Abstract
The Synechocystis Slr0642 protein and its plastidial Arabidopsis (Arabidopsis thaliana) ortholog At2g32040 belong to the folate-biopterin transporter (FBT) family within the major facilitator superfamily. Both proteins transport folates when expressed in Escherichia coli. Because the structural requirements for transport activity are not known for any FBT protein, we applied mutational analysis to identify residues that are critical to transport and interpreted the results using a comparative structural model based on E. coli lactose permease. Folate transport was assessed via the growth of an E. coli pabA abgT strain, which cannot synthesize or take up folates or p-aminobenzoylglutamate. In total, 47 residues were replaced with Cys or Ala. Mutations at 22 positions abolished folate uptake without affecting Slr0642 expression in membranes, whereas other mutations had no effect. Residues important for function mostly line the predicted central cavity and are concentrated in the core α-helices H1, H4, H7, and H10. The essential residue locations are consistent with a folate-binding site lying roughly equidistant from both faces of the transporter. Arabidopsis has eight FBT proteins besides At2g32040, often lacking conserved critical residues. When six of these proteins were expressed in E. coli or in Leishmania folate or pterin transporter mutants, none showed evidence of folate or pterin transport activity, and only At2g32040 was isolated by functional screening of Arabidopsis cDNA libraries in E. coli. Such negative data could reflect roles in transport of other substrates. These studies provide the first insights into the native structure and catalytic mechanism of FBT family carriers.
Keywords: Membrane/Proteins, Organisms/Bacteria, Organisms/Plant, Organisms/Protozoan, Transport, Transport/Folates
Introduction
Tetrahydrofolate and its one-carbon substituted derivatives (collectively termed folates) are essential cofactors in one-carbon metabolism (1, 2). They contain pterin, p-aminobenzoate (pABA),5 and glutamate moieties. Folates are made de novo by plants and most microorganisms but not by animals and certain protists, which must salvage exogenous folates (2–4).
Carrier-mediated folate transport into cells and between subcellular compartments is vital in organisms that require folates and is also vital in plants due to the way the folate synthesis pathway and folates are compartmented. Folates are made in mitochondria and exported to the cytosol, plastids, and vacuole (5–7). This implies the existence of several organellar folate transporters. Because plant cells can import folates, there must also be a transporter in the plasmalemma (8, 9).
Taking all organisms together, six types of folate transporter have been cloned so far. Five occur in mammals (10–12); of these, one (from the mitochondrial carrier family) occurs also in plants (13). The sixth type, the folate-biopterin transporter (FBT) family, also called the BT1 family, occurs in protists, cyanobacteria, and plants (3, 14, 15). The FBT family belongs to the major facilitator superfamily (MFS) (16). MFS proteins consist of a single polypeptide chain containing 12 transmembrane α-helices, with both N and C termini on the cytoplasmic side of the membrane (17). MFS transporters can have uniport, symport, or antiport mechanisms (18). Structures have now been determined for MFS proteins, including the lactose permease LacY, the glycerol-3-phosphate antiporter GlpT, and the multidrug transporter EmrD (19–21). These structures imply a general architecture for the superfamily, with substantial conservation of secondary and tertiary structure elements, and a common substrate translocation mechanism (17, 21).
FBT family members shown experimentally to transport folate include the FT1 and FT5 proteins from the protistan parasite Leishmania (22, 23), Slr0642 from the cyanobacterium Synechocystis, and its Arabidopsis ortholog At2g32040, which is located in the plastid envelope (15). Nothing is known for any of them about their structure in the membrane, the basis for substrate preference, or the catalytic mechanism, despite the centrality of FBTs in Leishmania to antifolate drug resistance (24).
The absence of information on the structure and mechanism of FBT carriers is due partly to the paucity of satisfactory eukaryotic or prokaryotic expression systems. Although Slr0642 and At2g32040 have both been functionally expressed in Escherichia coli, the experiments that could be done were limited by occasional spontaneous acquisition of folate uptake by the host cells (15). For example, the use of site-directed mutagenesis to probe the transport mechanism was precluded.
Besides At2g32040, the Arabidopsis genome encodes eight other FBT proteins, all phylogenetically more divergent from the cyanobacterial and Leishmania proteins than At2g32040 (15). Their function is unknown. They might a priori be supposed to transport folates, because folates (or pterins) are the only known substrates of the FBT family, and mitochondrial, vacuolar, and plasmalemmal folate carriers remain to be identified.
In this study, we first improved an E. coli expression system for FBT proteins. Using this system along with site-directed mutagenesis led to the definition of transport-critical residues in Slr0642. The expression system also enabled testing of more Arabidopsis FBT proteins for folate transport and functional screening of cDNA libraries. Last, we explored whether several FBT proteins could rescue the folate or biopterin growth requirements of Leishmania folate or biopterin transporter mutants.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
(6R,6S)-5-Formyl-5,6,7,8-tetrahydrofolate (5-FHTF) was from Schircks Laboratories (Jona, Switzerland), and pABA was from Sigma. Solutions were freshly prepared in water, filter-sterilized, and quantified spectrophotometrically using the molar extinction coefficient of 5-FHTF at 287 nm (31,500 m−1 cm−1) and of pABA at 267 nm (15,000 m−1 cm−1).
E. coli Strains and Culture Conditions
E. coli strain BN1143 (abg-1 abgT::kan zda-3061::Tn10 pabA1 rpsL704) (25) was obtained from B. P. Nichols (University of Illinois at Chicago). E. coli K-12 strain JW3323 (pabA::kan) was from the Keio collection (26). The kanamycin resistance cassette was removed from JW3323 via FLP-mediated recombination (27) to create K-12 ΔpabA with an in-frame deletion of the pabA gene. The abgT::kan locus from BN1143 was then introduced by P1 transduction into the K-12 ΔpabA strain to give the double mutant ΔpabA abgT::kan. Clones with the desired deletion or replacement were selected on LB plates containing 50 μg/ml kanamycin. The genotype of the double mutant was verified by PCR. Growth assays of the double mutant ΔpabA abgT::kan expressing the different FBT constructs were conducted on minimal medium as described (15).
