Abstract
Kainate receptors are widely expressed in the brain, and are present at pre- and postsynaptic sites where they play a prominent role in synaptic plasticity and the regulation of network activity. Within individual neurons, kainate receptors of different subunit compositions are targeted to various locations where they serve distinct functional roles. Despite this complex targeting, relatively little is known about the molecular mechanisms regulating kainate receptor subunit trafficking. Here we investigate the role of phosphorylation in the trafficking of the GluR6 kainate receptor subunit. We identify two specific residues on the GluR6 C terminus, Ser846 and Ser868, which are phosphorylated by protein kinase C (PKC) and dramatically regulate GluR6 surface expression. By using GluR6 containing phosphomimetic and nonphosphorylatable mutations for these sites expressed in heterologous cells or in neurons lacking endogenous GluR6, we show that phosphorylation of Ser846 or Ser868 regulates receptor trafficking through the biosynthetic pathway. Additionally, Ser846 phosphorylation dynamically regulates endocytosis of GluR6 at the plasma membrane. Our findings thus demonstrate that phosphorylation of PKC sites on GluR6 regulates surface expression of GluR6 at distinct intracellular trafficking pathways, providing potential molecular mechanisms for the PKC-dependent regulation of synaptic kainate receptor function observed during various forms of synaptic plasticity.
Keywords: Cell/Endocytosis, Cell/Trafficking, Phosphorylation, Receptors, Receptors/Endocytosis, Kainate Receptor, Surface Expression
Introduction
Kainate receptors are ionotropic glutamate receptors, which are expressed at both pre- and post-synaptic sites throughout the central nervous system. Postsynaptic kainate receptors mediate a slow EPSC (1–3), whereas presynaptic kainate receptors regulate neurotransmitter release (4–6). Due to their synaptic localization and regulation of both glutamatergic and GABAergic transmission, kainate receptors play a major role in defining network activity. For example, kainate receptors strongly influence the CA3 and CA1 networks in the hippocampus and are critical for development of hippocampal spontaneous rhythmic activity patterns (7, 8). Furthermore, pathologies such as epilepsy involve dysfunction of kainate receptors (9, 10). In addition, kainate receptors play a prominent role in long-term synaptic plasticity in the hippocampus (11–14) and neocortex (15, 16). Thus kainate receptors play vital roles in synaptic plasticity and network function.
Kainate receptors are tetramers made up of GluR5–7 and KA1–2 subunits. GluR5–7 can form functional homomers; in contrast, KA1–2 do not form functional ion channels unless assembled as heteromers with GluR5–7 (17–19). Kainate receptor subunits are widely expressed in the mammalian brain and their expression is developmentally and regionally regulated (20, 21). In addition, presynaptic and postsynaptic kainate receptors typically differ in their subunit composition. For example, in hippocampal CA3 pyramidal neurons, heteromeric GluR6/KA2 kainate receptors are targeted to dendrites (6, 10), whereas GluR5-containing kainate receptors are selectively targeted to axon terminals (4, 22, 23). Therefore, mechanisms exist for regulated and subunit-specific trafficking and targeting of kainate receptors. A prominent candidate for such a mechanism is receptor phosphorylation, which regulates trafficking of NMDARs (24–26) and AMPARs (27, 28). However, relatively little is known about the phosphorylation of kainate receptor subunits and the role it plays in kainate receptor trafficking in neurons.
Here we directly address the role of phosphorylation of GluR6, one of the most prominently expressed kainate receptor subunits in the brain. We show that the GluR6 C terminus is phosphorylated by PKC2 at two different sites, Ser846 and Ser868. Phosphorylation of these sites regulates GluR6 trafficking by at least two distinct mechanisms. First, phosphorylation of either Ser846 or Ser868 strongly reduces the ER exit of GluR6, thus decreasing surface expression of GluR6. Second, the phosphorylation of Ser846, but not Ser868, mediates a dynamic endocytosis of surface-expressed GluR6. Thus the direct phosphorylation of GluR6 on two residues plays distinct roles in regulating GluR6 trafficking and surface expression in neurons.
EXPERIMENTAL PROCEDURES
Cells
HeLa cells (ATCC) were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 1% l-glutamine. Hippocampal or cortical neurons were isolated from P0-1 mouse pups or E18 mouse embryos, respectively, and cultured according to the manufacturer's protocol (Invitrogen). Cultures were maintained in neurobasal medium (Invitrogen) supplemented with glutamine and B-27 supplement (Invitrogen) and analyzed at 14 days in vitro.
Antibodies and DNA Constructs
Anti-FLAG M2 antibody was purchased from Sigma. Antibody against GluR6/7 was purchased from Millipore. Green fluorescent protein (GFP)-Rab5 and Rab9 were obtained as a gift from Dr. Juan Bonifacino (NICHD, National Institutes of Health) and GFP-Rab11 was obtained as a gift from Dr. James Goldenring (Vanderbilt University, Nashville, TN). GluR6 in the pRK5 mammalian expression vector was obtained as a gift from Dr. P. Seeburg (Max Planck, Heidelberg, Germany). The FLAG epitope was inserted after the signal sequence of GluR6 by site-directed mutagenesis, and as with all other mutations in GluR6, it was engineered according to the QuikChange protocol (Stratagene). The sequences of all mutants were confirmed by automated sequence analysis. For fluorescence-activated cell sorting (FACS) analysis, an IRES-EGFP sequence was introduced into each mutant construct.
Immunostaining for Co-localization, Surface Expression, and Internalization in HeLa Cells and Neurons
Cells grown on glass coverslips were transfected with the cDNAs indicated as previously described (29) and analyzed 24 h later. Transfected live cells were washed once in PBS and labeled with anti-FLAG antibody for 30 min on ice for HeLa cells and at room temperature for neurons. For internalization assays, after being washed with PBS, cells were incubated at 37 °C for the indicated amount of time in medium containing leupeptin to allow the internalization of labeled GluR6. Cells were then fixed in 4% paraformaldehyde in PBS for 15 min at room temperature. After being washed with PBS, the cells were labeled with fluorescence-conjugated secondary antibody (Alexa 488-conjugated secondary antibody for surface expression analysis in HeLa cells or Alexa 568-conjugated secondary antibody for surface expression analysis in neurons and internalization assays; Molecular Probes) for 45 min at room temperature for visualizing surface-expressed GluR6 and then with anti-mouse IgG (1:50; Sigma) for 1 h at room temperature for staining the remaining unlabeled GluR6 on the cell surface. After being permeabilized in 0.2% Triton X-100 in PBS for 5 min, cells were incubated with fluorescence-conjugated secondary antibody (Alexa 488-conjugated secondary antibody for internalization assay or Alexa 568-conjugated secondary antibody for co-localization assay; Molecular Probes) for visualizing internalized GluR6. For the surface expression assay, the cells were again labeled with anti-FLAG antibody for 45 min at room temperature. After being washed with PBS, the cells were then labeled with fluorescence-conjugated secondary antibody (Alexa 568-conjugated secondary antibody; Molecular Probes) for visualizing intracellular GluR6. The coverslips were washed and then mounted with ProLong Antifade Kit (Molecular Probes). A series of optical sections were collected using a Zeiss LSM510 confocal microscope (Carl Zeiss MicroImaging). Figures show maximum projections. The fluorescence intensity of the surface expressed GluR6 was normalized with the fluorescence intensity of total GluR6 in each cell. The fluorescence intensity of internalized GluR6 was normalized with the fluorescence intensity of the surface expressing plus internalized GluR6 in each cell.
