Abstract
Huntington's disease (HD) is caused by expansion of the polymorphic polyglutamine segment in the huntingtin protein. Full-length huntingtin is thought to be a predominant HEAT repeat α-solenoid, implying a role as a facilitator of macromolecular complexes. Here we have investigated huntingtin's domain structure and potential intersection with epigenetic silencer polycomb repressive complex 2 (PRC2), suggested by shared embryonic deficiency phenotypes. Analysis of a set of full-length recombinant huntingtins, with different polyglutamine regions, demonstrated dramatic conformational flexibility, with an accessible hinge separating two large α-helical domains. Moreover, embryos lacking huntingtin exhibited impaired PRC2 regulation of Hox gene expression, trophoblast giant cell differentiation, paternal X chromosome inactivation and histone H3K27 tri-methylation, while full-length endogenous nuclear huntingtin in wild-type embryoid bodies (EBs) was associated with PRC2 subunits and was detected with trimethylated histone H3K27 at Hoxb9. Supporting a direct stimulatory role, full-length recombinant huntingtin significantly increased the histone H3K27 tri-methylase activity of reconstituted PRC2 in vitro, and structure–function analysis demonstrated that the polyglutamine region augmented full-length huntingtin PRC2 stimulation, both in HdhQ111 EBs and in vitro, with reconstituted PRC2. Knowledge of full-length huntingtin's α-helical organization and role as a facilitator of the multi-subunit PRC2 complex provides a novel starting point for studying PRC2 regulation, implicates this chromatin repressive complex in a neurodegenerative disorder and sets the stage for further study of huntingtin's molecular function and the impact of its modulatory polyglutamine region.
INTRODUCTION
In 1993, genetic studies identified the CAG trinucleotide repeat mutation that causes Huntington's disease (HD) (1). This dominantly inherited disorder is characterized by loss of brain neurons, especially in the striatum, and the inexorable onset of motor, cognitive and behavioral symptoms (2). The HD mutation comprises expanded versions of a polymorphic CAG repeat that elongate a variable polyglutamine segment in the huntingtin protein, from the normal range (8–37 residues) to 38 or more residues (1). This polymorphic polyglutamine segment is thought to confer a subtle structural alteration and gain of huntingtin function (3,4). Indeed, genotype–phenotype studies have demonstrated that the polyglutamine region normally modulates huntingtin function in cellular energy metabolism (3,5,6) and the expression of expanded polyglutamine tracts within the endogenous huntingtin protein is associated with dominant phenotypes in model systems (7,8). However, while polyglutamine, alone or embedded in short polypeptides, exhibits striking physical properties (9,10), typically measured in aggregation assays (11), little is known of the molecular impact of the polyglutamine region on the structure and function of the full-length huntingtin protein, though this information is needed to fully understand huntingtin biology and the trigger of HD pathogenesis.
Full-length huntingtin is now thought to comprise a large α-helical solenoid (repeated units arranged in a continuous superhelix/coil) (12) rather than a globular protein, as its ∼3144 amino acid length was predicted to be entirely spanned by loosely conserved HEAT/HEAT-like repeats (13). Indeed HEAT repeats, curling anti-parallel α-helical units, were first recognized within huntingtin (14) and were named for huntingtin and for elongation factor 3 component eIF3k, protein phosphatase 2A regulatory subunit PR65/A and target of rapamycin TOR1 (14), though this structural element now defines a larger class of proteins (15). HEAT and HEAT-like repeats may augment other motifs (e.g. eIF3k and TOR1) or may encompass the entire protein (e.g. PR65/A and importin-β), stacking to confer dramatic conformational flexibility and multi-contact protein interaction topologies suited to the role of these α/α-solenoid molecules as facilitators of dynamic multi-subunit complexes. PR65/A, for example, facilitates diverse phosphatase holoenzymes involved in cell signaling and metabolism (16), while importin-β serves distinct nuclear transport complexes that engage a variety of cargos (17,18).
The emerging molecular view of huntingtin as a predominant HEAT/HEAT-like α-helical facilitator protein is supported by the results of studies of the full-length protein. For example, endogenous full-length murine huntingtin exhibited distinct subcellular epitope patterns, implying multiple alternate conformations (8,19), and the first circular dichroism spectra of recombinant full-length human huntingtin denoted a predominantly α-helical molecule (20). Moreover, while the specific players in most cases remain to be identified, huntingtin is thought to be multi-functional, participating in diverse subcellular processes ranging from vesicle trafficking to energy metabolism and gene transcription (21).
As an approach to defining huntingtin's essential functional molecular interactions in a mammalian system, we, and others, are studying the consequences of targeted inactivation of the murine HD gene. Huntingtin was initially shown to be required in the extraembryonic tissue (22) to bypass a block early in embryonic development just before head-fold formation (23–25). Our subsequent analysis of huntingtin null embryos then revealed a constellation of other morphological and molecular phenotypes, including anterior streak and mesoderm patterning deficits, failure to properly silence growth (e.g. Nodal, Fgf8) and transcription (e.g. Evx1, T) factor genes, that also were reminiscent of embryos deficient in polycomb repressive complex 2 (PRC2), due to loss of core components Ezh2 (26), Suz12 (27) or Eed (28), thereby implying a possible intersection of huntingtin with this epigenetic silencer (29).
Here we have investigated the α-solenoid view of the HD protein, by determining the domain organization of a set of full-length recombinant human huntingtins and by utilizing these reagents, in conjunction with targeted mutations at the murine HD locus that probe the endogenous protein, to examine the specific hypothesis that huntingtin may assist PRC2. The results of our analysis nominate huntingtin as a novel α-solenoid stimulator of this multi-subunit histone H3 lysine 27 (H3K27) methyltransferase complex (30–33).
RESULTS
Recombinant full-length huntingtin hinged α-helical domain structure
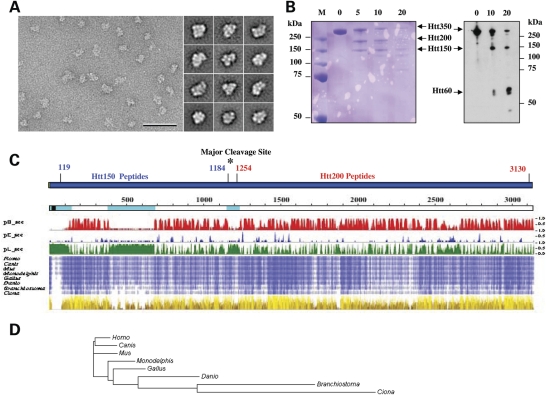
To determine whether full-length huntingtin might fulfill the flexible segmental organization of an α/α-solenoid protein, we analyzed a set of full-length (>3144 residues) recombinant human FLAG-tag huntingtins, with polyglutamine tracts of 23, 32 or 43 residues. The proteins were expressed in Sf9 insect cells from Baculovirus vectors, generated as described in Materials and Methods, and, though large, each recombinant protein was enriched by column chromatography from Sf9 insect cell extracts (Materials and Methods), yielding a sharp elution peak (see Materials and Methods, Supplementary Material, Fig. S1). As illustrated for Q23-huntingtin (Supplementary Material, Fig. S1), the enriched protein was full-length (∼350 kDa) and of high purity, yielding a single Coomassie blue stained band that immunoblot revealed was detected by N- and C-terminal huntingtin antibodies.