Site-directed Mutagenesis of Slr0642
Site-directed mutagenesis was performed using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The pLOI707HE vector containing the slr0642 cDNA plus a C-terminal hexahistidine tag sequence was used as template. Synthetic oligonucleotides used for mutagenesis are listed in supplemental Table S1. Mutations were confirmed by sequencing.
Membrane Isolation and Western Blotting
E. coli double mutant (ΔpabA abgT::kan) cells harboring pLOI707HE alone or containing wild-type or mutated slr0642 cDNA (hexahistidine-tagged) were cultured in LB medium at 37 °C until A600 reached 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mm, and incubation was continued for 5 h at 22 °C. After harvesting (6000 × g, 10 min), cells were resuspended in 100 mm potassium phosphate, pH 7.0, 10 mm β-mercaptoethanol and disrupted in a Mini- BeadBeater (Biospec, Bartlesville, OK) using zirconia beads. The lysate was centrifuged at 6,000 × g and 10,000 × g for 10 min each to remove debris and finally at 100,000 × g for 1 h to pellet membranes. Membrane proteins were quantified using the BCA protein assay kit (Pierce). For electrophoresis, 25 μg of proteins were mixed with 0.2 m Tris-HCl, pH 6.5, 8% (w/v) SDS, 8% (v/v) β-mercaptoethanol, 40% (v/v) glycerol, and 0.04% (w/v) bromphenol blue and incubated at 37 °C for 30 min. Proteins were separated by SDS-PAGE using 10% (w/v) polyacrylamide gels and electrotransferred (44 mA, 2 h) to Immobilon-P membranes (Millipore, Bedford, MA). Blotted membranes were incubated overnight in TBS (20 mm Tris-HCl, 150 mm NaCl, pH 7.5) containing 5% (w/v) nonfat milk powder, washed with TBS containing 0.1% (v/v) Tween 20, and incubated for 1 h with anti-pentahistidine antibody (1:2000) (Qiagen, Valencia, CA) in TBS containing 5% (w/v) nonfat milk powder. Membranes were then washed in TBS containing 0.1% (v/v) Tween 20 and incubated with alkaline phosphatase-conjugated goat anti-mouse IgG (1:3000) (Bio-Rad) in TBS containing 5% (w/v) nonfat milk powder. Alkaline phosphatase activity was detected by incubating for 10 min in 100 mm Tris-HCl, pH 9.5, 100 mm NaCl, 5 mm MgCl2, 0.04% (w/v) nitro blue tetrazolium, 0.02% (w/v) 5-bromo-4-chloroindolyl phosphate.
Arabidopsis FBT Expression in E. coli
Full-length cDNAs encoding At5g25040, At5g25050, At1g79710, At5g10820, At5g54860, and At1g04570 were isolated by PCR using cDNA prepared from Arabidopsis rosette leaves as a template and the primers listed in supplemental Table S1. The SuperScriptTM III first-strand synthesis system (Invitrogen) was used for reverse transcription of mRNA. For expression in E. coli, full-length cDNAs were cloned into pLOI707HE as described (15). Chimeric constructs (with FBT sequences truncated just before the first predicted transmembrane α-helix and fused to the N-terminal 27 residues of Slr0642) were also generated as described (15), using primers listed in supplemental Table S1. Constructs were made in E. coli strain DH5α, sequence-verified, and then introduced into the E. coli double mutant ΔpabA abgT::kan, harboring pACYC-RIL (Stratagene, La Jolla, CA), which contains three rare E. coli tRNA genes and the cat marker. Transformants were grown on LB plates containing 10 μg/ml tetracycline, 50 μg/ml kanamycin, and 20 μg/ml chloramphenicol.
Arabidopsis FBT Expression in Leishmania donovani Mutants
Two versions of At2g32040 and the protein-coding regions of At5g25040, At5g25050, At1g79710, At5g10820, and At5g54860 were cloned into the unique BglII site of pIR1SAT, inserting a Kozak sequence (CCACC) just before the start codon. pIR1SAT is an integrating Leishmania expression vector containing streptothricin acetyltransferase as a selectable marker. Transfection into Leishmania of the linear targeting fragment of the vector and selection for streptothricin resistance yields parasites where the construct has replaced a copy of the ribosomal small subunit gene and is transcribed at high levels from the polI promoter.6 All sequences were amplified by PCR using primers to add 5′ and 3′ BglII, BclI, or BamHI sites and were digested with the respective enzyme before ligating into pIR1SAT. For At1g79710, which has a BglII site at bp 81, the first 69 bp were replaced by a start codon using a primer that ablated the BglII site; this deleted about half the region before the first transmembrane domain. One At2g32040 construct was truncated to remove the plastid targeting sequence by replacing the first 309 bp with a start codon. For the other, the first 357 bp were replaced by the first 26 codons of Leishmania major BT1, in which 12 G or C residues were changed to A or T (to eliminate a GC-rich region), and a BspEI site was introduced to join the fragments. All constructs were sequence-verified. The plasmids were linearized with SwaI and transfected into bt1− or MTXA5 mutant L. donovani promastigotes, and transformants were isolated by plating on semisolid M199 medium containing 100 μg/ml nourseothricin, using published methods (28, 29). A positive control episomal add-back line was generated by the same procedure for the MTXA5 null mutant by introducing the L. major FT1 gene in the pXGNEO vector (B2437) and plating on medium containing 10 μg/ml G418 (3, 30). Episomal add-back lines for the bt1− null mutants were as described (22). Growth of wild-type L. donovani and bt1− and MTXA5 mutants and transformants under semidefined conditions used folate-free RPMI medium (Invitrogen) supplemented with 30 mm HEPES, pH 7.4, 63 μm adenosine, 5 μg/ml hemin, and 1% (v/v) heat-inactivated fetal calf serum. The growth of each transport-deficient parental line was compared with that of its episomal add-back and the Arabidopsis FBT transformants. The growth of the MTXA5 lines was examined over three passages in media with no additions or with the addition of 8.4 μm biopterin, 25 μm folic acid, or 8.4 μm biopterin plus 25 μm folic acid. Constructs were taken to complement the MTXA5 mutant if they allowed growth to continue in the presence of biopterin but the absence of exogenous folate. Similarly, growth of the bt1−/− lines was examined over 13 passages with no additions and with 0.1 or 0.2 μm biopterin. Constructs were taken to complement the bt1−/− mutant if they allowed continued growth in the presence of 0.1 μm biopterin.