Glycosidase Treatment and Immunoblotting
HeLa cells were transfected with the cDNAs indicated. After 24 h (±indicated treatment), cells were lysed in lysis buffer (TBS: 2 mm EDTA, 0.1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 μg/ml of leupeptin, and 5 mm iodoacetamide). The lysates were incubated for 1 h at 4 °C and centrifuged at 20,000 × g for 20 min. Fifty μg of supernatants of each sample were treated in three ways: (i) untreated; (ii) endoglycosidase Hf (endo Hf) (1,500 units); or (iii) PNGase F (750 units). All samples were incubated at 37 °C for at least 4 h and then resolved by SDS-PAGE, transferred onto polyvinylidene difluoride membranes, and probed with anti-GluR6/7 (1:5,000) antibody. The blots were incubated with anti-rabbit horseradish peroxidase secondary antibodies (1:5,000; Amersham Biosciences), followed by detection with ECL reagents (Pierce).
FACS Analysis of Cell Surface Protein
HeLa cells grown in 6-cm dishes were transiently transfected with FLAG-GluR6-IRES-EGFP (WT, S846A, S846D, S868A, and S868D) by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfected live HeLa cells were rinsed twice with ice-cold PBS containing 5 mm EDTA (PBS/EDTA) and then incubated with ice-cold PBS/EDTA for 10 min on ice. Detached 1 × 106 cells were transferred to polystyrene test tubes (VWR Scientific) and labeled with anti-FLAG antibody for 30 min on ice. Cells were then washed 3 times with ice-cold PBS and incubated with phycoerythrin-conjugated anti-mouse IgG1 secondary antibody (Caltag) for 30 min on ice. Cells were washed 3 times and suspended in ice-cold PBS. The cells were immediately analyzed using a dual-laser FACSVantage SE flow cytometer (BD Biosciences). Cell Quest Acquisition and Analysis software (BD Biosciences) was used to acquire and quantify the fluorescence signal intensities and graph the data as bivariate dot density plots. Fluorescence emissions of individual fluorophores were initially corrected for spectral overlap using “simple” single color controls and appropriate electronic compensation. The background fluorescence was determined using HeLa cells transfected with empty vector and stained as described above. In parallel, the total expression level of GluR6 was analyzed by FACS. Briefly, harvested cells as described above were first fixed with 4% paraformaldehyde in PBS for 15 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min. The fixed cells were then incubated with anti-FLAG antibody for 30 min at room temperature. After being washed with PBS, cells were incubated with phycoerythrin-conjugated anti-mouse IgG1 secondary antibody for 30 min at room temperature and then the total expression level of GluR6 was analyzed by FACS as described above.
Fusion Protein Production and in Vitro Phosphorylation
GST fusion proteins for GluR6 wild type (WT), S846A, and S868A were purified as previously described (30). The purified fusion proteins were phosphorylated in 20 mm HEPES, pH 7.4, 1.67 mm CaCl2, 1 mm dithiothreitol, 10 mm MgCl2, 200 mm cold ATP, 1 pmol of [γ-32P]ATP (3000 Ci/mmol) with 25 ng of purified PKC (Promega) at 30 °C for 30 min. The reactions were stopped by adding SDS-PAGE sample buffer, and the samples were boiled for 5 min. The phosphorylated proteins were resolved by SDS-PAGE and transferred to nitrocellulose membrane (Schleicher & Schuell). The bands were visualized by autoradiography and excised for in vitro phosphopeptide mapping.
Two-dimensional Phosphopeptide Mapping
Peptide mapping was performed as previously described (30). Briefly, phosphorylated proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The relevant bands were excised and soaked for 1 h in tubes containing 1 ml of 1% polyvinylpyrrolidone-40 in 100 mm acetic acid. After washing with 0.4% NH4HCO3, the proteins on the membrane were digested with trypsin overnight at 37 °C. Supernatants containing the tryptic digestion products were dried in a SpeedVac, washed twice with 900 μl of H2O with repeated drying, and resuspended in 5 μl of H2O. One or two μl of the dissolved phosphopeptides were spotted onto a cellulose thin layer chromatography plate (Merck). The phosphopeptides were resolved in the first dimension by electrophoresis in buffer containing 2.5% formic acid and 7.8% acetic acid. Separation by ascending chromatography in the second dimension was performed using buffer containing 62.5% isobutyric acid, 4.8% pyridine, 1.9% butanol, and 2.9% acetic acid. The thin layer chromatography plate was air-dried, and the peptide map was visualized by PhosphorImager analysis.
In Vitro Phosphorylation of Immunoprecipitated GluR6
Cortical neurons (14 days in vitro) were treated with the PKC activator, 13-O-acetylphorbol 12-myristate (PMA), or vehicle (dimethyl sulfoxide) for 10 min. Cell lysates were prepared as described above. GluR6 was immunoprecipitated with anti-GluR6 antibody using Protein G-Sepharose beads. The beads were washed with lysis buffer 3 times, and with PKC phosphorylation buffer twice. The immunoprecipitates were in vitro phosphorylated as described above and visualized by PhosphorImager analysis.
RESULTS
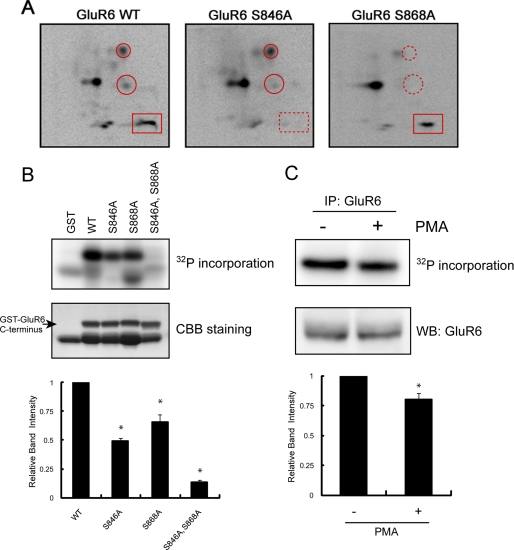
Because PKC activity regulates GluR6-containing kainate receptors (31), we began to characterize the direct phosphorylation of GluR6 by PKC. Like all ionotropic glutamate receptors, GluR6 has an intracellular C-terminal domain, which is accessible to protein kinases. In addition, the GluR6 C terminus can be phosphorylated by PKC in vitro (32). To define specific phosphorylation sites in this region, we generated a GST fusion protein containing the GluR6 intracellular C-terminal domain (GST-GluR6) and performed in vitro phosphorylation by incubating fusion protein with [γ-32P]ATP and purified PKC. We subjected the phosphorylated fusion proteins to SDS-PAGE, digested the proteins with trypsin, and resolved the resulting peptides in two dimensions. We visualized the resulting phosphopeptide map (Fig. 1A), which revealed that WT GluR6 contained multiple phosphopeptides or spots (Fig. 1A, left panel), consistent with the GluR6 C terminus having several PKC phosphorylation sites. To define the precise residues, we generated point mutations of several candidate residues, which are part of PKC consensus motifs based on the NetPhos program, including Ser846 and Ser868. Phosphopeptide maps of GST-GluR6 S846A and GST-GluR6 S868A each showed the disappearance of one or two phosphopeptides compared with maps of GST-GluR6 WT (Fig. 1A), indicating that these two residues are directly phosphorylated by PKC. In addition, we performed mass spectrometry of PKC-phosphorylated GST-GluR6, which identified Ser868 as a PKC phosphorylation site (supplemental Fig. S1). All of the other spots described above in Fig. 1A are thought to be phosphorylated peptides of the GST-GluR6 fusion protein.
