Abstract
A recent explosion of work surrounds the interactions between Sir3p (Silent Information Regulator 3) and chromatin. We review here the Sir3p functions related to its role in silencing in Saccharomyces cerevisiae. This unusual protein, which is absolutely required for silencing, is distantly related to the highly conserved replication initiator Orc1p, but is itself phylogenetically limited to “post-genome-duplicated” budding yeasts. Several recent studies revise earlier models for Sir3p action. Specifically, the N-terminal bromo-adjacent homology (BAH) domain plays a now well-defined role in silencing, and a picture is emerging in which both termini of Sir3p bind two locations on the nucleosome: (1) the loss of ribosomal DNA silencing (LRS) surface in the nucleosome core, and (2) the N-terminal histone tails for effective silencing at telomeres. We relate Sir3p structure and function, and summarize recent molecular studies of Sir3p/chromatin binding, Sir3p/Dot1p competition, and the possible role of O-Acetyl ADP ribose (O-AADPR) in Sir3p/chromatin binding. We emphasize recent genetic data that provide important new insights and settle controversies created by in vitro work. Finally, we synthesize these ideas to revise the model for how Sir3p mediates silent chromatin formation in yeast, in part through its affinity for the LRS region of the nucleosome, which must be “just right.”
Keywords: SIR3, silencing, chromatin, telomere, SIR2, SIR4
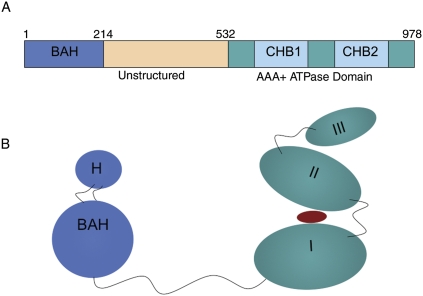
Classical silencing is typified by silent chromatin in Saccharomyces cerevisiae. Its heterochromatin-like structure occurs in three locus types: (1) silent mating-type cassettes HML and HMR, (2) telomeres, and (3) ribosomal DNA (rDNA) repeats (for review, see Rusche et al. 2003). All three share requirements for post-translational modification of the histone tails and core, as well as a Sir2p (Silent Information Regulator 2)/partner proteins “silencing complex.” Most mechanistic data on silent chromatin formation come from studies on telomeric and HM locus silencing. The Sir complex at these loci consists of Sir1, Sir2, Sir3, and Sir4p, with Sir1p playing a significant role in the establishment of HM locus silencing, variable roles at native telomeres, and only an indirect role at artificial truncated telomeres often used to study silencing. Sir2p is a histone deacetylase, while Sir1, Sir3, and Sir4p are thought to play more “structural” roles. It is interesting to note in this context that the Sir3p protein contains an AAA ATPase (“ATPases associated with a variety of cellular activities” domain) motif (see Fig. 1).
Figure 1.
Domain structure of Sir3. (A) The primary domain structure of Sir3p with important domains highlighted in different colors. The first 214 amino acids comprise the BAH domain in blue; the region from amino acids 214 to 532 (tan) is largely unstructured (McBryant et al. 2006); and from amino acids 532 to 978 (green), Sir3p is similar to the CDC6 subfamily of the AAA ATPase domain. CHB1 and CHB2 (blue) indicate the C-terminal histone-binding (CHB) activity associated with this portion of Sir3p. The BAH domain also contains histone-binding activity. (B) A cartoon version of the tertiary structure of Sir3 based on the crystal structures of the BAH domain 2FL7 (Hou et al. 2006) and the Archaeal cdc6 protein ortholog 1FNN (chain A) (Liu et al. 2000). The BAH domain is colored in blue, and the C-terminal domain is colored in teal. The red oval placed between domains I and II of the C-terminal region of Sir3 indicates a possible nucleotide moiety such as O-AADPR that could bind to Sir3p.
In the current model for telomeric silencing, Rap1 and Ku proteins bind directly to telomere chromatin, followed by recruitment of the Sir2/3/4p complex. Iterative cycles of Sir2p-mediated deacetylation of histone tail lysines, specifically at H4 K16, create high-affinity sites for the Sir2/3/4p complex in adjacent nucleosomes, allowing spreading of silent chromatin (Hoppe et al. 2002; Luo et al. 2002; Rusche et al. 2002; Tanny et al. 2004; Rudner et al. 2005). Encroachment of silent chromatin from silent loci into neighboring euchromatin is prevented by redundant competing mechanisms: anti-silencers or “boundary elements,” the inherent instability of the silencing complex antagonizing the silencing-promoting role of Sir2/3/4p, and other cis- and trans-acting factors (Lynch and Rusche 2009).
