Abstract
Upstream transcription factor 1 (USF1) has been associated with familial combined hyperlipidemia, the metabolic syndrome, and related conditions, but the mechanisms involved are unknown. In this study, we report validation of Usf1 as a causal gene of cholesterol homeostasis, insulin sensitivity and body composition in mouse models using several complementary approaches and identify associated pathways and gene expression network modules. Over-expression of human USF1 in both transgenic mice and mice with transient liver-specific over-expression influenced metabolic trait phenotypes, including obesity, total cholesterol level, LDL/VLDL cholesterol and glucose/insulin ratio. Additional analyses of trait and hepatic gene expression data from an F2 population derived from C57BL/6J and C3H/HeJ strains in which there is a naturally occurring variation in Usf1 expression supported a causal role for Usf1 for relevant metabolic traits. Gene network and pathway analyses of the liver gene expression signatures in the F2 population and the hepatic over-expression model suggested the involvement of Usf1 in immune responses and metabolism, including an Igfbp2-centered module. In all three mouse model settings, notable sex specificity was observed, consistent with human studies showing differences in association with USF1 gene polymorphisms between sexes.
INTRODUCTION
Upstream transcription factor 1 (USF1) is a ubiquitously expressed transcription factor that has been implicated in the genetic control of metabolic and vascular diseases. Prominent among these is familial combined hyperlipidemia (FCHL; MIM 144250), characterized by elevated levels of total cholesterol, triglyceride, or both, with other component traits including low HDL (high-density lipoprotein) cholesterol, small dense LDL (low-density lipoprotein) particles, elevated apolipoprotein B and/or free fatty acid. FCHL predisposes to premature coronary heart disease, and is the most common familial dyslipidemia, with a population prevalence of 1–6% in Western populations (1–3). Whole-genome scans and candidate gene studies performed in FCHL families in different populations have identified several putative loci for FCHL (4). A linkage peak on 1q21–23 was identified as a major locus for FCHL in Finns and subsequently replicated (5–9), which led to the identification of USF1 as the first positionally cloned gene for FCHL (10). In addition to FCHL, genetic variants of USF1 have been associated in independent studies with various complex disorders, including metabolic syndrome and related traits (11–15) and, in some studies, risk of type 2 diabetes (16–18) and cardiovascular disease (19–22). Interestingly, many of the clinical metabolic features of FCHL also represent trait components of type 2 diabetes and metabolic syndrome, including dyslipidemia and insulin resistance, both of which are associated with increased risk of cardiovascular diseases (23). However, the picture is not entirely clear, as not all studies have identified associations of these phenotypes with USF1 variants (16,17,22,24,25), and positively identified associations were not always with the same variant (13,15,21). Additionally, a frequent observation was the occurrence of sex differences in the presence or strength of associations (13,14,17). There are limited data regarding USF1 transcript levels in human tissues. In two studies of adipose tissue, USF1 transcript levels did not differ in relation to USF1 alleles (10,26). Direct correlations of USF1 transcript levels with phenotypes have not been reported.
USF1 is a member of the basic helix–loop–helix leucine zipper family of transcription factors. It can form homodimers or heterodimers with USF2, which recognize E-box regulatory sequences and lead to transcription activation and/or enhanced gene expression (27). Biochemical and molecular analyses in cells have shown that USF1 regulates multiple genes, including apolipoproteins, lipases and various enzymes participating in glucose and lipid metabolism (4,28,29). Taken together, these genetic and biochemical data suggest that USF1 may play an important role in regulating metabolic traits in vivo. However, there is limited direct evidence in animal models to confirm such a role (30,31).
Recently, using an integrative genetics analysis of an F2 mouse population, we identified Usf1 as one of a number of genes supported as having a causal role for relevant metabolic traits (32). These included HDL cholesterol, free fatty acid and insulin levels in plasma for both males and females. Sex differences were also noted, as in males, Usf1 was additionally a causal gene for adiposity, total cholesterol and unesterified cholesterol, whereas in females, for plasma triglyceride level and fat pad size. In this study, we report validation of Usf1 as a causal gene of cholesterol homeostasis, insulin sensitivity and body composition in the mouse using over-expression of USF1 in both transgenic (tg) mice and mice with transient liver-specific over-expression. Network and pathway analyses of the liver gene expression signatures in the hepatic over-expression model and the F2 population were also performed to further characterize possible mechanisms and effects of Usf1.