FIGURE 1.
Identification of Ser846 and Ser868 as PKC phosphorylation sites in the GluR6 C terminus. A, phosphopeptide map analysis shows that the GluR6 C terminus is phosphorylated by PKC on multiple peptides (left panel) including Ser846 and Ser868, as indicated by the disappearance of phosphopeptides on maps of the GluR6 mutants (middle and right panels). GST-GluR6 (WT, S846A, and S868A) fusion proteins were phosphorylated with purified PKC in vitro using [γ-32P]ATP and then subjected to two-dimensional phosphopeptide mapping as described under “Experimental Procedures.” A rectangle surrounds the phosphopeptide that includes phosphorylated Ser846. Circles surround the phosphopeptides that include phosphorylated Ser868. An X in each panel indicates the origin where the peptides were initially spotted. Representative phosphopeptide maps are shown of 3–4 independent experiments. B, Ser846 and Ser868 are the major PKC phosphorylation sites in the GluR6 C terminus. GST, GST-GluR6 WT, GST-GluR6 S846A, GST-GluR6 S868A, and GST-GluR6 S846A,S868A were phosphorylated by PKC in vitro and analyzed by PhosphorImager. The loaded amount of GST fusion protein was visualized by protein staining. The lower band in the WT lane is a degradation product of GST-GluR6. Data represent mean ± S.E.M. (error bars) of normalized relative band intensity. *, p < 0.05, Student's t test. CCB, Coomassie Brilliant Blue. C, pretreatment of neurons with PMA reduced PKC phosphorylation of GluR6 in vitro. GluR6 was immunoprecipitated from PMA or vehicle-treated neurons, phosphorylated by PKC in vitro, and analyzed by PhosphorImager. The loaded amount of GluR6 was visualized by Western blotting using anti-GluR6 antibody. Data represent mean ± S.E.M. of normalized relative band intensity. *, p < 0.05, Student's t test.
We next examined the phosphorylation efficiency of each identified site. As shown in Fig. 1B, phosphorylation of GluR6 S846A and GluR6 S868A was less efficient than GluR6 WT. Furthermore, GluR6 S846A,S868A showed a profound reduction (87%) in PKC phosphorylation. These results indicate that Ser846 and Ser868 are the major phosphorylation sites within the GluR6 C terminus. To study the phosphorylation of endogenous GluR6, we performed back phosphorylation of GluR6 in neurons. Specifically, we treated cultured cortical neurons with PMA (or vehicle) for 10 min to stimulate PKC phosphorylation of endogenous GluR6. Receptors were immunoprecipitated and phosphorylated in vitro with purified PKC. Phosphorylation of GluR6 isolated from PMA-treated neurons was less robust than phosphorylation of GluR6 from untreated neurons (Fig. 1C). This result indicates that endogenous GluR6 is phosphorylated by PKC in neurons.
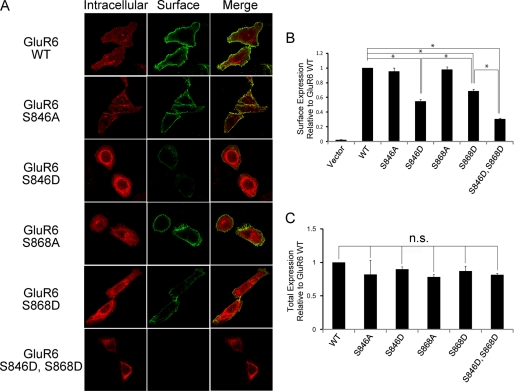
PKC activity is known to regulate GluR6 trafficking (31); therefore, we next examined the role of Ser846 and Ser868 in regulating GluR6 surface expression. Because the GluR6 Ser846 and Ser868 phosphospecific antibodies were not suitable for immunofluorescence microscopy, we used a combination of phosphomimetic (GluR6 S846D or GluR6 S868D) and nonphosphorylatable (GluR6 S846A or GluR6 S868A) mutations to evaluate the importance of these sites in GluR6 trafficking. We generated GluR6 constructs containing an N-terminal FLAG tag to allow specific labeling of the surface-expressed receptor. We incubated live HeLa cells expressing full-length GluR6 (WT, S846A, S846D, S868A, S868D, or S846D,S868D) with FLAG antibodies to label surface-expressed receptor, which were subsequently stained with Alexa 488-conjugated (green) secondary antibodies. Following fixation and permeabilization, we incubated the cells with FLAG antibodies and stained the cells with Alexa 568-conjugated (red) antibodies to detect intracellular GluR6. Using confocal imaging to compare surface and internal pools of GluR6, we observed a dramatic reduction in the surface-expressed receptor when Ser846 and Ser868 were mutated to aspartic acid, with the double mutation being the most profound (Fig. 2A). In contrast, surface expression of GluR6 S846A or GluR6 S868A was indistinguishable from GluR6 WT (Fig. 2A).
FIGURE 2.
GluR6 surface expression in HeLa cells was reduced when either Ser846 or Ser868 was mutated to mimic phosphorylation. A, GluR6 surface expression was evaluated in HeLa cells expressing FLAG-GluR6 (WT, S846A, S846D, S868A, S868D, or S846D,S868D) by immunofluorescence using confocal microscopy as described under “Experimental Procedures.” B, quantitative FACS analysis shows significantly decreased surface expression of GluR6 S846D and GluR6 S868D compared with GluR6 WT. HeLa cells were transfected with FLAG-GluR6 IRES-EGFP (GluR6 WT, S846A, S846D, S868A, S868D, or S846D,S868D). GluR6 surface expression was analyzed by FACS as described under “Experimental Procedures.” GluR6 S846D surface expression was decreased by 45%, GluR6 S868D by 31%, and GluR6 S846D,S868D by 69% compared with GluR6 WT. Data represent mean ± S.E.M. of the fold-increase in surface expression compared with GluR6 based on the fluorescence intensity of surface-expressed GluR6 in transfected cells. *, p < 0.01, Student's t test. C, quantitative FACS analysis shows that phosphomimetic mutations of GluR6 do not affect GluR6 total expression level. Data represent mean ± S.E.M. of the fold-increase in total expression compared with GluR6 WT based on the fluorescence intensity of total GluR6 in transfected cells.