Sir3p
A most striking feature of Sir3p is its ability to condense chromatin even when the Sir2/4 complex is depleted, suggesting that some intrinsic properties of Sir3p allow it to bind to and condense nucleosomes (Hecht et al. 1996; Strahl-Bolsinger et al. 1997). Sir3p structure enables its role in silent chromatin. It has three distinct structural domains (Fig. 1): the N-terminal bromo-adjacent homology (BAH) domain, followed by a 300-residue stretch of intrinsically disordered amino acid residues, and then a C-terminal AAA ATPase that is highly conserved among diverse protein types (Callebaut et al. 1999; Duina and Winston 2004; Connelly et al. 2006; Hou et al. 2006; McBryant et al. 2006).
BAH domain
In Sir3p, the BAH domain consists minimally of amino acids 1–214 (see Figs. 1, 2; Callebaut et al. 1999; Connelly et al. 2006; Hou et al. 2006; Sampath et al. 2009). The BAH domain's role in silencing has been underappreciated until recently for at least two reasons: Both recombinant protein production in Escherichia coli and N-terminal tags disrupt the N-terminal acetylation critical for both binding chromatin and Sir3p stability (Wang et al. 2004; Onishi et al. 2007; van Welsem et al. 2008). Only once Sir3p was expressed with a C-terminal tag was its autonomous ability to bind nucleosomes recognized.
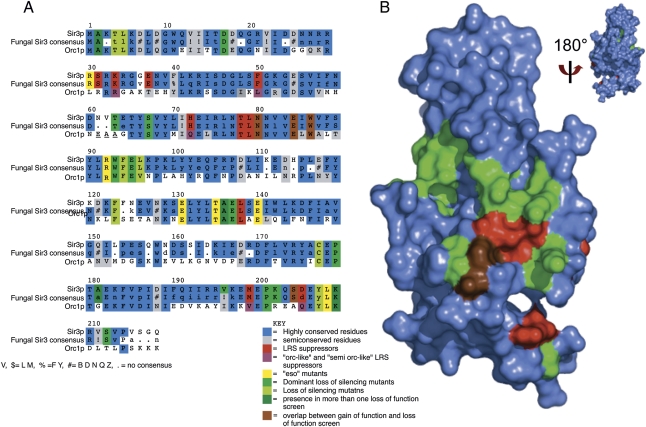
Figure 2.
Critical binding surface of the BAH domain revealed by multiple genetic screens. (A) Multiple sequence alignment of SIR3 BAH domain from S. cerevisiae, consensus sequence for SIR3 BAH domain from post-genome duplication yeast strains, and BAH domain from ORC1. The side chains identified in the eso, DNG, and lfm screens are highlighted in yellow-green, medium green, and light green, respectively; side chains from the slr screen are highlighted in red; residues overlapping between the slr screens and other loss-of-function screens are highlighted in brown; residues overlapping among the loss-of-function screens DNG, eso, and lfm are highlighted in dark green. Mutations in the slr screen that introduce an “Orc-like” residue are highlighted in purple. Conserved residues are highlighted in blue, and semiconserved residues are highlighted in gray. (B) Surface representation of the crystal structure 2FL7 (Hou et al. 2006). The color coding is similar to A, but simplified; the DNG, eso, and lfm screens are highlighted in green; the slr screen is highlighted in red; and the overlap between the two groups is highlighted in brown. The structure is rotated 180° around the X-axis to demonstrate that all of the side chains identified by the various screens are found on one face of the BAH domain.
The first clue that this domain regulated Sir3p function came from suppressor studies of the H4 K16Q nonmating phenotype. In the presence of sir3-D205N or sir3-W86R, the H4 K16Q yeast strain regained mating ability (Johnson et al. 1990). Interestingly enough, this same BAH domain mutation, sir3-D205N, subsequently appeared in several screens, including suppression of a rap1-s allele (missing the Rap1p C-terminal Sir3p interaction domain) and mutations in the nucleosome core (Thompson et al. 2003; Norris et al. 2008). Why would one mutation suppress so many loss-of-silencing mutations? As we suggest below, getting the Sir3p BAH domain to the nucleosome is not only the crux of Sir3p–nucleosome interactions, but it is also necessary that this interaction is “just right.” [An explanation of “Goldilocks” for nonnative English speakers. In the modern version of the fairy tale “Goldilocks and the Three Bears,” mutated somewhat from the original by Southey (1834), the fair maiden Goldilocks visits the house of the bears and tries out their three bowls of porridge (hot, medium, and cool), their three chairs (large, medium, and small), etc., and concludes in each case that the one in the middle is “just right” for her.]
