Abstract
Retinoic acid receptor-α (RARα) is a known estrogen target gene in breast cancer cells. The consequence of RARα induction by estrogen was previously unknown. We now show that RARα is required for efficient estrogen receptor-α (ER)-mediated transcription and cell proliferation. RARα can interact with ER-binding sites, but this occurs in an ER-dependent manner, providing a novel role for RARα that is independent of its classic role. We show, on a genome-wide scale, that RARα and ER can co-occupy regulatory regions together within the chromatin. This transcriptionally active co-occupancy and dependency occurs when exposed to the predominant breast cancer hormone, estrogen—an interaction that is promoted by the estrogen–ER induction of RARα. These findings implicate RARα as an essential component of the ER complex, potentially by maintaining ER–cofactor interactions, and suggest that different nuclear receptors can cooperate for effective transcriptional activity in breast cancer cells.
Keywords: Breast cancer, estrogen receptor, retinoic acid receptor-α, transcription, chromatin
Breast cancer is one of the most common cancers in women worldwide. About 70% of breast cancers are positive for the nuclear receptor estrogen receptor-α (ERα, from here on called ER) and require estrogen for cell proliferation. Over the past years, many ER cofactors have been identified that contribute to changes in chromatin structure and gene regulation (Glass and Rosenfeld 2000; Metivier et al. 2003; Hall and McDonnell 2005). Gene expression studies and transcription factor-binding maps have revealed that a number of estrogen-regulated genes are subsequently used by the ER complex for efficient gene transcription (Carroll et al. 2005; Eeckhoute et al. 2006, 2007; Lupien et al. 2008). Examples of this include the transcription factors FoxA1 (HNF3α) and GATA3, both of which are regulated in response to estrogen treatment and have been shown to be crucial for ERα to bind to the chromatin and activate transcription of its target genes (Carroll et al. 2005; Eeckhoute et al. 2006, 2007; Lupien et al. 2008). As such, estrogen–ER can transcribe genes that ultimately contribute to ER function. Given this, we explored the transcription factor retinoic acid (RA) receptor-α (RARα), an additional estrogen-induced gene (Laganiere et al. 2005a).
RARα is a nuclear receptor and classically binds to RA response elements (RAREs) as a heterodimer, often using retinoid X receptor (RXR) as a binding partner. RARα can bind to chromatin in the absence or presence of its natural ligands—all-trans RA (ATRA) or 9-cis RA (Clarke et al. 2004)—and recruits cofactors to activate or repress its target genes (Dilworth and Chambon 2001; Niederreither and Dolle 2008). In addition, RARα can bind to other nuclear receptors, such as the thyroid hormone receptor (Glass et al. 1989), and cross-talk between RARα and ER pathways in breast cancer cells have been suggested (Rousseau et al. 2003).
RARα is an estrogen-induced gene, and its expression in breast tumors has been shown to correlate with ER expression (Roman et al. 1993). Recently, the binding events of a tagged version of RARα were mapped, and RARα and ER were shown to share a subset of binding regions within the genome (Hua et al. 2009). Hua et al. (2009) concluded that RARα in the presence of its ligand can antagonize estrogen–ER function, and vice versa. Their hypothesis is that RARα and ER can, in some cases, share common cis-regulatory elements, and that the two nuclear receptors compete for transcriptional activity (Hua et al. 2009). For the first time, we map endogenous RARα-binding events in proliferating breast cancer cells in the presence of estrogen, and show that RARα does in fact share very similar binding profiles to ER. Furthermore, under conditions where breast cancer cells are stimulated with estrogen, RARα and ER form part of the same transcriptional complex. We show that RARα is required for estrogen-mediated transcription and cell proliferation. These findings suggest that the co-occupancy of these distinct nuclear receptors can occur in a cooperative manner.
Results
RARα is an estrogen-induced gene that is required for estrogen-mediated growth
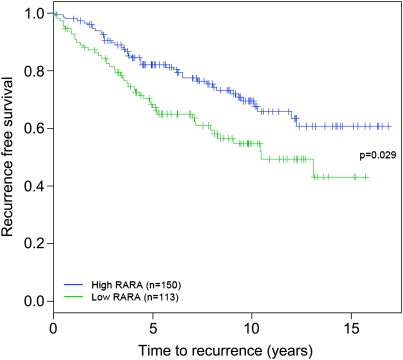
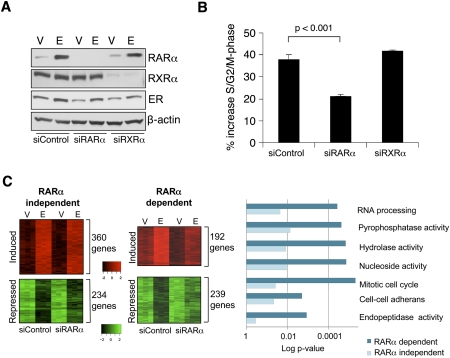
RARα is a well-documented estrogen target gene (Laganiere et al. 2005a), and the ER-binding event responsible for estrogen induction of RARα has been identified by ER chromatin immunoprecipitation (ChIP)-on-chip experiments (Laganiere et al. 2005a). In a cohort of 263 ER-positive breast cancer patients, all of whom received endocrine therapy, RARα was shown to predict positive clinical outcome (Fig. 1), supporting the hypothesis that it is an important ER target gene. Given the fact that a number of estrogen-regulated genes, including FoxA1, c-Myc, and cyclin D1, are essential for estrogen-mediated cell division (Prall et al. 1998; Laganiere et al. 2005b; Musgrove et al. 2008), we assessed whether RARα was also an important target gene for estrogen-mediated cell cycle progression by specifically silencing RARα using siRNA. As RARα classically binds as a heterodimer, we included siRNA targeting RXRα, one of its common binding partners, to determine if any role for RARα was dependent on RXRα. Western blot analysis confirmed effective silencing of both RARα and RXRα (Fig. 2A), and this silencing had negligible effects on ER protein levels (Fig. 2A).
Figure 1.
RARα predicts the outcome in breast cancer patients. RARα expression levels in 263 breast cancer patients, all of whom received endocrine therapy, were used to predict positive clinical outcome (recurrence-free survival).
Figure 2.
RARα is required for estrogen-mediated transcription and proliferation. (A) Western blot analysis of nuclear extracts of MCF-7 cells transfected with control siRNA or siRNA to RARα or RXRα and treated with vehicle (V) or estrogen (E). (B) Control siRNA or siRNA to RARα or RXRα was transfected into hormone-deprived cells, followed by estrogen treatment and assessment of percentage of S/G2/M phase. The data are the average of three independent replicates, and the error bars represent standard deviation (SD). (C) siControl- or siRARα-transfected cells were treated with vehicle or estrogen, and gene expression profiling was performed. Examples of estrogen-induced or estrogen-repressed genes are shown based on their dependency on RARα. GO category enrichments of the differentially estrogen-regulated genes (those dependent on RARα vs. those independent) are shown.