We next quantified the surface expression of GluR6 WT compared with GluR6 phosphorylation mutants using FACS analysis. We used a bicistronic expression construct containing the coding sequence of FLAG-tagged GluR6 and EGFP (FLAG-GluR6-IRES-EGFP) to monitor GluR6-transfected cells (GFP positive cells) and then performed FACS analysis to measure surface expression of FLAG-GluR6 in live HeLa cells expressing GFP. We found that surface expression of GluR6 S846D was strongly decreased compared with GluR6 WT (Fig. 2B). We also observed a profound decrease in GluR6 S868D surface expression compared with GluR6 WT. In addition, the simultaneous mutation of both of these serines to aspartic acid (GluR6 S846D,S868D) resulted in the lowest surface expression. However, there was no change in overall receptor expression for GluR6 S846D, GluR6 S868D, or GluR6 S846D,S868D compared with GluR6 WT (Fig. 2C). For both of these PKC sites, the robust reduction in surface expression was consistently observed only for the phosphomimetic mutations (GluR6 S846D or GluR6 S868D). In contrast, the non-phosphorylatable mutations (GluR6 S846A or GluR6 S868A) resulted in little or no change in GluR6 surface expression.
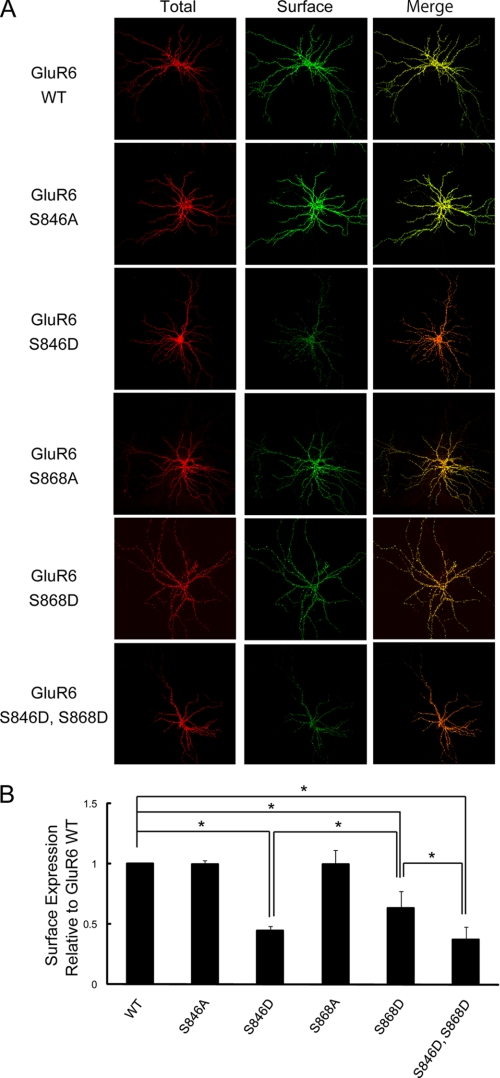
We also examined GluR6 surface expression in neurons. To specifically monitor surface expression of GluR6 WT versus GluR6 containing mutations of the PKC phosphorylation sites, we expressed exogenous GluR6 in neurons prepared from GluR6 knock-out (KO) mice. Thus, we could avoid potential oligomerization of recombinant GluR6 containing mutations of the critical phosphorylation sites with endogenous GluR6 WT. We imaged surface-expressed receptor using immunofluorescence confocal microscopy and observed a significant decrease in GluR6 surface expression when Ser846 and/or Ser868 was mutated to aspartic acid (Fig. 3, A and B). However, surface expression of GluR6 was unchanged when either Ser846 or Ser868 was mutated to alanine to eliminate phosphorylation of those residues. These findings support a specific role for phosphorylation of these two PKC sites in regulating GluR6 surface expression in neurons just as we found for GluR6 expressed in heterologous cells (Fig. 2).
FIGURE 3.
GluR6 surface expression in neurons was reduced when either Ser846 or Ser868 was mutated to mimic phosphorylation. A, cultured hippocampal neurons derived from GluR6 KO mice were transfected with FLAG-GluR6 (WT, S846A, S846D, S868A, S868D, or S846D,S868D). Surface-expressed and total GluR6 were visualized by immunostaining as described under “Experimental Procedures.” B, quantification of surface-expressed GluR6 in GluR6 KO neurons using Metamorph software. The surface expression of GluR6 S846D, GluR6 S868D, and GluR6 S846D,S868D were significantly decreased compared with GluR6 WT (60% reduction for GluR6 S846D and 35% reduction for GluR6 S868D). Data represent mean ± S.E.M. of the fold-increase in surface expression compared with GluR6 WT based on the relative fluorescence intensity of surface-expressed GluR6 to that of total expressed GluR6. *, p < 0.01, Student's t test.
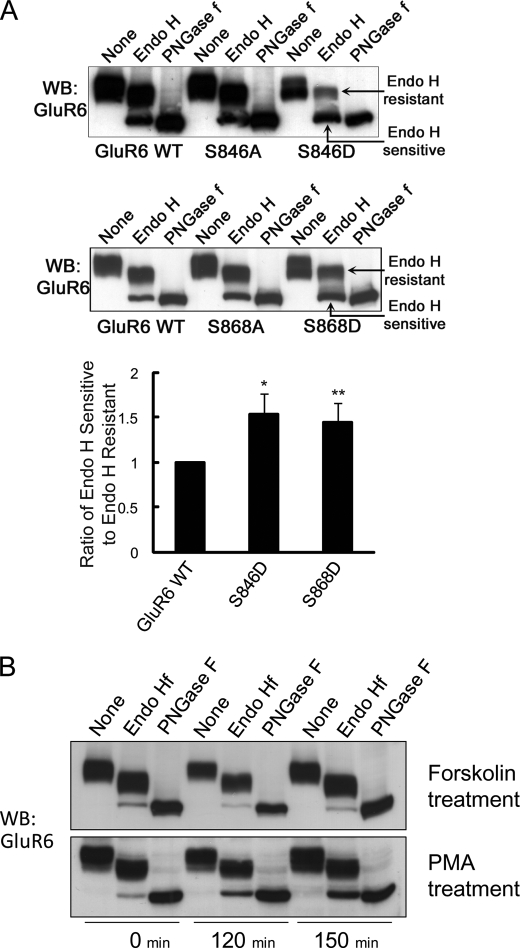
Receptor surface expression is determined by a balance of distinct trafficking events, which include protein transport through the biosynthetic pathway and exo-/endocytosis at the plasma membrane. To examine if PKC phosphorylation plays a role in regulating GluR6 trafficking through the ER/cis-Golgi, we utilized a glycosidase assay to test if GluR6 phosphorylation alters ER egress. Lysates from HeLa cells expressing GluR6 (WT, S846A, S846D, S868A, or S868D) were collected, treated with glycosidases (endo H or PNGase F), and proteins were resolved by SDS-PAGE and immunoblotted with GluR6/7 antibody. The majority of GluR6 WT was endo H resistant, as detected by the predominant higher molecular weight band following endo H treatment (Fig. 4A). However, PMA, but not forskolin, treatment of the cells increased the amount of the endo H-sensitive fraction of GluR6 (Fig. 4B). Furthermore, both GluR6 S846D and GluR6 S868D showed a dramatic increase in endo H sensitivity as detected by a decrease in the ratio of mature receptor (upper, endo H resistant band) to immature receptor (lower, endo H sensitive band), consistent with increased ER retention compared with GluR6 WT (1.53-fold increase for S846D, p < 0.05 (n = 4); 1.44-fold increase for S868D, p < 0.01 (n = 5), Fig. 4A). However, GluR6 containing non-phosphorylatable mutations (GluR6 S846A or GluR6 S868A) showed no change in trafficking through the biosynthetic pathway, with the ratio of immature to mature receptor being the same as with GluR6 WT (Fig. 4A). These results suggest that PKC phosphorylation of GluR6 retains GluR6 in the ER.