Screening of Arabidopsis cDNA Libraries
Arabidopsis cDNA expression libraries from 3-day-old hypocotyls (31) were obtained from the Arabidopsis Biological Resource Center (stock numbers CD4-14, CD4-15, and CD4-16). Mass excision was made using the ExAssist® Interference-Resistance Helper Phage kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Briefly, a portion of the λ bacteriophage libraries was combined with E. coli XL1-blue MRF′ cells and ExAssist helper phage at a 1:10:100 ratio, incubated 15 min at 37 °C, mixed with LB for a 3-h incubation at 37 °C with shaking, heated at 70 °C for 20 min, and centrifuged for 10 min at 1,000 × g. A portion of the resulting supernatant (excised phagemids) was incubated with E. coli SOLR cells for 20 min at 37 °C, plated onto 10 15-cm LB-ampicillin (100 μg/ml) agar plates, and incubated overnight at 37 °C. The resulting clones (∼106) were harvested in liquid LB and pooled for large scale plasmid DNA isolation by alkaline lysis. E. coli double mutant (ΔpabA abgT::kan) cells were transformed with 5 μg of plasmid DNA and plated on selective medium (M9 minimal medium containing 0.5 mm IPTG, 100 μg/ml ampicillin, 50 μg/ml kanamycin, 20 μg/ml chloramphenicol, 2 μg/ml sulfathiazole, and 11 μm 5-FTHF) for screening.
Bioinformatics
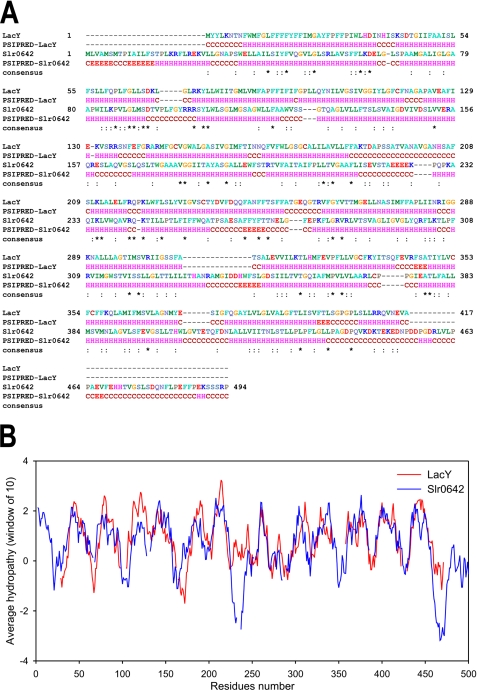
Because Slr0642 belongs to the MFS superfamily of transporters (16), a homology model of the transporter was built using the known structure of the lactose permease LacY (19) as template. However, the sequence homology between LacY and Slr0642 is only 9.5%, precluding an accurate alignment based on sequence identity alone. Therefore, additional criteria were applied to obtain a more reliable alignment, because the biophysical and chemical properties of related membrane proteins are often better conserved than the sequence identity. First, the secondary structure elements were determined using PSIPRED (32) to obtain a coarse alignment of the α-helices (Fig. 1A). Second, hydropathy profiles of LacY and Slr0642 were used to improve the alignment, because they are sensitive measures of the underlying structural similarity even when sequence similarity is virtually absent (33) (Fig. 1B). The orientation of the α-helices of Slr0642 was then improved in an iterative way by applying two further principles: (a) that residues that interact with the alkyl chains of the lipid bilayer are hydrophobic, whereas those in protein interactions or in the water phase are hydrophilic; and (b) that residues that interact with the membrane are less conserved than those involved in substrate and protein interactions, because there is less selective pressure on hydrophobic interactions (34). A hydropathy score was assigned to each residue by using the hydrophobicity scale of Kyte and Doolittle (35). A conservation score was assigned by ConSurf (36) by using an alignment of Slr0642 orthologs. For the iterative process, first a structure was built based on the alignment by using Swiss-Model (37). Second, the conservation and hydropathy scores were assigned to residues by using a color scale. Third, the structure was inspected visually with PyMol (38), and the alignment was altered to correct the inconsistencies. In total, 18 rounds of refinement were applied to generate a model that was most consistent with these criteria (see supplemental Fig. S1 and its interactive version, supplemental Fig. S2, A and B). The final pairwise alignment was used to calculate a comparative model of Slr0642 with MODELLER (39) by using the LacY structure (19) as template. The highest ranking model was analyzed with MOLPROBITY (40) to check the ψ, ϕ, and Cβ angles of the residues.
FIGURE 1.
Secondary structure predictions and hydropathy plots of the folate transporter Slr0642 and the lactose permease LacY. A, alignments of the amino acid sequences and the secondary structure predictions by PSIPRED. The asterisks and colons indicate identical and similar residues, respectively. B, hydropathy plots after refinement of the alignment by amphipathy and conservation analysis. The window was 20 amino acid residues, and the Kyte and Doolittle hydrophobicity scale was used.
RESULTS
Construction and Validation of an Improved E. coli pabA abgT Strain Suitable for Folate Uptake Studies
Because wild-type E. coli cells cannot take up folates, the functional expression of plasmid-encoded folate transporters can be detected in a pabA host strain (unable to synthesize the folate precursor pABA) by monitoring growth on medium containing a folate, 5-FTHF (15). However, this assay suffers interference from the occasional appearance of cells that spontaneously acquire the ability to grow on 5-FHTF (15), presumably due to sporadic manifestation of a latent capacity for uptake of folate or folate breakdown products that give rise to pABA.
