Abstract
The activity of uracil DNA glycosylases (UDGs), which recognize and excise uracil bases from DNA, has been well characterized on naked DNA substrates but less is known about activity in chromatin. We therefore prepared a set of model nucleosome substrates in which single thymidine residues were replaced with uracil at specific locations and a second set of nucleosomes in which uracils were randomly substituted for all thymidines. We found that UDG efficiently removes uracil from internal locations in the nucleosome where the DNA backbone is oriented away from the surface of the histone octamer, without significant disruption of histone-DNA interactions. However, uracils at sites oriented toward the histone octamer surface were excised at much slower rates, consistent with a mechanism requiring spontaneous DNA unwrapping from the nucleosome. In contrast to the nucleosome core, UDG activity on DNA outside the core DNA region was similar to that of naked DNA. Association of linker histone reduced activity of UDG at selected sites near where the globular domain of H1 is proposed to bind to the nucleosome as well as within the extra-core DNA. Our results indicate that some sites within the nucleosome core and the extra-core (linker) DNA regions represent hot spots for repair that could influence critical biological processes.
Keywords: Chromatin, Chromatin/Structure, DNA/Enzymes, DNA/Repair, Histones, Linker Histone
Introduction
DNA in the cell is constantly damaged from both exogenous and endogenous sources, which can result in deleterious consequences such as mutation or cell death (1–5). The cell employs a variety of mechanisms to counteract DNA damage and maintain genomic integrity including base excision repair (BER),4 which repairs damage to DNA bases that does not cause large distortions to the DNA helix (4, 6). BER typically is initiated by a DNA glycosylase that is specific to a type of damaged or misincorporated base. A prototypical family of DNA glycosylases are the Family 1 uracil DNA glycosylases (UDGs), which specifically remove uracil in DNA that results from either misincorporation during replication, leading to A-U matches, or deamination of cytosine, leading to G-U mismatches (7, 8). Aberrant appearance of uracil in DNA is estimated to occur hundreds of times per cell per day and can cause harmful effects resulting from G:C to A:T transitions, cytotoxic/mutagenic abasic (AP) sites, or inhibition of DNA methylation (9–13). In addition, enzymatic cytosine deamination to produce G:U mismatches by activation-induced deaminases in B cells and subsequent glycosylase-dependent U excision are critical to the mechanism of immunoglobulin gene maturation and rearrangement (14, 15).
UDG base excision involves flipping the uracil out of the DNA helix stack to allow hydrolytic cleavage of the glycosidic bond (16–19), creating an apurinic/apyrimidinic (AP) site in the DNA with release of a free uracil base (5, 20–22). The backbone at the AP site is then cleaved by an AP endonuclease activity, which leaves a normal 3′-hydroxyl end and a 5′-deoxyribose phosphate, which can then undergo either a short-patch or long-patch repair process. Both of these pathways result in a nick that is ligated by a DNA ligase to complete repair (3, 5, 23).
BER has been extensively studied on free DNA, however, in eukaryotic cells the substrate for repair is not bare DNA but chromatin, in which the genome is associated with histones and other non-histone chromosomal proteins. The basic repeating subunit of chromatin is the nucleosome, consisting of a nucleosome core in which ∼147 bp of DNA is wrapped ∼1.7 times around a protein spool consisting of two copies each the four core histones H2A, H2B, H3, and H4 and 10–80 bp of linker DNA that links cores together (24, 25). Each nucleosome repeat is bound by one linker histone (e.g. H10, H1, and H5) and non-histone chromosomal proteins that promote folding and condensation of nucleosomes into the 30-nm chromatin fiber and other higher order structures present in the cell nucleus (25, 26). The assembly of DNA into nucleosomes greatly restricts the accessibility of most DNA-binding factors, including those involved in DNA repair. For example, the activities of factors involved in nucleotide excision repair are inhibited by chromatin and evidence indicates that nucleotide excision repair is linked to chromatin remodeling activities (27–32). Likewise, the activity of the short-patch BER-associated polymerase β appears to be totally inhibited by the presence of a nucleosome (33, 34). However, some BER-associated enzymes have been found to exhibit significant activity on nucleosome substrates in vitro. For example, FEN1 and DNA ligase 1 are able to carry out their respective activities sequentially on the same nucleosome substrate without significant disruption of histone-DNA interactions (35, 36). This suggests that the final steps of some BER events may not require chromatin remodeling activities.
The ability of several DNA glycosylases, either alone or in conjunction with other BER factors, to process targets buried in nucleosomes has been investigated, with somewhat conflicting results. Recently, Menoni et al. (37) showed that OGG1, which removes 8-oxoG from DNA is inhibited ∼100-fold at a site 10 bp from the nucleosome dyad, whereas AP endonuclease 1 exhibited no activity on nucleosomes. However, the glycosylase NTH1 was shown to efficiently process thymine glycol lesions oriented away from the histone octamer surface at a site about 50 bp from the nucleosome dyad but sites oriented toward the histone octamer or closer to the dyad were removed at a reduced efficiency compared with naked DNA (38). The activity of the glycosylase MBD4 was found to be diminished but not abolished for T/G mismatches within a reconstituted nucleosome in a manner apparently not dependent on position with respect to the dyad, whereas histone hyperacetylation increased the efficiency with which the bases were excised (39). Likewise, two groups (33, 34, 40) have investigated the activity of human UDGs, SMUG1 and UNG2, in conjunction with AP endonuclease 1, and DNA polymerase β on nucleosomal substrates. Both groups found that the DNA glycosylases and AP endonuclease exhibited marginally reduced activities on nucleosomal substrates but determined somewhat different extents of inhibition compared with free DNA substrates and came to different conclusions regarding the effect of rotational orientation (33, 34, 40). A possible reason for these differences may be that the two groups assayed the enzymes' activities using different DNA sequences. Importantly all studies of glycosylase activity have been carried out at a single or limited number of sites within the nucleosome.
To address differences in these results and obtain a more comprehensive view of glycosylase activity in chromatin, we carried out a systematic examination of UDG activity throughout a nucleosome substrate. We find that UDG excision within the nucleosome is significantly decreased compared with free DNA, consistent with previously published data. However, UDG activity on nucleosomal DNA is dependent on both rotational and translational positioning of the uracil, resulting in significant variations in the proficiency of this first step of repair. UDG activity on DNA outside the core region is commensurate with that of naked DNA, whereas H1 association reduces UDG at specific sites with the nucleosome core and in the linker DNA.