FIGURE 4.
GluR6 S846D and GluR6 S868D are more highly retained in the ER than GluR6 WT. HeLa cells were transfected with FLAG-GluR6 (WT, S846A, S846D, S868A, or S868D). Cell lysates were prepared and subjected to a glycosidase assay as described under “Experimental Procedures.” Proteins were immunoblotted with GluR6 antibody. A, GluR6 S846D and GluR6 S868D were more endo H sensitive than GluR6 WT, GluR6 S846A, or GluR6 S868A. Endo H-sensitive fractions were quantitated by measuring the band intensity of the endo H-sensitive fraction compared with the band intensity of endo H-resistant fraction using NIH Image software. Error bars indicate S.E.M. *, p < 0.05, Student's t test. B, PKC activation increases ER retention of GluR6. Twenty-four hours after transfection, cells were treated with PMA (1 μm) and MG132 (20 μm) for the indicated time. GluR6 from PMA-treated cells was more endo H sensitive, as detected by a change in the ratio of immature GluR6 (lower band) to mature GluR6 (upper band) following endo H treatment. WB, Western blot.
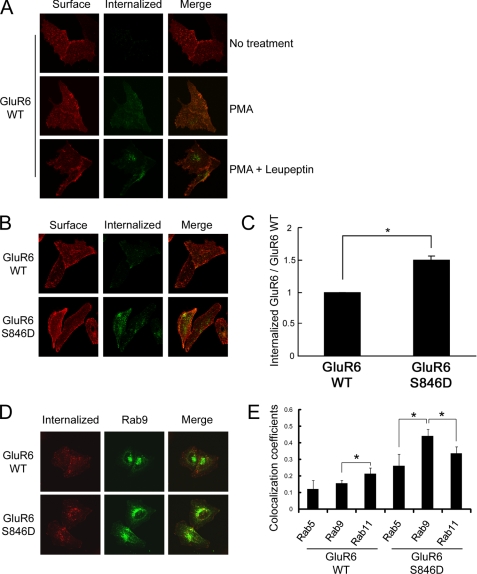
Although phosphomimetic mutations of Ser846 or Ser868 decrease ER egress of GluR6, there is still a substantial pool of surface-expressed receptor. Because PKC phosphorylation is implicated in kainate receptor endocytosis (31), we specifically evaluated GluR6 endocytosis. First, using immunofluorescence microscopy, we found that GluR6 endocytosis is increased following PMA treatment (Fig. 5A). In addition, treatment with leupeptin, a lysosomal inhibitor, increased the accumulation of internalized GluR6 (Fig. 5A). We also treated cells with PMA combined with MG132, a proteasome inhibitor, and evaluated GluR6 internalization and obtained similar results (data not shown). To examine the role of the two PKC phosphorylation sites on GluR6 in receptor endocytosis, we next performed internalization assays of GluR6 WT compared with GluR6 containing phosphomimetic mutations of Ser846 or Ser868. We found that GluR6 S846D underwent robust endocytosis compared with GluR6 WT (Fig. 5, B and C). In contrast, we observed no change in GluR6 S868D endocytosis compared with WT (data not shown). To determine the post-endocytic trafficking pathway of GluR6 S846D, we co-expressed GFP-Rab9 as a marker for late endosomes. We observed colocalization of GluR6 S846D with the late endosomal marker, Rab9, after 60 min of internalization (Fig. 5, D and E). From all of these data, we conclude that a portion of GluR6 traffics to the degradation pathway following Ser846 phosphorylation-induced internalization.
FIGURE 5.
Phosphorylation of Ser846 accelerates GluR6 internalization in HeLa cells. A, PMA treatment increases GluR6 internalization. GluR6 internalization was measured as described under “Experimental Procedures.” After surface labeling with anti-FLAG antibody, cells were incubated at 37 °C to allow labeled GluR6 to internalize for 60 min. Cells were fixed and incubated with fluorescence-conjugated secondary antibody for visualizing surface-expressed GluR6. After permeabilization, the cells were labeled with fluorescence-conjugated secondary antibody for visualizing internalized receptors. PMA treatment with leupeptin increased the accumulation of internalized GluR6. B, constitutive endocytosis of GluR6 S846D was more robust than GluR6 WT without PMA treatment. The brightness was adjusted equally across all panels in this figure to allow a direct comparison between GluR6 WT and GluR6 S846D. C, quantification of internalized GluR6 in HeLa cells was conducted using Metamorph software. GluR6 S846D showed a 1.5-fold increase in the normalized amount of internalized GluR6 compared with GluR6 WT. Error bars indicate S.E.M. *, p < 0.01. D, internalized GluR6 S846D was highly co-localized with a late endosomal marker, Rab9 in HeLa cells. HeLa cells were transfected with FLAG-GluR6 (WT or S846D) together with GFP-Rab9. Internalization assay of GluR6 (WT or S846D) in HeLa cells was performed following leupeptin treatment (60 min), and co-localization of internalized GluR6 with GFP-Rab9 was analyzed by immunocytochemistry. E, colocalization analysis was performed using LSM510 software (Zeiss). Values represent the mean ± S.E.M. of Pearson's correlation. *, p < 0.05.
