Abstract
Polypurine/polypyrimidine (pPU/pPY) tracts, which exist in the promoter regions of many growth-related genes, have been proposed to be very dynamic in their conformation. In this chapter, we describe a detailed protocol for DNase I and S1 nuclease footprinting experiments with supercoiled plasmid DNA containing such the promoter regions to probe whether there are conformational transitions to B-type DNA, melted DNA and G-quadruplex structures within this tract. This is demonstrated with the proximal promoter region of the human vascular endothelial growth factor (VEGF) gene, which also contains multiple binding sites for Sp1 and Egr-1 transcription factors.
Keywords: Plasmid footprinting, DNA Secondary structure, G-quadruplex, VEGF
1. INTRODUCTION
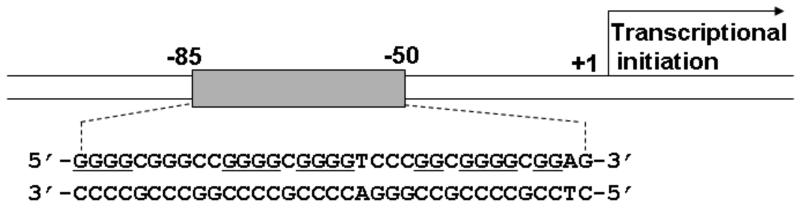
Polypurine/polypyrimidine tracts are known to exist at multiple sites in mammalian genomes, particularly in the proximal promoter regions of growth-related genes (1-7), including VEGF (Fig. 1). These cis-regulatory elements contain multiple Sp1 binding sites and several, studies have independently reported the presence of DNase I- or S1 nuclease-hypersensitive sites within the regions of DNA harboring this tract in both chromatin and negatively supercoiled plasmid DNA, suggesting that this tract is structurally dynamic and easily converted into alternative conformations different from the typical B-DNA structure (8-11). In general, the structural transition of B-DNA to alternative secondary structures is preceded by the local melting or unwinding of duplex DNA, which is facilitated by a negative supercoiling stress naturally generated behind the translocating RNA polymerase complex during the transcription of the genes (12-14). Thus, the structural transition of B-DNA to alternative non-B-conformations is believed to temporarily relieve a negative supercoiling stress generated under normal physiological conditions (15,16). The proximal region of the VEGF promoter contains such a pPu/pPy tract (Fig. 1) and this chapter uses this as an example for examining the structural dynamics, determining whether it can assume a number of different topological forms.
Fig. 1.
Schematic diagram showing the location of the pPu/pPy tract in the proximal promoter region of the VEGF gene.
1.1 VEGF
Most primary solid tumors go through a dormant state in which the maximum attainable size is about 1-2 mm in diameter when the tumor cells use only pre-existing host blood vessels (17,18). However, the growth of new blood vessels from pre-existing vessels by a process called angiogenesis allows the tumor cells to progressively expand and disseminate to distant organs (17,18). Therefore, angiogenesis represents an essential step for tumor growth and metastasis by providing not only oxygen and nutrients to proliferating tumor cells, but also escape routes for metastatic tumor cells (17,18). The switch to an angiogenic phenotype is mediated by a number of key regulators, such as fibroblast growth factors (FGFs), vascular endothelial growth factors (VEGFs) and angiopoietins, semaphorin, ephrin, Notch/Delta, and the roundabout/slit families of proteins (17,18). Among them, VEGF (or VEGF-A) has been considered to be the key mediator of tumor angiogenesis by stimulating proliferation, migration, survival, and permeability of endothelial cells (19-21).
