Abstract
p53 is a pleiotropic transcription factor driving a flexible transcriptional program that mediates disparate cellular responses to stress, including cell cycle arrest and apoptosis. The mechanisms by which p53 differentially regulates its diverse target genes remain poorly understood. In this issue of Genes & Development, Morachis and colleagues (pp. 135–147) demonstrate the critical role of core promoter elements at p53 target loci, in that they dictate differential RNA polymerase II recruitment and activity in a p53-autonomous fashion.
Keywords: p53, RNA polymerase II, transcription, core promoters, p21, Fas/APO1, NF-Y
Selective p53 target gene expression: many paths to diversity
p53 acts as a signaling node within a vast gene network that suppresses cancer development. In response to potentially oncogenic signals such as DNA damage or oncogene hyperactivation, p53 participates in diverse anti-tumoral cellular responses including cell cycle arrest, senescence, and apoptosis (Vousden and Prives 2009). The mechanisms defining which specific cellular response is adopted upon p53 activation are not fully understood. This ignorance hampers the development of therapies that could harness the apoptotic potential of p53 for the selective elimination of cancer cells. Accordingly, much effort over the past decade has been focused on delineating the mechanisms underlying context-dependent gene expression upon p53 activation. This research has yielded many potential models, but none seem to hold a universal truth.
At one end of the spectrum, p53-centric models postulate that differential cell fate choice results from modulation of the many activities of the p53 molecule itself. This regulation could be achieved by p53-binding proteins or p53 post-translational modifications capable of altering the transcriptional activity of p53 in a gene-specific manner. For example, the p53-interacting protein HZF (hematopoietic zinc finger) has been shown to stimulate p53 binding to the response elements found on cell cycle arrest genes while preventing binding to apoptotic genes (Das et al. 2007). In contrast, the ASPP (apoptosis-stimulating protein of p53) proteins promote p53 binding to certain apoptotic genes (Slee and Lu 2003). Similarly, p53 phosphorylation on Ser46 or acetylation on Lys120 was shown to favor transactivation of specific apoptotic genes (Oda et al. 2000; Sykes et al. 2006; Tang et al. 2006). At the other end of the spectrum, p53-autonomous models postulate that p53 transactivation potential is fairly invariant, and that the impact of p53 activation on the cellular transcriptome is determined by regulatory events acting independently of p53 modifications, p53 binding to DNA, or p53-interacting proteins. For example, hCAS (human cellular apoptosis susceptibility protein) associates with select p53 target genes independently of p53 to promote the apoptotic response (Tanaka et al. 2007). Mechanistically, hCAS seems to be required to generate a permissive chromatin landscape at specific p53 target loci by a process involving histone lysine demethylation. Research into the mechanisms driving stress-specific expression of p21 (CDKN1A), a key mediator of p53-dependent cell cycle arrest, has revealed a wealth of regulatory events modulating the transcriptional activity of this locus at steps subsequent to p53 binding and without any correlation to p53 post-translational modification status (Espinosa et al. 2003; Gomes et al. 2006; Donner et al. 2007; Mattia et al. 2007; Beckerman et al. 2009). Similarly, cell-type-specific expression of the p53 target genes p21, 14–3–3σ, and miR-34a was found to be defined by unequal p21 mRNA turnover, 14–3–3σ promoter DNA methylation, and processing of the miR-34a pri-mRNA among different cell types (Paris et al. 2008).
It is within this framework that the report by Morachis et al. (2010) illuminates a new mode of gene-specific regulation within the p53 network. Their work clearly demonstrates that the core promoter architecture of p53 target genes plays a major role in regulating their expression, and indicates that hard-wired, gene-specific p53-autonomous mechanisms may be more prevalent than previously appreciated.
Not all p53 target gene promoters are created (evolved) equally
A great deal of attention has been devoted to the fact that p53 binds to DNA sites that are highly variable in sequence, affinity, and topology. It is possible that the flexible nature of the p53 response element (p53RE) creates a reservoir of regulatory diversity, but this putative code has not been deciphered yet (Resnick-Silverman et al. 1998; Horvath et al. 2007). An equally important but underappreciated collection of DNA features capable of delivering gene-specific regulation within the p53 network are the core promoter elements (CPEs) found at its target loci. The core promoter of a gene is defined as the minimal DNA sequence required for accurate transcription initiation by RNA polymerase II (RNAPII), and is composed of a combinatorial arrangement of CPEs such as the TATA box, the TFIIB response element (TBRE), the downstream core promoter element (DPE), GC-rich islands, the initiator motif (Inr), or the motif ten element (MTE) (Juven-Gershon et al. 2008). These sequences mediate recruitment of general transcription factors (GTFs) to core promoters, the first step in the assembly of preinitiation complexes (PICs). Importantly, although PIC formation is a universal prerequisite for gene activation, different genes carry distinct combinations of CPEs. For example, the TATA box is bound by TBP (TATA-binding protein), a component of the GTF TFIID, but TATA boxes are present at only a small fraction of promoters. In the absence of a TATA box, TFIID can still be recruited via the interaction of its other subunits with Inr or DPE elements (for review, see Smale and Kadonaga 2003). It is not fully understood how different core promoter architectures may affect the rate of PIC assembly or PIC stability, as well as other steps of the transcription cycle such as promoter escape and early elongation. Interestingly, work from the Kadonaga laboratory (Butler and Kadonaga 2001) has demonstrated clearly that variations in CPEs alter the response of a given promoter to distal enhancers, suggesting that CPEs constitute a key variable within the regulatory circuit of a gene and should not be considered as mere scaffolding elements. The p53 transcriptional network is a good example of a collection of genes that share a common transcriptional activator but harbor highly diverse core promoters. A brief analysis of a few common CPEs at canonical p53 target genes demonstrates a wide range of architectures (Fig. 1). Does this CPE diversity contribute to the highly pleiotropic nature of the p53 transcriptional program?