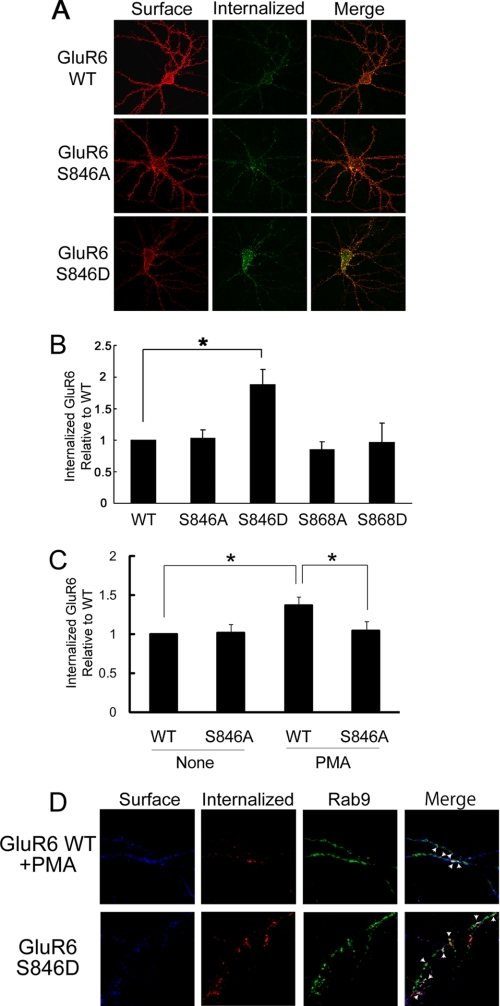
To evaluate the role of Ser846 in regulating endocytosis of surface-expressed GluR6 in neurons, we expressed exogenous GluR6 in cultured neurons derived from GluR6 KO mice. Briefly, we labeled surface-expressed receptors (GluR6 WT, GluR6 S846A, or GluR6 S846D) with FLAG antibodies and then an incubation at 37 °C for 20 min to allow internalization of the labeled GluR6. The cells were labeled with Alexa 568-conjugated secondary antibody (red) for visualizing surface-expressed GluR6. After being permeabilized, cells were incubated with Alexa 488-conjugated secondary antibody (green) for visualizing internalized GluR6. We found that GluR6 S846D, which mimics phosphorylation of GluR6 at this residue, had a dramatic increase in endocytosis compared with GluR6 WT or the unphosphorylatable mutant GluR6 S846A (Fig. 6, A and B). We next examined PMA-induced internalization of GluR6 in neurons and found that PMA treatment stimulated internalization of GluR6 WT, but not the unphosphorylatable mutant, GluR6 S846A (Fig. 6C). These findings were consistent with the phosphorylation of GluR6 on Ser846 facilitating receptor endocytosis in neurons.
FIGURE 6.
Phosphorylation of Ser846 mediates GluR6 internalization in neurons. A, mutation of Ser846 to aspartate accelerates GluR6 internalization. Internalization of GluR6 (WT, S846A, and S846D) was evaluated in GluR6 KO neurons as described in the legend to Fig. 5A. The brightness was adjusted equally across all panels in this figure to allow direct comparison between GluR6 WT and S846D. B, quantification of internalized GluR6 was conducted using Metamorph software. GluR6 S846D showed an almost 2-fold increase in the normalized amount of internalized GluR6 compared with GluR6 WT. Error bars indicate S.E.M. *, p < 0.01. C, mutation of Ser846 to alanine blocks PMA-induced GluR6 internalization. Quantification of internalized GluR6 was conducted using Metamorph software. Error bars indicate S.E.M. *, p < 0.05. D, internalized GluR6 S846D was highly co-localized with the late endosomal marker Rab9 in neurons. Neurons were transfected with FLAG-GluR6 (WT or S846D) together with GFP-Rab9. The co-localization of internalized GluR6 WT with GFP-Rab9 was analyzed with immunofluorescence microscopy. Arrows indicate areas of colocalization.
We also investigated the post-endocytic itinerary of internalized GluR6 in neurons. We found that internalized GluR6 (WT and S846D) colocalized with the late endosomal marker Rab9 in neurons (Fig. 6D), consistent with trafficking to late endosomes and similar to trafficking of internalized GluR6 in heterologous cells.
DISCUSSION
GluR6-containing kainate receptors are expressed throughout the brain and their function and surface expression dynamically regulated by synaptic activity. Although GluR6 endocytosis is regulated by PKC activity, it is not clear if direct phosphorylation of GluR6 mediates receptor trafficking. We now show that PKC phosphorylates the GluR6 C terminus on multiple residues including Ser846 and Ser868. We find that phosphorylation of either Ser846 or Ser868 decreases GluR6 surface expression in heterologous cells and neurons. In contrast, the non-phosphorylatable mutations (GluR6 S846A or GluR6 S868A) resulted in little or no change in GluR6 surface expression. It is possible that GluR6 basal phosphorylation is very low. Normally GluR6 is very efficiently expressed on the cell surface (45), which could explain why under basal conditions, we do not observe a difference between GluR6 WT and unphosphorylatable mutants. By studying the effects of mutations of these sites we further demonstrate that phosphomimetic mutations of either Ser846 or Ser868 reduce ER egress. We also show that phosphorylation of GluR6 on Ser846 plays a specific additional role in promoting GluR6 endocytosis from the cell surface. The ER egress of AMPA, NMDA, and kainate receptor subunits is regulated by a number of mechanisms. For example, ER exit of the AMPA receptor subunit GluR2 is regulated by the RNA-editing state of the receptor (33). Edited GluR2 contains a critical arginine in the pore lining region of the subunit, which restricts Ca2+ permeability. Only edited GluR2 can efficiently traffic out of the ER, resulting in surface expression of Ca2+-impermeable heteromeric AMPARs. The regulation of ER exit of NMDA and kainate receptors is determined by specific ER-retention motifs. For NMDA receptors, an ER-retention motif prevents ER exit of NR1 subunits unless they are co-assembled with NR2 subunits (24, 34). Similarly, the kainate receptor subunit KA2 has ER-retention motifs that prevent ER egress unless this subunit co-assembles with GluR5–7 (19, 29, 35). ER retention of individual subunits ensures that only functional heteromeric NMDA and kainate receptors are allowed to traffic to the plasma membrane. Interestingly, ER retention of the NR1 subunit of NMDA receptors is also regulated by phosphorylation. Specifically, PKC and PKA-dependent phosphorylation of NR1 on critical residues overrides the ER-retention motif (24, 25). We now show that phosphorylation of specific residues on the C terminus of GluR6 increases ER retention of kainate receptors. Our data support a model in which PKC phosphorylation of either Ser846 or Ser868 increases ER retention of GluR6. Thus PKC activation can arrest forward trafficking of GluR6, thereby causing a reduction of surface GluR6 levels. Moreover, this mechanism also enables the selective trafficking of GluR6 subunits dependent on their phosphorylation state.
Glutamate receptor endocytosis is regulated by a number of mechanisms. AMPA, NMDA, and kainate receptors undergo clathrin-dependent endocytosis (35–39). For both AMPA and NMDA receptors, there is evidence that direct phosphorylation of receptor subunits regulates endocytosis (40–42). Although there are several studies implicating phosphorylation as a key event in regulating kainate receptor trafficking, it has been unclear whether direct phosphorylation of receptor subunits regulates endocytosis. Recent studies show that GluR5 and GluR6 kainate receptor subunits can undergo endocytosis that is dependent on PKC (31, 43). In the present study we now identify specific sites on GluR6 as targets of PKC phosphorylation and show that phosphorylation of one particular site, Ser846, specifically increases endocytosis of kainate receptors. Recent work shows that the C-terminal splice variants, GluR6a, GluR6b, couple to distinct synaptic signaling cascades (44) and exhibit differential regulation of their surface expression (45). Of the two sites that we have identified as regulating GluR6 trafficking, Ser846 is present in both splice variants, but Ser868 exists only in GluR6a. This raises the interesting possibility that differences in GluR6a and GluR6b splice isoform trafficking and signal transduction are dependent upon differential regulation by phosphorylation at these two sites.