C-terminal domain
Long thought to be its “business end,” the Sir3p C terminus dominates its known cellular roles (Fig. 1), including Rap1p, histone tail, and self-interactions (Johnson et al. 1990; Cockell et al. 1995; Hecht et al. 1995; Liu and Lustig 1996; Park et al. 1998; Luo et al. 2002). C-terminal residues 623–762 and 799–910 can bind H3 and H4 tails in vitro (Hecht et al. 1995). Sir3p's anti-parallel dimerization activity maps to residues 464–728 and 832–978 (King et al. 2006). The 464–728 region is also the minimal domain for Sir4 interaction (Chang et al. 2003; King et al. 2006). A crystal structure of the complex of an Archaeal ORC1 homolog with DNA demonstrates that this domain can bind ATP/ADP and DNA nonspecifically (Gaudier et al. 2007), suggesting that the C terminus of Sir3p may bind both DNA and proteins. Based on combined structure and sequence alignment data (Liu et al. 2000), the Sir3p C terminus can be divided into three domains typical of this family: Domain I consists of Sir3p residues ∼532–726, domain II consists of residues from 727–831, and domain III consists of residues from 832–978 (see Fig. 1; Liaw and Lustig 2006; A Norris and JD Boeke, unpubl.). In the AAA+ family, the ATP-binding pocket lies between domains I and II, which contain several conserved motifs important for nucleotide binding. Sir3p is enigmatic because its AAA+ domain lacks many residues important for ADP/ATP binding, but secondary structure algorithms predict a similar overall topology to the AAA+ family (Neuwald et al. 1999; Iyer et al. 2004; Gaudier et al. 2007). Domain III is the most variable among this class of proteins, and is probably important for auxiliary factor binding (Neuwald et al. 1999; Iyer et al. 2004; Gaudier et al. 2007). Because SIR3 is a duplicated gene, it is free to evolve faster and “neofunctionalize” (Ohno 1970) than a gene required for viability, like Orc1p. The lack of conservation of the Sir3p C-terminal ATP-binding pocket could suggest that Sir3p binds a related nucleotide such as O-acetyl ADP ribose (O-AADPR) for activity (see below). Tethering studies employing Sir3-LexA fusions, recognizing binding sites adjacent to a telomeric URA3 reporter, have mapped the functional regions of Sir3p required post-recruitment to the last 144 amino acids (Lustig et al. 1996; Park et al. 1998; Liaw and Lustig 2006).
Interaction with chromatin
Initial in vitro experiments mapped the necessary and sufficient Sir3p–nucleosome interaction domains to the C-terminal residue blocks 623–762 and 799–910 of Sir3p and the histone tails of H3 and H4 (Hecht et al. 1995). Subsequent studies demonstrated that the nucleosome core was also important, as was the N terminus of Sir3p for nucleosome–Sir3p interaction. Surface plasmon resonance (SPR) experiments show that Sir3p can bind the H3–H4 tetramer in vitro, as can the BAH domain alone, albeit with lower affinity, demonstrating that Sir3p can bind to nucleosomes independently of DNA (Onishi et al. 2007). Two recent elegant studies that reconstitute chromatin in vitro demonstrate the importance of both H4 K16 (tail) and H3 K79 (core) for Sir3 binding to nucleosome arrays (Johnson et al. 2009; Martino et al. 2009). Martino et al. (2009) purified Sir2, Sir3, and Sir4p from Sf9 cells and recombinant tailless Xenopus trinucleosome arrays and demonstrated that Sir3p could bind independently of histone tails, albeit with diminished affinity. The same study showed that Sir2,3,4p also bound to naked DNA, an activity attributed to Sir4 (Martino et al. 2009). Johnson et al. (2009) used Sir2, Sir3, and Sir4p from yeast extracts with recombinant nucleosomes and the HMR locus as a DNA template, creating a longer nucleosomal array. This study demonstrated that Sir3p can bind to nucleosome arrays without the Sir2/4 complex, but does so with much less affinity. Additionally, the Sir3p nucleosome interaction is disrupted by acetylation or mutation of H4 K16, mutation of H3 K79, or introduction of Htz1. All of these aberrations have much less effect on Sir3p binding to nucleosome arrays if Sir2/4 is added, at which point Sir2/3/4 binds irrespective of the status of H4 K16, while H3 K79A of Htz1 only slightly reduces Sir3p nucleosome binding (Johnson et al. 2009). The interaction of Sir3p with modified nucleosomal arrays depends on the coiled-coil domain of Sir4 that binds Sir3, suggesting that Sir2/4 bridges Sir3 to modified chromatin (Johnson et al. 2009). Collectively, these results show that Sir3p binds both tail and nontail regions of the nucleosome in vitro, and that the Sir2/4 complex not only increases Sir3p's affinity for the nucleosome, but also makes it less sensitive to H4 K16 modification. This difference in affinity most likely reflects the initial recruitment and stable binding steps in silent chromatin assembly, and suggests that it may be Sir3p sensing the acetylation status of H4 K16 and hence responsible for preventing further spread of the silencing complex.