EXPERIMENTAL PROCEDURES
DNA Templates, 154-bp Single U Substitution Substrates
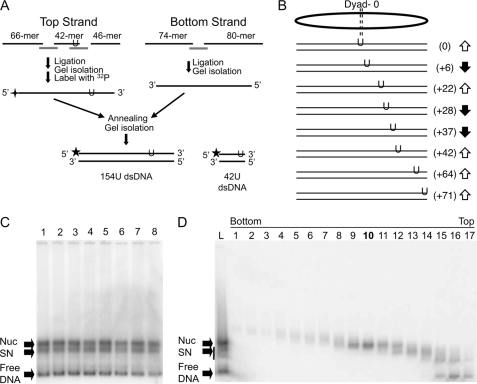
Eight 154-bp DNA fragments each containing different single T → U substitutions were generated based on the nucleosome-positioning element within the Xenopus borealis somatic-type 5 S RNA gene repeat (26). Fragments encompassed positions −78 to +76 with respect to the 5 S transcription start site on the top strand, however, in this work base positions are identified relative to the nucleosome dyad, located 2 bp upstream from the transcription start site (Fig. 1, A and B). The top and bottom strands of the DNA fragment were generated separately. For the top strand, 10 μg each of a 42-nucleotide (nt) and a 46-nt oligomer (see Fig. 1A and supplemental Table S1) were 5′-phosphorylated with ATP and T4 polynucleotide kinase (New England BioLabs). These oligos and the 66-nt upstream oligo were annealed with the 66 + 42 and the 42 + 46 complementary “splints” (see Fig. 1A and supplemental Table S1) and ligated by addition of 2400 units of T4 DNA ligase (New England BioLabs) in appropriate buffer, and incubation at 37 °C overnight. Full-length single-stranded 154-mers were isolated on 6% denaturing polyacrylamide gels containing 8 m urea, bands were identified by staining with ethidium bromide, and the DNA purified from the gel. For the bottom strand a 74-nt oligomer was 5′-phosphorylated with ATP and T4 polynucleotide kinase (New England BioLabs) as above, then annealed with the 74 + 80 splint and an 80-nt oligomer (Fig. 1A and supplemental Table S2). The oligos were ligated with T4 DNA ligase (New England BioLabs) and the 154-nt bottom strand was purified as described above. The top strands were radioactively 5′ end-labeled with [γ-32P]ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (New England BioLabs), then annealed to the bottom strand by heating to 95 °C for 10 min, turning off the heating block, and allowing the solutions to cool down slowly. The labeled double-stranded DNAs were isolated on non-denaturing 6% polyacrylamide gels, the wet gels were exposed to x-ray film, and the DNA eluted from the gel slice by crushing and soaking in 10 mm Tris, pH 8.0, and 1 mm EDTA (TE) overnight. A 42-bp fragment was also prepared and used as a naked DNA reference in the reactions by first radioactively end labeling the 42 + 22 U oligo (see Fig. 1A and supplemental Table S3) as described above, annealing with a complimentary “bottom strand” oligomer, and gel isolation as described above. In all cases, complete annealing was confirmed by complete restriction enzyme cleavage of the fragments.
FIGURE 1.
Assembly of nucleosomes containing single uracil residues. A, scheme of template preparation. Oligos used are indicated by black lines and denoted by length. Sequences of oligos are listed in supplemental Tables S1–S3. Gray lines indicate splints and are identified in the tables according to oligos they overlap. Note the single uracil (U) is located in either the 42- or 46-mer (see supplemental Table S1). Stars indicate positions of the radioactive phosphate label. B, positions of uracil substitutions. Note all single U substitutions are located in the top strand of the DNA fragments used. Arrows indicate predicted orientation of the phosphodiester backbone at the site of substitution with respect to the surface of the histone octamer. C, reconstitution of U-substitution templates. Products of reconstitution were separated on 0.7% agarose nucleoprotein gels, the gels were dried and analyzed by phosphorimager. Lanes 1–8 correspond to templates shown in B, from top to bottom, respectively. Bands corresponding to free 154-bp DNA (Free DNA), nucleosome (Nuc), and subnucleosome (SN) products are indicated by the arrows. D, sucrose gradient purification of reconstituted nucleosomes. Nucleosomes reconstituted with the 154(+22U) template (shown in C, lane 3) were purified by sedimentation through a sucrose gradient and fractions were analyzed on agarose nucleoprotein gels as described under “Experimental Procedures.” Numbers on top of gel correspond to fractions taken from the bottom to top of the gradient, as indicated. In this case fraction 10 was chosen for further study. Arrows indicate bands as in C. Fractionation of the remaining nucleosomes was carried out in an identical fashion. dsDNA, double-stranded DNA.
Random U Substitution Templates
A collection of 227-bp DNA fragments containing random T residues substituted with Us were generated by PCR. Either the top strand (ACCATGATTACGAATTCGAGC) or bottom strand (GAATGGCAAAAGTGCAAAAGC) primer was radioactively end-labeled with [γ-32P]ATP (PerkinElmer Life Sciences) T4 polynucleotide kinase (New England BioLabs) by standard methods. PCR contained 1 μg of each primer, 10 μl of 10× Taq polymerase buffer, 8 μl of 50 mm MgCl2, 5 μg of pXP-10 plasmid (41), 1 μl (5 units) of Taq polymerase (New England BioLabs), and either 8 μl of a stock containing 10 mm of each of the four dNTPs or 6 μl containing 10 mm dATP, dCTP, and dGTP and 2 μl of dUTP/dTTP, in a 5/95% ratio in a 100-μl reaction. The labeled double-stranded DNA fragments were purified on 6% polyacrylamide gels as described. The 227-bp products encompassed residues −90 to +137 within the 5 S rDNA sequence from pXP-10, renumbered as −88 to +139 in accordance with the dyad being fixed as position 0 (see above).
Nucleosome Reconstitution
Nucleosomes were reconstituted onto DNA substrates by standard salt-step dialysis (41). Equal amounts of H3/H4 and H2A/H2B purified from chicken erythrocytes were mixed with 18 μg of unlabeled calf thymus DNA and ∼200 ng (0.5 × 106 cpm) of one of the radiolabeled DNA fragments in 2 m NaCl, 10 mm Tris, pH 8.0, 1 mm EDTA, and 10 mm DTT. Samples were dialyzed against TE buffer containing 10 mm 2-mercaptoethanol and 1.2, 1.0, 0.8, and 0.6 m NaCl for 1.5 h each at 4 °C. A final dialysis against TE buffer only was carried out overnight. Proper formation of nucleosomes was confirmed by running samples of the reconstitutions on 0.7% agarose gels (41). Nucleosomes reconstituted with 154-bp templates were purified on 10-ml 5–20% sucrose gradients containing 10 mm Tris, pH 8.0, 0.1 mm EDTA and centrifuged for 15–18 h at 4 °C and 34,000 × g in a SW41 ultracentrifuge rotor (Beckman Coulter). Fractions were analyzed by running samples on 0.7% agarose nucleoprotein gels (41).
Hydroxyl Radical Footprinting
Nucleosome substrates were analyzed using hydroxyl radical footprinting as described (42). The free DNA and corresponding nucleosome were incubated in the presence or absence of hydroxyl radicals and the reactions were stopped by adjusting the solution to 5% glycerol. The DNAs were then purified by ethanol precipitation, resuspended in loading buffer, and run on 6% polyacrylamide DNA sequencing gels. The gels were then dried down, exposed, and quantified by phosphorimaging (GE Healthcare).