Synaptic kainate receptors are regulated during development and also acutely during certain forms of long-term synaptic plasticity. In the neonatal barrel cortex both pre- and postsynaptic kainate receptors are expressed at thalamocortical inputs to layer 4 and are developmentally down-regulated during the first postnatal week (15, 46). In addition, postsynaptic kainate receptors at thalamocortical synapses are rapidly down-regulated during the induction of long term potentiation (15) in a mechanism that requires PKC (47). In perirhinal cortex layer 2/3 pyramidal neurons, a form of long term depression induced by kainate receptor activation is expressed as a rapid reduction in kainate receptor-mediated synaptic transmission, and, interestingly, this form of plasticity also requires PKC (16). At CA1 synapses in neonatal hippocampus, long term potentiation is associated with a rapid regulation of presynaptic kainate receptor function, switching presynaptic kainate receptors from a high affinity receptor to a low affinity receptor and producing a change in the dynamic properties of CA1 inputs (14). Interestingly at this period of development, CA1 long term potentiation is also dependent upon PKC (48). These studies therefore indicate that PKC activation can lead to the regulation of kainate receptor function during synaptic plasticity. Our present data provide a candidate mechanism for this PKC-dependent down-regulation of kainate receptor function: the PKC phosphorylation of the ubiquitously expressed GluR6 subunit leading to an increase in kainate receptor endocytosis.
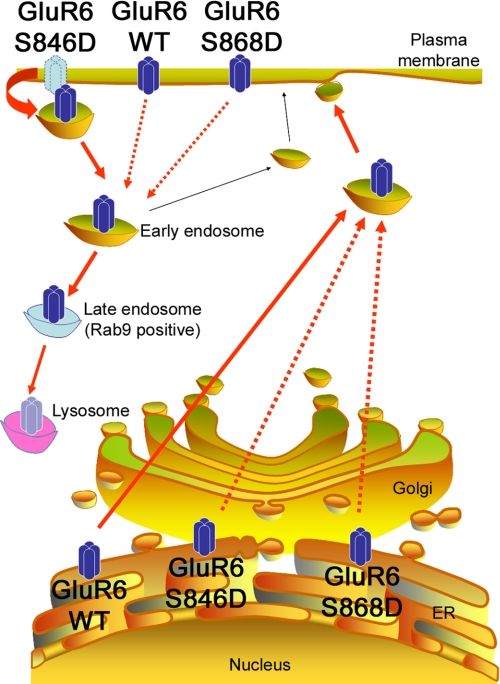
In this study we have identified a role for direct phosphorylation of GluR6 by PKC in the regulation of two distinct trafficking pathways (Fig. 7). Our findings indicate that PKC-dependent phosphorylation of Ser846 or Ser868 on the GluR6 C terminus promotes ER retention of kainate receptors. However, Ser846 phosphorylation plays an additional specific role in promoting GluR6 endocytosis that may underlie the PKC-dependent regulation of synaptic kainate receptors in certain forms of synaptic plasticity. Thus there appear to be multiple mechanisms by which phosphorylation of kainate receptor subunits lead to the specific regulation of receptor trafficking. The rapid regulation of kainate receptor trafficking allows neurons to selectively target receptors of different subunit compositions to different neuronal compartments where they serve distinct functional roles (6). It will be of interest to determine whether additional phosphorylation sites exist on other kainate receptor subunits that regulate their trafficking and whether such mechanisms can act in concert to selectively regulate kainate receptor trafficking and targeting that is dependent on receptor subunit composition.
FIGURE 7.
Phosphorylation of GluR6 at Ser846 or Ser868 inhibits GluR6 exit from the ER and traffic to the cell surface. In addition, phosphorylation of GluR6 at Ser846 accelerates GluR6 endocytosis from the plasma membrane and traffic to late endosomes.
Supplementary Material
Acknowledgments
We thank Dr. D. Maric (NINDS FACS Facility) for advice and expertise in the analysis of FACS data, Dr. C. L. Smith (NINDS Light Imaging Facility) for assistance with confocal microscopy, and Dr. S. Heinemann (Salk Institute) for making the GluR6 KO mice available. In addition, we acknowledge J. Nagle and D. Kauffman (NINDS Sequencing Facility) for DNA sequencing.
This work was supported, in whole or in part, by the National Institutes of Health NINDS Intramural Research Program and a Japan Society for the Promotion of Science Research Fellowship for Japanese Biomedical and Behavioral Researchers at National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PKC
- protein kinase C
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- FACS
- fluorescence-activated cell sorting
- PBS
- phosphate-buffered saline
- GST
- glutathione S-transferase
- PMA
- 13-O-acetylphorbol 12-myristate
- WT
- wild-type
- KO
- knock-out
- NMDA
- N-methyl-d-aspartic acid
- AMPAR
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors.