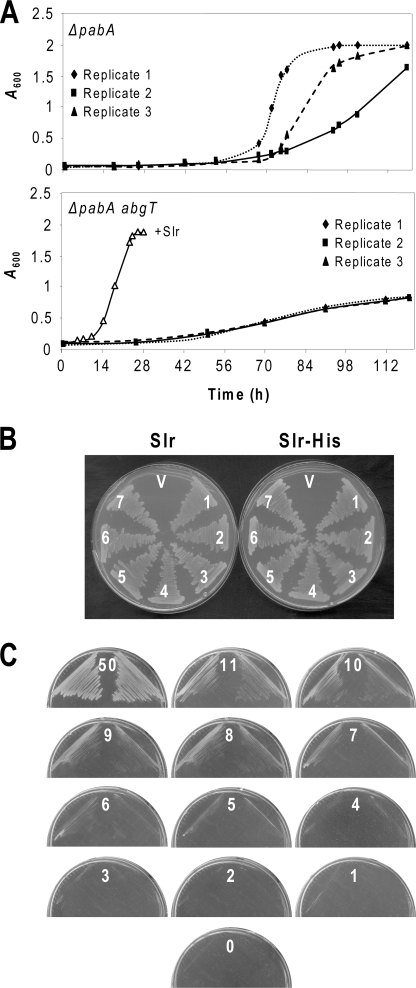
The E. coli abgT gene encodes a cryptic transporter of pABA-glutamate (a folate breakdown product) that is activated by point mutations in its promoter region (25, 41, 42), making it a prime suspect to explain the interference problem. We therefore constructed a pabA abgT strain and compared its propensity to grow in liquid minimal medium plus 5-FHTF with that of the parent pabA strain (Fig. 2A). A strain harboring the folate transporter Slr0642 was included for comparison. In triplicate observations, the double mutant cultures showed only slight and uniform growth, most likely sustained by traces of pABA produced by 5-FHTF breakdown in the medium. In contrast, all of the single mutant cultures began to grow exponentially after various lag times. This result was verified by streaking the double mutant on minimal medium plates containing 5-FHTF; in multiple trials, no colonies appeared. We therefore concluded that AbgT expression is responsible for the interference problem and used the pabA abgT mutant for all further work.
FIGURE 2.
Validation of the E. coli expression system. A, suppression of the spontaneous acquisition of folate uptake by inactivation of abgT. Triplicate cultures of the pabA and pabA abgT mutants were inoculated in minimal medium containing 0.5 mm IPTG and 11 μm 5-FTHF and incubated for 120 h at 37 °C, monitoring growth by absorbance at 600 nm. The growth of the pabA abgT mutant expressing Slr0642 (+Slr) is shown for comparison. No growth occurred in any cultures when 5-FTHF was omitted (not shown). B, lack of effect of a C-terminal His tag on Slr0642 function. Seven independent clones (1–7) of the pabA abgT strain harboring native (Slr) or His-tagged (Slr-His) versions of Slr0642 were plated on minimal medium containing 0.5 mm IPTG and 11 μm 5-FTHF. Single clones harboring vector alone (V) were included as controls. Clones were streaked three times in succession and photographed 2 days after the last streaking. C, growth of the pabA abgT strain harboring Slr0642 on minimal medium containing 0.5 mm IPTG and the standard 5-FTHF concentration (11 μm) as well as higher (50 μm) and lower (0–10 μm) concentrations. Duplicate clones were streaked three times in succession and photographed 2 days after the last streaking. Note that growth is not seen at 5-FTHF concentrations below 5 μm.
Functionality of a His-tagged Slr0642 Protein and Vector Choice
Synechocystis Slr0642 was chosen for site-directed mutagenesis because it is more readily expressed in E. coli than the At2g32040 protein (15). A C-terminal His tag was added to allow immunodetection, which did not alter performance; the tagged and untagged versions of Slr0642 gave identical growth of the pabA abgT mutant on 5-FHTF medium (Fig. 2B). In this and subsequent work, we retained the pLOI707HE vector used previously (15). This is a 13-kb, medium copy plasmid with a tac promoter and a copy of the lacIq repressor gene to ensure tight control of expression by IPTG (43). The high copy, less tightly IPTG-regulated pBluescript plasmid gave poorer results; colonies grew only half as fast as when pLOI707HE was used. Others have noted that high copy plasmids can be unsuitable for membrane protein expression in E. coli (44).
Sensitivity of the Growth Assay
To enhance the sensitivity of the growth assay to reductions of transport activity in mutant Slr0642 proteins, it was carried out at a 5-FTHF concentration determined to be subsaturating (11 μm). Thus, after a standard 2-day incubation, growth was markedly less at 11 μm than at 50 μm 5-FTHF (Fig. 2C). When 5-FTHF concentration was lowered in 1-μm steps from 11 μm, growth ceased to be evident at a concentration of 4 μm (Fig. 2C). If the 5-FTHF concentration is limiting for growth, it follows that transporter activity is likewise limiting and that if a concentration drop from 11 to 4 μm (∼60%) suffices to halt growth, then a similar activity drop would also do so. That a mutant protein retaining as much as 40% of wild type activity would fail to support growth makes the assay a responsive one.
Mutagenesis Round 1
Seventeen potential active site residues were selected (Table 1) based on their predicted location in transmembrane α-helices, their chemistry, and their pattern of conservation among FBT family members shown to transport folates. Polar residues were emphasized because these could in principle face the hydrophilic cavity of the carrier and interact with the folate substrate. Twelve of the residues are conserved or conservatively replaced in all of the folate-transporting FBT proteins and five others are conserved between Slr0642 and At2g32040. Two non-conserved Arg residues (Arg-158 and Arg-301) were included for comparison. All residues were replaced by Cys except for Cys-371, where Ala was used. Cys substitution mutants have been extensively used to investigate the structure and function of transporters (e.g. see Ref. 45).