Uracil DNA Glycosylase Kinetic Assays, Single U Substitution Substrates
Escherichia coli uracil DNA glycosylase (2 units/μl; ∼0.1 μg/μl) was obtained from New England Biolabs and judged to be pure based on observation of a single band on a SDS-PAGE gel (data not shown). The bacterial enzyme was used based on commercial availability and conservation with the human enzyme (see “Discussion”). For UDG cleavage reactions typically 200 μl of a 154-mer nucleosome purified by sucrose gradient (104 cpm, ∼10 ng labeled nucleosomes) was mixed with an equal amount (cpm) of the 42-mer single U substitution-free DNA in UDG reaction buffer (10 mm Tris, pH 8.0, 1 mm EDTA, 1 mm DTT, and 100 μg/μl of bovine serum albumin) in a total volume of 350 μl. UDG in dilution buffer (20 mm Tris, pH 8.0, 50 mm NaCl, 1 mm EDTA, 1 mm DTT, 20% glycerol, and 100 μg/μl of bovine serum albumin) was added in amounts indicated in the figure legends and the reaction was incubated at 37 °C. At each time point 35 μl was removed to a new 1.5-ml tube and an equal volume of 1% SDS was added to stop the reaction. DNA was recovered by ethanol precipitation and resuspended in 90 μl of ultrapure water. Piperidine (10 μl) was added to each tube and base elimination was initiated by heating the tubes to 95 °C for 30 min. The reactions were dried to completion in a SpeedVac (Savant), resuspended in 30 μl of TE, and scintillation counted using an LSC 3500 (Beckman). Equivalent amounts per radioactivity from each reaction were run on 6% polyacrylamide DNA sequencing gels containing 1× TBE and 8 m urea. A G-reaction of the same DNA fragments, where the DNA has been modified with dimethyl sulfate and undergone base elimination as described in Ref. 43, was used as a marker. The gels were dried down and exposed in GE Healthcare PhosphorImager cassettes. Images were analyzed using ImageQuant and the data fitted to a first-order exponential kinetic profile using GraphPad Prism. All rate determinations were repeated at least three times.
Uracil DNA Glycosylase Assays with Global U Substitution Substrates
Forty μl of nucleosomes reconstituted with the 227-bp global U substitution substrates (10 pmol of total nucleosomes, 0.1 pmol of radiolabel 227-bp nucleosomes, ∼50 × 103 cpm) were incubated in UDG reaction buffer (10 mm Tris, pH 8.0, 1 mm EDTA, 1 mm DTT, and 100 μg/μl of bovine serum albumin) with 0.05 units of UDG (New England BioLabs) in a 50-μl total volume for various times at 37 °C. The samples were adjusted to 5% glycerol, loaded onto 0.7% agarose gels, and run at 120 V for 2 h. The gels were covered in plastic wrap and exposed to x-ray film overnight. Bands corresponding to nucleosome and free DNA for each time point were excised, the gel crushed, and DNA eluted passively by soaking in TE and 1% SDS overnight with constant agitation. Gel fragments were removed by centrifugation through Spin-X filters (Costar) and the solution volume reduced by drying in a SpeedVac (Savant). The DNA was ethanol precipitated and resuspended in 90 μl of ultrapure water. Base elimination was carried out, the DNA was resuspended in 30 μl of TE, and ∼1,000 cpm of each reaction run on sequencing gels, as described above.
H1 Preparation and Binding Assays
H1 was expressed in bacterial cells from the plasmid pET3dH1oa, containing the coding sequence for Xenopus H1o and purified as described (44). The ratio of H1 to nucleosome was determined from binding assays in which 10 μl of the 227-mer global U substitution nucleosome reconstitution (about 2.5 pmol of total nucleosomes, 0.025 pmol of radiolabeled 227-bp nucleosomes) was incubated in the presence of various amounts of H1 in binding buffer (50 mm NaCl, 5% glycerol, 10 mm Tris, pH 8.0, 1 mm EDTA, 1 mm DTT) for 30 min at 37 °C. The resulting complexes were analyzed on 0.7% agarose nucleoprotein gels to determine the optimal amount of H1 to achieve nucleosome binding with no binding to free DNA. The effect of H1 on UDG activity was determined by incubating 80 μl (∼105 cpm) of the 227-mer nucleosome reconstitution in the absence or presence of the selected amounts of H1 in the binding buffer for 30 min at 37 °C, followed by addition of 0.1 unit of total UDG. Naked DNA, nucleosomes, and H1-bound nucleosomes were isolated on preparative 0.7% agarose nucleoprotein gels and DNA was recovered and subjected to base elimination and cleavage patterns were analyzed on 6% polyacrylamide sequencing gels as described above.
RESULTS
To examine the activity of UDG at specific sites within a nucleosome, single thymine residues were substituted for uracil to generate U:A base pairs at specific locations within a 154-bp DNA fragment containing the Xenopus 5 S nucleosome-positioning element (Fig. 1, A and B). Sites were chosen for substitution based on previous characterization of the translational and rotational orientation of 5 S DNA assembled into a nucleosome and correspond to locations where the DNA backbone is oriented approximately toward or away from the surface of the histone octamer (42, 45). All of the U for T substitutions were in the “top” strand of the DNA fragment, which was specifically radiolabeled at the 5′ end before annealing with the complementary strand (Fig. 1A). We also prepared a 42-bp DNA fragment containing a single U substitution to include in the reactions as an internal naked DNA reference (Fig. 1A). UDG cleavage of the glycosidic bond of U residues was followed by base-catalyzed elimination of the AP site and breakage of the DNA backbone so that cleavage could be detected by DNA sequencing gel electrophoresis. Control experiments demonstrated that both the presence of a U in the DNA and the UDG enzyme were necessary to observe cleavage products and that UDG cleavage of Us within the 154- and 42-bp free DNA substrates occurred at equivalent rates (not shown, see below).
The 154-bp U-containing DNA templates were then reconstituted into nucleosomes via standard salt dialysis with purified histones. Analysis of nucleosomes reconstituted with native and U-containing DNA fragments on 0.7% native agarose gels indicated that the presence of uracil did not detectably alter the efficiency of reconstitution (Fig. 1C). Remaining naked DNA and subnucleosomal products from the reconstitution were purified away from the nucleosome products by sedimentation through sucrose gradients (Fig. 1D and not shown). Hydroxyl radical footprinting of purified U-containing nucleosomes and naked DNA showed that the latter is cleaved uniformly throughout, the nucleosome exhibits the expected wave-like pattern for a properly rotationally and translationally positioned 5 S nucleosome (supplemental Fig. S1 and results not shown) (42). Thus, the single U substitutions do not detectably alter nucleosome positioning or rotational orientation of 5 S DNA on the histone surface as expected, given the rather modest change in structure (loss of one C5 methyl group) upon single U substitution within the DNA fragment.