REFERENCES
- 1.Lerma J., Paternain A. V., Rodríguez-Moreno A., López-García J. C. (2001) Physiol. Rev. 81, 971–998 [DOI] [PubMed] [Google Scholar]
- 2.Vignes M., Collingridge G. L. (1997) Nature 388, 179–182 [DOI] [PubMed] [Google Scholar]
- 3.Castillo P. E., Malenka R. C., Nicoll R. A. (1997) Nature 388, 182–186 [DOI] [PubMed] [Google Scholar]
- 4.Chittajallu R., Vignes M., Dev K. K., Barnes J. M., Collingridge G. L., Henley J. M. (1996) Nature 379, 78–81 [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez-Moreno A., Herreras O., Lerma J. (1997) Neuron 19, 893–901 [DOI] [PubMed] [Google Scholar]
- 6.Isaac J. T., Mellor J., Hurtado D., Roche K. W. (2004) Pharmacol. Ther. 104, 163–172 [DOI] [PubMed] [Google Scholar]
- 7.Lauri S. E., Segerstråle M., Vesikansa A., Maingret F., Mulle C., Collingridge G. L., Isaac J. T., Taira T. (2005) J. Neurosci. 25, 4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisahn A., Contractor A., Traub R. D., Buhl E. H., Heinemann S. F., McBain C. J. (2004) J. Neurosci. 24, 9658–9668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smolders I., Bortolotto Z. A., Clarke V. R., Warre R., Khan G. M., O'Neill M. J., Ornstein P. L., Bleakman D., Ogden A., Weiss B., Stables J. P., Ho K. H., Ebinger G., Collingridge G. L., Lodge D., Michotte Y. (2002) Nat. Neurosci. 5, 796–804 [DOI] [PubMed] [Google Scholar]
- 10.Mulle C., Sailer A., Pérez-Otaño I., Dickinson-Anson H., Castillo P. E., Bureau I., Maron C., Gage F. H., Mann J. R., Bettler B., Heinemann S. F. (1998) Nature 392, 601–605 [DOI] [PubMed] [Google Scholar]
- 11.Bortolotto Z. A., Clarke V. R., Delany C. M., Parry M. C., Smolders I., Vignes M., Ho K. H., Miu P., Brinton B. T., Fantaske R., Ogden A., Gates M., Ornstein P. L., Lodge D., Bleakman D., Collingridge G. L. (1999) Nature 402, 297–301 [DOI] [PubMed] [Google Scholar]
- 12.Contractor A., Swanson G., Heinemann S. F. (2001) Neuron 29, 209–216 [DOI] [PubMed] [Google Scholar]
- 13.Schmitz D., Mellor J., Breustedt J., Nicoll R. A. (2003) Nat. Neurosci. 6, 1058–1063 [DOI] [PubMed] [Google Scholar]
- 14.Lauri S. E., Vesikansa A., Segerstråle M., Collingridge G. L., Isaac J. T., Taira T. (2006) Neuron 50, 415–429 [DOI] [PubMed] [Google Scholar]
- 15.Kidd F. L., Isaac J. T. (1999) Nature 400, 569–573 [DOI] [PubMed] [Google Scholar]
- 16.Park Y., Jo J., Isaac J. T., Cho K. (2006) Neuron 49, 95–106 [DOI] [PubMed] [Google Scholar]
- 17.Herb A., Burnashev N., Werner P., Sakmann B., Wisden W., Seeburg P. H. (1992) Neuron 8, 775–785 [DOI] [PubMed] [Google Scholar]
- 18.Werner P., Voigt M., Keinänen K., Wisden W., Seeburg P. H. (1991) Nature 351, 742–744 [DOI] [PubMed] [Google Scholar]
- 19.Nasu-Nishimura Y., Hurtado D., Braud S., Tang T. T., Isaac J. T., Roche K. W. (2006) J. Neurosci. 26, 7014–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wisden W., Seeburg P. H. (1993) J. Neurosci. 13, 3582–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahn S., Volk B., Wisden W. (1994) J. Neurosci. 14, 5525–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vignes M., Clarke V. R., Parry M. J., Bleakman D., Lodge D., Ornstein P. L., Collingridge G. L. (1998) Neuropharmacology 37, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 23.Clarke V. R., Collingridge G. L. (2002) Neuropharmacology 42, 889–902 [DOI] [PubMed] [Google Scholar]
- 24.Scott D. B., Blanpied T. A., Swanson G. T., Zhang C., Ehlers M. D. (2001) J. Neurosci. 21, 3063–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott D. B., Blanpied T. A., Ehlers M. D. (2003) Neuropharmacology 45, 755–767 [DOI] [PubMed] [Google Scholar]
- 26.Chung H. J., Huang Y. H., Lau L. F., Huganir R. L. (2004) J. Neurosci. 24, 10248–10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche K. W., O'Brien R. J., Mammen A. L., Bernhardt J., Huganir R. L. (1996) Neuron 16, 1179–1188 [DOI] [PubMed] [Google Scholar]
- 28.Lee H. K. (2006) Pharmacol. Ther. 112, 810–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes D. M., Braud S., Hurtado D. E., McCallum J., Standley S., Isaac J. T., Roche K. W. (2003) Biochem. Biophys. Res. Commun. 310, 8–13 [DOI] [PubMed] [Google Scholar]
- 30.Kim C. H., Braud S., Isaac J. T., Roche K. W. (2005) J. Biol. Chem. 280, 25409–25415 [DOI] [PubMed] [Google Scholar]
- 31.Martin S., Henley J. M. (2004) EMBO J. 23, 4749–4759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirbec H., Francis J. C., Lauri S. E., Braithwaite S. P., Coussen F., Mulle C., Dev K. K., Coutinho V., Meyer G., Isaac J. T., Collingridge G. L., Henley J. M., Couthino V. (2003) Neuron 37, 625–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greger I. H., Khatri L., Ziff E. B. (2002) Neuron 34, 759–772 [DOI] [PubMed] [Google Scholar]
- 34.Standley S., Roche K. W., McCallum J., Sans N., Wenthold R. J. (2000) Neuron 28, 887–898 [DOI] [PubMed] [Google Scholar]
- 35.Ren Z., Riley N. J., Garcia E. P., Sanders J. M., Swanson G. T., Marshall J. (2003) J. Neurosci. 23, 6608–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wenthold R. J., Prybylowski K., Standley S., Sans N., Petralia R. S. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 335–358 [DOI] [PubMed] [Google Scholar]
- 37.Roche K. W., Standley S., McCallum J., Dune Ly C., Ehlers M. D., Wenthold R. J. (2001) Nat. Neurosci. 4, 794–802 [DOI] [PubMed] [Google Scholar]
- 38.Man H. Y., Lin J. W., Ju W. H., Ahmadian G., Liu L., Becker L. E., Sheng M., Wang Y. T. (2000) Neuron 25, 649–662 [DOI] [PubMed] [Google Scholar]
- 39.Lin J. W., Ju W., Foster K., Lee S. H., Ahmadian G., Wyszynski M., Wang Y. T., Sheng M. (2000) Nat. Neurosci. 3, 1282–1290 [DOI] [PubMed] [Google Scholar]
- 40.Lavezzari G., McCallum J., Dewey C. M., Roche K. W. (2004) J. Neurosci. 24, 6383–6391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavezzari G., McCallum J., Lee R., Roche K. W. (2003) Neuropharmacology 45, 729–737 [DOI] [PubMed] [Google Scholar]
- 42.Ahmadian G., Ju W., Liu L., Wyszynski M., Lee S. H., Dunah A. W., Taghibiglou C., Wang Y., Lu J., Wong T. P., Sheng M., Wang Y. T. (2004) EMBO J. 23, 1040–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rivera R., Rozas J. L., Lerma J. (2007) EMBO J. 26, 4359–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coussen F., Perrais D., Jaskolski F., Sachidhanandam S., Normand E., Bockaert J., Marin P., Mulle C. (2005) Neuron 47, 555–566 [DOI] [PubMed] [Google Scholar]
- 45.Jaskolski F., Coussen F., Nagarajan N., Normand E., Rosenmund C., Mulle C. (2004) J. Neurosci. 24, 2506–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kidd F. L., Coumis U., Collingridge G. L., Crabtree J. W., Isaac J. T. (2002) Neuron 34, 635–646 [DOI] [PubMed] [Google Scholar]
- 47.Scott H. L., Braud S., Bannister N. J., Isaac J. T. (2007) Neuropharmacology 52, 185–192 [DOI] [PubMed] [Google Scholar]
- 48.Wikström M. A., Matthews P., Roberts D., Collingridge G. L., Bortolotto Z. A. (2003) Neuropharmacology 45, 828–836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.