VEGF expression is mainly regulated at the transcriptional level, and its expression is induced by a variety of factors, including hypoxia, pH, activated oncogenes, inactivated tumor suppressor genes, and growth factors (22-29). VEGF is frequently overexpressed in many types of cancer and the stable expression of VEGF appears to arise from increased VEGF promoter activity (20-30). The VEGF promoter region contains binding sites for several putative transcription factors, such as HIF-1, AP-1, AP-2, Egr-1, Sp1, and many others, suggesting that they may be involved in VEGF transcriptional regulation (3,22-29). Functional analysis of the human VEGF promoter using the full-length VEGF promoter reporter revealed that the proximal 36-bp region (−85 to −50 relative to transcription initiation site) is essential for basal or inducible VEGF promoter activity in several human cancer cells (3).
1.2 Probes for unusual DNA structures
The presence of the pPu/pPy tract within the proximal region of the VEGF promoter (Fig. 1) led us to speculate that this region might be structurally dynamic and could potentially assume a number of different topological forms. Since our previous studies demonstrated that oligonucleotides representing the coding strands of this tract could adopt G-quadruplex structures (30), we further investigated the structural dynamics and the overall forms of the pPu/pPy tract within the promoter of the VEGF gene. We employed in vitro footprinting analysis utilizing a negatively supercoiled plasmid DNA containing this region in order to mimic the in vivo situation, where negative supercoils prevail due to the active DNA transaction (12-16,30). The footprinting agents used in this study include DNase I, and S1 nuclease, since the reactivity of these probes is very sensitive to the conformation of DNA molecules (31-32). These reagents have been utilized in many previous studies to probe structural transitions from B-DNA to non-B-type DNA structures, such as melted DNA, hairpin structures, G-quadruplex structures, and others (9-11). While DNase I preferentially cleaves locally unwound or normal duplex regions over single-stranded regions, S1 nuclease preferentially cleaves single-stranded regions of DNA over duplex DNA (9-11). However, both enzymes show the lowest cleavage activity toward highly organized secondary structures such as hairpins or G-quadruplex structures (9-11, 31,32). For these reasons, the combined use of both nucleases in in vitro footprinting experiments reveals pertinent information about unusual structural features of defined elements within the global region of DNA duplex molecules (31,32).
2. MATERIALS
2.1. Labeling 5′-termini of nucleic acids with [32P]
T4 polynucleotide kinase (Fermentas).
Kinase buffer (10X): 500 mM Tris-HCl (pH 7.6), 100 mM MgCl2, 50 mM DTT, 1 mM spermidine, and 1 mM EDTA.
Adenosine 5′-gamma 32P triphosphate (γ-32P ATP), triethylammonium salt (6000 Ci/mmole, 10 mCi/mL, GE, Healthcare).
Micro Bio-Spin™ 30 Columns (Bio-Rad).
2.2. Digestion of plasmid DNA with nucleases
2.3. Radioactive cycle sequencing and linear amplification
Thermo Sequenase DNA Polymerase (USB); 4 units/μl, 0.0006 units/μl Thermoplasma acidophilum inorganic pyrophosphatase; 50 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 0.5% Tween®-20, 0.5% Igepal™ CA-630, 50% glycerol.
Reaction Buffer (concentrate): 260 mM Tris-HCl, pH 9.5, 65mM MgCl2.
Telomestatin (see Note 3).
Gene-specific primer: 0.5 pmol/μl; (such as VEGF) 5′-CCCAGCGCCACGACCTCCGAGCTACC -3′ (see Notes 4-5).
dNTP (10 mM) solution: 10 mM each dATP, dGTP, dCTP, dTTP (see Note 6).
ddG Termination Mix: 150μM each dATP, dCTP, 7-deaza-dGTP, dTTP; 1.5μM ddGTP.
ddA Termination Mix: 150 μM each dATP, dCTP, 7-deaza-dGTP, dTTP; 1.5 μM ddATP.
ddT Termination Mix: 150 μM each dATP, dCTP, 7-deaza-dGTP, dTTP; 1.5 μM ddTTP.
ddC Termination Mix: 150 μM each dATP, dCTP, 7-deaza-dGTP, dTTP; 1.5 μM ddCTP.
Stop Solution: 95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol.