To determine the effect on cell cycle progression, we hormone-depleted MCF-7 cells for 3 d to induce cell cycle arrest and transfected cells with siRNA targeting RARα, RXRα, or control siRNA, followed by estrogen treatment for 24 h. Flow cytometry-based cell cycle analysis demonstrated that siRNA to RARα, but not RXRα, affected the estrogen-mediated cell cycle progression (Fig. 2B). The requirement for RARα on ER-mediated cell proliferation was confirmed using independent approaches for determining cell growth (Supplemental Fig. 1). This suggests that RARα plays an essential role in ER-mediated proliferation. No decrease in cell proliferation was observed after depleting RXRα protein levels, confirming that the decrease was specific to RARα.
RARα is a rate-limiting factor for ER transcriptional activity
We sought to determine the requirement for RARα in the ER-mediated transcriptional response. We hormone-depleted MCF7 cells and transfected them with siRNA to control or RARα. We subsequently treated cells with vehicle or estrogen for 12 h, collected total RNA, and performed gene expression profiling using Illumina bead arrays. Triplicate experiments were performed, and analysis revealed a total of 552 genes that were estrogen-induced and 473 genes that were estrogen-repressed. Under conditions where RARα was specifically silenced, we observed that a significant number of these estrogen-regulated genes were not statistically regulated (Fig. 2C). About a third (192 genes) of all estrogen-induced genes were no longer induced in the absence of RARα, and approximately half of the estrogen-repressed genes (239 genes) were no longer repressed. Gene ontology (GO) category enrichment analysis of the genes differentially regulated in the presence or absence of RARα confirmed that genes involved in cell cycle were enriched in the RARα-dependent, estrogen-regulated genes (Fig. 2C).
RARα can bind to ER-binding regions in vitro and in vivo, and this is dependent on the presence of ER
To directly determine whether RARα could bind to ER-binding loci in a chromatin context, we performed ChIP of endogenous RARα in proliferating MCF-7 cells, where RARα protein levels are abundant. RARα was shown to bind to all five tested ER-binding loci in asynchronous MCF-7 cells (Supplemental Fig. 2). This was also shown in two other ER-positive breast cancer cell lines; namely, T-47D and ZR75-1 (Supplemental Fig. 3). We could confirm that this RARα binding was dependent on the presence of ER, since treatment with ICI 182780 (an ER down-regulator) inhibited RARα binding to the chromatin. As a control, RARα binding to a known classic binding site (RARβ) was not altered by treatment with ICI 182780 (Supplemental Fig. 2), confirming that decreased RARα binding was not due to changes in RARα protein levels. Furthermore, to confirm that RARα can bind to the ER-binding events in hormone-deprived media, where RARα ligands are depleted, we repeated the RARα ChIP in hormone-depleted cells that were subsequently treated with estrogen or vehicle for 16 h. ChIP confirmed that RARα is recruited to the ER-binding sites tested after estrogen treatment, showing an estrogen-dependent enrichment of RARα binding to the ER cis-regulatory sites (Supplemental Fig. 4A). No increased binding of RARα to the known target promoter of RARβ was observed, showing that the increased RARα binding was specific to ER-binding events.
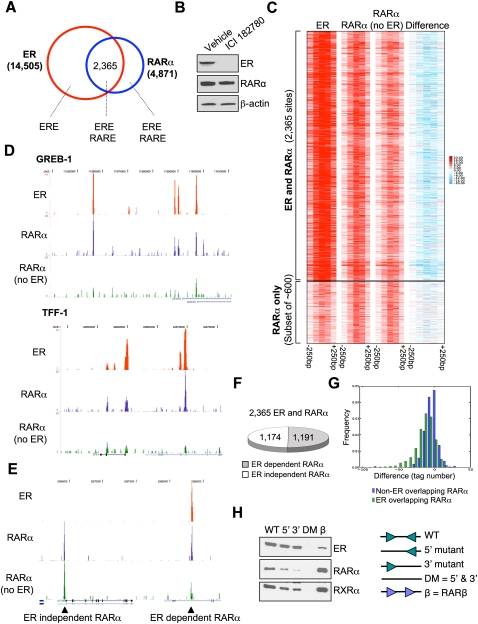
We extended these findings to map ER- and RARα-binding events on a global scale by ChIP-sequencing (ChIP-seq). We performed this experiment in asynchronous MCF-7 cells grown in complete media in order to capture RARα-binding events under conditions where RARα protein is abundant. This is particularly important given the difficulty in performing RARα ChIP, where enrichment is poor. We acquired a minimum of ∼30 million aligned reads for RARα ChIP-seq and, after using stringent cutoffs, we found 4871 RARα-binding events across the genome (tabulated Solexa data are shown in Supplemental Fig. 5A). Analysis of the genomic location of the RARα-binding events showed a similar pattern to that seen with other nuclear receptors (Supplemental Fig. 5C). RARα was rarely found at promoter regions (∼5%), and most RARα-binding events occur in intergenic regions.
ER ChIP-seq was also performed in asynchronous cells, and peaks were called using stringent thresholds, resulting in 14,505 ER-binding events. This ER-binding list had excellent representation (>85%) of a previous ER-binding list (Carroll et al. 2006). We found that 2365 of the ER- and RARα-binding sites overlapped, representing 48.6% of the RARα-binding sites (Fig. 3A). Within the overlapping sites, we find enrichments for both estrogen response element (ERE) and RARE motifs, as expected. Importantly, when looking at all ER- and RARα-binding sites (Supplemental Fig. 5), we find that, on the whole, the RARα sites that are not ER-binding events do in fact have weak (below the threshold used to call peaks) ER binding. Similarly, the ER-binding events that were not considered to be RARα-binding events do have weak RARα binding. This suggests that the overlap in ER and RARα binding may be significantly higher than the 48.6% reported.
Figure 3.