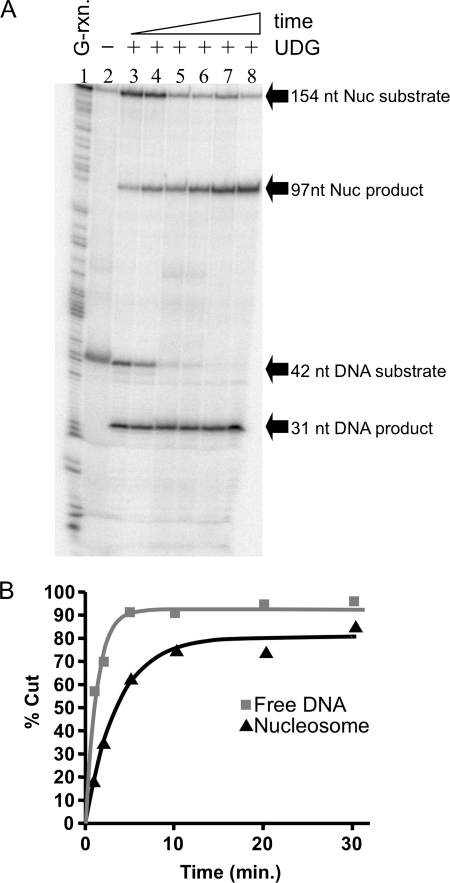
We first investigated the activity of UDG on a U located at position +22 within the 154(+22U) nucleosome (numbers indicate distance from the nucleosome dyad in base pairs), where the DNA backbone is oriented away from the core histone surface. Equivalent moles and specific activities of the nucleosome and free DNA substrates were incubated together with 0.05 units of UDG at 37 °C. At each time point an aliquot was removed, the reaction was stopped by addition of SDS and strand cleavage induced by base treatment. The products of the reactions were analyzed on sequencing gels and the fraction of substrate cleaved by UDG was determined and plotted versus reaction time. The results of these experiments revealed that UDG clearly excised a significant fraction of the uracil from the +22 site in the 154-bp nucleosome (Fig. 2A). A plot of product produced versus time is fit well by a single exponential for both the nucleosome and naked DNA substrates and indicates that 80–90% of each substrate was digested during the course of these reactions (Fig. 2B). Comparison of apparent first-order rate constants obtained from the fits showed that the rate of UDGs cleavage at the “outward facing” +22 site in the nucleosome substrate was about 6-fold slower than the rate of cleavage of the naked DNA substrate (see Table 1).
FIGURE 2.
Uracil located at an outward facing site within the nucleosome is readily excised by UDG. Nucleosomes reconstituted with the 154(+22U) DNA template were combined with the 42-bp reference DNA fragment (∼104 cpm each) and incubated with 0.05 units of UDG and the extent of cleavage analyzed on sequencing gels as described in the text. A, phosphorimage of a sequencing gel showing UDG-dependent cleavage of the 154-bp nucleosome (Nuc) and the 42-bp naked DNA (DNA) substrates to produce 97- and 31-bp products, respectively. Lane 1, G-reaction marker; lane 2, no UDG; lanes 3–8, substrates incubated with UDG for 1, 2, 5, 10, 20, and 30 min, respectively. B, plot of data taken from the gel shown in A. The amount of cleavage was quantified as described and points were fitted to a single-phase exponential curve to obtain apparent first-order rate constants. Cleavage data for the DNA (gray squares) and nucleosome (black triangles) are shown.
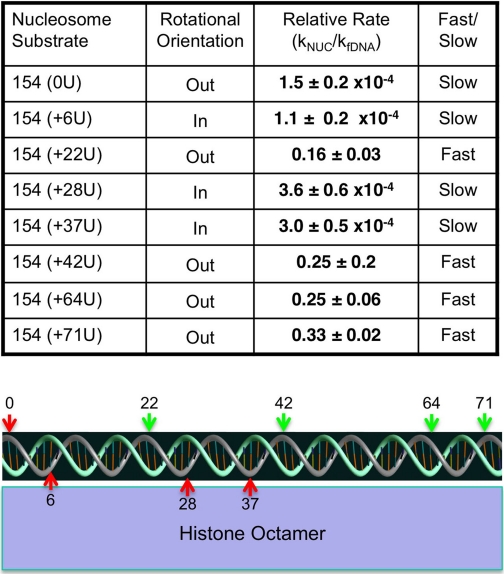
TABLE 1.
Single U kinetic assay results
Nucleosome substrates and rotational orientation of the uracils with respect to the histone surface are as described in the text. The relative rate for each site was calculated based on the major kinetic component in nucleosome assays (see text) compared with the internal naked DNA control, corrected for enzyme activity. Errors shown are mean ± S.E. from n ≥ 3 independent determinations for each site. Rates at each site are categorized as either fast or slow relative to the rate of spontaneous DNA unwrapping (see Ref. 52). The graphic depicts the rotational orientation of the 5 S DNA in the downstream half of the nucleosome (dyad indicated by “0”) and location of sites used for analysis relative to the histone octamer surface. All uracils are located within the top (grey) strand. Green and red arrows indicate fast or slow kinetics of UDG cleavage corresponding to the last column in the table.
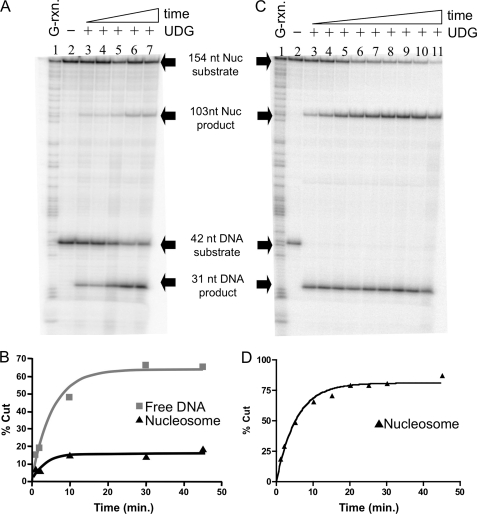
We next investigated a neighboring “inward facing” site, in which a uracil was located at position +28 within the 154(+28U) nucleosome substrate (Figs. 1B and 3A). Interestingly, at the same enzyme concentration used in the experiment shown in Fig. 2A, the digestion kinetics for the nucleosome appeared biphasic, with about 10% of the substrate digested at rates approximately equivalent to that of naked DNA, whereas additional substrate was cleaved much more slowly, such that the bulk of the substrate remained undigested at the longest time points investigated (Fig. 3B). We thus increased the amount of enzyme several hundredfold and repeated the assays (Fig. 3, C and D). At the increased enzyme concentration the rapid phase was over before the first time point and a second, slower phase was now apparent that included the majority of the nucleosome substrate. We assume the more slowly digesting material, which we estimate encompasses >80% of the nucleosome substrate, represents bona fide nucleosomes positioned at the expected location, whereas the ∼10% of rapidly digesting material may represent a small amount of dissociated or alternatively positioned templates, not uncommon for nucleosome preparations as demonstrated by restriction enzyme digests of similar samples (46) (see “Discussion”). Thus a small amount (10%) of nucleosome sample digests as free DNA, whereas the bulk of the nucleosome substrate bearing inward facing U digests with kinetics significantly slower than the naked DNA. Quantification showed UDG digests the inward facing U at position +28 in the nucleosome almost 3,000 times more slowly than the uracil in the 42-bp naked DNA fragment (Table 1).
FIGURE 3.
Uracil at an inward facing site within the nucleosome is excised very slowly by UDG. Nucleosomes reconstituted with the 154(+28U) DNA template were combined with the 42-bp reference DNA fragment before incubation with UDG and analysis of cleavage as described in the text. A, nucleosomes and naked DNA were incubated with 0.05 units of UDG. Lane 1, G-reaction marker; lane 2, substrates incubated in the absence of UDG; lanes 3–7, substrates incubated in the presence of UDG for 1, 2, 10, 20, and 30 min, respectively. Substrates and products are indicated as described in the legend to Fig. 2A. B, quantification of data shown in A. C, nucleosomes and naked DNA cleaved with 23.4 units of UDG. Lane 1, G-reaction marker; lane 2, substrates incubated in the absence of UDG; lanes 3–11, substrates incubated for 1, 2, 5, 10, 15, 20, 25, 30, and 45 min, respectively. D, quantification of data shown in C. Data were fitted to a single exponential as described in the legend to Fig. 2B.