2.4. Denaturing PAGE
TBE electrophoresis buffer (10X): 0.89 M Tris-HCl (pH 8.0), 0.89 M boric acid, 20 mM EDTA. Store at room temperature.
Sixteen percent acrylamide/bisacrylamide (29:1 with 3.3% C) with 8 M urea and N,N,N′,N′- TEMED, Bio-Rad, Hercules, CA.
Ammonium persulfate: prepare 10% solution in water. Store at 4 °C up to 1 month.
3. METHODS
In order to test if the pPu/pPy tract of a promoter region is structurally dynamic and can easily adopt a non-B-DNA conformation under physiological conditions, in vitro footprinting of the with DNase I and S1 nuclease is performed with a supercoiled form of a plasmid (such as pGL3-V789, which contains the VEGF promoter region from −727 to +50). This plasmid is incubated in the absence of any salt, or in the presence of 100 mM KCl to facilitate the evolution of the secondary structures from the pPu/pPy region. To test the binding of the G-quadruplex interactive agent to the secondary structures, the plasmid DNA is incubated with and without 1 μM telomestatin (33) for 1 hr at 37°C, and then treated with DNase I or S1 nuclease for 2 min. To map S1 and DNase I cleavage sites, linear amplification by PCR was performed with 32P-labeled-gene specific primers to amplify the top strand of both nuclease treated plasmid DNA.
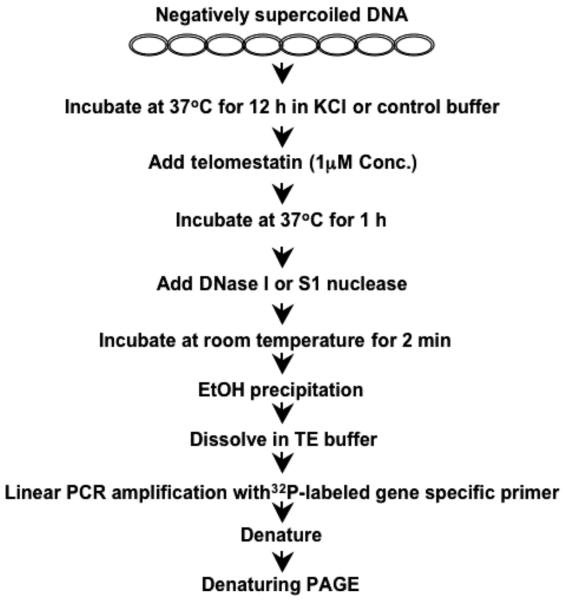
An overall strategy to perform in vitro footprinting of the wild-type VEGF promoter contained in a supercoiled plasmid in the presence of K+ and G-quadruplex-interactive compounds is shown schematically in Fig. 2.
Fig. 2.
Flowchart of DNase I and S1 nuclease footprinting experiments of the VEGF promoter region in a supercoiled plasmid.
3.1. Isolation of supercoiled plasmids
The supercoiled plasmids are isolated from transformed E. coli strain DH5α using the QIAGEN Plasmid Maxi Kit. This method is based on modified SDS-alkaline lysis of bacterial cells in combination with selective binding of the DNA to silica beads in the presence of certain salts (see Note 7).
3.2. Treatment plasmid DNA with nuclease
In an empty tube, supercoiled plasmid (2 μg) and 2.5 μl of 10X KCl or control buffer are mixed and brought to 25 μl with the addition of DDW (see Note 8).
The incubation proceeds at 37°C for over 12 h or overnight to allow the secondary structures to evolve from the pPu/pPy region.
For testing the drug binding to the secondary structures, add 1 μl of diluted drug solution in KCl or control buffer (e.g., 25 μM telomestatin in KCl or control buffer) to 25 μL DNA solution and mix them by vortexing and centrifuge briefly.
Incubate the reaction mixture at 37°C for 1h.