Importantly, studies in yeast cell extracts demonstrated that full-length Sir3p or ectopically expressed Sir3p BAH domain coimmunoprecipitate histone H3, and that this interaction is dependent on H3 residue 79 of the loss of rDNA silencing (LRS) surface and H4 tail residues 16, 17, and 19. Sir3p prefers hypomethylated H3 K79 and hypoacetylated H4 K16 for its interaction with the nucleosome, as indicated by an increase in H3 coimmunoprecipitations (co-IPs) with Sir3p in dot1Δ and sas2Δ cells (Onishi et al. 2007). These co-IP studies are corroborated by chromatin immunoprecipitation (ChIP) studies in which Sir3p does not chromatin-immunoprecipitate to telomeric DNA in an LRS mutant H3 A75V, or in tail mutants H4 K16Q or R17/19 A (Altaf et al. 2007; Fingerman et al. 2007; Norris et al. 2008). These studies suggest that the H4 tail and the LRS domain represent two surfaces important for Sir3p interaction with chromatin and, given that the H4 tail is flexible, they could represent a single contiguous surface stabilized in silent chromatin. Until recently, some controversy surrounded the exact region of Sir3p important for this interaction. Recombinant Sir3p C-terminal domain immunoprecipitates GST-tagged H4 peptides and an H3 peptide encompassing residues 68–83 (Hecht et al. 1995; Altaf et al. 2007). The interactions with the tail and the H3 residues surrounding H3 K79 are modification-dependent, confirming the in vivo importance of the modification status of these two regions (Altaf et al. 2007). However, an ectopically expressed BAH domain can more efficiently coimmunoprecipitate H3 from a yeast whole-cell extract than does full-length Sir3p, whereas Sir3p lacking the BAH domain fails to coimmunoprecipitate any H3, suggesting that the C terminus may inhibit the BAH domain to some extent (Onishi et al. 2007).
Genetics seals the deal
Whereas in vitro binding studies and whole-cell extract co-IPs can give a sense of the possible, genetics fills in the gaps in vivo. In the case of Sir3p, where in vitro interactions can be masked by requirements for nucleosome and Sir3p modification, recent genetic studies directly implicate the BAH domain as playing a critically important regulatory role in telomeric silencing. Four different screens isolated one surface of the SIR3 BAH domain as important for silencing in yeast in loss-of-function, dominant loss-of-function, and gain-of-function screens related to both telomeric and HM locus silencing. A summary of these screens is represented in Figure 2 and Supplemental Table 1. For the sake of clarity in this publication, we classified the various screens as summarized below. The screen done by Stone et al. (2000) generated eso (for enhancer of sir one alleles). The screen done by Buchberger et al. (2008) produced DNG (dominant-negative alleles), the alleles isolated by Sampath et al. (2009) are named lfm (for loss of function missense), and the alleles isolated by Norris et al. (2008) are named slr (for supressors of LRS).
The eso alleles enhance the mating defect of a sir1Δ mutation. Normally, sir1Δ yeast mate at a very low but detectable level; the sir3 eso mutants can also mate, but abolish all mating in the sir1Δ strain (Stone et al. 2000). All mutations but one (S813A) localized to the BAH domain. The DNG screen necessarily isolated dominant loss of telomeric silencing SIR3 mutations, as it was performed in a SIR3+ background. The screen isolated mutants that were somewhat defective in binding to telomeric chromatin but much more defective in spreading (Buchberger et al. 2008). This screen isolated many mutations clustered on the BAH domain and, as in the eso screen, isolated a single distinct mutation in the C-terminal region (L738P) that was the weakest in terms of its effects on telomeric silencing. While these screens revealed that the BAH domain is crucial for silencing, the slr screen as well as mutagenesis by Sampath et al. (2009) revealed a genetic link between the BAH domain and the LRS region of the nucleosome. In the slr screen, SIR3 alleles D205N and L79I, along with histone alleles H3 D77N and H4 H75Y, were isolated as LRS suppressors. The same histone alleles as well as SIR3 D205N were identified as suppressors of the sir3 A2T loss-of-silencing mutation (Norris et al. 2008; Sampath et al. 2009). Additionally, the subset of the lfm and DNG alleles failed to coimmunoprecipitate histone H3 from whole-cell extract or to chromatin-immunoprecipitate telomeric DNA, demonstrating that BAH is crucial for chromatin interaction. It is interesting that these loss-of-function alleles were dominant; presumably, the dominance reflects essential Sir3p multimerization, with loss of function leading to a poisoned subunit. The slr screen differed in that it isolated alleles of the BAH domain that suppressed histone LRS alleles’ loss of telomeric silencing. Indeed, these mutations suppressed multiple LRS histone alleles, but not other nucleosome mutations that interfered with telomeric silencing. Informatively, the slr alleles resulted in paradoxically decreased telomeric silencing in the presence of wild-type histones, despite a significant increase in ChIP signal to telomeric DNA compared with wild-type Sir3p (Norris et al. 2008). Similarly, H3 K56Q mutations disrupt telomeric silencing but do not decrease the ability of the SIRs to chromatin-immunoprecipitate to telomeric DNA (Xu et al. 2007). These results suggest that SIR binding is a prerequisite for silencing but is not sufficient. The question remains: Why would “wild-type” histones lose silencing in the presence of SIR3 suppressor alleles D205N and L79I, but still chromatin-immunoprecipitate ample Sir3p to telomeric DNA? This seeming conundrum was recapitulated by studies with Orc1p–Sir3p hybrids. Whereas an ectopically expressed Orc1p BAH domain coimmunoprecipitates more histone H3 than Sir3p BAH, Orc1p BAH does not substitute well for Sir3p BAH