RESULTS
Generation and characterization of USF1 tg mice
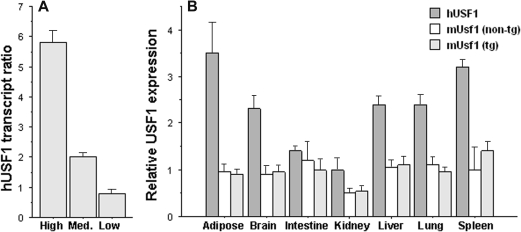
In order to maintain the endogenous regulatory regions of USF1, we used a bacterial artificial chromosome (BAC) clone to construct tg mice over-expressing human USF1. The BAC clone contained the entire coding region of USF1 as well as two flanking genes, PVRL4 and ARGHAP30 (Supplementary Material, Fig. S1A). Efforts to isolate a BAC fragment with the USF1 gene alone were unsuccessful and could have resulted in the removal of regulatory elements. The potential role of the additional genes on phenotype was addressed using transient over-expression studies described in what follows. Three independent lines of USF1 tg mice were generated as described in Materials and Methods and maintained in the heterozygous state on the FVB strain background. On the basis of the expression level, the three tg lines were designated as being high, medium and low expressers of USF1, where a 5:2:1 ratio of human USF1 transgene expression was observed, respectively (Fig. 1A). As assessed by quantitative RT–PCR, significant expression of the human USF1 transgene was observed in a number of tissues, including the liver, adipose, skeletal muscle, spleen, intestine, lung and whole brain, similar to the endogenous mouse Usf1 gene in both tg and non-tg mice (Fig. 1B). USF1 tg mice developed and reproduced normally. No behavioral abnormalities or anatomical defects were observed in any major organs.
Figure 1.
(A) Hepatic hUSF1 expression in three independent tg lines, shown as the ratio of expression levels in the three lines relative to the average of the low expresser line. (B) Survey of human USF1 (hUSF1) and endogenous mouse Usf1 (mUsf1) gene expression in USF1 tg mice (high expresser line) and in non-tg littermates. Graphs display mean ± 1 SEM of transcript levels normalized to beta-actin.
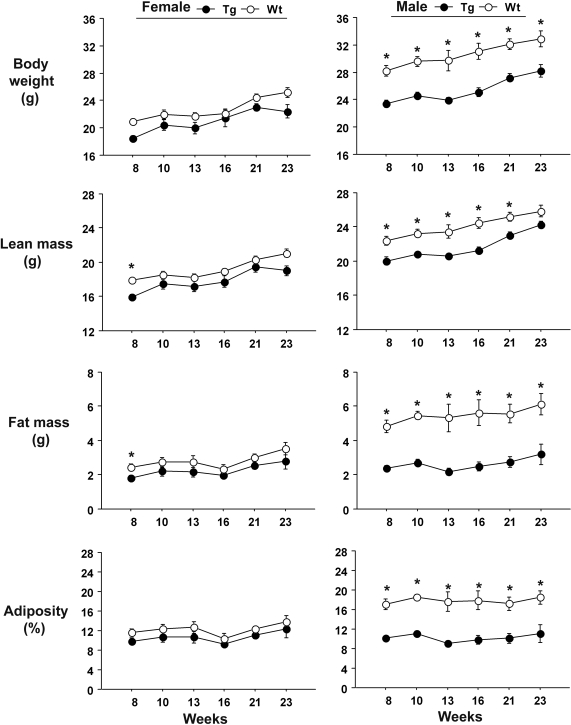
High expresser male USF1 tg mice showed significant decreases in total body weight, fat mass and overall adiposity, compared with their wild-type (wt) littermates (Fig. 2). We noted that the differences appeared throughout the entire observation period, between week 8 and week 23 after birth. However, in females, the differences as the mice grew older were less clear. Follow-up studies showed that differences in these traits for both sexes were not observed at the immediate post-weaning time, indicating that the effects were not developmental in origin (data not shown). Food consumption was assessed in 8-week-old female mice of the high expresser line and found to be greater in transgenics compared with controls (P < 0.05; Supplementary Material, Fig. S2), suggesting that the transgenics had a higher metabolic rate or level of activity. Body temperatures did not differ.