Add 2 μL of diluted DNase I (0.2 U) or 200 U of S1 nuclease to the tube, mix gently by pipetting up and down several times, cap the tubes and centrifuge briefly. After 1 minute digestion, add 100 μL of 0.3 M sodium acetate to the reactions followed by DNA precipitation with 2 volumes 100% ethanol and placement at −20°C overnight.
Spin in microfuge 30 min and allow pellet to air dry.
Resuspend the dried pellet completely in 25 μL of TE buffer.
3.3. Labeling 5′-termini gene-specific primers with [32P]
Prepare a reaction mixture (25 μL), containing oligonucleotide (4 μM), 3 μL γ-32P ATP (6000 Ci/mmole, 10 mCi/mL), T4 polynucleotide kinase (10 U), 2.5 μL 10× kinase buffer, and water.
Incubate the reaction mixture at 37 °C for 1 h in water bath for labeling 5′-termini of oligonucleotides with γ-[32P]-ATP.
After completion of the reaction, use Micro Bio-Spin™ 30 Columns (Bio-Rad) to remove unincorporated radioactive γ-32P ATP (6000 Ci/mmole, 10 mCi/mL) from labeled DNA. The instructions for use of Bio-Spin™ 30 Columns are based on recommendations from the manufacturer. In brief, the reaction mixture (25 μL) is loaded at the top of the column after centrifuging the column at 1,000 × g for 4 min in a swinging bucket and removing the packing buffer. The column is then centrifuged for 4 min at 1,000 × g to collect the purified 5′-end-labeled oligonucleotide in water (see Note 9).
3.4. Radiolabeled Primer Cycle Sequencing
Label four tubes representing G, A, T and C.
Place 4 μl of the ddGTP termination mix in the tube labeled G. Similarly fill the A, T and C tubes with 4 μl of the ddATP, ddTTP and ddCTP termination mixes respectively.
In a separate microcentrifuge tube, combine the following: 1 μL of plasmid DNA (20 pmole), 0.5 μL of concentrated reaction buffer, 1.0 μL of labeled primer, 1.0 μL of distilled water and 0.5 μL of Thermo Sequenase DNA Polymerase (see Note 10).
Transfer it to the PCR tube (from step 1), mix gently by pipetting up and down several times, cap the tubes and place them in the thermal cycler (see Note 11).
Carry out PCR using cycling conditions consisting of an initial 10-min denaturation step at 94°C, 1 min at 60°C, and 1 min at 72°C, for a total of 42 cycles (see Note 12).
Add 4 μl of stop solution to each of the termination reactions, mix thoroughly and centrifuge briefly.
3.5. Linear amplification of the plasmid DNA digested with DNase I or S1 nuclease using 32P-labeled primers
Place 4 μl of the plasmid DNA digested with DNase I or S1 nuclease from step 3.2 in each PCR tube.
In a separate microcentrifuge tube, combine the following: 0.5 μL of 10 mM dNTP solution, 0.5 μL of concentrated reaction buffer, 0.5 μL of labeled primer, 2.0 μL of distilled water and 0.5 μL of Thermo Sequenase DNA Polymerase (see Note 9).
Transfer it to the PCR tube (from step 1), mix gently by pipetting up and down several times, cap the tubes and place them in the thermal cycler (see Note 13).
Carry out PCR using cycling conditions consisting of an initial 10-min denaturation step at 94°C, 1 min at 60°C, and 1 min at 72°C, for a total of 42 cycles (see Note 14).
3.6. Separation of PCR products on denaturing PAGE
Set up a denaturing 10% polyacrylamide gel of 30 cm × 30 cm × 0.4 mm.
Prepare 60 mL of gel solution by mixing 6 mL TBE buffer (10X), 15 mL of 40% acrylamide/bisacrylamide (29:1), and 30 g urea and adding water to 60 mL. After adding 100 μL ammonium persulfate solution and 20 μL TEMED, pour the gel and insert the comb.