Analysis of the recombinant protein by circular dichroism produced spectra that confirmed a predominant α-helical structure (data not shown), as previously reported (20). To investigate the protein's overall domain organization, FLAG-tag huntingtin was examined by negative stain electron microscopy (EM). Analysis of about 10 000 interactively selected Q23-huntingtin particles revealed 100 structurally distinguishable classes (Supplementary Material, Fig. S2A), illustrated by the representative class averages shown in Figure 1A. These structural classes were also similar for Q32 huntingtin and Q43 huntingtin (Supplementary Material, Fig. S2B), revealing that the polyglutamine region did not overtly alter huntingtin's remarkable conformational variability. Consistent with a flexible segmental α-helical domain organization, limited tryptic digestion of the recombinant protein readily yielded two large domains (∼150 kDa and ∼200 kDa fragments) (Fig. 1B) and mass spectrometry (Supplementary Material, Tables S1 and S2) located a major accessible hinge region between residues 1184–1254. Notably, the smaller fragments produced by continued digestion (Fig. 1B) revealed that the extreme N-terminus, with its polyglutamine segment, was buried within the ∼150 kDa domain, on an initially inaccessible ∼60 kDa subdomain. This was consistent with the results of CHOP, for structural domains (34,35), which predicted residues 124–971 in a super-helical architecture, with significant similarity (2.5e−20) to PR65/A (PDB I.D. 1b3u).
Figure 1.
Huntingtin conformational flexibility and domain organization. (A) Representative area of an electron micrograph of negatively stained FLAG-Q23 huntingtin (scale bar 50 nm) (left) and representative class averages (side length of panels is 28.8 nm) (right) showing the structural variability of huntingtin. (B) Timed proteolysis of Q23 FLAG-huntingtin (Htt350) with trypsin (0, 5, 10, 20 min) yielded two major Coomassie Blue stained products at ∼150 kDa (Htt150) and ∼200 kDa (Htt200) (left), though immunoblot probed with anti-FLAG detected the ∼150 kDa fragment and a smaller ∼60 kDa product (right). (C) Schematic of human huntingtin (blue bar), with FLAG-tag (green) and locations of Htt150 and Htt250 mass spectrometry peptides (Supplementary Material, Tables S1 and S2) and major trypsin cleavage site (asterisk). Below this, huntingtin (open line) is depicted with the polyglutamine tract (black block) and amino acid coordinates (above), NORSp predicted disordered regions (light blue), matching the predictions of PROF where pHsec, pEsec and pLsec represent the probability (1 = high, 0 = low) for helix (red), strand (blue) and neither helix nor strand (green). Below this, compressed multiple sequence alignment of huntingtin from human and seven chordates with increasing intensity of blue shading for residues identical in 4 to 8 organisms and physico-chemical properties (from Jalview) conserved for each amino acid position, from dark brown (least) to bright yellow (most), with height corresponding to increasing conservation. (D) Phylogram based upon alignment of huntingtin homologues for the eight representative chordates, with branch lengths proportional to the inferred evolutionary change.
Multiple sequence alignment of eight representative chordate huntingtins (sea squirt to human) (see Materials and Methods), summarized in Figure 1C and the phylogram in Figure 1D, revealed that the conservation of the unstructured hinge segment and pattern of α-helical structure were conserved through 500 million years of evolution, strongly implying that huntingtin's overall modular organization and conformational flexibility are critical for its biological function.
Impaired PRC2 function in the absence of huntingtin
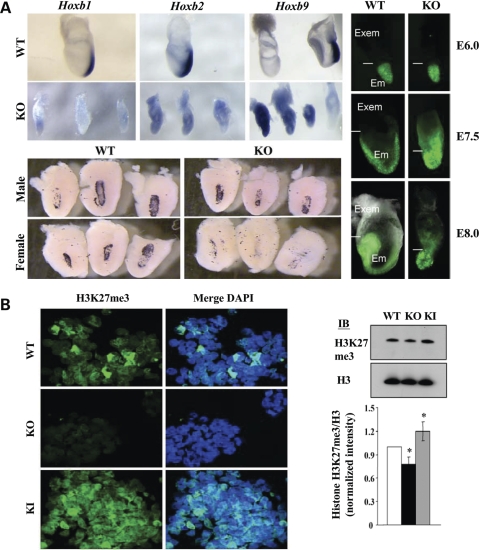
The hypothesis that huntingtin may facilitate PRC2 was first explored in vivo by examining huntingtin null Hdhex4/5 homozygote embryos and cultured EBs for molecular epigenetic phenotypes reported in PRC2-deficient embryos (26–28,36). Though Hdhex4/5 homozygote embryos at embryonic stage E7.5 have previously been shown to properly localize stage-appropriate markers such as Otx2, Hnf3β and Hesx1 (29), the results of whole mount in situ hybridization, in Figure 2A, revealed that huntingtin null embryos failed to properly repress PRC2 regulated Hox gene expression, as evidenced by ectopic Hoxb1, Hoxb2 and Hoxb9 mRNA. Moreover, female huntingtin null embryos exhibited decreased differentiation of trophoblast giant cells, marked by PL-1 mRNA expression, which normally requires proper obligatory silencing of the paternally inherited X chromosome. Indeed, in the absence of huntingtin, female embryos inheriting a paternally transmitted X chromosome marked by a GFP-transgene, displayed inappropriate reactivation of GFP-signal in cells of the extra-embryonic tissue, though random X chromosome inactivation in the embryro proper appeared to be normal.
Figure 2.