RARα binds to ER-binding regions in chromatin. (A) ER- and RARα-binding events were mapped in asynchronous MCF-7 cells. The ERE and RARE motifs that were enriched within each of the categories are shown. (B) Cells were treated with vehicle or ICI 182780, an ER down-regulator, for 3 h. Total protein was analyzed using Western blot analysis. (C) Clustering of ER and RARα binding ±250 bp. Signal based on tag count within all ER-binding events and corresponding RARα binding in full media or ICI 182780-treated cells. The difference in tag number within RARα-binding events between full media and ICI 182780 treatments is shown. As a control, ∼600 RARα-binding events (but not ER-binding events) are shown, and these are not influenced by the presence of ICI 182780. (D) Examples of ER- and RARα-binding events adjacent to the TFF-1 and GREB-1 genes. In both cases, the down-regulation of ER results in significant decreases in RARα binding. (E) A control region showing two RARα-binding events: one that does not overlap with ER binding and is ER-independent, and the adjacent site that is ER-dependent. (F) Graph representing the percentage of RARα-binding events that require ER. (G) Frequency histogram plotting the difference values (RARα in ICI 182780-treated cells minus RARα signal in full media) comparing RARα-binding events that overlap with ER sites versus non-ER-overlapping RARα-binding events. (H) An in vitro oligonucleotide pull-down was performed using a biotinylated 38-bp double-stranded oligonucleotide of an experimentally verified ER-binding region containing a perfect ERE. Various mutants lacking the 5′ half-site, the 3′ half-site, or both half-sites were included. Also included was a positive RARα/RXRα-binding site (RARβ promoter). Asynchronous MCF-7 cell lysate was collected and incubated with the various oligonucleotides, and purified protein was subjected to Western blot analysis.
When we compare our RARα-binding sites with the recent RARα-binding sites mapped by an EGFP-RARα fusion (generating a total of 7346 RARα-binding events) (Hua et al. 2009), we find an overlap of only 1332 binding events (27%). This low overlap may be due to a number of factors: different conditions (RARα ligand treatment in hormone-free conditions vs. binding in full media with abundant estrogen levels), different platforms (Affymetrix tiling microarrays vs. Solexa sequencing), or, potentially, the presence of the EGFP tag may affect RARα-binding properties. Our current RARα-binding profile represents endogenous RARα binding in the presence of estrogen, and is a distinct data set from the recently published EGFP-RARα binding following treatment with the RARα ligand (Hua et al. 2009).
We aimed to confirm that the RARα-binding events were not antagonizing ER (Hua et al. 2009), but were in fact acting in a cooperative manner with ER. We repeated the RARα ChIP-seq but pretreated asynchronous cells with ICI 182780, an ER down-regulator (McClelland et al. 1996), for 3 h. Importantly, ICI 182780 is a rapid and specific down-regulator of ER, allowing for acute inhibition of ER levels. At this 3-h time point, ER protein levels have decreased substantially (Fig. 3B), as well as ER binding (Supplemental Fig. 6), but RARα levels have not changed (Fig. 3B).
We acquired a minimum of ∼30 million aligned reads for RARα ChIP-seq in ICI 182780-treated cells and found that a number of the RARα-binding events had decreased binding in the absence of ER (Fig. 3C). As examples, RARα binding at the GREB1 and TFF1 regulatory regions are shown in the presence and absence of ER (Fig. 3D). RARα and ER co-occupy similar cis-regulatory elements, but the absence of ER results in loss of RARα binding to these regions. Also shown is a control region showing adjacent RARα-binding events: one that overlaps with an ER-binding event and is dependent on the presence of ER, and another RARα-binding event that is not altered by the loss of ER (Fig. 3E). More than half (50.4%) of the RARα-binding events were significantly decreased when ER was removed by ICI 182780 treatment (Fig. 3F), such that they were not called as binding events, despite the fact that RARα protein levels did not change. This was specific to the RARα sites that overlapped with ER (Fig. 3G). As such, approximately half of the RARα sites that overlap with ER sites are dependent on the presence of ER, confirming a cooperative and dependent interaction between ER and RARα.
We used an in vitro oligonucleotide pull-down approach to confirm that RARα could bind to an ERE motif. We designed biotin-labeled double-stranded 38-nucleotide oligonucleotides containing a perfect ERE (GGTCAnnnTGACC). This motif and the flanking sequence were derived from an experimentally identified ER-binding site (Carroll et al. 2006). Included were 5′ and 3′ mutants lacking one of the ERE half-sites, and a double mutant (DM) lacking both ERE half-sites. Also included was a double-stranded oligonucleotide containing part of the RARβ gene promoter, which contains a well-characterized RARα- and RXRα-binding site (de The et al. 1990). Total protein from proliferating MCF-7 cells was incubated with the various double-stranded oligonucleotides. Streptavidin purification of the proteins associated with each oligonucleotide was performed, and Western blots for ER, RARα, and RXRα were performed. The results obtained (Fig. 3H) show that ER can bind to the sequence and only requires one of the ERE half-sites. RARα was shown to bind to the perfect ERE, but RARα binding was decreased significantly when one of the ERE half-sites was deleted. We could recapitulate the same findings on a different ER-binding region that also contained a perfect ERE (but with different flanking sequence) where RARα binding was shown to occur, but, again, only in the presence of the ERE half-site (data not shown). This in vitro assay shows that the RARα binding to ER-binding sites requires the presence of the ERE half-site.
RARα and ER co-occupy the DNA simultaneously
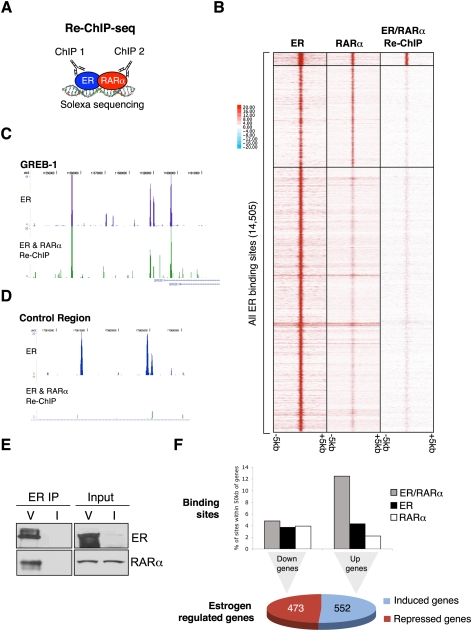
To directly implicate RARα in the ER transcriptional complex, we performed an ER ChIP, followed by “release” of the ER-enriched chromatin and a subsequent RARα ChIP (termed Re-ChIP). This shows simultaneous chromatin co-occupancy of both proteins. As a control, an ER ChIP was performed followed by an IgG ChIP. Real-time PCR of a number of known ER-binding loci revealed clear enrichment of ER/RARα-bound ER-binding events, but not the classic RARα-binding site in the RARβ promoter (Supplemental Fig. 4B). This Re-ChIP experiment provides definitive evidence that ER and RARα co-occupy the same cis-regulatory elements on the chromatin. As controls, ER ChIP followed by IgG Re-ChIP was used to normalize the data. Similarly, IgG ChIP followed by RARα Re-ChIP did not result in any enrichment of the ER-binding loci (data not shown).
