the expression of other FGF family members, such as FGF23, which is secreted to act locally or systemically in a paracrine or endocrine manner. In support of this hypothesis, we observed increased FGF23 expression in osteoblasts as well as increased serum FGF23 in TgHMW mice.
The skeletal phenotype of the TgHMW mice is similar to that of the FGF23 transgenic mouse (10), the Hyp mouse (11), and the Dmp1 null mouse (12). In addition, all three models have high levels of circulating FGF23 as well as increased osteoblast/osteocyte FGF23 expression. There are other similarities and differences between TgHMW and these mouse models. For example, similar to FGF23 Tg mice (35), long bones of TgHMW have increased mRNA for alkaline phosphatase, an early marker of osteoblast differentiation that has been shown to be elevated in human rickets and osteomalacia (36). In contrast to the FGF23Tg mice, there were significant reductions in α1(I) collagen and increased expression of osteopontin mRNA in long bones of TgHMW compared with TgVector mice. Both TgHMW and FGF23Tg mice had elevated serum concentrations of CTX, a marker of increased bone degradation. However, in contrast to the FGF23Tg mice, histomorphometry demonstrated increased osteoclast number and activity in the TgHMW mice. Interestingly, we did not observe any significant differences in mRNA for receptor activator of NF-κB ligand (RANKL) or osteoprotegerin (OPG) or the RANKL/OPG ratio that is important in osteoclastogenesis (supplemental Fig. 2). These results are in contrast to the reported significant increase in RANKL/OPG ratio reported in bones of FGF23Tg mice (35).
Because hypophosphatemia is a major defect in these rachitic models, we assessed the effect of a high phosphate diet in rescuing the TgHMW phenotype. There was reversal of the dwarfism and partial rescue of the osteomalacia similar to that obtained in high phosphate rescue diets in the Hyp (37) and Dmp1 null mice (9) as well as in humans with vitamin D-resistant rickets (38). The failure to fully correct the osteomalacia in these models suggests a direct effect for FGF23 on mineralization. Similarly, lack of Dmp1 resulted in defective mineralization, and a high phosphate diet did not fully rescue the osteomalacia, suggesting that the lack of functional DMP1 in osteocytes was responsible for osteomalacia (9). However, we did not observe a decrease in Dmp1 mRNA in flushed bones of HMW mice. As a matter of fact, Dmp1 mRNA was significantly increased in flushed bones from TgHMW mice (supplemental Fig. 3).
It was originally proposed that PHEX and FGF23, which are both produced by bone, are part of a bone-kidney axis regulating phosphate homeostasis and mineralization (39). This hypothesis was supported by the finding of increased FGF23 in the setting of inactivating PHEX mutations in osteocytes (40–42). However, Phex deficiency was not the cause of the TgHMW phenotype because its mRNA was significantly increased in the bones of these mice. This observation is similar to that reported for Dmp1 null mice in which Phex mRNA was also increased (43).
Several studies have shown that FGF23 modulates phosphate homeostasis via FGFRs and KLOTHO in kidney to affect downstream signaling pathways, such as the MAPK/ERK pathway, resulting in down-regulation of the Na-Pi co-transporter Npt2a (44–46). Our studies support FGF23 signaling via FGFR1c and/or FGFR3c as well as KLOTHO and ERK to reduce expression of Npt2a in kidneys from TgHMW mice. The importance of KLOTHO in phosphate homeostasis was first shown in Klotho null mice, a model of senescence with hyperphosphatemia (47). KLOTHO was shown to function as a co-receptor for FGF23 with several subtypes of FGFRs (44, 48, 46). Controversy also exists as to the specific FGF receptor that mediates the action of FGF23 in kidneys. There are four FGFR genes and several splice variants, resulting in multiple FGFR subtypes (46, 49). Interestingly, a recent immunohistochemical study demonstrated increased FGFR3 but not FGFR1, FGFR2, or FGFR4 in the proximal tubule of Hyp mice (50). However, crossing the Hyp mouse with an FGFR3 null mouse or FGFR4 mouse did not correct any of the metabolic abnormalities of the Hyp mouse (50).