We repeated the experiment with nucleosomes containing single Us substituted for Ts at six other sites throughout the 5 S sequence (Fig. 1B). In general, we found that the outward facing sites were cleaved only marginally more slowly than equivalent sites in free DNA (3–6-fold slower), whereas inward facing sites exhibited several thousand-fold slower cleavage rates compared with both free DNA and the outward facing sites (see Table 1). An exception was the one outward facing site located at the nucleosome dyad, which is cleaved by UDG with much slower kinetics compared with other outward facing sites.
Although the single U substitution templates allowed accurate determination of rates of UDG cleavage within nucleosome and naked DNA substrates, we also wished to assess the activity of UDG more globally throughout the nucleosome. Thus we used a PCR technique to generate a 227-bp DNA template based on the Xenopus 5 S nucleosome positioning sequence that contained random U for T substitutions throughout the DNA sequence. This template allowed us to investigate the rate of UDG activity on core versus extra-core (linker) DNA, and to study the effects of linker histone H1 on the activity of UDG. We determined that replacement of 5% of the dTTP in the PCR with dUTP resulted in about one uracil incorporated per strand on average within the nucleosome templates (results not shown). Labeling of one of the two primers allowed analysis of UDG cleavage throughout each DNA strand independently (see “Experimental Procedures”). Nucleoprotein gels showed that the DNAs were efficiently reconstituted into nucleosomes (results not shown).
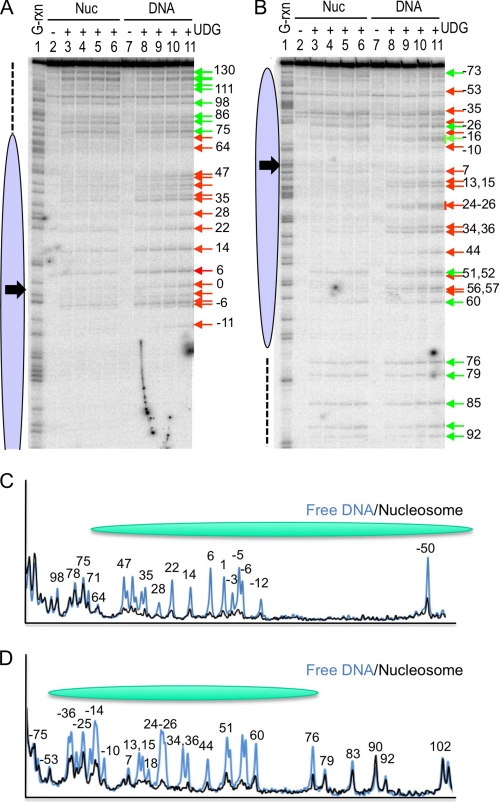
The 227-bp global U substitution nucleosomes were incubated with UDG for increasing amounts of time, the reactions were terminated, and nucleosomes and free DNA were isolated on preparative nucleoprotein gels. In these experiments we employed a concentration of enzyme sufficient to just cut naked DNA to completion over the time course to provide better contrast between exposed and protected regions, thus most internal nucleosome sites are not appreciably cleaved. DNA was isolated from the naked DNA and nucleosome bands and UDG activity analyzed as for the single-U assays. As expected from the single-site assays, we found activity of UDG was significantly decreased throughout the region encompassed by the nucleosome core (−73 to +73, top strand) on both strands (Fig. 4, A and B, ovals). However, DNA outside of the nucleosome core region was cut to an extent equal to that of the naked DNA (dashed line). Densitometric analysis of the cleavage patterns confirms the patterns of protection (Fig. 4, C and D). Although statistically reliable rate data cannot be derived for the core region from these gels due to the low extent of cleavage, it is clear that a few sites within the nucleosome core are appreciably cleaved, specifically positions −26, −16, +51, and +60 on the bottom strand, whereas nearby sites at +44 and +56/57 are protected from cleavage (Fig. 4B). It is interesting to note that all cleaved sites occur where the DNA backbone is predicted to be oriented away from the core histone octamer in the nucleosome, complementing our single-site analysis (see “Discussion”).
FIGURE 4.
Global analysis of UDG activity within nucleosome core and linker DNA regions. Nucleosomes reconstituted with the 227-bp DNA fragments containing randomly incorporated uracil residues were incubated with UDG and cleavage analyzed as described under “Experimental Procedures.” A and B, cleavage within the top and bottom strands of nucleosomes assembled with the 227-bp template, respectively. Lanes 2 and 7, 3 and 8, 4 and 9, 5 and 10, and 6 and 11 were incubated 0, 1, 5, 15, or 30 min, respectively. The positions of residues substituted for U are marked with arrows, red for positions blocked by the nucleosome and green for positions in which cleavage appears approximately commensurate with that of the naked DNA control. Black arrows indicate location of nucleosome dyad. C and D, densitometric scans of nucleosome and naked DNA cleavage patterns for the top and bottom strands, respectively. Lanes 4 and 9 were scanned in gels A and B to produce traces in C and D, respectively.
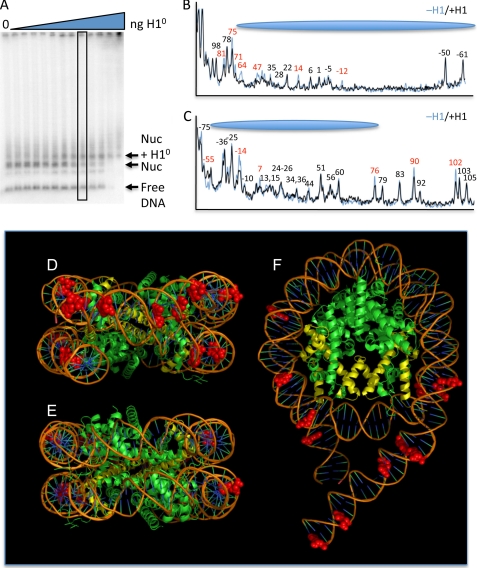
It has been previously demonstrated that linker histones bind preferentially and in a 1:1 ratio to nucleosomes over free DNA fragments (47), despite the propensity of these proteins to bind strongly and cooperatively to naked DNA (48). To investigate whether binding of a linker histone to the nucleosome affects UDG activity, we first titrated increasing amounts of purified H1 with the 227-bp global U nucleosomes to determine the optimal ratio that provided a discernable nucleosome + H1 gel shift band but no binding to naked DNA to ensure a single H1 bound to each nucleosome in the shifted band (Fig. 5A) (see “Experimental Procedures”). The effect of H1 on the activity of UDG was assayed by incubating the complexes with enzyme, and separating free DNA, nucleosome, and nucleosome + H1 complexes by preparative nucleoprotein gels (not shown). DNA was extracted from the gel slices and the UDG cleavage pattern resolved on sequencing gels, as before. Densitometric comparisons of the nucleosome and nucleosome + H1 cleavage patterns show that in the presence of H1, UDG cleavage within the nucleosome core region was reduced at positions +71, +64, +47, +14, and −12 on the top DNA strand and −55, −14, +7 on the bottom strand (Fig. 5, B and C, red numbers), whereas other residues were cleaved to identical extents in the presence and absence of H1 (black numbers). Interestingly, when mapped onto a model of the nucleosome (Fig. 5, D–F), we find that these sites are primarily located in the “front” of the nucleosome, near where the nucleosome dyad passes through the DNA. H1 binding also reduced cleavage at selected sites within the “linker” DNA outside of the core region. For example, UDG cleavages at sites +81 and +75 on the top strand and +76, +90, and +102 on the bottom strand are reduced in the presence of H1, whereas cleavages at nearby sites (e.g. +79, +83, +92, +103, and +105 on the bottom strand) are unchanged (Fig. 5, B and C).