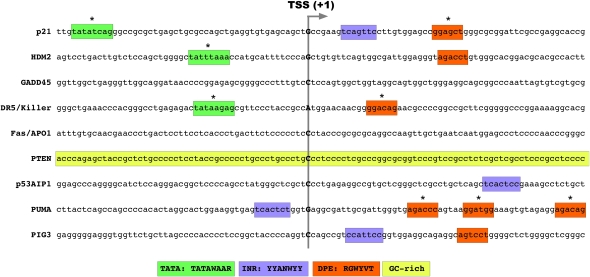
Figure 1.
Diverse core promoter architectures among p53 target genes. A search for TATA boxes, Inr elements, and DPEs reveals multiple CPE arrangements at p53 target loci. Sequences were aligned according to the first nucleotide (+1) of the most common EST for each gene, as found in GenBank. Notice that these putative transcription start sites (TSSs) rarely colocalize with the identified Inr motifs, which supposedly carry the +1 as defined by in vitro transcription assays. Likewise, the location of the TATA boxes and DPEs marked here do not always match with the consensus positions (−30 for the first nucleotide of the TATA box and +28 for the first nucleotide of the DPE). The motifs marked with asterisks are imperfect matches to the consensus. Even when employing these loose criteria, our search did not identify obvious CPEs at the GADD45a and FAS promoters. Beyond the study by Morachis et al. (2010) analyzing the core promoters of p21 and FAS, there is little information about which, if any, of these putative CPEs is involved in the regulation of their corresponding p53 target genes.
Differential kinetics of p53 target gene expression are driven by CPEs
Early microarray expression studies demonstrated that subsets of p53 target genes are activated within different time frames, regardless of stress type, with a general trend of cell cycle arrest genes displaying early induction in response to p53 activation, while proapoptotic genes show relatively more delayed expression (Zhao et al. 2000). One plausible explanation is that cell cycle arrest genes tend to carry high-affinity p53REs as compared with apoptotic genes, which may enable the former to respond to lower levels of nuclear p53. However, this correlation is not absolute, with some proapoptotic p53 target genes harboring relatively high-affinity binding sites (Szak et al. 2001). Furthermore, a cause–effect relationship between binding site affinity and kinetics of activation in vivo has not been formally established. A different hypothesis is borne out by the observations that cell cycle arrest genes constitutively harbor high levels of RNAPII at their core promoters, while proapoptotic genes do not (Espinosa et al. 2003). These observations suggest that cell cycle arrest genes may be intrinsically set up to be the rapid first responders to cellular stress, while proapoptotic genes wait in reserve. Differential RNAPII preloading has been associated with the timing of activation in other transcriptional networks (Hargreaves et al. 2009), but, once again, a formal proof that RNAPII preloading is responsible for differential expression kinetics is missing.
To investigate these issues more deeply, Morachis et al. (2010) employed in vitro transcription reactions in a side-by-side comparison of two functionally distinct p53 target promoters. They used short DNA fragments carrying the core promoters of the cell cycle arrest gene p21 and the proapoptotic gene FAS. Of note, these DNA fragments were devoid of p53-binding sites, and their transcription reactions were carried out with nuclear extracts lacking active p53. Their selection was based on previous in vivo experiments demonstrating that the p21 promoter carries large amounts of preloaded RNAPII, whereas the FAS promoter does not (Espinosa et al. 2003). In elegant time-course experiments, Morachis et al. (2010) monitored both the velocity and persistence of PIC assembly at each core promoter. Akin to a 100-m Olympic sprinter, the p21 core promoter undergoes rapid PIC assembly and transcription initiation, but it reinitiates very poorly, with rapid decay in transcriptional competence being apparent. On the other hand, the FAS promoter behaves more like a marathon runner, as it undergoes slow but sustained PIC formation, and, once it becomes transcriptionally engaged, it delivers multiple rounds of efficient reinitiation. One additional apoptotic p53 target gene, APAF1, showed a similar behavior to FAS, with slow PIC formation but enhanced reinitiation capacity.