The Fgf2 null mouse allowed us to examine whether endogenous FGF2 played a role in mediating the effects of HMW FGF2 isoforms to regulate FGF23 and PHEX. The specificity of the regulation of FGF23 by the HMW FGF2 is clearly demonstrated in FGF2 null osteoblasts transfected with an empty vector or the cDNA that encodes only for HMW FGF2, whereas Fgf2 null cells transfected with the cDNA encoding LMW 18-kDa FGF2 had no increase in Fgf23 mRNA (supplemental Fig. 4).
The availability of the Hyp mouse a mouse homolog of X-linked hypophosphatemic rickets/osteomalacia allowed us to determine the potential relevance of HMW FGF2 in phosphate wasting disorders. The observation that FGF2 mRNA and protein was increased in bones/osteoblasts and that FGF23 and HMW FGF2 protein colocalized in osteocytes from the Hyp mouse confirms the importance of this new mouse model for the study of phosphate homeostasis.
In summary, this study describes a novel Pi-regulating system in mice, which involves a cascade of signals involving HMW FGF2, FGF23, and other phosphate regulators in bone, which is then able to modulate renal phosphate transport. Specifically, FGF2 HMW isoforms differentially regulate bone phosphate metabolism and renal phosphate transport through an FGF23-dependent pathway. These new observations in the TgHMW and Hyp mice indicate a potentially important pathogenic role of FGF2 isoforms in human phosphaturic disorders and merit further exploration.
Supplementary Material
Acknowledgments
We thank the University of Connecticut Health Center microcomputed tomography facility for performing the micro-CT studies. The retroviral construct was a gift from D. Dorsky and R. Kosher (University of Connecticut Health Center). We thank B. Koeppen for reviewing the kidney histology and L. G. Raisz (University of Connecticut Health Center) for helpful critique of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AG021189 (to M. M. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- FGF
- fibroblast growth factor
- FGFR
- FGF receptor
- HMW
- high molecular weight
- LMW
- low molecular weight
- IRES
- internal ribosome entry site
- GFP
- green fluorescent protein
- BMD
- bone mineral density
- BMC
- bone mineral content
- micro-CT
- microcomputed tomography
- BV
- bone volume
- TV
- total volume
- BS
- bone surface
- PBS
- phosphate-buffered saline
- CTX
- C-terminal telopeptide of type 1 collagen
- RT
- reverse transcription
- OB
- osteoblast
- TRITC
- tetramethylrhodamine isothiocyanate
- BMSC
- bone marrow stromal cell
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- eGFP
- enhanced green fluorescent protein
- CMV
- cytomegalovirus
- L3
- third lumbar
- PTH
- parathyroid hormone
- Pp44/42
- phospho-p44/42
- RANKL
- receptor activator of NF-κB ligand
- OPG
- osteoprotegerin
- Tg
- transgenic
- hFGF2
- human FGF2
- KO
- knock-out
- WT
- wild type.
REFERENCES
- 1.Hurley M. M., Marie P. J., Florkiewicz R. Z. (2002) in Principles of Bone Biology (Bilezikian J. P., Raisz L. G., Rodan G. A. eds) pp. 825–851, Academic Press, Inc., San Diego, CA [Google Scholar]
- 2.Muenke M., Schell U. (1995) Trends. Genet. 11, 308–313 [DOI] [PubMed] [Google Scholar]
- 3.De Moerlooze L., Dickson C. (1997) Curr. Opin. Genet. Dev. 7, 378–385 [DOI] [PubMed] [Google Scholar]
- 4.Coffin J. D., Florkiewicz R. Z., Neumann J., Mort-Hopkins T., Dorn G. W., 2nd, Lightfoot P., German R., Howles P. N., Kier A., O'Toole B. A., Sasse J., Gonzalez A. M., Baird A., Doetschman T. (1995) Mol. Biol. Cell. 6, 1861–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X., White K. E. (2005) Cytokine Growth Factor Rev. 16, 221–232 [DOI] [PubMed] [Google Scholar]