FIGURE 5.
H1 association reduces UDG activity in both core and linker DNA. A, optimization of the H1:nucleosome ratio. H1 was incubated with nucleosomes reconstituted with the 227-bp DNA fragment containing uracil randomly substituted for thymine and complexes analyzed by agarose nucleoprotein gels and phosphorimaging. Lanes contain 0, 1, 2, 5, 10, 25, 50, 100, 200, 400, 800, 1,600, and 2,400 ng of H1 incubated with ∼10 pmol of total nucleosomes (see “Experimental Procedures”). The position of the free DNA, nucleosome, and H1-bound nucleosomes bands are indicated. Note that excess H1 results in aggregation of the sample and slowly migrating species. B and C, effect of H1 on UDG cleavage of nucleosome DNA. The 227-bp global U-substitution nucleosomes were incubated with H1. H1-bound and unbound species were isolated from preparative versions of the gel shown in A. DNA isolated from these samples was separated on sequencing gels and the cleavage patterns analyzed by densitometric analysis. Patterns for the top and bottom strands are shown in B and C, respectively. Numbers correspond to distance from the dyad of the nucleosome. Positions protected upon H1 association are indicated in red. D–F, molecular model of the 227-bp nucleosome, showing front, back, and top views, respectively. Base positions for which UDG cleavage is reduced upon H1 binding are indicated as red space-filling structures. H3 is shown in yellow for orientation. The model was adapted from Ref. 54 using MacPyMOL.
DISCUSSION
In this work we show that UDG activity on nucleosome DNA is dependent on both the rotational and translational positions of the target. We find sites within nucleosomes where UDG is virtually blocked from cleavage as well as sites where cleavage occurs at rates only a few-fold less than that observed with naked DNA. Generally, outward facing sites within the nucleosome core region are cleaved at significantly faster rates than inward facing sites, and cleavage is more rapid for sites further from the nucleosome dyad. Interestingly linker DNA outside of the core region exhibits cleavage kinetics similar to free DNA, whereas binding of linker histone restricts UDG activity at specific sites near the proposed binding site of the H1 globular domain (49–51) as well as at selected sites within the linker DNA. These results have significant biological implications for UDG activity and DNA repair in chromatin.
We find that sites oriented toward the histone octamer are cleaved at rates several thousand times slower than analogous sites on naked DNA. Nevertheless, cleavage of these sites involving a majority of the nucleosome substrate can be observed at high enzyme concentrations and occurs at rates indicative of a process limited by spontaneous unwrapping of DNA from the surface of the nucleosome to expose the free sites (52). As originally demonstrated by Widom and colleagues (52, 53) by monitoring the activity of restriction enzymes, nucleosome DNA undergoes spontaneous unwrapping to expose interior sites with a probability of about 1 in 3000–5000, depending on location within the nucleosome, thus limiting the rate of processes requiring fully exposed DNA by the same factor. The rates we measure for inward facing sites are entirely consistent with this model. Moreover, our global substitution experiments indicate that sites oriented intermediately between inward and outward facing are also cleaved with very slow kinetics. Thus our rate measurements indicate that UDG requires DNA unwrapping for activity at sites not oriented maximally away from the surface of the histone octamer.
On the other hand, we measured rates of cleavage for outward facing sites that are significantly faster than that dictated by spontaneous DNA unwrapping. These rates, only 3–6-fold slower than the rate for naked DNA, indicate that UDG can recognize an appropriately positioned uracil within nucleosome DNA and cleave the glycosidic bond without significant disruption of histone-DNA interactions. Indeed, the energy penalty for breakage of only one histone-DNA contact is likely to translate into a several hundredfold reduction in the rate for an associated process (52). Thus, our data indicate that UDG can accommodate both occlusion of one side of the DNA helix by the core histone proteins and the large extent of bending of the DNA within the nucleosome. Interestingly, a co-crystal structure of UDG bound to a DNA substrate indicates that the substrate is bent away from the main body of the enzyme at an angle approximating the extent of bending required for nucleosome wrapping (18). Thus, the natural shape of the DNA substrate when bound to the UDG active site is consistent with activity of this enzyme for outward facing sites on the nucleosome.
Previous studies investigating the ability of uracil DNA glycosylases to excise targets from nucleosomal DNA showed that U was removed from nucleosomes 3–10 times slower than free DNA, but in these studies there appeared to be little effect of rotational orientation of the U within the nucleosome (34, 40). Although these numbers are in good agreement with what we have determined for most of the outward facing sites in the nucleosome, they are not consistent with our results for the inward facing sites where we find UDG is inhibited several thousand-fold on nucleosomes compared with free DNA. The basis for this discrepancy is not clear. We note that in our nucleosome preparations a small fraction of the substrate exhibited much more rapid cleavage kinetics than the bulk. Given the well known propensity of nucleosomes to undergo disproportionation or dissociation, especially at low concentrations (55, 56), we believe that the rapidly digesting component in our experiments represents a non-nucleosomal contaminant that can dominate the kinetic profile of a more slowly digesting component at low enzyme concentrations. This is a critical issue as standard enzyme kinetic assays typically examine the first few percent of substrate processed, something clearly impossible for nucleosome samples. Indeed, previous studies corrected for a fraction of the nucleosome sample that behaved like free DNA (40) or used restriction enzyme cleavage to remove naked DNA contaminants (34). In our kinetic profiles we defined a uniformly digesting component representing the bulk (>80%) of the nucleosome observed substrate at drastically increased enzyme levels due to the low probability of DNA site exposure for inward facing sites (see above). In addition, it is known that a small fraction of 5 S nucleosomes adopt alternative positions on the DNA (57), which may exhibit drastically different kinetics compared with the main translational position. Thus it is possible that, despite attempts to correct for non-nucleosomal contaminants, previous measurements of UDG kinetics did not effectively separate out fast-digesting components and thus yielded aberrantly fast kinetics for inward facing sites.
We note that this study employed commercially available E. coli UDG, whereas previous studies have used Homo sapiens UDG. However, we believe it is unlikely that these enzymes would behave significantly different in our assays due to extreme conservation within the ∼220-amino acid residue catalytic domain of this family of enzymes (7, 58). Although H. sapiens UDG also has an 84-amino acid residue domain N-terminal to the catalytic domain, this region interacts with other proteins, such as proliferating cell nuclear antigen and replication protein A and is not part of the catalytic core of the enzyme (58). Indeed, due to alternative splicing, this domain differs between the nuclear and mitochondrial forms of the human enzyme to provide appropriate targeting to these subcellular organelles (59). The catalytic cores of human and E. coli UDG are highly conserved, exhibiting 57% sequence identity and 71% similarity, with no gaps. In addition, the bacterial and human enzymes (PDB codes 1EUG and 1AKZ, respectively) are very similar in overall structure, with a root mean square deviation of 0.9 Å for all Cα positions (60). Moreover, residues comprising the active sites of the two enzymes are strictly conserved and exhibit a root mean square deviation of 0.35 Å (60). Not surprisingly, the two enzymes are found to have virtually identical catalytic mechanisms. Thus, it is highly unlikely that significant differences would be obtained with the human enzyme in our assays.