Huntingtin stimulated PRC2 during development. (A) Whole mount in situ hybridization Hoxb1, Hoxb2, Hoxb9 mRNA expression in E7.5 Hdhex/4/5/Hdhex/4/5 (KO) huntingtin null embryos was not properly silenced and restricted to the posterior, as observed for wild-type Hdh+/Hdh+ (WT) embryos (top). Decreased placental lactogen (PL-1) mRNA signal (purple) in bisected decidua revealed fewer giant trophoblast cells in female embryos lacking huntingtin (KO), compared with null male (KO) or wild-type (WT) embryos of either gender (bottom). Female embryos, at the indicated developmental stages (ectoplacental cone, Reichert's membrane removed), show aberrant Xp GFP-transgene reactivation in the Hdhex/4/5/Hdhex/4/5 (KO) extraembryonic tissue (Exem), compared with Hdh+/Hdh+ (WT) tissue, while random inactivation in the embryo proper (Em) appeared normal (right). (B) Confocal images of immunostained sections of day 4 Hdh+/Hdh+ (WT) and Hdhex/4/5/Hdhex/4/5 (KO) huntingtin null EBs revealed decreased histone H3K27me3 signal in the latter, whereas knock-in Hdh+/HdhQ111 (KI) EBs, expressing huntingtin with a 111 residue polyglutamine segment exhibit increased histone H3K27me3 signal. Immunoblot of nuclear extracts and plot of histone H3K27me3/H3 band intensity ratio, normalized to wild-type (white bar), confirmed significantly (P < 0.016) decreased histone H3K27me3 in Hdhex/4/5/Hdhex/4/5 (KO) extracts (black bar) and significantly (P < 0.044) increased histone H3K27me3 in knock-in Hdh+/HdhQ111 (KI) extract (grey bar), compared with wild-type (n = 4).
Investigation of histone H3K27 methylation in Hdhex4/5 null EBs, developing in cell culture from embryonic stem cells, demonstrated that huntingtin was required for efficient re-establishment of global tri-methylated histone H3K27. The level of tri-methylated histone H3K27 was decreased at day 2 (data not shown) and day 4, as illustrated by immunostaining and immunoblot analysis, in Figure 2B, though was normalized by days 6–11 (Supplementary Material, Fig. S3A). Notably, global levels of di-methylated histone H3K27 were not altered in huntingtin null EBs (Supplementary Material, Fig. S3B), indicating that lack of huntingtin specifically affected H3K27 tri-methylation.
The phenotypes that indicated impaired PRC2 function in huntingtin-null embryos were less severe than complete loss of PRC2 methyltransferase activity, due to lack of the Ezh2 catalytic subunit, and appeared milder than phenotypes due to loss of Eed. This is consistent with a role for huntingtin, not as a core PRC2 component, but as an essential, potentially dynamic, facilitator of PRC2 activity during development.
Full-length huntingtin stimulated PRC2 tri-methyltransferase activity
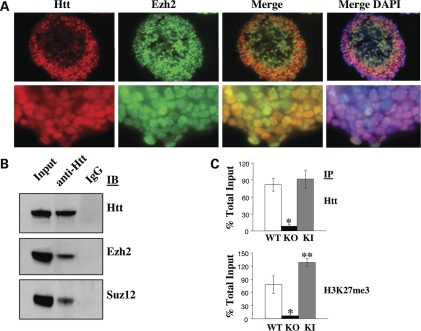
Developing day 4 EBs were chosen as a system amenable to biochemical analysis, to evaluate the possibility that full-length huntingtin might intersect with PRC2 in the nucleus. Full-length huntingtin was detected by immunoblot analysis in nuclear, as well as cytoplasmic extracts (Supplementary Material, Fig. S3C), and antibody reagent AP194, which immunostained nuclear though not cytoplasmic conformations of full-length huntingtin (8), revealed huntingtin in the nuclei of cells in all three germ-layers, with Ezh2-stain, especially in the outermost endodermal cells (Fig. 3A). Furthermore, the results of analysis of nuclear extracts by gel filtration chromatography demonstrated that full-length huntingtin was co-eluted with PRC2 subunits Ezh2 and Suz12 (Supplementary Material, Fig. S3D). Analysis by co-immunoprecipitation, with specific antibody reagents, yielded a proportion of full-length huntingtin, with Ezh2 and Suz12, as revealed by immunoblot analysis of the precipitated proteins shown in Figure 3B. In addition, as summarized in Figure 3C, chromatin immunoprecipitation (ChIP), with anti-huntingtin or anti-histone H3K27me3, enriched Hoxb9 sequences from wild-type, though not from huntingtin null day 4 EB nuclei, thereby placing huntingtin at Hoxb9 chromatin in wild-type cells and supporting a functional role for huntingtin in stimulating histone H3K27 trimethylation.
Figure 3.
Huntingtin associated with PRC2 in the nucleus. (A) Fluorescent microscope images of an immunostained section of a day 4 Hdh+/Hdh+ wild-type (WT) EB, demonstrating the nuclear conformation of huntingtin (Htt), detected by AP194 antibody (8), and anti-Ezh2-signal in DAPI-stained nuclei (merge). (B) Immunoblot showing co-immunoprecipitation of Ezh2 and Suz12 with huntingtin (Htt), from nuclear extracts of day 4 Hdh+/Hdh+ (WT) EBs. (C) Plot of the results of chromatin immunoprecipitation analysis of Hoxb9, demonstrating enrichment of huntingtin (Htt) and trimethylated histone H3K27 (H3K27me3) in day 4 Hdh+/Hdh+ wild-type (WT) EBs, that is not apparent in the absence of huntingtin in Hdhex4/5/Hdhex4/5 (KO) EBs, but is increased in Hdh+/HdhQ111 knock-in (KI) EBs, expressing 111-glutamine huntingtin (n = 3; anti-Htt KO versus WT P < 0.0004; anti-histone H3K27me3 KO versus WT P < 0.0037; anti-Htt KI versus WT P < 0.4267; anti-histone H3K27me3 KI versus WT P < 0.0157).
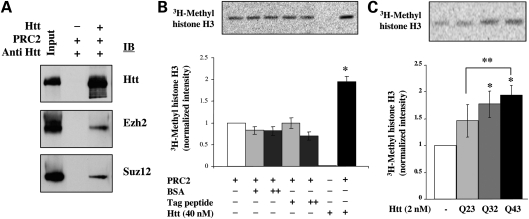
This interpretation was confirmed by the results of in vitro experiments, to determine whether recombinant full-length human huntingtin would interact with and alter the histone H3K27 methyltransferase activity of reconstituted PRC2 in a previously reported in vitro assay (30–33) (see Materials and Methods). As shown in the immunoblot in Figure 4A, full-length FLAG-tag Q23 huntingtin added to recombinant PRC2 was co-immunoprecipitated with Ezh2 and Suz12, and, as illustrated in Figure 4B, the recombinant protein significantly increased PRC2-specific histone H3K27 methylation, as judged by the intensity of bands of incorporated tritium. The stimulatory effect of full-length huntingtin, compared with reactions without huntingtin or with control peptides, was observed over a range of huntingtin (Supplementary Material, Fig. S4A) and nucleosomal array (Supplementary Material, Fig. S4B) concentrations. Furthermore, consistent with the finding that huntingtin was needed to stimulate tri- but not di-methylation of histone H3K27 in vivo (Fig. 2B, Supplementary Material, Fig. S3B), the results of immunoblot analysis of the in vitro PRC2 reaction products demonstrated that recombinant huntingtin specifically enhanced histone H3K27 tri-methylation but not di-methylation (Supplementary Material, Fig. S4C).