TABLE 1.
Residues of the Synechocystis Slr0642 folate carrier mutagenized in round 1
Strictly conserved residues are indicated by a plus sign, conservatively replaced residues by “c”, and non-conserved residues by a minus sign.
| Residue | Conservation pattern in proven folate carriers |
Critical | |||
|---|---|---|---|---|---|
| Slr0642 | At2g32040 | FT1 | FT5 | ||
| Fully conserved | |||||
| Trp-82 | + | + | + | + | Yes |
| Lys-85 | + | + | + | + | Yes |
| Asp-93 | + | + | + | + | Yes |
| Asp-145 | + | + | + | + | Yes |
| Asp-149 | + | + | c | c | Yes |
| Ser-169 | + | + | + | + | No |
| Trp-172 | + | + | + | + | No |
| Glu-263 | + | c | c | c | Yes |
| Arg-309 | + | + | + | + | Yes |
| Met-361 | + | + | + | + | No |
| Arg-369 | + | + | + | + | Yes |
| Cys-371 | + | + | + | + | Yes |
| Partly conserved | |||||
| Arg-53 | + | + | − | − | Yes |
| Glu-154 | + | + | − | − | No |
| Arg-155 | + | + | − | − | Yes |
| Lys-304 | + | + | − | − | No |
| Asn-388 | + | + | − | + | Yes |
| Non-conserved | |||||
| Arg-158 | + | − | − | − | No |
| Arg-301 | + | − | − | − | No |
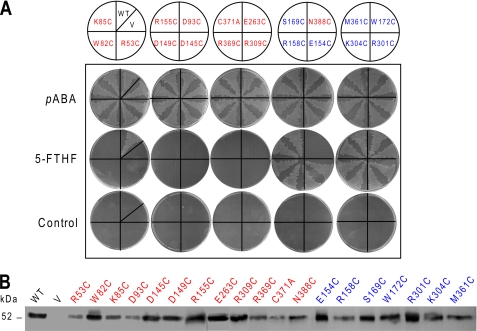
Of the 19 mutations tested, 12 were unable to support growth of the pabA abgT mutant on 5-FTHF medium, whereas the others had no noticeable effect (Fig. 3A). Western blotting of membrane fractions from the cells harboring the mutant constructs showed that Slr0642 protein was always expressed, albeit at varying levels (Fig. 3B). However, it varied about as much among the constructs that were functional as among those that were not, making it unlikely that the inactive mutant phenotype arose from insufficient protein. Although the protein expression results do not exclude the possibility that the deleterious mutations resulted in incorrect insertion into the membrane, not inhibition of transport per se, aberrant insertion is the less economical explanation and is inconsistent with the high incidence of criticality.
FIGURE 3.
First round of site-directed mutagenesis of the Slr0642 protein. A, in vivo folate uptake assays. The E. coli pabA abgT strain was transformed with pLOI707HE with no insert (V) or containing native slr0642 (WT) or 19 mutants thereof. Two independent clones of each mutant construct were streaked on minimal medium plus 0.5 mm IPTG alone (control) or containing 3.6 μm pABA or 11 μm 5-FTHF. Clones were streaked once on pABA-supplemented plates and three times in succession on the others. The pABA plates were photographed 1 day after streaking, and others were photographed after 2 days. Red, transport-critical residues; blue, non-critical residues. B, Western blot analysis of Slr0642 protein levels in membrane preparations from the above clones. Membrane proteins (25 μg/track) were separated by SDS-PAGE. The position of the 52-kDa molecular size marker is shown.
Of the 12 target amino acids conserved in known FBT folate transporters, nine were critical; of the five less conserved residues only three were critical; and of the two non-conserved residues, neither was critical (Table 1). There is thus a general correlation between conservation and importance for transport function.
Mutagenesis Rounds 2 and 3
A second set of 16 residues (Table 2) was selected based on (a) a high degree of conservation among cyanobacterial and plant orthologs of Slr0642, (b) a predicted location in the aqueous cavity, and (c) a polar or charged nature (or commonly found to be interacting with folate in other proteins of known structure) (supplemental Fig. S3). All were changed to Cys. Of the 16 residues, seven were important for transport (supplemental Fig. S4), six of which were predicted to face the central cavity.
TABLE 2.
Residues of Synechocystis Slr0642 mutagenized in round 2
Strictly conserved residues are indicated by a plus sign, conservatively replaced residues by “c”, and non-conserved residues by a minus sign.
| Residue | Conservation pattern in proven folate carriers |
Critical | |||
|---|---|---|---|---|---|
| Slr0642 | At2g32040 | FT1 | FT5 | ||
| Tyr-43 | + | + | − | c | No |
| Gln-46 | + | + | − | − | No |
| Lys-61 | + | + | − | − | Yes |
| Asp-62 | + | + | − | − | Yes |
| Glu-159 | + | + | − | − | No |
| Gln-168 | + | + | − | − | No |
| Phe-252 | + | + | − | − | Yes |
| Trp-256 | + | + | − | − | No |
| Phe-267 | + | + | + | + | Yes |
| Gln-300 | + | c | c | c | No |
| Asp-348 | + | + | + | + | Yes |
| Gln-357 | + | + | − | − | Yes |
| Phe-360 | + | + | + | c | No |
| Glu-376 | + | + | + | + | No |
| Phe-380 | + | + | c | c | Yes |
| Glu-396 | + | − | − | − | No |
Collectively, the results of rounds 1 and 2 provided good support for the overall structure of the comparative model because a high proportion of the residues that are important for transport line the predicted central cavity. In the model, α-helices H1 and H7 are dominant in forming the cavity, followed by H4 and H10, and together these four α-helices accounted for two-thirds of the critical residues. The third round of mutagenesis therefore focused mainly on these α-helices and sought to confirm their orientation and to better define the extent of the folate binding site within the cavity (Table 3). Of the 12 residues mutated (all to Cys), only three were critical (supplemental Fig. S5).