We also note that our nucleosome assays with lower concentrations (0.05 units) of UDG contained double-stranded DNA substrate in ∼10-fold excess over enzyme (9), thus our assays report multiturnover kinetics. Free enzyme concentrations also could be reduced due to the presence of about 10-fold (w/w) excess “carrier” DNA over radiolabeled uracil-containing template in the single-U nucleosome reactions. However, we found that DNA fragments lacking uracil are a very poor competitive inhibitor of UDG activity compared with fragments containing uracil (data not shown).
We did find one outward facing site in the single U substitution experiments, positioned exactly at the dyad axis of the nucleosome, which was cleaved much more slowly than other outward facing sites. We previously reported that hDNA ligase I activity was highly inhibited at a site near the nucleosome dyad in nucleosome cores but that this inhibition was relieved when a nucleosome substrate containing linker DNA was used, presumably due to linker DNA-dependent repositioning of the core histone tail domains (36, 61). Our experiments with nucleosomes assembled onto the 227-bp DNA fragments do not show a significant increase in accessibility near the dyad, however, we have not carried out quantitative single-site assays for the dyad site with a longer DNA fragment. We note that others have shown that hyperacetylation of histone tails or proteolytic removal of the tail domains apparently does not significantly alter UDG activity on individual nucleosomes (33, 40).
Interestingly, we find that the UDG excision of uracil located immediately outside of the nucleosome core region occurred at rates commensurate with that of naked DNA. These results are entirely consistent with a recent examination of UDG activity within a nucleosomal array containing chemically generated U:G base pairs (62). The Smerdon group (62) showed that UDG cleavage occurred in a facile manner, primarily within the linker regions between nucleosomes, even in conditions in which the arrays are folded into higher order secondary structures. In addition, these authors also detected limited sites within nucleosome core regions amenable to robust cleavage by UDG, consistent with our global analysis.
Linker histones bind to the exterior of nucleosomes and stabilize folding and condensation of nucleosome arrays by association of the highly positively charged C-terminal domain with linker DNA (25, 63, 64). Specific binding is driven by the so-called globular domain of the protein, which associates with the nucleosome near the dyad and where the DNA exits/enters the structure (49, 50, 65). However, the exact modes and locations of binding of the globular and C-terminal domains within the nucleosome have not been defined. Interestingly, we find H1 binding reduces UDG activity exclusively at sites clustered in the front of the nucleosome core, near where the dyad passes through the DNA, whereas no protection is found for sites on the back side of the nucleosome (Fig. 5, D–F). In addition, linker histones protect 10 additional base pairs of “chromatosome” DNA on either side of the core region from nuclease digestion and may organize linker DNA into a stem structure (66–68). Consistent with this idea, we found that H1 association exerts subtle effects on UDG cleavage at specific sites throughout the linker DNA, even at sites beyond the 10-bp chromatosome regions at each edge of the nucleosome core. Overall the effects of H1 binding on cleavage are subtle in our single nucleosome assays but may be much more significant in stably folded arrays of nucleosomes (62). Nevertheless, these results indicate that association of the C-terminal tail and reorganization of the linker DNA upon H1 binding can affect UDG access to the DNA.
In general, our data are consistent with data showing that complex DNA repair systems are affected enormously by assembly of the DNA substrate into chromatin (69). Uracil left undetected by UDG due to chromatin could have a variety of mutagenic consequences (11, 12, 70). In addition, differential UDG activity within the nucleosome may lead to selective repair of uracils and provide a mechanism by which translational heterogeneity of nucleosomes could contribute to diversity in antibody production (15). Activation-induced cytidine deamination converts cytidines to uracils during transcription of immunoglobulin loci, leading to somatic hypermutation, initiation of class switch recombination, and gene conversion (71). Several mechanisms are believed to be linked to the initial conversion of C to U, including glycosylase-dependent error prone DNA repair in B cells (15, 71). If nucleosome reformation after transcription is more rapid than initiation of the repair process, natural cell to cell variations in positioning of nucleosomes coupled with the observation of “hot spots” for UDG activity (this work) would contribute to selective repair of a subset of Us, and thus greater diversity in sequence outcome. Moreover, recent work shows that nucleosomes can greatly influence the initial activation-induced cytidine deamination activity (72). It will be interesting in the future to examine rates of nucleosome formation, repair, and roles of other activities such as AP endonuclease on these processes.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant RO1GM52426 and American Cancer Society Grant RPG GMC-99742.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1–S3.
- BER
- base excision repair
- UDG
- uracil DNA glycosylase
- AP
- apurinic/apyrimidinic
- nt
- nucleotide
- DTT
- dithiothreitol.
REFERENCES
- 1.Friedberg E. C. (1995) Trends Biochem. Sci. 20, 381. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers J. H. (2001) Nature 411, 366–374 [DOI] [PubMed] [Google Scholar]
- 3.Wood R. D. (1996) Annu. Rev. Biochem. 65, 135–167 [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T., Karran P., Wood R. D. (1997) Curr. Opin. Genet. Dev. 7, 158–169 [DOI] [PubMed] [Google Scholar]
- 5.Memisoglu A., Samson L. (2000) Mutat. Res. 451, 39–51 [DOI] [PubMed] [Google Scholar]
- 6.Annunziato A. T., Hansen J. C. (2000) Gene Expr. 9, 37–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearl L. H. (2000) Mutat. Res. 460, 165–181 [DOI] [PubMed] [Google Scholar]
- 8.Liu P., Burdzy A., Sowers L. C. (2002) Chem. Res. Toxicol. 15, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 9.Mosbaugh D. W., Bennett S. E. (1994) Prog. Nucleic Acid Res. Mol. Biol. 48, 315–370 [DOI] [PubMed] [Google Scholar]
- 10.Krokan H. E., Standal R., Slupphaug G. (1997) Biochem. J. 325, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl T. (1993) Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 12.Duncan B. K., Weiss B. (1982) J. Bacteriol. 151, 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otterlei M., Warbrick E., Nagelhus T. A., Haug T., Slupphaug G., Akbari M., Aas P. A., Steinsbekk K., Bakke O., Krokan H. E. (1999) EMBO J. 18, 3834–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krokan H. E., Nilsen H., Skorpen F., Otterlei M., Slupphaug G. (2000) FEBS Lett. 476, 73–77 [DOI] [PubMed] [Google Scholar]