Based on the observation that ER and RARα form part of the same complex at specific tested ER-binding loci, we extended this on a genome-wide level (Fig. 4A) by performing ER/RARα Re-ChIP in full media followed by Solexa sequencing (Re-ChIP-seq). More than 14 million aligned reads were acquired for both Re-ChIP-seq replicates. As a control, ER then ER Re-ChIP-seq and ER then IgG Re-ChIP-seq were performed (Supplementary Fig. 7). We subtracted the ER/IgG Re-ChIP signal from the specific ER/RARα Re-ChIP-seq data to show the difference in binding. After this subtraction, we found 352 high-confidence ER/RARα Re-ChIP sites. These ER/RARα Re-ChIP sites represent ∼15% of the 2365 ER and RARα overlapping binding sites (Fig. 4B). Given the technical difficulties with this assay, more ER/RARα Re-ChIP binding events would likely be gained with additional sequencing.
Figure 4.
Genome-wide Re-ChIP-seq shows that ER and RARα co-occupy the chromatin together. (A) A schematic showing the Re-ChIP-seq protocol, where ER ChIP is performed, followed by RARα ChIP and Solexa sequencing. (B) Clustered binding signal from all ER-binding events (±5 kb) is shown. The corresponding RARα-binding events as well as the ER/RARα Re-ChIP binding signal are shown. The signal from the ER/RARα Re-ChIP sites are after the ER/IgG signal has been subtracted. (C) An example of a region around the GREB-1 gene showing the ER/RARα Re-ChIP signal. (D) A control region showing a strong ER-binding events but no ER/RARα Re-ChIP signal. (E) Coimmunoprecipitation showing direct ER and RARα interactions in asynchronous cells treated with vehicle (V). As a control, cells were treated with ICI 182780 (I). Also shown are total input levels. (F) The ER/RARα Re-ChIP binding events, the ER-binding events (not Re-ChIP sites), and the RARα-binding events (not Re-ChIP sites) were assessed relative to estrogen-induced or estrogen-repressed genes. The percentage of binding sites within 50 kb of the transcription start sites of the estrogen-regulated genes are shown.
As an example of ER and RARα co-occupancy, the Re-ChIP regions adjacent to the GREB1 gene are shown (Fig. 4C). Equally strong ER-binding sites were not detected as ER/RARα co-occupied regions (an example is shown in Fig. 4D), confirming specificity in the regions cobound by ER and RARα. These 352 ER/RARα Re-ChIP binding events clearly confirm that ER and RARα co-occupy the chromatin together, supporting a cooperative role. To show that ER and RARα form direct interactions and are not simply tethered together by regions of DNA, we performed an ER immunoprecipitation followed by an RARα Western blot. The data (Fig. 4E) confirmed that ER and RARα could be coimmunoprecipitated and are therefore part of the same protein complex.
We combined the ER/RARα Re-ChIP sites (vs. the ER- and RARα-binding sites that are not Re-ChIP sites) with the estrogen-mediated gene expression data. We found that the ER/RARα co-occupied Re-ChIP binding events were more likely to be adjacent to estrogen-induced genes when compared with ER- or RARα-binding events that were not Re-ChIP sites (Fig. 4F; Supplemental Fig. 8). This provides evidence that the regions simultaneously co-occupied by ER and RARα are transcriptionally active and biased toward genes that are estrogen-induced. Although a number of estrogen-repressed genes were dependent on RARα (Fig. 2C), no bias was observed near down-regulated genes, as has been shown previously (Carroll et al. 2006). Similarly, we did not see enrichment of ER/RARα Re-ChIP binding events adjacent to a random set of control genes (data not shown).
RARα can induce estrogen-mediated transcription
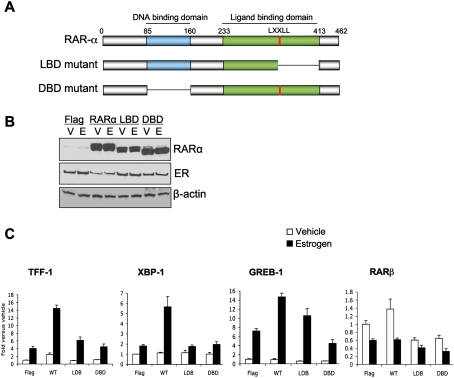
Our data suggested that specific silencing of RARα could negatively affect estrogen–ER-mediated transcription. We also aimed to determine if RARα was sufficient to promote estrogen–ER transcription. Hormone-depleted cells were transfected with an RARα-expressing construct (the Flag-RARα vector was a kind gift from Shinobu Tsuzuki, Institute of Cancer Research, London) expressing full-length RARα, or mutant versions lacking either a core part of the ligand-binding domain (LBD; deleted amino acids 351–412) or the DNA-binding domain (DBD; deleted amino acids 85–160) (Fig. 5A). We expressed empty vector (Flag) as a control. Under hormone-deprived conditions, when RARα protein levels are low, we expressed the vectors and treated cells with vehicle or 1 nM estrogen for 5 h. We collected protein and performed Western blot analysis to ensure effective expression of RARα and the mutants (Fig. 5B). We also collected RNA after vehicle or estrogen treatment and assessed expression of a number of classic estrogen-regulated genes, all of which have ER/RARα-binding sites adjacent to them. As shown in Figure 5C, expression of full-length RARα resulted in increased expression of all assessed estrogen target genes, confirming that the addition of RARα could potentiate ER-mediated transcription. Interestingly, when either the LBD or the DBD of RARα was deleted, the RARα potentiation of ER-mediated transcription was inhibited (Fig. 5C). We did not see any changes in control genes, including RARβ (Fig. 5C). This suggested that RARα is a rate-limiting component of the ER machinery, and that intact LBD and DBD are required for effective RARα–ER interactions.
Figure 5.
Expression of RARα can potentiate estrogen–ER-mediated transcription. (A) Schematic representation of the RARα mutants generated. (B) Western blot analysis of nuclear fraction of transfected cells after vehicle (V) or estrogen (E) treatment. (C) Hormone-depleted cells were transfected with control (Flag), RARα (WT), or the LBD and DBD mutants of RARα, and cells were subsequently treated with vehicle or estrogen. Real-time RT–PCR was conducted of TFF-1, XBP-1, and GREB-1 (all estrogen-regulated genes). RARβ (a RARα target gene) was included as a control gene.