in silencing (Stone et al. 2000; Onishi et al. 2007). Hence, we propose that Sir3p follows the “Goldilocks principle” (Southey 1834): Just as in the case of the Orc1p BAH domain, when Sir3p binds too strongly to chromatin, it is counterproductive for telomeric silencing (Norris et al. 2008). Sir3p may need to either bind and release telomeric DNA for proper silencing or, alternatively, bind in more than one conformation, one of which involves the interaction of both BAH and LRS surfaces. Following the presumed gene duplication event that gave rise to SIR3, and during its “neofunctionalization” (Ohno 1970), Sir3p's BAH may have evolved a lower affinity for the LRS surface, consistent with the fact that Sir3p seems to have at least one additional distinct chromatin-binding domain (Hecht et al. 1995; Altaf et al. 2007; Onishi et al. 2007; Buchberger et al. 2008). Both the DNG and eso screens isolated single mutations in the C terminus of Sir3p, L738P, and S813A, respectively. Using GST-tagged H4 peptides, the L738P mutation interacted with H4 peptides more strongly than wild type, but, unlike all of the other DNG mutations, L738P was not defective in co-IP of H3, suggesting strong binding of the C terminus to H4 tails could also be deleterious to silencing (Buchberger et al. 2008). A picture emerges in which a balance needs to be struck between Sir3p BAH and C-terminal domains for binding to the nucleosome; this balance is crucial for effective silent chromatin formation.
Electrostatic repulsion dominates the interaction
The slr screen shows an electrostatic trend in the Sir3p BAH suppressors isolated, in that they tend to increase the positive charge of the BAH domain. Additionally, the DNG screen also isolated two positively charged lysines on the BAH surface that lead to loss of silencing. This trend is strikingly similar to a complementary electrostatic trend on the LRS surface of the nucleosome, in which mutations that increase positive charge tend to increase telomeric silencing, whereas mutations that decrease positive charge tend to lose silencing. The simplest explanation for these data is that the Sir3p BAH domain makes direct contact with the LRS nucleosomal domain, and electrostatic repulsion dominates this interaction. The nucleosome and Sir3p normally repel each other in wild-type yeast to a certain extent, and it is the slightly unstable nature of this interaction that determines the strength of silencing (Norris et al. 2008). Thus, these slr mutants decrease potential repulsion, presumably by increasing nucleosome-binding affinity. In contrast, charge-neutralizing substitutions in the two lysines K202 and K209 on α helix 8 of Sir3p that, by this electrostatic hypothesis, should make positive contributions to Sir3p nucleosome binding were isolated in both the DNG and lfm screens (Buchberger et al. 2008; Sampath et al. 2009). Furthermore, mutations on the LRS surface of the nucleosome that decrease the charge repulsion of D77 also suppress the loss of silencing of the SIR3 A2T or the LRS allele H3 A75V. Therefore, the crux of the interaction between Sir3p and the nucleosome is getting the BAH domain of Sir3p to the LRS surface of the nucleosome.
The collective interpretation of the phenotypes of these mutants is that (1) there is a functional surface on the BAH domain that interacts with the LRS surface of the nucleosome, and (2) the D205 residue makes an important repulsive contribution that limits nucleosome-binding affinity. In situations where Sir3p is able to bind all over the genome, as occurs in dot1Δ or sas2Δ cells, telomere silencing is lost; hence, it is important to (1) get Sir3p to the right place and (2) balance its affinity for the nucleosome. Like Goldilocks, the Sir3p BAH domain needs its affinity for the LRS surface of the nucleosome to be “just right.”
Furthermore, weakening BAH-binding affinity might facilitate a role requiring alternating binding and release of chromatin or allowing for conformational changes that might be required for silencing. The simplest explanation is that Sir3p requires a distinct “sweet spot” in between a stable LRS–BAH interaction and one that is sufficiently weak to allow the necessary dynamic structural changes.
Competition with Dot1p
Dot1p methylation in euchromatin helps keep silent chromatin from spreading and the Sir2,3,4p complex properly localized (van Leeuwen and Gottschling 2002; van Leeuwen et al. 2002). By ChIP analysis, both deletion and overexpression of DOT1 led to mislocalization of the Sir2,3,4p complex. Sir3p coimmunoprecipitates more H3 in both dot1 deletion and H4K16R strains in a Sir3p BAH domain-dependent manner (Onishi et al. 2007). However, the Sir3p C terminus inhibits Dot1p in vitro methylation reactions of H3 (Altaf et al. 2007; Fingerman et al. 2007), possibly by competing for the Dot1p-binding site. While Dot1p most certainly binds the LRS region, residues 17, 18, and 19 of the H4 tail are also important for the methylation reaction (Fingerman et al. 2007). The C terminus of Sir3p was known to interact in vitro with the H4 tail, and hence could be competing with that region for binding (Hecht et al. 1995). Considering that the H4 tail emerges from a nearby nucleosome surface and is flexible and short, it is entirely possible that it can form a contiguous surface with the LRS domain for binding of Sir3p, Dot1p, or both. A related study found that the Sir3p C terminus could bind a peptide consisting of residues 68–83 of H3, and this binding was methylation-sensitive (Altaf et al. 2007). It appears that both the BAH C-terminal domains have the potential to bind the LRS region and the H4 tail, and Dot1p competes with both domains of Sir3p for this surface. While Dot1p is insensitive to H3 K79 methylation status, Sir3p binds only to the unmethylated surface; this competition between Sir3p and Dot1p helps dictate whether chromatin is silenced or expressed.