TABLE 3.
Residues of Synechocystis Slr0642 mutagenized in round 3
Strictly conserved residues are indicated by a plus sign, conservatively replaced residues by “c”, and non-conserved residues by a minus sign.
| Residue | Conservation pattern in proven folate carriers |
Critical | |||
|---|---|---|---|---|---|
| Slr0642 | At2g32040 | FT1 | FT5 | ||
| Leu-54 | + | + | − | − | No |
| Val-56 | + | + | − | − | No |
| Ser-57 | + | + | c | − | No |
| Phe-58 | + | + | c | − | No |
| Leu-60 | + | + | c | c | No |
| Ser-150 | + | + | − | − | No |
| Thr-259 | + | + | c | − | Yes |
| Ser-264 | + | + | c | + | No |
| Thr-271 | + | + | + | − | No |
| Asn-272 | + | + | − | − | No |
| Arg-335 | + | + | − | − | Yes |
| Met-384 | + | − | c | c | Yes |
Prediction and Testing of Folate Transport Activity of Other Arabidopsis FBT Proteins
The above analysis identified seven residues that are not only important to Slr0642 function but also strictly conserved in known folate transporters as well as in cyanobacterial and plant orthologs of Slr0642 (Tables 1 and 2). These residues are Trp-82, Lys-85, Asp-93, Asp-145, Phe-267, Asp-348, and Cys-371. The absence of any of the seven might thus suggest lack of folate transport activity. Inspection of the aligned sequences of the eight unstudied Arabidopsis FBT proteins (15) showed that five of them (At5g10820, At5g54860, At2g33280, At1g04570, and At1g64890) lack up to four of the seven residues and that only three (At5g25040 and At5g25050, which are 88% similar, and At1g79710) have the whole set.
Folate transport activity was tested for five of these proteins (At5g25050, At1g79710, At5g10820, At5g54860, and At1g04570) by expressing them in the pabA abgT strain and monitoring growth on 5-FTHF medium. Two types of expression construct were used: native sequences and sequences truncated before the first predicted transmembrane α-helix and fused to the N-terminal 27 residues of Slr0642. A strategy similar to the latter was used to express At2g32040 in E. coli (15). Neither version of any of the five proteins conferred growth on 5-FTHF, although positive controls with At2g32040 did so (data not shown). Negative results were also obtained from functional complementation tests in Leishmania folate and biopterin transport mutants with At5g25040, At5g25050, At1g79710, At5g10820, and At5g54860 proteins (data not shown). It should be noted, however, that constructs of the known folate transporter At2g32040 gave no complementation in the Leishmania tests, although positive Leishmania FT1 and BT1 controls behaved as expected.
As an indirect way to further test Arabidopsis FBT proteins for folate transport activity, we screened three cDNA expression libraries for functional complementation of the pabA abgT mutant on 5-FHTF medium. The only folate transporter found by screening 107 recombinants from each library was At2g32040, which was recovered three times from the same library. All three clones were truncated at residue 43, close to the predicted start of the mature protein.
DISCUSSION
This study was made possible by the development of a robust E. coli expression system for FBT carriers, comprising a pabA abgT double mutant host strain, and the medium copy, tightly IPTG-regulated pLOI707HE plasmid (43). This expression system could potentially be used for other FBT proteins and has already been used for another class of folate transporter (46). When used with a folate concentration that was subsaturating for growth, this system enabled a sensitive growth assay to detect transport defects in mutant proteins.
It should be said that extensive efforts were made to express Slr0642 in E. coli, Lactococcus lactis, or yeast at levels sufficient for transport assays with 3H-labeled folates. These efforts included recoding the gene, using specialized membrane protein expression strains, and modifying the N terminus to enhance membrane insertion. Protein expression in all systems was detectable immunologically, but none gave enough transport activity to measure on a time scale less than that of growth. We therefore measured growth itself.
In the course of this work, we tested other members of the Arabidopsis FBT family in our E. coli expression system and also in a heterologous eukaryotic system, employing Leishmania folate or pterin transporter mutants where only a low level of active protein expression is required to relieve folate or pterin auxotrophy. Although positive and negative controls were successful, no evidence of activity could be found in either system. These negative findings may have technical explanations, but other explanations cannot be excluded. Perhaps these transporters act on other substrates or function in compartments incompatible with the heterologous systems used. Further studies will be required to address these possibilities.
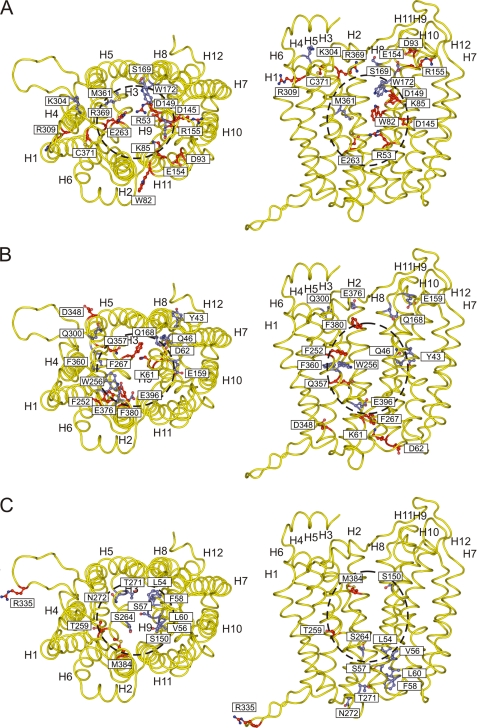
Our comparative structural model of Slr0642 is necessarily speculative due to the lack of close homology between FBT proteins and other MFS proteins whose three-dimensional structures have been determined. However, criteria beyond homology were carefully combined to guide model development, and the final result is quite consistent with the pattern of residue conservation and with the results of site-directed mutagenesis. Moreover, our mutagenesis data are consistent with the results of a recent study in which 10 conserved polar residues in the Leishmania FT1 transporter were changed to Leu or Val (47). Of the six residues common to both studies, five (Lys-85, Asp-93, Asp-145, Arg-309, and Asp-348 in Slr0642) were found to be essential or important to transport function in both. As summarized in Fig. 4 (and its interactive version, supplemental Fig. S6, A–C), the majority of critical residues line the predicted central cavity and are concentrated in α-helices H1, H4, H7, and H10, which form the cavity's core.