RARα is required for estrogen-induced gene transcription by facilitating ER–coactivator interactions
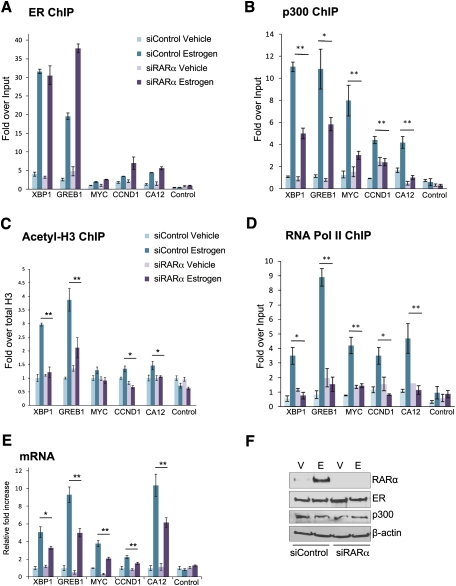
We sought to determine the functional role that RARα plays in mediating ER activity. We transfected cells with control siRNA or siRNA targeting RARα, treated cells with vehicle or estrogen, and performed ER ChIP after 6 h of estrogen treatment. ER binding was assessed at ER/RARα-cobound sites adjacent to well-established ER-regulated genes, including MYC, Cyclin D1, XBP-1, GREB-1, and CA12. ER binding at all of the tested sites was not decreased in the absence of RARα (Fig. 6A), and global chromatin-associated ER levels did not change (Supplemental Fig. 6). Therefore, ER binding to chromatin is not dependent on the presence of RARα.
Figure 6.
RARα is required for effective coactivator loading. MCF-7 cells were hormone-depleted and transfected with control siRNA or siRNA targeting RARα. ChIP of ER (A), p300 (B), or acetylated-H3 (C) was performed after 6 h of vehicle (V) or estrogen (E) treatment, followed by real-time PCR of a number of ER-binding regions. The acetylated-H3 ChIP was first normalized to total H3, then to Input. (D) Following transfection, RNA PolII ChIP was performed, and real-time PCR of the promoter regions of the estrogen-regulated genes was performed. (E) Changes in mRNA levels of the genes were assessed following silencing of RARα. (F) After silencing of control or RARα, total protein levels of ER, p300, and RARα were assessed. (*) P < 0.05; (**) P < 0.01. The data are representative of triplicate experiments, ±SD.
We hypothesized that RARα may function as a scaffold in the ER complex for maintaining cofactor interactions. We tested this by specifically silencing RARα and performing ChIP of p300, a well-known ER coactivator (Shang et al. 2000), at the ER-binding loci. We found a significant decrease in p300 binding at all of the ER-binding events in the absence of RARα (Fig. 6B). We could also confirm that binding of the Mediator subunit TRAP220 and the ER coactivator AIB-1 was decreased following specific silencing of RARα (Supplemental Fig. 9). These findings confirm that the decreased coactivator binding at the ER-binding events was specifically due to the absence of RARα. The changes in cofactor and Mediator binding following specific silencing of RARα also resulted in changes in chromatin structure, as measured by acetylated-H3 at some of the ER-binding regions (Fig. 6C).
To confirm that the specific silencing of RARα had an influence on the transcription of these genes, we performed ChIP of RNA polymerase II (PolII) followed by real-time PCR of the promoter regions of these genes. As expected, silencing of RARα resulted in a significant reduction in RNA PolII binding to the promoters of all of these genes, confirming RARα dependency for the estrogen-mediated transcription of these target genes (Fig. 6D). This was supported by data showing changes in mRNA levels of the genes tested (Fig. 6E). No significant changes in total protein levels were observed (Fig. 6F; Supplemental Fig. 9).
An estrogen–ER-dependent role for RARα can be inhibited by an RARα ligand
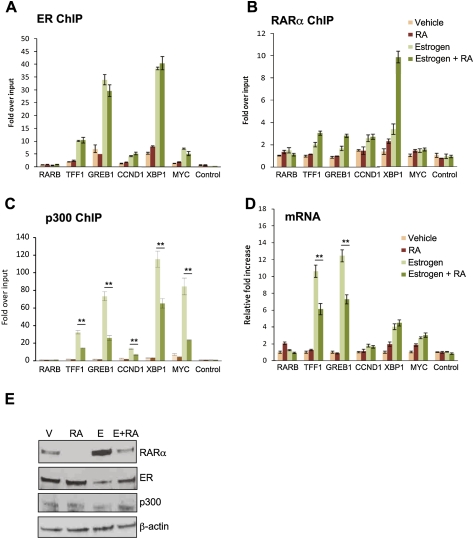
Our data show that estrogen–ER transcription involves RARα. However, a previous investigation (Hua et al. 2009) suggested that RARα and ER share similar binding regions, but that the addition of the RARα ligand ATRA could inhibit estrogen–ER function. Our data suggest that, when presented with estrogen, ER and RARα cooperate for efficient estrogen-mediated transcription. We aimed to reconcile this difference by determining if the presence of the RARα ligand could inhibit estrogen–ER-mediated activity. We hormone-depleted cells and treated cells with vehicle, estrogen, ATRA, or estrogen plus RA for 6 h, after which chromatin was collected for ChIP. We found that estrogen induced ER binding as expected, and cotreatment with RA did not decrease ER binding (Fig. 7A). ChIP of RARα also revealed that it can still bind in the presence of estrogen and RA and, in some cases, binding was increased (Fig. 7B). In addition, the estrogen-mediated recruitment of p300 (Fig. 7C) was inhibited by cotreatment of estrogen and RA. Similar results were observed when assessing binding of TRAP220 (Supplemental Fig. 9B). The cotreatment of RA with estrogen could inhibit the estrogen-mediated induction of some target genes while others were unaffected (Fig. 7D). These data confirm that RARα can have an antagonist effect on ER transcriptional activity when liganded with RA, as suggested previously (Hua et al. 2009); however, cotreatment with estrogen and RA did not perturb binding of either nuclear receptor.
Figure 7.
RARα ligands can inhibit estrogen–ER-mediated transcriptional activity. Hormone-depleted cells were treated with vehicle, estrogen, RA (ATRA), or estrogen + RA for 6 h, and ChIP was performed for ER (A), RARα (B), and p300 (C). Real-time PCR of a number of ER-binding regions was assessed. (D) Changes in mRNA levels of several regulated genes. (E) Western blot of total protein levels in treated cells. The data are the average of three independent experiments, ±SD. (*) P < 0.05; (**) P < 0.01.
Discussion
RARα is a well-studied nuclear receptor transcription factor that plays an important role during development and in normal physiological functioning, where it can regulate a number of cellular processes, including proliferation and differentiation. RARα forms heterodimers with members of the RXR family, and these heterodimers can form in the absence or presence of ligand and can bind to the chromatin (Dilworth and Chambon 2001). RARα can also form fusion genes with a number of genes; notably, PML (Borrow et al. 1990; Goddard et al. 1991), which is the defining translocation in acute promyelocytic leukemia. A role for RARα in breast cancer has been postulated, and RARα ligands have been shown to be effective in breast cancer treatment (Darro et al. 1998), although the mechanisms of this are unknown.