O-AADPR
One very tantalizing idea is that Sir3p does not bind ADP or ATP, but has evolved to bind the related molecule O-AADPR, a specific product of the NAD+-dependent Sir2p-mediated deacetylation reaction (Tanner et al. 2000; Sauve et al. 2001). There is striking evidence both for and against this idea. When O-AADPR is added to purified Sir2, Sir3, Sir4, and nucleosomes, filaments visible in the electron microscope (EM) are formed (Liou et al. 2005; Onishi et al. 2007). The most striking evidence for O-AADPR binding is that Sir2,3,4 complexes or Sir3p alone purified from baculovirus-infected cell extract have an increased affinity for trinucleosomes, as measured by gel shift when O-AADPR is added but not when analogs such as AADPR, ADP, ATP, or nonhydrolyzable ATP analogs are added (Martino et al. 2009). The effect was dramatic with the Sir2,3,4p complex, but is also quite significant with Sir3p alone, suggesting that it is the key target (Martino et al. 2009). The evidence against this hypothesis is based on a Sir3p-Hos3 fusion protein used to demonstrate that telomeric silencing can occur in the absence of O-AADPR (Chou et al. 2008). Hos3 is an NAD+-independent deacetylase that does not produce O-AADPR, nor does it normally deacetylate histones. When fused in-frame to the C terminus of Sir3p, it deacetylates histones at silent loci as measured by ChIP, and this effectively silences DNA (Chou et al. 2008). However, while it does not silence as well as wild-type yeast, it does silence as well as a Sir3–Sir2p fusion protein (Chou et al. 2008). Taken together, these experiments suggest that O-AADPR may indeed increase the affinity of Sir3p in the Sir2,3,4p complex for chromatin and increase silencing, but the small molecule is not absolutely required for silencing. It would be interesting to know whether O-AADPR is required for the type of silencing that occurs when Sir3p is overexpressed, leading to regions of silent DNA depleted of Sir2p and Sir4p, but enriched for Sir3p. Additionally, a crystal structure of the C terminus would shed light on whether Sir3p really does have a similar structure as the AAA+ family, as has been predicted sequence alignments, and whether it can bind a nucleotide.
Model for Sir3p and telomeric silent chromatin formation
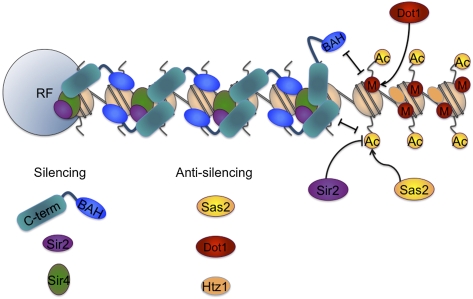
Silencing is a dynamic feature of telomeric DNA; there are silencing-promoting activities in the cell that are balanced with destabilizing activities that ultimately separate the silent regions from the expressed regions of the genome. The Sir3p BAH and C-terminal regions play critical roles in this balance (see Fig. 3). Establishment of silencing occurs through Rap1p and the Ku complex binding to nonnucleosomal telomeric repeats at the chromosome tips. Rap1p then recruits Sir3p and the Sir2/4p complex. The deacetylation activity of Sir2p then creates a high-affinity site for the SIR complex, allowing the SIR complex and silencing to spread. Several actions inhibit silencing: One is the inherent instability of the SIR complex, and the second is the competition between Sir2p and Sas2p for the acetylation state of histone H4 K16. The inherent instability can be exacerbated or counteracted by poorly defined cis-acting sequences, presumably Rap1-, Abf1-, and/or Orc1-binding sites; ARS sequences; and other sequences in the X core and Y′ domains of the telomeres (Boscheron et al. 1996; Fourel et al. 1999, 2002; Pryde and Louis 1999; Cheng and Gartenberg 2000; Mondoux and Zakian 2007). Acetylation of H4 K16 leads to deposition of histone variant HTZ1 and decreased affinity of the SIR complex for the nucleosomes, and especially for that of Sir3p and the nucleosome. This loss of affinity is reinforced by the competition between Dot1p and Sir3p for the methylation status of histone H3 K79, as well as forces that act genome-wide such as HTZ1 deposition and H3 K4 methylation (Venkatasubrahmanyam et al. 2007). Mutations such as H3 K79R may disrupt some of the Dot1p anti-silencing