Once the gel is polymerized, carefully remove the comb, and wash the well with TBE buffer (1×) using a pasture pipette.
Attach the gel plates to the electrophoresis apparatus, and fill both reservoirs of the electrophoresis tank with 1× TBE. Pre-run and warm the gel for a least 30 minutes at 1400 V (constant voltage) using a DC power supply.
Heat the samples and sequencing ladders at 95 °C for 3 min, and chill the sample on ice before loading. Run the gel at about 1400 V.
After the desired resolution is obtained, detach the gel plates from the electrophoresis apparatus, and carefully separate both plates, leaving the gel attached to one plate.
Place a piece of thin chromatography paper (DE81) on top of the gel, and slowly pull back on the paper to transfer gels to the paper.
Place a piece of Whatman paper (3MM) underneath, and cover the wet gel with plastic wrap on top.
Put the gel sandwich in a dryer between a plastic fiber mat and clear plastic sheet, and dry the gel at 80 °C for at least 1 h with a vacuum.
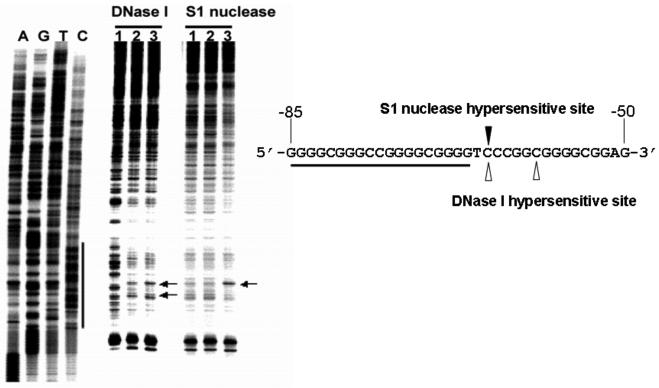
Place the dried gel in an X-ray film cassette. Obtain an autoradiogram by exposing the X-ray film to the dried gel. Alternatively, the image can be obtained by exposing the dried gel to the phosphor screen for an appropriate time and scanning the phosphor screen. Fig. 3 is an example of an autoradiogram of a 10% polyacrylamide sequencing gel, showing the results of S1 and DNase I footprinting experiments carried out with a supercoiled pGL3-V789 plasmid.
Fig. 3.
In vitro footprinting of the VEGF promoter region with DNase I, S1 nuclease or DMS. Autoradiograms showing S1 nuclease and DNase I cleavage sites on the G-strand of a supercoiled pGL3-V789 plasmid. This plasmid was incubated in control (lane 1), or in KCl buffer without (lane 2) and with 1 μM telomestatin (lane 3) at 37°C for 1 h before digesting with nucleases. Arrows indicate the hypersensitive cleavage sites to nucleases. The primer extension reaction revealed a long protected region at approximately −53 to −123 bp, including the G-rich sequences, when a supercoiled pGL3-V789 plasmid was incubated with 100 mM KCl and digested with DNase I (compare lanes 1 and 2). This indicates a possible transition from B-DNA to a G-quadruplex structure in the VEGF promoter region, which is consequently resistant to DNase I digestion. Significantly, a striking hypersensitivity was found in the presence of KCl and telomestatin at a cytosine located at the 3′-side of the G-quadruplex-forming region (underlined sequence), which is the junction site separating the putative G-quadruplex from the adjacent normal B-DNA (see arrow “A” in lane 3). The reactivity of S1 nuclease at the VEGF proximal promoter region was also moderately reduced in the presence of telomestatin and KCl, and the hypersensitivity site observed with S1 nuclease corresponds to one of those obtained with DNase I in the presence of telomestatin and KCl (lane 3) (Figure modified from ref. 30).
Acknowledgements
This research was supported by grants from the National Institutes of Health (CA109069). We are grateful to Drs. Allison Hays and Keith Fox for proofreading and editing the final version of the manuscript and figures. We also thank Drs Keping Xie and Kazuo Shin-ya for providing pGL3-V789 and telomestatin, respectively, for this study.