FIGURE 4.
The results of the mutagenesis study mapped onto the comparative model of Slr0642. A–C, first, second, and third rounds of mutagenesis, respectively. The data were taken from Tables 1–3, respectively. Shown are the extracellular view (left) and lateral view from the membrane (right) with the backbone of the model shown in a schematic diagram (yellow) and the mutated residues in a stick representation. Residues shown in red are critical for function, whereas the blue ones are not. The approximate position of the substrate binding site is indicated by the dashed circle. Supplemental Fig. S6, A–C, shows interactive versions of the three parts of this figure.
Slr0642 most probably operates via an alternate access mechanism in which the transporter has a central substrate binding site and two gates on either side of the central cavity that regulate access to the binding site in an alternating fashion. Since the model is only an approximation of the real structure, it is not possible to carry out substrate docking or molecular dynamics studies to obtain the binding conformation of the substrate. However, the substrate binding site is likely to be in the middle of the central cavity, corresponding to the area for binding of galactopyranosylthiogalactopyranoside in the structure of LacY (19). Several conserved residues that are found in the central cavity are critical for transport: Arg-53, Lys-85, Asp-145, Asp-149, Phe-252, Thr-259, Glu-263, Gln-357, Phe-380, Met-384, and Asn-388. The interactions of folate with the side chains of amino acids were inspected (using the Protein Data Bank) to understand the chemistry of folate binding (supplemental Fig. S3). Generally, the carboxyl groups of the glutamate moiety interact with Lys or Arg, the N3 and amine group of the pterin moiety interact with Asp or Glu, the ring structure of the pterin moiety interacts with the aromatic Phe or Tyr or hydrophobic Val or Ala, and the pABA interacts with the hydrophobic Leu or Ile. These interactions are to be expected based on the chemical properties of the substrate and side chains. On this basis, Asp-145 and/or Asp-149 could be interacting with N3 and the amine group of the pterin moiety, and Arg-53 and/or Lys-85 could have electrostatic interactions with the carboxyl groups of the glutamate group of folate. The aromatic Phe-252 and/or Phe-380 could have aromatic stacking interactions with the pterin ring, and the hydrophobic residue Met-384 could interact with the pABA or the pterin ring. The polar residues Thr-259, Gln-357, and Asn-388 could be interacting with the amino, carboxylate, or keto groups.
Several other residues could be involved in the gating aspects of the mechanism, and they line the interface between the two domains. On the cytoplasmic side are residues Glu-263 and Arg-53, which might form a salt bridge interaction, linking the two domains. Other residues in the interface are Lys-61, Asp-62, and Phe-267, which have no obvious interaction partners. Facing the outside of the cell is residue Asp-93, which is in the interface of the domains and is conserved not just in all FBT proteins (15) but in all other members of the major facilitator superfamily, pointing to a universal catalytic or structural role. In addition, there are Arg-369 and Phe-380, which could be important in gating. Trp-82 and Arg-309 may be closing the gaps between the two domains of the transporter, whereas the roles of Arg-155, Thr-259, Cys-371, Arg-309, and Asp-348 are unclear. Arg-335 is a conserved residue present in a long loop that does not exist in LacY.
Finally, the identification of conserved, transport-critical residues is a step toward prediction of substrate preference for FBT proteins. In this respect, it is encouraging that of the seven residues that are essential in Slr0642 and strictly conserved in known folate transporters, two are missing from the BT1 transporter of Leishmania, which strongly prefers biopterin (22, 24).
Supplementary Material
Acknowledgments
We thank Dr. B. P. Nichols for strain BN1143 and for advice, Dr. V. de Crécy-Lagard for help with E. coli genetics, Dr. A. Robinson for help with the Slr0642 model, and M. J. Ziemak for technical support.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM071382 and AI21903, by the Medical Research Council UK, by European Molecular Biology Organization Fellowship ALTF106-2005, and by an endowment from the C. V. Griffin, Sr. Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
T. J. Vickers and S. M. Beverley unpublished data.
- pABA
- p-aminobenzoic acid
- FBT
- folate-biopterin transporter
- MFS
- major facilitator superfamily
- 5-FHTF
- (6R,6S)-5-formyl-5,6,7,8-tetrahydrofolate
- IPTG
- isopropyl-β-d-thiogalactopyranoside
- TBS
- Tris-buffered saline.