- 15.Kavli B., Otterlei M., Slupphaug G., Krokan H. E. (2007) DNA Repair 6, 505–516 [DOI] [PubMed] [Google Scholar]
- 16.Savva R., McAuley-Hecht K., Brown T., Pearl L. (1995) Nature 373, 487–493 [DOI] [PubMed] [Google Scholar]
- 17.Mol C. D., Arvai A. S., Slupphaug G., Kavli B., Alseth I., Krokan H. E., Tainer J. A. (1995) Cell 80, 869–878 [DOI] [PubMed] [Google Scholar]
- 18.Slupphaug G., Mol C. D., Kavli B., Arvai A. S., Krokan H. E., Tainer J. A. (1996) Nature 384, 87–92 [DOI] [PubMed] [Google Scholar]
- 19.Barrett T. E., Savva R., Panayotou G., Barlow T., Brown T., Jiricny J., Pearl L. H. (1998) Cell 92, 117–129 [DOI] [PubMed] [Google Scholar]
- 20.Dodson M. L., Michaels M. L., Lloyd R. S. (1994) J. Biol. Chem. 269, 32709–32712 [PubMed] [Google Scholar]
- 21.Parikh S. S., Walcher G., Jones G. D., Slupphaug G., Krokan H. E., Blackburn G. M., Tainer J. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5083–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stivers J. T., Drohat A. C. (2001) Arch. Biochem. Biophys. 396, 1–9 [DOI] [PubMed] [Google Scholar]
- 23.Lindahl T., Wood R. D. (1999) Science 286, 1897–1905 [DOI] [PubMed] [Google Scholar]
- 24.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 25.van Holde K. E. (1989) Chromatin, Springer Verlag, New York [Google Scholar]
- 26.Wolffe A. P. (1998) Chromatin Structure and Function, 3rd Ed., Academic Press, San Diego, CA [Google Scholar]
- 27.Ura K., Hayes J. J. (2002) Eur. J. Biochem. 269, 2288–2293 [DOI] [PubMed] [Google Scholar]
- 28.Golding A., Chandler S., Ballestar E., Wolffe A. P., Schlissel M. S. (1999) EMBO J. 18, 3712–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R. E., Hoeijmakers J. H., Vermeulen W. (2000) Mol. Cell. Biol. 20, 7643–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong F., Fahy D., Smerdon M. J. (2006) Nat. Struct. Mol. Biol. 13, 902–907 [DOI] [PubMed] [Google Scholar]
- 31.Gong F., Fahy D., Liu H., Wang W., Smerdon M. J. (2008) Cell Cycle 7, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson C. L., Côté J. (2004) Genes Dev. 18, 602–616 [DOI] [PubMed] [Google Scholar]
- 33.Beard B. C., Stevenson J. J., Wilson S. H., Smerdon M. J. (2005) DNA Repair 4, 203–209 [DOI] [PubMed] [Google Scholar]
- 34.Beard B. C., Wilson S. H., Smerdon M. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7465–7470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huggins C. F., Chafin D. R., Aoyagi S., Henricksen L. A., Bambara R. A., Hayes J. J. (2002) Mol. Cell 10, 1201–1211 [DOI] [PubMed] [Google Scholar]
- 36.Chafin D. R., Vitolo J. M., Henricksen L. A., Bambara R. A., Hayes J. J. (2000) EMBO J. 19, 5492–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menoni H., Gasparutto D., Hamiche A., Cadet J., Dimitrov S., Bouvet P., Angelov D. (2007) Mol. Cell. Biol. 27, 5949–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasad A., Wallace S. S., Pederson D. S. (2007) Mol. Cell. Biol. 27, 8442–8453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishibashi T., So K., Cupples C. G., Ausió J. (2008) Mol. Cell. Biol. 28, 4734–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilsen H., Lindahl T., Verreault A. (2002) EMBO J. 21, 5943–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayes J. J., Lee K. M. (1997) Methods 12, 2–9 [DOI] [PubMed] [Google Scholar]
- 42.Thiriet C., Hayes J. J. (1998) J. Biol. Chem. 273, 21352–21358 [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J., Fritsch O., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 44.Hayes J. J. (1996) Biochemistry 35, 11931–11937 [DOI] [PubMed] [Google Scholar]
- 45.Hayes J. J., Tullius T. D., Wolffe A. P. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 7405–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aoyagi S., Hayes J. J. (2002) Mol. Cell. Biol. 22, 7484–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayes J. J., Wolffe A. P. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 6415–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark D. J., Thomas J. O. (1986) J. Mol. Biol. 187, 569–580 [DOI] [PubMed] [Google Scholar]
- 49.Crane-Robinson C. (1997) Trends Biochem. Sci. 22, 75–77 [DOI] [PubMed] [Google Scholar]
- 50.Brown D. T., Izard T., Misteli T. (2006) Nat. Struct. Mol. Biol. 13, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan L., Roberts V. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8384–8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polach K. J., Widom J. (1995) J. Mol. Biol. 254, 130–149 [DOI] [PubMed] [Google Scholar]
- 53.Anderson J. D., Thåström A., Widom J. (2002) Mol. Cell. Biol. 22, 7147–7157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schalch T., Duda S., Sargent D. F., Richmond T. J. (2005) Nature 436, 138–141 [DOI] [PubMed] [Google Scholar]
- 55.Godde J. S., Wolffe A. P. (1995) J. Biol. Chem. 270, 27399–27402 [DOI] [PubMed] [Google Scholar]
- 56.Claudet C., Angelov D., Bouvet P., Dimitrov S., Bednar J. (2005) J. Biol. Chem. 280, 19958–19965 [DOI] [PubMed] [Google Scholar]
- 57.Yang Z., Zheng C., Hayes J. J. (2007) J. Biol. Chem. 282, 7930–7938 [DOI] [PubMed] [Google Scholar]
- 58.Parikh S. S., Putnam C. D., Tainer J. A. (2000) Mutat. Res. 460, 183–199 [DOI] [PubMed] [Google Scholar]
- 59.Otterlei M., Haug T., Nagelhus T. A., Slupphaug G., Lindmo T., Krokan H. E. (1998) Nucleic Acids Res. 26, 4611–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao G., Tordova M., Jagadeesh J., Drohat A. C., Stivers J. T., Gilliland G. L. (1999) Proteins 35, 13–24 [PubMed] [Google Scholar]
- 61.Angelov D., Vitolo J. M., Mutskov V., Dimitrov S., Hayes J. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6599–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakanishi S., Prasad R., Wilson S. H., Smerdon M. (2007) Nucleic Acids Res. 35, 4313–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clark D. J., Kimura T. (1990) J. Mol. Biol. 211, 883–896 [DOI] [PubMed] [Google Scholar]
- 64.Carruthers L. M., Bednar J., Woodcock C. L., Hansen J. C. (1998) Biochemistry 37, 14776–14787 [DOI] [PubMed] [Google Scholar]
- 65.Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. (1986) J. Mol. Biol. 187, 591–601 [DOI] [PubMed] [Google Scholar]
- 66.Furrer P., Bednar J., Dubochet J., Hamiche A., Prunell A. (1995) J. Struct. Biol. 114, 177–183 [DOI] [PubMed] [Google Scholar]
- 67.Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. (1980) Nature 288, 675–679 [DOI] [PubMed] [Google Scholar]
- 68.Simpson R. T. (1978) Biochemistry 17, 5524–5531 [DOI] [PubMed] [Google Scholar]
- 69.Smerdon M. J., Conconi A. (1999) Prog. Nucleic Acid Res. Mol. Biol. 62, 227–255 [DOI] [PubMed] [Google Scholar]
- 70.Otterlei M., Kavli B., Standal R., Skjelbred C., Bharati S., Krokan H. E. (2000) EMBO J. 19, 5542–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu M., Schatz D. G. (2009) Trends Immunol. 30, 173–181 [DOI] [PubMed] [Google Scholar]
- 72.Shen H. M., Poirier M. G., Allen M. J., North J., Lal R., Widom J., Storb U. (2009) J. Exp. Med. 206, 1057–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.