Figure 4.
Huntingtin stimulated recombinant PRC2 in vitro. (A) Immunoblot showing co-immunoprecipitation of FLAG-huntingtin with recombinant Ezh2 and Suz12 members of reconstituted PRC2 to which huntingtin (Htt) was added (+) or was not added (−). (B) Autoradiogram of bands of 3H-methyl histone H3 produced by reconstituted PRC2, and below the plot of the quantified band intensities, demonstrating that addition of 40 nm recombinant Q23 huntingtin (Htt), but not bovine serum albumin (BSA), or tag-peptide (Tag peptide), significantly stimulated PRC2 activity. (n = 3; *P < 0.05). (C) Autoradiogram showing bands of 3H-methyl histone H3 produced by reconstituted PRC2 in the absence (−) and presence of 2 nm recombinant huntingtins, with different polyglutamine sizes, and below a plot of quantified band intensities, relative to baseline PRC2 activity, demonstrating a progressive increase in huntingtin's stimulation of PRC2 as polyglutamine size is increased (n = 3; Q43Htt or Q32 versus no Htt *P < 0.017; Q43Htt versus Q23Htt **P < 0.045).
The polyglutamine region modulated huntingtin PRC2 stimulation
Functional interactions involving globular proteins are typically validated by structure–function experiments that entail targeted disruption of a single point-to-point protein interaction motif. However, consistent with previous evidence of striking conformational variability in vivo, our analysis of recombinant huntingtin revealed a flexible, non-globular HEAT repeat α-helical domain organization (Fig. 1, Supplementary Material, Fig. S2). For other HEAT solenoids, protein-interaction entailed dramatic conformational switches and complex multiple points of contact along the idiosyncratic contours formed by HEAT repeat packing, not accurately mapped by the methods that determine the sites of docking-interactions between globular proteins (37).
Therefore, in the absence of detailed knowledge of huntingtin's likely complex interactions with PRC2, and perhaps its chromatin substrate, we assessed the potential impact of huntingtin's only known naturally occurring functional polymorphism. The polyglutamine region has been shown to subtly but significantly modulate the consequences of endogenous full-length murine and human huntingtin in vivo (5,6). Moreover, our analysis of recombinant full-length huntingtins demonstrated that the polymorphism, even into the expanded HD range, did not overtly alter the protein's overall structure (Supplementary Material, Fig. S2), consistent with the finding that this modulatory region did not impair (38) and indeed was not needed for huntingtin's early developmental activity (5).
The potential effect of the polyglutamine region on PRC2 activity was first assessed by analysis of day 4 EBs expressing endogenous murine huntingtin with 111-glutamines, from the previously described HdhQ111 knock-in allele (19,39). Compared with wild-type EBs, and in contrast to huntingtin-null EBs, the cells of HdhQ111/Q7 EBs exhibited elevated levels of tri-methylated histone H3K27, by immunostaining and immunoblot analysis, as shown in Figure 2B, though not di-methylated H3K27 (Supplementary Material, Fig. S3B). Moreover, ChIP analysis revealed increased enrichment of huntingtin and tri-methylated histone H3K27 at Hoxb9, as summarized in Figure 3C. Consistent with these findings, as shown in Figure 4C, in the in vitro assay with reconstituted PRC2, full-length recombinant human proteins with polyglutamine segments longer than 23-residues (32- and 43-residues) progressively increased huntingtin stimulation of histone H3K27 methylation. Thus, in both the cell-based and the molecularly-defined structure–function experiments, the impact of the polyglutamine modulatory region confirmed huntingtin's role in facilitating the PRC2 histone H3K27 methyltransferase complex.
DISCUSSION
The expansion of the polymorphic polyglutamine region in the huntingtin HEAT repeat protein is the root genetic cause of HD pathogenesis. Despite this compelling reason, and though this ancient protein is of interest because it is the founding member of a growing class of HEAT repeat proteins, huntingtin's molecular organization and function have attracted relatively little attention. The results of our genetic structure–function experiments extend the single report of native huntingtin's predominant α-helical nature (20), by demonstrating features consistent with a flexible α-solenoid organization and by providing strong empirical support for huntingtin as a dynamic facilitator of at least one multi-functional macromolecular complex, the PRC2 methyltransferase.
Probing the structural organization of full-length native recombinant human huntingtin by circular dichroism spectroscopy, we confirmed the protein's previously reported predominant α-helical nature (20). However, in contrast to that report, our purification strategy yielded a sharp chromatographic peak comprising only full-length huntingtin, without the reported ∼220 kDa piece of huntingtin (starting at residue 622), which we speculate may have arisen from proteolysis of partially denatured protein produced by the harsher purification conditions utilized in that study. Limited proteolysis of the native full-length recombinant huntingtin initially yielded two major products, an N-terminal ∼150 kDa segment (ending at residue 1184) and an ∼200 kDa C-terminal segment (starting at residue 1254), implying a domain organization comprising two large nearly equal sized α-helical arms, separated by an accessible hinge region. Continued digestion of these domains yielded additional bands, including an ∼60 kDa N-terminal fragment bearing the polyglutamine region that implied cleavage within an unstructured region located at ∼residue 500. Thus, though it may be susceptible to proteolytic cleavage when huntingtin is denatured, this sub-domain was buried within the 150 kDa N-terminal arm of the native protein, likely because this domain may assume a super-helical structure resembling the PR65/A α-solenoid (PDB I.D. 1b3u).
The segmental α-helical domain organization and the conformational flexibility of full-length huntingtin, revealed by negative stain EM, are general structural features expected of a predominant HEAT repeat α/α-solenoid protein. However, high-resolution analysis will be needed to prove the continuous helical structure of the molecule. Indeed, though the elongated and often curved shapes formed by stacking of adjacent HEAT and HEAT-like repeats are similar for different HEAT repeat proteins, the precise contours are determined by the amino acid sequences of these degenerate structural elements. To date, crystal structures for the first 60 amino acids of huntingtin (encoded by exon 1) have been analyzed, directly demonstrating the α-helical secondary structure of the first 17 amino acids, the structural variability of the abutting 17 residue polyglutamine segment and helical arrangement of the adjacent polyproline rich segment (40). Knowledge of the domain organization of native huntingtin, which was not accurately predicted using various pieces of the protein (41), should now spur efforts to determine the higher-order structure of the functional protein.