RARα was shown recently to be a direct ER target gene that is induced upon estrogen treatment of breast cancer cells (Laganiere et al. 2005a). Other estrogen-regulated transcription factors include FoxA1 (HNF3-α) and GATA3, both of which are markers of ER-positive luminal breast cancer (Sorlie et al. 2003), and both of which are subsequently used by ER to bind chromatin and regulate gene transcription (Carroll et al. 2005; Laganiere et al. 2005b; Eeckhoute et al. 2006, 2007). We hypothesized that RARα might function in a similar role. In fact, cross-talk between RARα and ER has been shown previously (Rousseau et al. 2003), but the basis of this was unclear. Proliferation of ER-positive breast cancer tumors is inhibited by the RARα agonist ATRA (Dawson et al. 1995) and, interestingly, the RARα antagonist Ro 41-5253 also results in decreased cell growth and induction of apoptosis in ER-positive cell lines (Toma et al. 1998).
We now show that RARα is part of the ER transcription complex, where it functions in an essential role to regulate estrogen-mediated gene expression. Specific silencing of RARα resulted in inhibition of estrogen-regulated target genes and inhibition of estrogen-mediated proliferation. We could show that RARα and ER form complexes on the chromatin and that RARα binding is dependent on the presence of ER. We hypothesize that RARα can have two independent functions: (1) a classic role as a heterodimeric partner of RXR proteins that respond to natural ligands such as RA, and (2) as an ER-associated protein required for maintaining cofactor interactions during estrogen-mediated gene transcription. As such, any shift between the classic and novel pathways may influence ER function in breast cancer cells. This is supported by recent data showing that RA can inhibit estrogen target genes (Hua et al. 2009), therefore promoting the classic RARα role, at the cost of estrogen–ER function. This potentially explains why RARα ligands are effective treatments for breast cancer (Darro et al. 1998), and why both agonists and antagonists (Dawson et al. 1995; Toma et al. 1998) inhibit breast cancer cell growth; activation of the classic RARα pathways can deplete RARα from its essential role as a component of the estrogen–ER pathway in breast cancer cells.
Our data show that, in the presence of estrogen, ER and RARα can co-occupy the same regions in the genome. Understanding the mechanisms required for these proteins to interact with the genome remains to be fully defined. ER and RARα can bind to the same half-site—namely, GGTCA (Klinge 2001; Balmer and Blomhoff 2005)—yet the spacing and orientation between the motifs is different. RAR–RXR bind to two direct repeat motifs with 1-, 2-, or 5-base-pair (bp) spacers (Bastien and Rochette-Egly 2004; Balmer and Blomhoff 2005), whereas ER binds to inverted half-sites with a 3-bp spacer (Klinge 2001). However, more recently, it has been observed in ChIP-on-chip and ChIP-seq data that, in a chromatin context, various nuclear receptors, including ER and RARα, can bind to a single half-site (Carroll et al. 2005; Lin et al. 2007; Massie et al. 2007; Wang et al. 2007) or widely spaced half-sites (Kato et al. 1995), or via other factors. As such, the classic paradigms of nuclear receptor–chromatin interactions may need to be re-evaluated.
These data confirm an integral role for RARα as an ER-associated factor. Such an interaction is distinct from the classic role of RARα and from an antagonist role between ER and RARα (Hua et al. 2009). Given the fact that the predominant hormone in breast cancer biology is estrogen, it is important that the interactions between ER and RARα be determined under these specific conditions. This is of utmost importance, considering that RARα is induced by the estrogen–ER complex in breast cancer cells (Laganiere et al. 2005a). By focusing on the interactions between ER and RARα under these clinically relevant conditions, we can now show that different nuclear receptors can interact to positively regulate gene transcription in breast cancer cells.
Materials and methods
Cell culture
MCF-7, T-47D, and ZR75-1 human cell lines were grown as described previously (Neve et al. 2006). Unless stated, in all experiments, estrogen was added at a final concentration of 100 nM and ICI 182780 (Fulvestrant) was added at a final concentration of 10 nM. ATRA was added at a final concentration of 1 μM.
siRNA
Cells were grown in phenol red-free DMEM supplemented with 5% charcoal-dextran-treated serum for at least 3 d. Cells were transfected with siRNA using Lipofectamine2000 (Invitrogen) on day 2. siGENOME SMARTpool from Dharmacon (M-003437-02) was used to target RARα. An independent siRNA targeting RARα was used to confirm the results. The sequences of this siRNA were 5′-GCAAAUACACUACGAACAA-3′ (sense) and 5′-UUGUUCGUAGUGUAUUUGC-3′ (antisense). The sequences of the other siRNAs used were as follows: RXRα siRNA, 5′-GGGAGAAGGUCUAUGCGUC-3′ (sense), 5′-GACGCAUAGACCUUCUCCC-3′ (antisense). AllStars Negative Control siRNA (Qiagen) was used as a negative control.
Western blot analysis
Whole-cell lysate or nuclear protein was harvested. Antibodies used were anti-ER (sc-543), anti-RARα (sc-551), anti-RXRα (sc-553), anti-p300 (sc-585), anti-TRAP220 (sc-8998), and anti-AIB-1 (sc-9119) from Santa Cruz Biotechnologies, and anti-β-actin (ab6276) from Abcam.
Cell cycle analysis
Cells were plated at equal confluence, deprived of hormones, and transfected using siRNA as described above. Total cells were harvested for flow cytometry analysis, using propidium iodide staining.
Oligonucleotide pull-down assay
Protein binding to synthesized double-stranded oligonucleotides was performed as described previously (Hata et al. 2000). A double-stranded oligonucleotide sequence was derived from an experimentally validated ER-binding event containing a perfect ERE: sense, 5′-biotin-CCTTGGTGTCGCGGGTCATAATGACCGGAGCTTTTCCC-3′, and antisense, 5′-biotin-GGGAAAAGCTCCGGTCATTATGACCCGCGACACCAAGG-3′. The various mutants were as follows: 5′ ERE mutant, sense, 5′-biotin-CCTTGGTGTCGCGATCTTTAATGACCGGAGCTTTTCCC-3′, and antisense, 5′-biotin-GGGAAAAGCTCCGGTCATTAAAGATCGCGACACCAAGG-3′; 3′ ERE mutant, sense, 5′-biotin-CCTTGGTGTCGCGGGTCATAAGCTTCGGAGCTTTTCCC-3′, and antisense, 5′-biotin-GGGAAAAGCTCCGAAGCTTATGACCCGCGACACCAAGG-3′; double ERE mutant, sense, 5′-biotin-CCTTGGTGTCGCGATCTTTAAGCTTCGGAGCTTTTCCC-3′, and antisense, 5′-biotin-GGGAAAAGCTCCGAAGCTTAAAGATCGCGACACCAAGG-3′. The ERE half-sites are underlined. A double-stranded oligonucleotide containing the RARE present in the RARβ promoter was included as a positive control for RARα and RXRα binding. The sequence of this oligonucleotide was sense, 5′-biotin-AGCAGCCCGGGTAGGGTTCACCGAAAGTTCACTCGCATATAT-3′, and antisense, 5′-biotin-ATATATGCGAGTGAACTTTCGGTGAACCCTACCCGGGATGCT-3′. The RAREs have been underlined.