- 6.Autosomal Dominant Hypophosphatemic Rickets Consortium (2000) Nat. Genet. 26, 345–348 [DOI] [PubMed] [Google Scholar]
- 7.Shimada T., Mizutani S., Muto T., Yoneya T., Hino R., Takeda S., Takeuchi Y., Fujita T., Fukumoto S., Yamashita T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6500–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hypophosphatemic Rickets Consortium (1995) Nat. Genet. 11, 130–136 [DOI] [PubMed] [Google Scholar]
- 9.Feng J. Q., Ward L. M., Liu S., Lu Y., Xie Y., Yuan B., Yu X., Rauch F., Davis S. I., Zhang S., Rios H., Drezner M. K., Quarles L. D., Bonewald L. F., White K. E. (2006) Nat. Genet. 38, 1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsson T., Marsell R., Schipani E., Ohlsson C., Ljunggren O., Tenenhouse H. S., Jüppner H., Jonsson K. B. (2004) Endocrinology. 145, 3087–3094 [DOI] [PubMed] [Google Scholar]
- 11.Beck L., Soumounou Y., Martel J., Krishnamurthy G., Gauthier C., Goodyer C. G., Tenenhouse H. S. (1997) J. Clin. Invest. 99, 1200–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorenz-Depiereux B., Bastepe M., Benet-Pagès A., Amyere M., Wagenstaller J., Müller-Barth U., Badenhoop K., Kaiser S. M., Rittmaster R. S., Shlossberg A. H., Olivares J. L., Loris C., Ramos F. J., Glorieux F., Vikkula M., Jüppner H., Strom T. M. (2006) Nat. Genet. 38, 1248–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Globus R. K., Plouet J., Gospodarowicz D. (1989) Endocrinology 124, 1539–1547 [DOI] [PubMed] [Google Scholar]
- 14.Wang J. S. (1996) Acta Orthop. Scand. Suppl. Rev. 269, 1–33 [DOI] [PubMed] [Google Scholar]
- 15.Florkiewicz R. Z., Sommer A. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 3978–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arese M., Chen Y., Florkiewicz R. Z., Gualandris A., Shen B., Rifkin D. B. (1999) Mol. Biol. Cell. 10, 1429–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delrieu I. (2000) FEBS Lett. 468, 6–10 [DOI] [PubMed] [Google Scholar]
- 18.Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A. C., Vagner S. (2003) Biol. Cell 95, 169–178 [DOI] [PubMed] [Google Scholar]
- 19.Ma X., Dang X., Claus P., Hirst C., Fandrich R. R., Jin Y., Grothe C., Kirshenbaum L. A., Cattini P. A., Kardami E. (2007) J. Cell. Physiol. 213, 690–698 [DOI] [PubMed] [Google Scholar]
- 20.Yu P. J., Ferrari G., Galloway A. C., Mignatti P., Pintucci G. (2007) J. Cell. Biochem. 100, 1100–1108 [DOI] [PubMed] [Google Scholar]
- 21.Liao S., Porter D., Scott A., Newman G., Doetschman T., Schultz Jel J. (2007) J. Mol. Cell. Cardiol. 42, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Z. S., Jeyaraman M., Wen G. B., Fandrich R. R., Dixon I. M., Cattini P. A., Kardami E. (2007) J. Mol. Cell. Cardiol. 42, 222–233 [DOI] [PubMed] [Google Scholar]
- 23.Sobue T., Naganawa T., Xiao L., Okada Y., Tanaka Y., Ito M., Okimoto N., Nakamura T., Coffin J. D., Hurley M. M. (2005) J. Cell. Biochem. 95, 83–94 [DOI] [PubMed] [Google Scholar]
- 24.Montero A., Okada Y., Tomita M., Ito M., Tsurukami H., Nakamura T., Doetschman T., Coffin J. D., Hurley M. M. (2000) J. Clin. Invest. 105, 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao L., Liu P., Li X., Doetschman T., Coffin J. D., Drissi H., Hurley M. M. (2009) J. Biol. Chem. 284, 3170–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Florkiewicz R. Z., Shibata F., Barankiewicz T., Baird A., Gonzalez A. M., Florkiewicz E., Shah N. (1991) Ann. N.Y. Acad. Sci. 638, 109–126 [DOI] [PubMed] [Google Scholar]