REFERENCES
- 1.Lucock M. (2000) Mol. Genet. Metab. 71, 121–138 [DOI] [PubMed] [Google Scholar]
- 2.Scott J., Rébeillé F., Fletcher J. (2000) J. Sci. Food Agric. 80, 795–824 [Google Scholar]
- 3.Cunningham M. L., Beverley S. M. (2001) Mol. Biochem. Parasitol. 113, 199–213 [DOI] [PubMed] [Google Scholar]
- 4.de Crécy-Lagard V., El Yacoubi B., de la Garza R. D., Noiriel A., Hanson A. D. (2007) BMC Genomics 8, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bekaert S., Storozhenko S., Mehrshahi P., Bennett M. J., Lambert W., Gregory J. F., 3rd, Schubert K., Hugenholtz J., Van Der Straeten D., Hanson A. D. (2008) Trends Plant Sci. 13, 28–35 [DOI] [PubMed] [Google Scholar]
- 6.Jabrin S., Ravanel S., Gambonnet B., Douce R., Rébeillé F. (2003) Plant Physiol. 131, 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orsomando G., de la Garza R. D., Green B. J., Peng M., Rea P. A., Ryan T. J., Gregory J. F., 3rd, Hanson A. D. (2005) J. Biol. Chem. 280, 28877–28884 [DOI] [PubMed] [Google Scholar]
- 8.Crosti P., Malerba M., Bianchetti R. (1993) Plant Sci. 88, 97–106 [Google Scholar]
- 9.Prabhu V., Chatson K. B., Lui H., Abrams G. D., King J. (1998) Plant Physiol. 116, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Titus S. A., Moran R. G. (2000) J. Biol. Chem. 275, 36811–36817 [DOI] [PubMed] [Google Scholar]
- 11.Matherly L. H., Goldman D. I. (2003) Vitam. Horm. 66, 403–456 [DOI] [PubMed] [Google Scholar]
- 12.Qiu A., Jansen M., Sakaris A., Min S. H., Chattopadhyay S., Tsai E., Sandoval C., Zhao R., Akabas M. H., Goldman I. D. (2006) Cell 127, 917–928 [DOI] [PubMed] [Google Scholar]
- 13.Bedhomme M., Hoffmann M., McCarthy E. A., Gambonnet B., Moran R. G., Rébeillé F., Ravanel S. (2005) J. Biol. Chem. 280, 34823–34831 [DOI] [PubMed] [Google Scholar]
- 14.Massimine K. M., Doan L. T., Atreya C. A., Stedman T. T., Anderson K. S., Joiner K. A., Coppens I. (2005) Mol. Biochem. Parasitol. 144, 44–54 [DOI] [PubMed] [Google Scholar]
- 15.Klaus S. M., Kunji E. R., Bozzo G. G., Noiriel A., de la Garza R. D., Basset G. J., Ravanel S., Rébeillé F., Gregory J. F., 3rd, Hanson A. D. (2005) J. Biol. Chem. 280, 38457–38463 [DOI] [PubMed] [Google Scholar]
- 16.Marger M. D., Saier M. H., Jr. (1993) Trends Biochem. Sci. 18, 13–20 [DOI] [PubMed] [Google Scholar]
- 17.Abramson J., Kaback H. R., Iwata S. (2004) Curr. Opin. Struct. Biol. 14, 413–419 [DOI] [PubMed] [Google Scholar]
- 18.Saier M. H., Jr., Tran C. V., Barabote R. D. (2006) Nucleic Acids Res. 34, D181–D186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. (2003) Science 301, 610–615 [DOI] [PubMed] [Google Scholar]
- 20.Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 21.Yin Y., He X., Szewczyk P., Nguyen T., Chang G. (2006) Science 312, 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemley C., Yan S., Dole V. S., Madhubala R., Cunningham M. L., Beverley S. M., Myler P. J., Stuart K. D. (1999) Mol. Biochem. Parasitol. 104, 93–105 [DOI] [PubMed] [Google Scholar]
- 23.Richard D., Leprohon P., Drummelsmith J., Ouellette M. (2004) J. Biol. Chem. 279, 54494–54501 [DOI] [PubMed] [Google Scholar]
- 24.Ouellette M., Drummelsmith J., El-Fadili A., Kündig C., Richard D., Roy G. (2002) Int. J. Parasitol. 32, 385–398 [DOI] [PubMed] [Google Scholar]
- 25.Hussein M. J., Green J. M., Nichols B. P. (1998) J. Bacteriol. 180, 6260–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapler G. M., Coburn C. M., Beverley S. M. (1990) Mol. Cell. Biol. 10, 1084–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bello A. R., Nare B., Freedman D., Hardy L., Beverley S. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 11442–11446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ha D. S., Schwarz J. K., Turco S. J., Beverley S. M. (1996) Mol. Biochem. Parasitol. 77, 57–64 [DOI] [PubMed] [Google Scholar]
- 31.Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., Ecker J. R. (1993) Cell 72, 427–441 [DOI] [PubMed] [Google Scholar]
- 32.Bryson K., McGuffin L. J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. (2005) Nucleic Acids Res. 33, W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lolkema J. S, Slotboom D. J. (1998) Mol. Membr. Biol. 15, 33–42 [DOI] [PubMed] [Google Scholar]
- 34.Baldwin J. M., Schertler G. F., Unger V. M. (1997) J. Mol. Biol. 272, 144–164 [DOI] [PubMed] [Google Scholar]
- 35.Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 36.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) Nucleic Acids Res. 33, W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 38.DeLano W. L. (2002) Curr. Opin. Struct. Biol. 12, 14–20 [DOI] [PubMed] [Google Scholar]
- 39.Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. (2008) Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 40.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green J. M., Nichols B. P. (2002) in Chemistry and Biology of Pteridines and Folates (Milstien S., Kapatos G., Levine R. A., Shane B. eds) pp. 631–635, Kluwer, Dordrecht [Google Scholar]
- 42.Carter E. L., Jager L., Gardner L., Hall C. C., Willis S., Green J. M. (2007) J. Bacteriol. 189, 3329–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arfman N., Worrell V., Ingram L. O. (1992) J. Bacteriol. 174, 7370–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bibi E., Edgar R., Béjà O. (1998) Methods Enzymol. 292, 370–382 [DOI] [PubMed] [Google Scholar]
- 45.Cappello A. R., Miniero D. V., Curcio R., Ludovico A., Daddabbo L., Stipani I., Robinson A. J., Kunji E. R., Palmieri F. (2007) J. Mol. Biol. 369, 400–412 [DOI] [PubMed] [Google Scholar]
- 46.Rodionov D. A., Hebbeln P., Eudes A., ter Beek J., Rodionova I. A., Erkens G. B., Slotboom D. J., Gelfand M. S., Osterman A. L., Hanson A. D., Eitinger T. (2009) J. Bacteriol. 191, 42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dridi L., Haimeur A., Ouellette M. (2010) Biochem. Pharmacol. 79, 30–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.