ChIP
ChIP experiments were performed as described previously (Carroll et al. 2005). Antibodies used were anti-ER (sc-543), anti-RARα (sc-551), anti-RXRα (sc-553), anti-p300 (sc-585), anti-TRAP220 (sc-8998), and anti-AIB-1 (sc-9119) from Santa Cruz Biotechnologies, and anti-RNA PolII (ab5408), anti-Histone H3 acetyl K9 + K14 + K18 + K23 + K27 (ab47915), and anti-Histone H3 (ab1791) from Abcam. Primer sequences are provided in Supplemental Table 1.
Re-ChIP
For the Re-ChIP experiments, an ER ChIP was performed as described in Carroll et al. (2005), with the exception that 25 μL of 10 mM DTT were used to elute the bound chromatin from the beads after the washes and incubated at 37°C with shaking for 30 min as described previously (Metivier et al. 2003). The supernatant was removed and diluted at least 70 times using ChIP dilution buffer. The second ChIP, using an antibody against either RARα, RXRα, or ER and IgG as negative controls, was then performed according to standard ChIP protocols (Carroll et al. 2005). All Re-ChIP data are shown relative to IgG control, which is a more rigorous control for Re-ChIP experiments.
Solexa sequencing and enrichment analysis
RNase A and proteinase K digestions were performed on the DNA from the ChIP or Re-ChIP experiments. The DNA was subsequently amplified using the Genomic DNA Sample Prep Kit from Illumina. Sequences generated by the Illumina genome analyzer were aligned against NCBI Build 36.3 of the human genome using MAQ (http://maq.sourceforge.net) with default parameters. Identification of enriched sites in the sequenced ChIP and Re-ChIP samples was performed using a consensus peak-calling technique. Only sequence reads with a MAQ mapping quality score ≥35 were used for the analysis. For each biological replicate (two ER ChIP replicates, two ER/RARα Re-ChIP replicates, two ER/IgG ReChIP replicates, ER–ER ReChIP, and RARα ChIPs), a corresponding set of Input sequence reads of similar size was obtained by random sampling from the full set of Input sequence reads. In addition, a set of Input read sequences was derived from the ER/IgG Re-ChIP for use with the ER/RARα Re-ChIP. These Input sequence read sets were used as control tracks for initial identification of enriched target sites, which was accomplished by using two publicly available peak callers. The first is the ChIP-seq Peak Finder based on that used in Johnson et al. (2007), with the fragment size parameter (maxSpacing) set to 75, the minimum number of sequences in a peak (minHits) set to 15, and the fold enrichment over Input parameter (minRatio) set to 5 for the ER ChIPs and 3 for the ER/RARα Re-ChIPs. The second peak caller used was the Model-based Analysis for ChIP-seq (MACS) (Zhang et al. 2008), run using default parameters (except mfold = 30); only peaks identified with a P-value of <6.3 × 10−7 were considered. Consensus peaks were determined by including only regions identified by both peak callers. Consensus peaks were merged into enriched regions by using the outer coordinates of the overlapping regions, as described in Schmidt et al. (2008). Once consensus peaks were determined for each biological replicate, consensus peaks were determined for each condition (ER and ER/RARα) by merging overlapping peaks from each replicate.
To generate the heat maps of the raw ChIP-seq data, ER, RARα, ER/RARα, ER/ER, and ER/IgG Re-ChIP binding peaks were used as targets to center each window. Each window was divided into five bins of 100 bp in size (Fig. 3C) or 100 bins of 100 bp in size (Fig. 4B; Supplemental Figs. 5, 7). An enrichment value was assigned to each bin, counting the number of sequencing reads in that bin. The ER/RARα Re-ChIP signal was further normalized by subtracting the background signal of an ER/IgG Re-ChIP experiment. For this purpose, the same number of sequencing reads of ER/RARα Re-ChIP and ER/IgG ReChIP were considered. The distribution of the sum of all five bins was used to plot the histogram of RARα-binding change of the ER-independent and ER-dependent RARα sites (subset of ∼600 and 2365 sites, respectively). The frequency was normalized to the number of binding events and bin size of the histogram. Data were visualized with Treeview (Saldanha 2004).
Coimmunoprecipitation
ER antibodies were cross-linked to agarose beads as described in the Abcam protocol. Proliferating MCF-7 cells were harvested and lysed in lysis buffer (50 mM HEPES at pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 1 mM DTT, protease inhibitors). Samples were immunoprecipitated for 2 h at 4°C, after which they were washed twice with lysis buffer containing 1 M NaCl and then twice with 50 mM HEPES (pH 7.5) containing 1 mM DTT. Bound protein was eluted by the addition of 1 M glycine (pH 3.0), neutralized using Tris (pH 9.0), and then analyzed by Western blot analysis.
Gene expression
MCF-7 cells were hormone-depleted for 3 d and treated with 100 nM estrogen for 12 h. Total RNA was collected, and RNA was hybridized to Illumina arrays. The Illumina BeadChip (HumanWG-6 version 3) bead-level data were preprocessed, log2-transformed, and quantile-normalized using the bead array package in Bioconductor. Differential expression analysis was performed using limma eBayes (Smyth 2004) with a Benjamini and Hochberg multiple test correction procedure (Benjamini and Hochberg 1995) to identify statistically significant differentially expressed genes (adjusted P-value, <0.01).
Motif and GO category analysis
The motif analysis was performed using the program CLOVER (Frith et al. 2004), which compares sets of DNA sequences to a library of transcription factor-binding motifs and identifies whether any of the motifs are statistically overrepresented or underrepresented in the sets. The JASPAR CORE (Sandelin et al. 2004) collection of transcription factor-binding patterns and the RARE probability weight matrix (Balmer and Blomhoff 2005) were used for the analysis. GO category enrichment was determined using GeneTrail (Backes et al. 2007).