Figure 2.
Body composition traits in high expresser line USF1 tg and wt mice. Graphs display mean ± 1 SEM at each time point. *P < 0.05 by unpaired t-test.
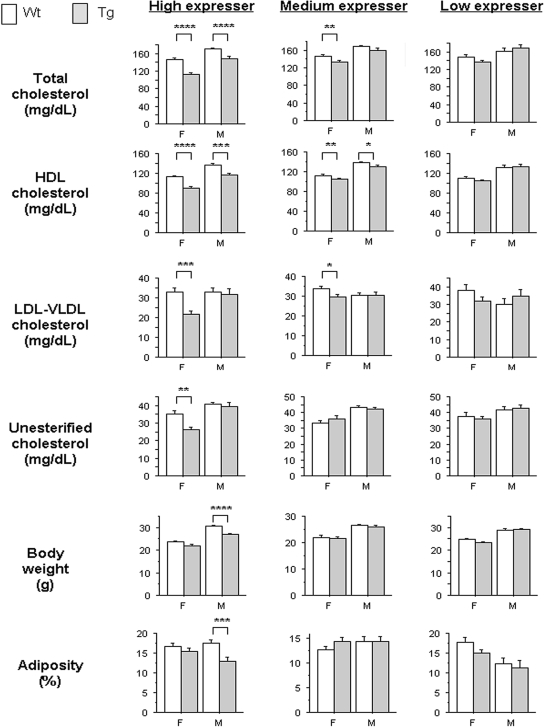
In addition to altered body composition, both male and female USF1 tg mice were observed to have significant decreases in total cholesterol and HDL cholesterol, compared with their wt littermates (Fig. 3). In female USF1 tg mice, significant reductions in LDL + VLDL, and unesterified cholesterol fractions, were also observed. Medium expresser mice showed comparable but less pronounced differences, whereas the low expresser transgenics did not differ from the non-tg littermates (Fig. 3). Plasma triglyceride levels were consistently higher in transgenics relative to controls, except in high expresser line females. However, there was substantial variability, and the differences were not significant (P > 0.05; Supplementary Material, Table S1).
Figure 3.
Plasma lipid levels, body weight and adiposity (at 10 weeks of age) observed in female (F) and male (M) USF1 tg mice (shaded bars) and corresponding wt littermates (open bars) for each of the three tg lines constructed (n≥ 4 in each group). Bars represent mean ± 1 SEM. Significant differences between tg and wt mice by unpaired t-test are indicated by *P< 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Metabolic effects of transient adenoviral delivered USF1 over-expression in liver
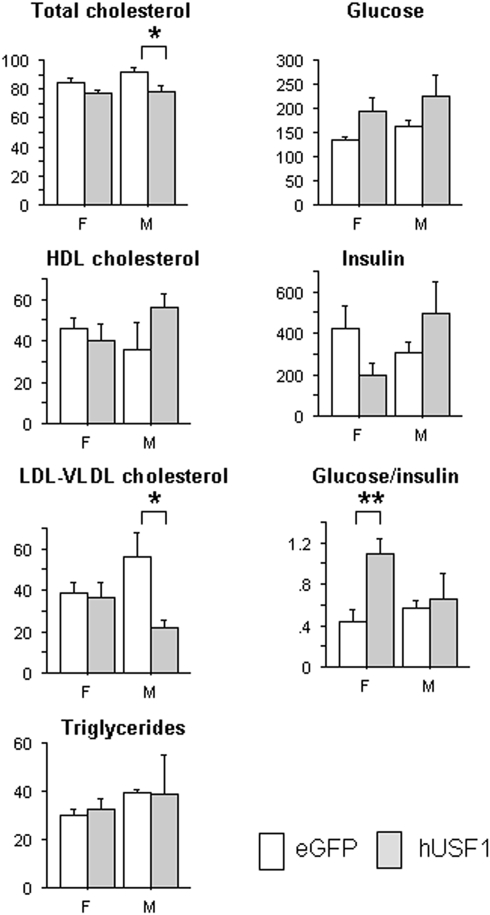
The effects of transient USF1 over-expression in the liver were studied using a USF1 recombinant adenoviral infection in C57BL/6J mice. The AdUSF1 construct (Supplementary Material, Fig. S1B) was delivered via tail vein injection, resulting in the liver being the major site for USF1 over-expression. Expression of both human USF1 mRNA and protein was confirmed, with mRNA levels being comparable with the high expresser tg line (Supplementary Material, Fig. S3). Analysis of plasma collected 5 days post-injection showed significant deceases in total cholesterol and LDL + VLDL cholesterol in AdUSF1 injected male mice compared with the corresponding control animals injected with AdGFP (Fig. 4). In female mice, there was a trend towards lower total cholesterol (P = 0.068), but no other differences in plasma lipids. However, in contrast to males, there was a significant increase in the glucose/insulin ratio and a trend towards lower insulin levels (P = 0.076). Plasma triglycerides did not differ for either sex. Apoa1 and Apob transcript levels were measured to see whether reduced levels of these might contribute to the plasma lipid changes observed here and in the transgenics. No significant difference of Apoa1 transcript was found, but that of Apob was mildly increased in the AdUSF1-transfected livers (P < 0.05; Supplementary Material, Fig. S4), suggesting that decreased synthesis was not the cause.