activity, but all of the other anti-silencing mechanisms are still in place, leading to only a minor defect in silencing. The Sir3p BAH domain binds the LRS surface of the nucleosome that encompasses H3 K79 and occludes its methylation by Dot1p; conversely, when H3 K79 is methylated, Sir3p binding is blocked. The C-terminal region may also play a secondary role in binding the LRS surface, but seems to be most important for the establishment of silencing. The C terminus has several important roles, especially in the initiation of silencing: It binds Rap1p for recruitment to the telomeric DNA; it recognizes the modification status of H4 K16; it binds Sir4p, which intensifies its affinity for chromatin; and, importantly for spreading, it contains the anti-parallel dimerization domain. If these prerequisites are not met, silencing is impaired. However, the crux of getting the right balance of silencing in the cell is getting the Sir3p BAH domain to the LRS surface of the nucleosome for which the affinity must be “just right”: Too tight and silencing is lost; too loose and silencing is also lost. Just how “too tight” is counterproductive for silencing in molecular terms remains to be determined. Two models have been put forth here: Sir3p may need to bind in more than one conformation or, alternatively, Sir3p may need to bind and release for proper chromatin formation. We favor the model where Sir3p binds in more than one conformation; this is based largely on the fact that Sir3p has multiple nucleosome interaction domains as well as its own dimerization domain. Additionally, Sir3p binds with a different affinity and stoichiometry, depending on whether it is binding a mono-, tri-, or multinucleosomal array. It will be interesting to find out more clearly how Sir3p mediates compaction of silent chromatin and what, if any, role a small molecule such as O-AADPR could play in that process.
Figure 3.
Competing actions for silencing followed by reinforcements. This figure is largely based on Altaf et al. (2007) and Buchberger et al. (2008). There are silencing-promoting actions such as Sir2p deacetylating the histone tails, which creates a high-affinity site for the SIR complex. Several actions inhibit silencing: One is the inherent instability of the SIR complex, and a second is the competition between Sir2p and Sas2p for the acetylation state of histone H4 K16. HTZ1 deposition also contributes to the destabilization of silencing (Venkatasubrahmanyam et al. 2007). Acetylation of H4 K16 leads to decreased affinity of the SIR complex for the nucleosomes and especially for that of Sir3 and the nucleosome; this loss of affinity is reinforced by the competition between Dot1p and Sir3p for the methylation status of histone H3 K79. Sir3p binds the LRS surface of the nucleosome, which encompasses H3 K79 and occludes its methylation by Dot1p; conversely, when H3 K79 is methylated, Sir3p binding is blocked. The orientation of Sir3p is meant to suggest that the N-terminal BAH and the C terminus of Sir3p may bind to both the LRS domain and the histone N-terminal tails, and that this dual binding may in fact be crucial to compaction of chromatin and silencing. Additionally, the Sir3 C terminus is shown to be most sensitive to the actelyation of H4 K16, which inhibits Sir3 “lockdown” onto the chromatin (Johnson et al. 2009), and hence spreading of the silent chromatin. Sir4p is depicted as binding to linker DNA as suggested in Martino et al. (2009), but this aspect of the model should be considered preliminary. The stoichiometry of the SIR complex is also preliminary, and is based primarily on Kimura and Horikoshi (2004) and Johnson et al. (2009).
Acknowledgments
We thank the Boeke laboratory for helpful discussions, and reviewers of this paper for pointing out important recent information. Work on silencing was supported by NIH Roadmap grant U54 RR020839 to J.D.B.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1865510.
Supplemental material is available at http://www.genesdev.org.