In support of huntingtin's role as a facilitator, implied by its predominant HEAT domain structure, the lack of huntingtin led to impaired PRC2 epigenetic gene and chromatin silencing function in embryos and impaired reestablishment of global histone H3K27 tri-methylation in developing EBs, whereas full-length recombinant human huntingtin specifically stimulated the tri-methyltransferase activity of reconstituted PRC2. Furthermore, as implied by co-immunoprecipitation of the full-length endogenous and recombinant proteins with core PRC2 members, huntingtin's direct role in stimulating PRC2, in vivo and in vitro, was confirmed by the progressive effect of huntingtin's polymorphic polyglutamine region, previously recognized as a modulatory segment of full-length endogenous murine (5) and human (6) huntingtin.
Though our data reveal that huntingtin's role in facilitating PRC2 tri-methyltransferase activity is important for normal murine embryonic development, the timing and duration of this interaction, as well as the subset of PRC2-target genes that may, like Hoxb9, be modulated, are areas that remain to be investigated. Though it is not clear exactly how huntingtin may interact with PRC2 and/or its nucleosomal histone substrate, it is unlikely to stimulate PRC2 in the same manner as PHF1, a recently reported globular PRC2 accessory protein that is thought to contact the Ezh2 catalytic subunit via its PHD finger domains (42,43). Indeed, as discussed earlier, full-length huntingtin's proposed α-helical solenoid structure promises to offer a novel mode of PRC2 regulation. It seems reasonable, from the striking alternate sub-cellular epitope patterns of the full-length endogenous protein, that this will entail dramatic conformational switches and complex contacts along the topological contours formed by stacking of the protein's adjacent HEAT/HEAT-like repeats, as reported for other predominant α-solenoid proteins (16–18). The availability of a system for purifying native full-length recombinant huntingtin, and empirical knowledge of its domain organization, should now enable high-resolution structural studies to determine the details of huntingtin's functional molecular interactions with PRC2/chromatin, including the potential structural role of the modulatory polyglutamine region.
In summary, the proposal that full-length huntingtin comprises a large hinged α-helical solenoid, which serves as a facilitator of the PRC2 complex, now provides a nuanced view of the molecule and its polymorphic polyglutamine region, thereby setting the stage for defining other functional complexes that full-length huntingtin may assist. Furthermore, it provides novel starting points for understanding the in vivo regulation of mammalian PRC2 and suggests that enhanced activity of this epigenetic regulator merits investigation as a potential contributor to HD neurodegeneration.
MATERIALS AND METHODS
Human FLAG-huntingtin insect vector expression clones
pFASTBAC1 vector (Invitrogen), with a FLAG-tag sequence adjacent to the unique BamHI site, was cleaved and a 10 kb EagI–BssHII fragment encoding human huntingtin (23 glutamines) (Genbank accession number L12392) was inserted. pFASTFLAGHttQ23 encodes FLAG-tag-KGERGAASRPEASGDCRAGRETA polypeptide in frame with the 3144 amino acid huntingtin sequence (FLAG-Q23 huntingtin). pALHDQ32, pALHDQ43 encoding full-length human FLAG-Q32 and -Q43 huntingtins, respectively, were generated in pFASTBAC1, modified to insert a polylinker containing FLAG, 6X histidine tag sequence and TEV protease recognition site, between the unique BamHI–KpnI sites. NcoI–XhoI HD cDNA fragments, encoding huntingtin amino acids 1–171 with different size polyglutamine tracts (Q32, Q43) (10,44) were inserted between the unique NcoI and XhoI sites, followed by in frame insertion of a 9046 bp human HD cDNA XhoI–SacII fragment, encoding human amino acids 172–3144 (10,44). All clones were verified by full DNA sequence analysis.
FLAG-huntingtin purification
FLAG-tag huntingtin was expressed from pFASTFLAGHttQ23 in the Bac-to-Bac Baculovirus Expression system (Invitrogen). The Sf9 cell lysate, generated by freeze/thawing in buffer A (50 mm Tris–HCl pH 8.0, 500 mm NaCl, 5% glycerol and complete protease inhibitors), was spun at 15 000 rpm (2 h). The supernatant was incubated with M2 anti-FLAG beads (Sigma) (2 h, 4°C). FLAG-huntingtin was eluted with buffer (50 mm Tris–HCl pH 8.0, 300 mm NaCl, 5% glycerol) containing 0.4 mg/ml FLAG peptide and loaded onto a calibrated Superose 6™ 10/300 column, equilibrated with 50 mm Tris–HCl pH 8.0, 150 mm NaCl. FLAG-huntingtin eluted discretely and was estimated to be at least 90% pure by Coomassie staining. Recombinant FLAG-Q32 and -Q43 huntingtins were purified in exactly the same manner. Comparisons of huntingtins with different polyglutamine sizes were performed with an equal amount of each protein, as judged by Bio-Rad DC protein assay and R-250 Coomassie blue staining of bands on 6% SDS–PAGE, which controlled for potential differences in purity and confirmed equal amounts of protein. The molarity for all huntingtins was calculated using a molecular weight of 350 kDa deduced from the human cDNA sequence (GenBank accession number L12392).
EM and image processing
Samples were prepared by negative staining with 0.75% (w/v) uranyl formate as described previously (45). Images were collected with a Tecnai T12 electron microscope (FEI, Hillsboro, OR) equipped with an LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded on imaging plates at a nominal magnification of 67 000x and a defocus value of −1.5 µm using low-dose procedures. Imaging plates were read out with a Ditabis micron imaging plate scanner (DITABIS Digital Biomedical Imaging System AG, Pforzheim, Germany) using a step size of 15 µm, a gain setting of 20 000 and a laser power setting of 30%. 2 × 2 pixels were averaged to yield a pixel size of 4.5 Å on the specimen level. Using BOXER display (EMAN software package) (46), 10 061 particles interactively selected from 53 images were windowed into 64 × 64 pixel images using the SPIDER software package (47), which was also used for all other image processing procedures. The particles were rotationally and translationally aligned and subjected to 10 cycles of multireference alignment, with K-means classification, specifying 100 output classes, after each round. The references used for the first multi-reference alignment were randomly chosen from the raw images.