As mentioned above, the differential regulation of p53 target genes observed in vivo could be explained by a myriad of factors, including p53 regulatory partners, p53REs, the chromatin context, and other variables. It is therefore an interesting surprise that some key aspects of this gene-specific regulation can be recapitulated in the minimal setting of an in vitro transcription assay using “naked” DNA templates. These results indicate that the CPEs within p53 target genes have evolved to regulate the speed of RNAPII engagement and the duration of its commitment. Cell cycle arrest genes engage quickly but commit poorly, while apoptotic genes engage more slowly but show sustained commitment. In the biological context, this would allow cells to mount an early but reversible prosurvival response, eventually followed by an irreversible lethal response.
Nuclear factor Y (NF-Y) is a gene-specific regulator of p53 target core promoters
Through extensive dissection of the cis-elements present at the p21 and FAS core promoters, Morachis et al. (2010) identified the key sequences responsible for these different behaviors. Perhaps not too surprisingly, the TATA box in the p21 promoter was found to be responsible for rapid PIC assembly. Furthermore, experiments using chimeric reporters showed that the TATA box could confer rapid PIC assembly when inserted into the otherwise TATA-less FAS promoter. Nicely, the artificial TATA-FAS construct displayed both rapid PIC assembly and sustained reinitiation, demonstrating that these core promoter properties are not mutually exclusive. Interestingly, analysis of the FAS promoter identified a novel sequence downstream from the transcription start site that was required for its transcriptional properties. Using this DNA sequence to purify interacting factors from nuclear extracts, Morachis et al. (2010) identified the NF-Y as a critical regulator of FAS expression. NF-Y is a heterotrimeric complex comprised of three subunits—A, B, and C—that has been reported previously to participate in regulation of other p53 target genes (Imbriano et al. 2005; Di Agostino et al. 2006; Peart and Prives 2006; Benatti et al. 2008). Of note, chimeric promoter constructs revealed a dual behavior for NF-Y. While it is a positive regulator of FAS transcription, insertion of the NF-Y-binding site into the p21 core promoter represses its activity and it cannot synergize with or replace the natural TATA box. This experiment reveals the topological constraints within core promoter architectures, where the precise combination and spatial ordering of CPEs are critical. Furthermore, Morachis et al. (2010) found that NF-Y binds exclusively to the FAS promoter in cells, and that NF-Y overexpression specifically increases FAS transcription.
Future perspectives
In sum, the data presented by Morachis et al. (2010) demonstrate a previously underappreciated role of core promoter architecture in the flexible character of the p53 transcriptional program. Their results should lead us to recognize that the intrinsic diversity of CPEs among p53 target promoters must and does play a significant role in dictating how and when these genes are expressed. Future studies will be required to integrate CPE-mediated effects with other regulatory events within the p53 network, and to define the overall impact of core promoter architecture on cell fate choice upon cellular stress. Ideally, this would be achieved by a combination of biochemical assays, as those developed by Morachis et al. (2010), and in vivo assays where the impact of CPEs could be tested in the natural context of the cell nucleus.
Acknowledgments
Work in the Espinosa laboratory is supported by grants from the NIH (CA117907) and NSF (MCB-0842974). J.M.E. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1893610.
References
- Beckerman R, Donner AJ, Mattia M, Peart MJ, Manley JL, Espinosa JM, Prives C. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes & Dev. 2009;23:1364–1377. doi: 10.1101/gad.1795709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatti P, Basile V, Merico D, Fantoni LI, Tagliafico E, Imbriano C. A balance between NF-Y and p53 governs the pro- and anti-apoptotic transcriptional response. Nucleic Acids Res. 2008;36:1415–1428. doi: 10.1093/nar/gkm1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes & Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes & Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: Cell cycle versus apoptosis. PLoS Genet. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R. Direct p53 transcriptional repression: In vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol. 2005;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter—The gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattia M, Gottifredi V, McKinney K, Prives C. p53-dependent p21 mRNA elongation is impaired when DNA replication is stalled. Mol Cell Biol. 2007;27:1309–1320. doi: 10.1128/MCB.01520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morachis JM, Murawsky CM, Emerson BM. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes & Dev. 2010 doi: 10.1101/gad.1856710. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- Paris R, Henry RE, Stephens SJ, McBryde M, Espinosa JM. Multiple p53-independent gene silencing mechanisms define the cellular response to p53 activation. Cell Cycle. 2008;7:2427–2433. doi: 10.4161/cc.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart MJ, Prives C. Mutant p53 gain of function: The NF-Y connection. Cancer Cell. 2006;10:173–174. doi: 10.1016/j.ccr.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Resnick-Silverman L, St Clair S, Maurer M, Zhao K, Manfredi JJ. Identification of a novel class of genomic DNA-binding sites suggests a mechanism for selectivity in target gene activation by the tumor suppressor protein p53. Genes & Dev. 1998;12:2102–2107. doi: 10.1101/gad.12.14.2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee EA, Lu X. The ASPP family: Deciding between life and death after DNA damage. Toxicol Lett. 2003;139:81–87. doi: 10.1016/s0378-4274(02)00421-6. [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak ST, Mays D, Pietenpol JA. Kinetics of p53 binding to promoter sites in vivo. Mol Cell Biol. 2001;21:3375–3386. doi: 10.1128/MCB.21.10.3375-3386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes & Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]