References
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheron C, Maillet L, Marcand S, Tsai-Pflugfelder M, Gasser SM, Gilson E. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 1996;15:2184–2195. [PMC free article] [PubMed] [Google Scholar]
- Buchberger JR, Onishi M, Li G, Seebacher J, Rudner AD, Gygi SP, Moazed D. Sir3–nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol. 2008;28:6903–6918. doi: 10.1128/MCB.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- Chang JF, Hall BE, Tanny JC, Moazed D, Filman D, Ellenberger T. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure. 2003;11:637–649. doi: 10.1016/s0969-2126(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Cheng TH, Gartenberg MR. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes & Dev. 2000;14:452–463. [PMC free article] [PubMed] [Google Scholar]
- Chou CC, Li YC, Gartenberg MR. Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing. Mol Cell. 2008;31:650–659. doi: 10.1016/j.molcel.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig AJ, Gasser SM. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: Evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JJ, Yuan P, Hsu HC, Li Z, Xu RM, Sternglanz R. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol Cell Biol. 2006;26:3256–3265. doi: 10.1128/MCB.26.8.3256-3265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina AA, Winston F. Analysis of a mutant histone H3 that perturbs the association of Swi/Snf with chromatin. Mol Cell Biol. 2004;24:561–572. doi: 10.1128/MCB.24.2.561-572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerman IM, Li HC, Briggs SD. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: Identification of a new trans-histone pathway. Genes & Dev. 2007;21:2018–2029. doi: 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Lebrun E, Gilson E. Protosilencers as building blocks for heterochromatin. Bioessays. 2002;24:828–835. doi: 10.1002/bies.10139. [DOI] [PubMed] [Google Scholar]
- Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Danzer JR, Fox CA, Keck JL. Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 A resolution. Protein Sci. 2006;15:1182–1186. doi: 10.1110/ps.052061006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol Cell. 2009;35:769–781. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Horikoshi M. Partition of distinct chromosomal regions: Negotiable border and fixed border. Genes Cells. 2004;9:499–508. doi: 10.1111/j.1356-9597.2004.00740.x. [DOI] [PubMed] [Google Scholar]
- King DA, Hall BE, Iwamoto MA, Win KZ, Chang JF, Ellenberger T. Domain structure and protein interactions of the silent information regulator Sir3 revealed by screening a nested deletion library of protein fragments. J Biol Chem. 2006;281:20107–20119. doi: 10.1074/jbc.M512588200. [DOI] [PubMed] [Google Scholar]
- Liaw H, Lustig AJ. Sir3 C-terminal domain involvement in the initiation and spreading of heterochromatin. Mol Cell Biol. 2006;26:7616–7631. doi: 10.1128/MCB.01082-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Liu C, Lustig AJ. Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics. 1996;143:81–93. doi: 10.1093/genetics/143.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM. Structure and function of Cdc6/Cdc18: Implications for origin recognition and checkpoint control. Mol Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- Luo K, Vega-Palas MA, Grunstein M. Rap1–Sir4 binding independent of other sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes & Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ, Liu C, Zhang C, Hanish JP. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2483–2495. doi: 10.1128/mcb.16.5.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch PJ, Rusche LN. A silencer promotes the assembly of silenced chromatin independently of recruitment. Mol Cell Biol. 2009;29:43–56. doi: 10.1128/MCB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM. Reconstitution of yeast silent chromatin: Multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell. 2009;33:323–334. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- McBryant SJ, Krause C, Hansen JC. Domain organization and quaternary structure of the Saccharomyces cerevisiae silent information regulator 3 protein, Sir3p. Biochemistry. 2006;45:15941–15948. doi: 10.1021/bi061693k. [DOI] [PubMed] [Google Scholar]
- Mondoux MA, Zakian VA. Subtelomeric elements influence but do not determine silencing levels at Saccharomyces cerevisiae telomeres. Genetics. 2007;177:2541–2546. doi: 10.1534/genetics.107.079806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Norris A, Bianchet MA, Boeke JD. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet. 2008;4:e1000301. doi: 10.1371/journal.pgen.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer; Berlin: 1970. [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Park Y, Hanish J, Lustig AJ. Sir3p domains involved in the initiation of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:977–986. doi: 10.1093/genetics/150.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Hall BE, Ellenberger T, Moazed D. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol Cell Biol. 2005;25:4514–4528. doi: 10.1128/MCB.25.11.4514-4528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Sampath V, Yuan P, Wang IX, Prugar E, van Leeuwen F, Sternglanz R. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol Cell Biol. 2009;29:2532–2545. doi: 10.1128/MCB.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: The mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- Southey R. The doctor. Harper, NY: 1834. [Google Scholar]
- Stone EM, Reifsnyder C, McVey M, Gazo B, Pillus L. Two classes of sir3 mutants enhance the sir1 mutant mating defect and abolish telomeric silencing in Saccharomyces cerevisiae. Genetics. 2000;155:509–522. doi: 10.1093/genetics/155.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D. Budding yeast silencing complexes and regulation of Sir2 activity by protein–protein interactions. Mol Cell Biol. 2004;24:6931–6946. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JS, Snow ML, Giles S, McPherson LE, Grunstein M. Identification of a functional domain within the essential core of histone H3 that is required for telomeric and HM silencing in Saccharomyces cerevisiae. Genetics. 2003;163:447–452. doi: 10.1093/genetics/163.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gottschling DE. Genome-wide histone modifications: Gaining specificity by preventing promiscuity. Curr Opin Cell Biol. 2002;14:756–762. doi: 10.1016/s0955-0674(02)00393-9. [DOI] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- van Welsem T, Frederiks F, Verzijlbergen KF, Faber AW, Nelson ZW, Egan DA, Gottschling DE, van Leeuwen F. Synthetic lethal screens identify gene silencing processes in yeast and implicate the acetylated amino terminus of Sir3 in recognition of the nucleosome core. Mol Cell Biol. 2008;28:3861–3872. doi: 10.1128/MCB.02050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci. 2007;104:16609–16614. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Connelly JJ, Wang CL, Sternglanz R. Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing. Genetics. 2004;168:547–551. doi: 10.1534/genetics.104.028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]