Huntingtin structure prediction and evolutionary conservation
Human huntingtin amino acid sequence (Homo sapiens; NP_002102) was analyzed for predicted secondary structure using: NORSp (48) and PROF (Profile network prediction HeiDelberg) (49) from the PredictProtein server (http://www.predictprotein.org/) (50). Huntingtin orthologues for the multiple sequence alignments: dog (Canis familiaris; XP_536221), mouse (Mus musculus; AAA89100), opossum (Monodelphis domestica; XP_001364862), chicken (Gallus gallus; XP_420822), zebrafish (Danio rerio; NP_571093); lancelet (Branchiostoma floridae; ABP04240) and sea squirt (Ciona intestinalis; NP_001119700), were aligned using ClustalW2 (European Bioinformatics Institute: http://www.ebi.ac.uk/Tools/clustalw2/) (51) and viewed and edited using Jalview 2.3 (http://www.jalview.org/) (52). The initial alignment used the ClustalW2 server default parameters (except iteration:tree and numiter:8). For secondary structure predictions, ‘extra’ sequences of greater than five amino acids relative to human huntingtin, were deleted from orthologues (apparent insertions of 23, 50 and 24 residues in zebrafish, lancelet and sea squirt at human 1051, 10 and 20 amino acids in sea squirt and lancelet at human 2145, 11 residues in lancelet at human 2195 and 26 residues in sea squirt at human 2642). After re-alignment, gaps of <5 residues in the human sequence were removed from any of the other sequences. The final ‘no-gap’ set was re-aligned producing the final multiple alignment. This was exported as an image file (PNG format) and compressed in PowerPoint. Jar files of the initial and final alignments are available on request. The physico-chemical properties conserved for each amino acid position were calculated in Jalview (53). CHOP (34,35) predicted tertiary structure, with searches of the CATH protein structure classification database, to identify potential structural domain homologues. FoldIndex was also used to predict unfolded human huntingtin structure (54).
Mice and embryos
Wild-type and Hdhex4/5/Hdhex4/5 embryos were obtained from timed matings of Hdhex4/5/Hdh+ heterozygote mice and genotyped by PCR assay as described (23). The day of plug was defined as E0.5. GFP X chromosome transgene mice were from The Jackson Laboratory (strain 003116).
In situ hybridization
Dissected embryos and decidua were fixed in 4% paraformaldehyde at 4°C brought through a sucrose gradient (15% sucrose, 30% sucrose), embedded in OCT and sectioned at 10 µm. RNA in situ hybridizations, performed as reported (55), were with antisense PL-1 probe synthesized with Sp6 and anti-sense Hox probes generated as described previously (56).
Embryonic stem cell and EB tissue culture
Wild-type and huntingitn null Hdhex4/5/Hdhex4/5 ES cells, reported previously (23), as well as Hdh CAG knock-in Hdh+/HdhQ111 ES cells (19) were cultured at 37°C in 5% CO2 in ESCM medium [DMEM, 15% FBS, penicillin streptomycin, glutamine, non-essential amino acids, 0.7% β-mercaptoethanol, 106 U/l ESGRO (Chemicon-Millipore)]. EB formation was in ESCM without ESGRO (LIF) on untreated polystyrene plates.
EB immunocytochemistry
EBs were fixed, embedded and sectioned as earlier. PBS washed sections, blocked with 10% goat serum in PBST (0.1% Triton) were incubated with primary antibody (overnight, 4°C) in PBST/0.1% goat serum. Secondary antibody was in PBST/0.1% goat serum. Mounting was in Vectashield containing DAPI (Vector labs). Primary antibodies: tri-methyl histone H3K27 (Upstate Biotechnology, Inc.), di-methyl histone H3K27 (Abcam), Ezh2 (BD Transduction labs) and huntingtin AP194 (Dr A. Sharp).
Immunoblots
Proteins were transferred from SDS–PAGE to nitrocellulose membrane (Schleicher & Schuell), blocked in 10% non-fat powered milk in TBS-T (50 mm Tris–HCl, 150 mm NaCl, pH 7.4, 0.1% Tween 20). Primary antibody incubation was overnight (4°C). Incubation with horseradish peroxidase-conjugated secondary antibody (Amersham Pharmacia Biotech) was 1 h (room temperature). Signal was detected by ECL chemiluminiscence (PerkinElmer) with autoradiographic film (Hyperfilm ECL; Amersham Bioscience). Additional primary antibodies were: huntingtin mAb2166 (Chemicon-Millipore), Suz12 (Upstate Biotechnology, Inc.), polyglutamine mAb 1F8 (10), histone H3 (Abcam), α-tubulin (Sigma) and fibrillarin (Santa Cruz Biotechnology, Inc.).
Subcellular fractionation
Subcellular fractions were prepared as reported (57). Washed cell pellets were homogenized in buffer [20 mm HEPES, pH 7.0, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 1 mm PMSF and complete protease inhibitors (Roche Applied Science)], using 10 strokes with a glass Dounce pestle. Supernatant from the 1000 g spin (10 min, 4°C) was the cytoplasmic fraction. Nuclear extract was generated from the pellet by resuspending in ELB buffer (250 mm NaCl, 0.1% NP-40, 50 mm HEPES, pH 7.0, 5 mm EDTA, 0.5 mm DTT, 1 mom PMSF, and complete protease inhibitors), sonicating (3 times, 10 s pulses), and saving the supernatant of a 14 000 g spin (10 min, 4°C). Protein concentration for this experiment and all others was measured with Bio-Rad DC (Detergent Compatible) (Bio-Rad).
Immunoprecipitation
Immunoprecipitation assays entailed: (i) incubation (RT for 2 h) of nuclear lysate (500 µg) and mAB2166 or control IgG from mouse serum (Sigma) or (ii) incubation (at 37°C for 2 h) of purified PRC2 complex (2.5 µg) and FLAG-huntingtin (1.25 µg) with mAb2166 or anti-Ezh2. Protein A Sepharose beads (Roche) were added for 1 h (4°C), washed (8 times) in ELB plus (250 mm NaCl, 0.1% NP-40, 50 mm HEPES, 5 mm EDTA) and boiled for 5 min in reducing SDS sample buffer to remove the complexes from the beads.
Chromatin immunoprecipitation and quantitative PCR
ChIP assays were performed using the Agilent mammalian ChIP-on-chip protocol as specified by the manufacturer (Agilent Technologies), except for huntingtin immunoprecipitation, where mAb 2166 was incubated with chromatin for 2 h at room temperature, to reduce non-specific background. Antibodies were anti-histone H3K27me3 (Abcam ab6002) antibody, anti-huntingtin (Millipore mAb 2166) and control IgG (Sigma).