Clinical expression data
Microarray data sets from 263 tamoxifen-treated patients were obtained from previous investigations (Sotiriou et al. 2006; Loi et al. 2007). All data had been normalized previously by RMA. Recursive partitioning (RP) in the R package Party was used on the normalized gene expression data. RP works by ordering the gene expression values and working down the values using each as a cutoff to create two groups. The difference in survival between the two groups is then calculated. The cutoff that provides the most significant difference between the two subsequent groups is chosen. Then the process is carried out recursively, until there are no more significant cutoffs in the data (Hothorn et al. 2006). A correction for multiple testing is applied.
Statistical analysis
All statistical analyses were performed using the two-tailed Students t-tests or the χ2 test using Excel. Only values with a P-value <0.05 were considered statistical. Error bars represent standard deviations.
Data depostion
The sequencing data are deposited at NCBI SRA study accession number SRA010193.
Acknowledgments
We thank Nik Matthews and James Hadfield for Solexa sequencing, and Gordon Brown and Mike Wilson for technical and bioinformatic help. We thank Shinobu Tsuzuki (Institute of Cancer Research, London) for the RARα plasmid. We acknowledge the support of The University of Cambridge, Cancer Research UK, and Hutchison Whampoa Limited. C.R.-I. is supported by a Commonwealth Scholarship, and J.S.C. is supported by an ERC Starting Grant.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.552910.
Supplemental material is available at http://www.genesdev.org.
References
- Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP. GeneTrail—Advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer JE, Blomhoff R. A robust characterization of retinoic acid response elements based on a comparison of sites in three species. J Steroid Biochem Mol Biol. 2005;96:347–354. doi: 10.1016/j.jsbmb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- Borrow J, Goddard AD, Sheer D, Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249:1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Clarke N, Germain P, Altucci L, Gronemeyer H. Retinoids: Potential in cancer prevention and therapy. Expert Rev Mol Med. 2004;6:1–23. doi: 10.1017/S1462399404008488. [DOI] [PubMed] [Google Scholar]
- Darro F, Cahen P, Vianna A, Decaestecker C, Nogaret JM, Leblond B, Chaboteaux C, Ramos C, Petein M, Budel V, et al. Growth inhibition of human in vitro and mouse in vitro and in vivo mammary tumor models by retinoids in comparison with tamoxifen and the RU-486 anti-progestagen. Breast Cancer Res Treat. 1998;51:39–55. doi: 10.1023/a:1006098124087. [DOI] [PubMed] [Google Scholar]
- Dawson MI, Chao WR, Pine P, Jong L, Hobbs PD, Rudd CK, Quick TC, Niles RM, Zhang XK, Lombardo A, et al. Correlation of retinoid binding affinity to retinoic acid receptor α with retinoid inhibition of growth of estrogen receptor-positive MCF-7 mammary carcinoma cells. Cancer Res. 1995;55:4446–4451. [PubMed] [Google Scholar]
- de The H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor β gene. Nature. 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–3054. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes & Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Krasnickas Keeton E, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor α expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes & Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Glass CK, Lipkin SM, Devary OV, Rosenfeld MG. Positive and negative regulation of gene transcription by a retinoic acid-thyroid hormone receptor heterodimer. Cell. 1989;59:697–708. doi: 10.1016/0092-8674(89)90016-0. [DOI] [PubMed] [Google Scholar]
- Goddard AD, Borrow J, Freemont PS, Solomon E. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP. Coregulators in nuclear estrogen receptor action: From concept to therapeutic targeting. Mol Interv. 2005;5:343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massague J. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: A conditional inference framework. J Comput Graph Statist. 2006;15:651–674. [Google Scholar]
- Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kato S, Sasaki H, Suzawa M, Masushige S, Tora L, Chambon P, Gronemeyer H. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol Cell Biol. 1995;15:5858–5867. doi: 10.1128/mcb.15.11.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Giguere V. Functional genomics identifies a mechanism for estrogen activation of the retinoic acid receptor α1 gene in breast cancer cells. Methods Enzymol. 2005a;19:1584–1592. doi: 10.1210/me.2005-0040. [DOI] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci. 2005b;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie CE, Adryan B, Barbosa-Morais NL, Lynch AG, Tran MG, Neal DE, Mills IG. New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep. 2007;8:871–878. doi: 10.1038/sj.embor.7401046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland RA, Gee JM, Francis AB, Robertson JF, Blamey RW, Wakeling AE, Nicholson RI. Short-term effects of pure anti-oestrogen ICI 182780 treatment on oestrogen receptor, epidermal growth factor receptor and transforming growth factor-α protein expression in human breast cancer. Eur J Cancer. 1996;32A:413–416. doi: 10.1016/0959-8049(95)00517-x. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Musgrove EA, Sergio CM, Loi S, Inman CK, Anderson LR, Alles MC, Pinese M, Caldon CE, Schutte J, Gardiner-Garden M, et al. Identification of functional networks of estrogen- and c-Myc-responsive genes and their relationship to response to tamoxifen therapy in breast cancer. PLoS One. 2008;3:e2987. doi: 10.1371/journal.pone.0002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Dolle P. Retinoic acid in development: Towards an integrated view. Nat Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Prall OW, Rogan EM, Musgrove EA, Watts CK, Sutherland RL. c-Myc or cyclin D1 mimics estrogen effects on cyclin E–Cdk2 activation and cell cycle reentry. Mol Cell Biol. 1998;18:4499–4508. doi: 10.1128/mcb.18.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman SD, Ormandy CJ, Manning DL, Blamey RW, Nicholson RI, Sutherland RL, Clarke CL. Estradiol induction of retinoic acid receptors in human breast cancer cells. Cancer Res. 1993;53:5940–5945. [PubMed] [Google Scholar]
- Rousseau C, Pettersson F, Couture MC, Paquin A, Galipeau J, Mader S, Miller WH., Jr The N-terminal of the estrogen receptor (ERα) mediates transcriptional cross-talk with the retinoic acid receptor in human breast cancer cells. J Steroid Biochem Mol Biol. 2003;86:1–14. doi: 10.1016/s0960-0760(03)00255-3. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Stark R, Wilson MD, Brown GD, Odom DT. Genome-scale validation of deep-sequencing libraries. PLoS One. 2008;3:e3713. doi: 10.1371/journal.pone.0003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. http://www.bepress.com/sagmb/vol3/iss1/art3. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Toma S, Isnardi L, Raffo P, Riccardi L, Dastoli G, Apfel C, LeMotte P, Bollag W. RARα antagonist Ro 41-5253 inhibits proliferation and induces apoptosis in breast-cancer cell lines. Int J Cancer. 1998;78:86–94. doi: 10.1002/(sici)1097-0215(19980925)78:1<86::aid-ijc14>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu X-S, Carroll JS, Janne OA, Krasnickas Keeton E, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]