- 27.Dacic S., Kalajzic I., Visnjic D., Lichtler A. C., Rowe D. W. (2001) J. Bone Miner. Res. 16, 1228–1236 [DOI] [PubMed] [Google Scholar]
- 28.Parfitt A. M., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., Recker R. R. (1987) J. Bone Miner. Res. 2, 595–610 [DOI] [PubMed] [Google Scholar]
- 29.Lizarraga G., Lichtler A., Upholt W. B., Kosher R. A. (2002) Dev. Biol. 243, 44–54 [DOI] [PubMed] [Google Scholar]
- 30.Terashima Y., Shimabukuro Y., Terashima H., Ozasa M., Terakura M., Ikezawa K., Hashikawa T., Takedachi M., Oohara H., Yamada S., Murakami S. (2008) J. Cell. Physiol. 216, 640–650 [DOI] [PubMed] [Google Scholar]
- 31.Suttamanatwong S., Franceschi R. T., Carlson A. E., Gopalakrishnan R. (2007) J. Cell. Biochem. 102, 496–505 [DOI] [PubMed] [Google Scholar]
- 32.Wamsley H. L., Iwaniec U. T., Wronski T. J. (2005) Toxicol. Pathol. 33, 577–583 [DOI] [PubMed] [Google Scholar]
- 33.Korpela J., Tiitinen S. L., Hiekkanen H., Halleen J. M., Selander K. S., Väänänen H. K., Suominen P., Helenius H., Salminen E. (2006) Anticancer. Res. 26, 3127–3132 [PubMed] [Google Scholar]
- 34.Razzaque M. S., Lanske B. (2007) J. Endocrinol. Rev. 194, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollberg K., Marsell R., Norgård M., Larsson T., Jonsson K. B., Andersson G. (2008) Bone 42, 1111–1121 [DOI] [PubMed] [Google Scholar]
- 36.Holick M. F. (2006) J. Clin. Invest. 116, 2062–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eicher E. M., Southard J. L., Scriver C. R., Glorieux F. H. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 4667–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marie P. J., Glorieux F. H. (1983) Calcif. Tissue. Int. 35, 443–448 [DOI] [PubMed] [Google Scholar]
- 39.Quarles L. D. (2003) J. Clin. Invest. 112, 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S., Guo R., Simpson L. G., Xiao Z. S., Burnham C. E., Quarles L. D. (2003) J. Biol. Chem. 278, 37419–374126 [DOI] [PubMed] [Google Scholar]
- 41.Weber T. J., Liu S., Indridason O. S., Quarles L. D. (2003) J. Bone. Miner. Res. 18, 1227–1234 [DOI] [PubMed] [Google Scholar]
- 42.Liu S., Tang W., Zhou J., Vierthaler L., Quarles L. D. (2007) Am. J. Physiol. Endocrinol. Metab. 293, E1636–E1644 [DOI] [PubMed] [Google Scholar]
- 43.Liu S., Zhou J., Tang W., Menard R., Feng J. Q., Quarles L. D. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E254–E261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urakawa I., Yamazaki Y., Shimada T., Iijima K., Hasegawa H., Okawa K., Fujita T., Fukumoto S., Yamashita T. (2006) Nature 444, 770–774 [DOI] [PubMed] [Google Scholar]
- 45.Yamashita T., Konishi M., Miyake A., Inui K., Itoh N. (2002) J. Biol. Chem. 277, 28265–28270 [DOI] [PubMed] [Google Scholar]
- 46.Fukumoto S. (2008) Intern. Med. 47, 337–343 [DOI] [PubMed] [Google Scholar]
- 47.Kuro-o M., Matsumura Y., Aizawa H., Kawaguchi H., Suga T., Utsugi T., Ohyama Y., Kurabayashi M., Kaname T., Kume E., Iwasaki H., Iida A., Shiraki-Iida T., Nishikawa S., Nagai R., Nabeshima Y. I. (1997) Nature 390, 45–51 [DOI] [PubMed] [Google Scholar]
- 48.Kurosu H., Ogawa Y., Miyoshi M., Yamamoto M., Nandi A., Rosenblatt K. P., Baum M. G., Schiavi S., Hu M. C., Moe O. W., Kuro-o M. (2006) J. Biol. Chem. 281, 6120–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Itoh N., Ornitz D. M. (2004) Trends Genet. 20, 563–569 [DOI] [PubMed] [Google Scholar]
- 50.Liu S., Vierthaler L., Tang W., Zhou J., Quarles L. D. (2008) J. Am. Soc. Nephrol. 19, 2342–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.