Footnotes
Plasmid pGL3-V789 was originally constructed by Dr. Keping Xie by subcloning a 789 bp fragment containing 5′ VEGF promoter sequences from −729 to +50 relative to the transcription initiation site into the KpnI and NheI sites of pGL3-basic (Promega, Madison, WI), which contains firefly luciferase coding sequences (3).
KCl Buffer provides optimum conditions for the formation of G-quadruplex structures from the single stranded DNA.
Telomestatin was kindly provided by Dr. Kazuo Shin-ya.
It is also a good idea to check the sequence of the primer for possible self-annealing (dimer formation could result) and for potential ‘hairpin’ formation, especially those involving the 3′ end of the primer.
Finally, check for possible sites of false priming in the vector or other known sequence if possible, again stressing matches which include the 3′ end of the primer.
All enclosed reagents should be stored frozen at −20°C and keep all reagents on ice once removed from storage for use.
The protocol is suitable for obtaining pure plasmid DNA up to 100μg from 30~100 ml bacterial culture grown in LB medium. The culture volume should be reduced to half or less when bacteria grown in rich medium are used.
It is best to prepare one large reaction mix and then aliquot 25 μl into each sample tube.
No further purification is required for most sequencing.
Use 1.0 pmol of fresh 32P-labeled primers, but 2-5-fold more primer can be used for shorter exposure times.
It is best to prepare one large reaction mix and then aliquot 4 μl into each sample tube.
The specific cycling parameters used will depend on the primer length and sequence and the amount and purity of the template DNA.
It is best to prepare one large reaction mix and then aliquot 4 μl into each sample tube.
If your gels seem to require longer exposures, and more template is not available, increase the number of PCR cycles.
References
- 1.McCarthy JG, Heywood SM. A long polypyrimidine/polypurine tract induces an altered DNA conformation on the 3′ coding region of the adjacent myosin heavy chain gene. Nucleic Acids Res. 1987;15:8069–8085. doi: 10.1093/nar/15.19.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Q, Le X, Abbruzzese JL, Peng Z, Qian CN, Tang H, et al. Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res. 2001;61:4143–4154. [PubMed] [Google Scholar]
- 4.Rustighi A, Tessari MA, Vascotto F, Sgarra R, Giancotti V, Manfioletti G. A polypyrimidine/polypurine tract within the Hmga2 minimal promoter: a common feature of many growth-related genes. Biochemistry. 2002;41:1229–1240. doi: 10.1021/bi011666o. [DOI] [PubMed] [Google Scholar]
- 5.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS protooncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 7.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pullner A, Mautner J, Albert T, Eick D. Nucleosomal structure of active and inactive c-myc genes. J Biol Chem. 1996;271:31452–31457. doi: 10.1074/jbc.271.49.31452. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Lin XH, Qiu QQ, Deuel TF. Modulation of transcription of the platelet-derived growth factor A-chain gene by a promoter region sensitive to S1 nuclease. J. Biol. Chem. 1992;267:17022–17031. [PubMed] [Google Scholar]
- 10.Siebenlist U, Henninghausen L, Battey J, Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984;37:381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- 11.Evans T, Efstratiadis A. Sequence-dependent S1 nuclease hypersensitivity of a heteronomous DNA duplex. J. Biol. Chem. 1986;261:14771–14780. [PubMed] [Google Scholar]
- 12.Benham CJ. Theoretical analysis of conformational equilibria in superhelical DNA. Ann. Rev. Biophys. Biophysical Chem. 1985;14:23–45. doi: 10.1146/annurev.bb.14.060185.000323. [DOI] [PubMed] [Google Scholar]