Purified input of chromatin (25 ng) or immunoprecipitated (IP) DNA (100 ng) from ChIP were used as a template in 50 µl reactions containing 25 µl of 2X SYBR Green Master Mix (Applied Biosystems) and 10 pmol of each primer. PCR reactions were performed with iCycler thermal cycler (Bio-Rad) and as follows: 50 cycles of 95°C for 15 s, 54°C for 15 s and 72°C for 15 s. All PCRs were performed in triplicate and threshold amplication cycle numbers (Tc) using iCycler software were used to calculate IP DNA quantities as percentages of corresponding inputs using the following equation: IP DNA as a percentage of input = 2(ΔTc) × 100, ΔTc = input DNA Tc − IP DNA Tc. Statistics were analyzed using Student's t-test. Hox B9 promoter primer sequences for q-PCR were; left primer 5′-TGGCCTTAGGCAGGCTATAA-3′ and right primer 5′-GGCTCTTCCCTTGATCCTTT-3′.
Acid precipitation of histones
Cells were lysed in 5–10 volumes of lysis buffer (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl), hydrochloric acid was added to 0.2 m final concentration, incubated (30 min, ice) and the extract spun at 11 000 g (10 min, 4°C). The supernatant was dialyzed twice against 20 volumes of 0.1 m acetic acid (1 h) and then dialyzed three times against 20 volumes of water. The acid precipitated proteins were loaded on 12% SDS–PAGE gel for immunoblot.
Size exclusion chromatography fractionation
Day 4 EB nuclear lysates were subjected to size exclusion chromatography on a pre-calibrated (with size standards listed in legend) Superose-6™ HR 16/60 column, equilibrated with 20 mm HEPES pH 7.5 containing 1 mm MgCl2, 150 mm NaCl and 0.5 mm DTT. Fractions (1.5 ml) were collected. Blue Dextran was used as void volume marker and was eluted at fraction number 26.
Cell-free assay for PRC2 histone H3 methyltransferase activity
Pre-assembled human PRC2 complex, FLAG-EED, EZH2, SUZ12 and RbAp48 proteins, was purified, from Sf9 cells co-infected with the cognate pFastBac1 constructs, by M2 anti-FLAG bead affinity column and Superose 6™ gel filtration chromatography (equilibrated with 50 mm Tris–HCl pH 8.0, 150 mm NaCl, 10% glycerol). The molarity was calculated using a MW of 270 kDa, which assumes one copy of each subunit per complex. The G5E4 nucleosomal array was assembled with Xenopus recombinant histones and G5E4 DNA fragments containing 12 nucleosomal positioning sequences, as reported (58,59).
The reconstituted PRC2 activity assay was optimized from a previous method (33). A reaction mixture of 15 µl comprised: 100 mm Tris–HCl pH 8.3, 1 mm DTT, 0.5 µm 3H-SAM, 4 nm PRC2 and 0.025–0.1 µm nucleosomal array. Incubations were at 30°C for 30 min, unless otherwise stated. For comparison of huntingtin's with different polyglutamine tracts, reactions were performed at 30°C with 2 nm of each protein for 30 min. Sample buffer was added to stop the reactions, which were subjected to 12% SDS–PAGE, transferred to Immobilon-PSQ membrane (Millipore), and exposed to phosphorimager screen. The 3H-H3K27 bands detected with Typhoon (ImageQuant as software, GE Healthcare Life Science) were quantified with Quantity One software (Bio-Rad). For the scaled up cold assay, a 60 µl reaction included: 100 mm Tris–HCl pH 8.3, 1 mm DTT, 2 µm SAM, 50 nm PRC2 and 0.1 µm nucleosomal array, with or without 40 nm FLAG-huntingtin (4 h, 30°C). Bands on immunoblots were scanned with GS-800 Calibrated Densitometer (Bio-Rad). All values were expressed as mean ± 1 SD. The statistical significance was determined by Student's t-test.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health NINDS grants NS32765, NS16367, NS49206; the Huntington's Disease Society of America Coalition for the Cure; and an anonymous donor. T.W. is an investigator of the Howard Hughes Medical Institute. J.M.W. received the Milton Wexler Postdoctoral Fellowship from the Hereditary Disease Foundation. J.-J.S. was a recipient of the Jane Coffin Childs Memorial Fund for Medical Research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Alan Sharp (Case Western Reserve University) for AP194 antibody, Dr Jay Cross (University of Calgary) for PL-1 plasmid, Joseph Garlick for insect cell culture work, the Taplin Mass spectrometry facility (Dr Steven Gygi's lab), Morgan Thompson and Dr Susan Cotman Director of the CHGR Confocal Resource. The molecular EM facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained by funds from National Institutes of Health grant GM62580 (S.C. Harrison).
Conflict of Interest statement. None declared.