- 13.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams DL, Kowalski D. Easily unwound DNA sequences and hairpin structures in the Epstein-Barr virus origin of plasmid replication. J. Virol. 1993;67:2707–2715. doi: 10.1128/jvi.67.5.2707-2715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front. Biosci. 2007;12:4409–4423. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 16.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 17.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan DC, Bicknell R. New molecular pathways in angiogenesis. Br. J. Cancer. 2003;89:228–231. doi: 10.1038/sj.bjc.6601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martiny-Baron G, Marme D. VEGF-mediated tumour angiogenesis: a new target for cancer therapy. Curr. Opin. Biotechnol. 1995;6:675–680. doi: 10.1016/0958-1669(95)80111-1. [DOI] [PubMed] [Google Scholar]
- 20.Goodsell DS. The molecular perspective: VEGF and angiogenesis. Stem Cells. 2003;21:118–119. doi: 10.1634/stemcells.21-1-118. [DOI] [PubMed] [Google Scholar]
- 21.Jain RK. Tumor angiogenesis and accessibility: role of vascular endothelial growth factor. Semin. Oncol. 2002;29:3–9. doi: 10.1053/sonc.2002.37265. [DOI] [PubMed] [Google Scholar]
- 22.Gunningham SP, Currie MJ, Han C, Turner K, Scott PA, Robinson BA, et al. Vascular endothelial growth factor-B and vascular endothelial growth factor-C expression in renal cell carcinomas: regulation by the von Hippel-Lindau gene and hypoxia. Cancer Res. 2001;61:3206–3211. [PubMed] [Google Scholar]
- 23.Schafer G, Cramer T, Suske G, Kemmner W, Wiedenmann B, Hocker M. Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J. Biol. Chem. 2003;278:8190–8198. doi: 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- 24.Maeno T, Tanaka T, Sando Y, Suga T, Maeno Y, Nakagawa J, et al. Stimulation of vascular endothelial growth factor gene transcription by all trans retinoic acid through Sp1 and Sp3 sites in human bronchioloalveolar carcinoma cells. Am. J. Respir. Cell Mol. Biol. 2002;26:246–253. doi: 10.1165/ajrcmb.26.2.4509. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Ye D, Xie X, Chen B, Lu W. VEGF, VEGFRs expressions and activated STATs in ovarian epithelial carcinoma. Gynecol. Oncol. 2004;94:630–635. doi: 10.1016/j.ygyno.2004.05.056. [DOI] [PubMed] [Google Scholar]
- 26.Pal S, Datta K, Khosravi-Far R, Mukhopadhyay D. Role of protein kinase Czeta in Ras-mediated transcriptional activation of vascular permeability factor/vascular endothelial growth factor expression. J. Biol. Chem. 2001;276:2395–2403. doi: 10.1074/jbc.M007818200. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka T, Kanai H, Sekiguchi K, Aihara Y, Yokoyama T, Arai M, et al. Induction of VEGF gene transcription by IL-1 beta is mediated through stress-activated MAP kinases and Sp1 sites in cardiac myocytes. J. Mol. Cell Cardiol. 2000;32:1955–1967. doi: 10.1006/jmcc.2000.1228. [DOI] [PubMed] [Google Scholar]
- 28.Finkenzeller G, Sparacio A, Technau A, Marme D, Siemeister G. Sp1 recognition sites in the proximal promoter of the human vascular endothelial growth factor gene are essential for platelet-derived growth factor-induced gene expression. Oncogene. 1997;15:669–676. doi: 10.1038/sj.onc.1201219. [DOI] [PubMed] [Google Scholar]
- 29.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saluz HP, Jost JP. Approaches to characterize protein-DNA interactions in vivo. Crit. Rev. Eukaryot. Gene Expr. 1993;3:1–29. [PubMed] [Google Scholar]
- 32.Dabrowiak JC, Goodisman J, Ward B. Quantitative DNA footprinting. Methods Mol. Biol. 1997;90:23–42. doi: 10.1385/0-89603-447-X:23. [DOI] [PubMed] [Google Scholar]
- 33.Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]