REFERENCES
- 1.The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Vonsattel J.P., DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Gusella J.F., Macdonald M.E. Huntington's disease: seeing the pathogenic process through a genetic lens. Trends Biochem. Sci. 2006;31:533–540. doi: 10.1016/j.tibs.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Gusella J.F., MacDonald M.E. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nat. Rev. Neurosci. 2000;1:109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- 5.Clabough E.B., Zeitlin S.O. Deletion of the triplet repeat encoding polyglutamine within the mouse Huntington's disease gene results in subtle behavioral/motor phenotypes in vivo and elevated levels of ATP with cellular senescence in vitro. Hum. Mol. Genet. 2006;15:607–623. doi: 10.1093/hmg/ddi477. [DOI] [PubMed] [Google Scholar]
- 6.Seong I.S., Ivanova E., Lee J.M., Choo Y.S., Fossale E., Anderson M., Gusella J.F., Laramie J.M., Myers R.H., Lesort M., et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 7.Menalled L.B. Knock-in mouse models of Huntington's disease. NeuroRx. 2005;2:465–470. doi: 10.1602/neurorx.2.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V.C., Sharp A.H., Persichetti F., Cattaneo E., MacDonald M.E. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 9.Li P., Huey-Tubman K.E., Gao T., Li X., West A.P., Jr, Bennett M.J., Bjorkman P.J. The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model. Nat. Struct. Mol. Biol. 2007;14:381–387. doi: 10.1038/nsmb1234. [DOI] [PubMed] [Google Scholar]
- 10.Persichetti F., Trettel F., Huang C.C., Fraefel C., Timmers H.T., Gusella J.F., MacDonald M.E. Mutant huntingtin forms in vivo complexes with distinct context-dependent conformations of the polyglutamine segment. Neurobiol. Dis. 1999;6:364–375. doi: 10.1006/nbdi.1999.0260. [DOI] [PubMed] [Google Scholar]
- 11.Klein F.A., Pastore A., Masino L., Zeder-Lutz G., Nierengarten H., Oulad-Abdelghani M., Altschuh D., Mandel J.L., Trottier Y. Pathogenic and non-pathogenic polyglutamine tracts have similar structural properties: towards a length-dependent toxicity gradient. J. Mol. Biol. 2007;371:235–244. doi: 10.1016/j.jmb.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Kobe B., Kajava A.V. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem. Sci. 2000;25:509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- 13.Takano H., Gusella J.F. The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/Rel/dorsal family transcription factor. BMC Neurosci. 2002;3:15. doi: 10.1186/1471-2202-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrade M.A., Bork P. HEAT repeats in the Huntington's disease protein. Nat. Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 15.Andrade M.A., Petosa C., O'Donoghue S.I., Muller C.W., Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y., Xing Y., Chen Y., Chao Y., Lin Z., Fan E., Yu J.W., Strack S., Jeffrey P.D., Shi Y. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 17.Cingolani G., Petosa C., Weis K., Muller C.W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 18.Lee S.J., Imamoto N., Sakai H., Nakagawa A., Kose S., Koike M., Yamamoto M., Kumasaka T., Yoneda Y., Tsukihara T. The adoption of a twisted structure of importin-beta is essential for the protein-protein interaction required for nuclear transport. J. Mol. Biol. 2000;302:251–264. doi: 10.1006/jmbi.2000.4055. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler V.C., White J.K., Gutekunst C.A., Vrbanac V., Weaver M., Li X.J., Li S.H., Yi H., Vonsattel J.P., Gusella J.F., et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 20.Li W., Serpell L.C., Carter W.J., Rubinsztein D.C., Huntington J.A. Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J. Biol. Chem. 2006;281:15916–15922. doi: 10.1074/jbc.M511007200. [DOI] [PubMed] [Google Scholar]
- 21.Bernards A. Huntingtin interactors. available at http://chgr.mgh.harvard.edu/genomesrus .
- 22.Dragatsis I., Efstratiadis A., Zeitlin S. Mouse mutant embryos lacking huntingtin are rescued from lethality by wild-type extraembryonic tissues. Development. 1998;125:1529–1539. doi: 10.1242/dev.125.8.1529. [DOI] [PubMed] [Google Scholar]
- 23.Duyao M.P., Auerbach A.B., Ryan A., Persichetti F., Barnes G.T., McNeil S.M., Ge P., Vonsattel J.P., Gusella J.F., Joyner A.L., et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 24.Nasir J., Floresco S.B., O'Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 25.Zeitlin S., Liu J.P., Chapman D.L., Papaioannou V.E., Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat. Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 26.O'Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shumacher A., Faust C., Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 29.Woda J.M., Calzonetti T., Hilditch-Maguire P., Duyao M.P., Conlon R.A., MacDonald M.E. Inactivation of the Huntington's disease gene (Hdh) impairs anterior streak formation and early patterning of the mouse embryo. BMC Dev. Biol. 2005;5:17. doi: 10.1186/1471-213X-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 31.Cao R., Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O'Connor M.B., Kingston R.E., Simon J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 34.Liu J., Rost B. CHOP proteins into structural domain-like fragments. Proteins. 2004;55:678–688. doi: 10.1002/prot.20095. [DOI] [PubMed] [Google Scholar]
- 35.Liu J., Rost B. CHOP: parsing proteins into structural domains. Nucleic Acids Res. 2004;32:W569–W571. doi: 10.1093/nar/gkh481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hemberger M. Epigenetic landscape required for placental development. Cell. Mol. Life Sci. 2007;64:2422–2436. doi: 10.1007/s00018-007-7113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobe B., Gleichmann T., Horne J., Jennings I.G., Scotney P.D., Teh T. Turn up the HEAT. Structure. 1999;7:R91–R97. doi: 10.1016/s0969-2126(99)80060-4. [DOI] [PubMed] [Google Scholar]
- 38.White J.K., Auerbach W., Duyao M.P., Vonsattel J.P., Gusella J.F., Joyner A.L., MacDonald M.E. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat. Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 39.Wheeler V.C., Auerbach W., White J.K., Srinidhi J., Auerbach A., Ryan A., Duyao M.P., Vrbanac V., Weaver M., Gusella J.F., et al. Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum. Mol. Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 40.Kim M.W., Chelliah Y., Kim S.W., Otwinowski Z., Bezprozvanny I. Secondary structure of Huntingtin amino-terminal region. Structure. 2009;17:1205–1212. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palidwor G.A., Shcherbinin S., Huska M.R., Rasko T., Stelzl U., Arumughan A., Foulle R., Porras P., Sanchez-Pulido L., Wanker E.E., et al. Detection of alpha-rod protein repeats using a neural network and application to huntingtin. PLoS Comput. Biol. 2009;5:e1000304. doi: 10.1371/journal.pcbi.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao R., Wang H., He J., Erdjument-Bromage H., Tempst P., Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol. Cell. Biol. 2008;28:1862–1872. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarma K., Margueron R., Ivanov A., Pirrotta V., Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol. Cell. Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faber P.W., Barnes G.T., Srinidhi J., Chen J., Gusella J.F., MacDonald M.E. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 45.Ohi M., Li Y., Cheng Y., Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludtke S.J., Baldwin P.R., Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 47.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 48.Liu J., Rost B. NORSp: Predictions of long regions without regular secondary structure. Nucleic Acids Res. 2003;31:3833–3835. doi: 10.1093/nar/gkg515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rost B., Sander C., Schneider R. PHD–an automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 50.Rost B., Yachdav G., Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 53.Livingstone C.D., Barton G.J. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 54.Prilusky J., Felder C.E., Zeev-Ben-Mordehai T., Rydberg E.H., Man O., Beckmann J.S., Silman I., Sussman J.L. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 55.Conlon R.A., Herrmann B.G. Detection of messenger RNA by in situ hybridization to postimplantation embryo whole mounts. Methods Enzymol. 1993;225:373–383. doi: 10.1016/0076-6879(93)25026-x. [DOI] [PubMed] [Google Scholar]
- 56.Conlon R.A., Rossant J. Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development. 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- 57.Sewalt R.G., Lachner M., Vargas M., Hamer K.M., den Blaauwen J.L., Hendrix T., Melcher M., Schweizer D., Jenuwein T., Otte A.P. Selective interactions between vertebrate polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of Polycomb group proteins. Mol. Cell. Biol. 2002;22:5539–5553. doi: 10.1128/MCB.22.15.5539-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikeda K., Steger D.J., Eberharter A., Workman J.L. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polach K.J., Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.