Figure 4.
Differences in plasma lipid and glucose/insulin traits observed in adenovirus vector-injected animals at day 5, comparing those receiving control (open bars) and USF1 (shaded bars) containing vectors (n= 5 and 4, respectively). Mean ± 1 SEM depicted; *P<0.05; **P < 0.01.
Identification of genes induced by USF1 over-expression in vivo
In order to explore mechanisms underlying the physiological effects of USF1 over-expression, we profiled liver tissue collected from mice injected with AdUSF1 or the corresponding control viral particles (AdGFP) and identified differentially expressed genes (USF1-induced gene sets). At P < 0.05 as determined by Student's t-test, and with fold changes ≥ 1.5 or ≤ 0.67, 345 and 265 genes were observed as differentially expressed, in males and females, respectively (Table 1; Supplementary Material, Table S2). The false discovery rates (FDRs) at this P-value cutoff were 0.15 and 0.22, respectively. The high FDR values in the profiling experiments are likely due to the modest number of mice profiled (n = 3 for each group). A significant sex effect was present, as only 56 (∼20%) of differentially expressed genes were found in common between the sexes (highlighted in Supplementary Material, Table S2). Among these 56, the direction of mRNA level change was consistent between males and females.
Table 1.
USF1 hepatic gene expression signature sets data summary
| Ad-USF1 F | Ad-USF1 M | F2 ApoE F | F2 ApoE M | |
|---|---|---|---|---|
| Signature size (P < 0.05) | 265 | 345 | 2802 | 2613 |
| FDR at P< 0.05 | 0.22 | 0.15 | 0.30 | 0.30 |
| Overlap Ad-USF1 F (P-value) | 1.64 × 10−34 | >0.05 | 7.11 × 10−3 | |
| Overlap Ad-USF1 M (P-value) | 1.53 × 10−34 | >0.05 | 1.03 × 10−7 | |
| Enrichment | MEMN seta | MEMN seta | MEMN seta | MEMN seta |
| Obesity setb | Obesity setb | Obesity setb | Immune response | |
| Amino acid metabolism | Response to biotic stimulus | Intracellular organelle | Defense response | |
| Oxidoreductase activity | Defense response | Cytoplasm | Response to biotic stimulus | |
| Organic acid metabolism | Immune response | Cellular physiological processes | ||
| Carboxylic acid metabolism | Antigen presentation | Metabolism | ||
| Nitrogen compound metabolism | Antigen processing | Mitochondrion | ||
| Catalytic activity | Immunological synapse | Catalytic activity | ||
| Monoxygenase activity | Cytoplasm | Cellular metabolism | ||
| Immune response | Cytosol | |||
| Response to biotic stimulus |
Enrichment of GO and KEGG categories are significant at P < 0.05, after Benjamini correction for multiple comparisons. A detailed listing of the enrichment analysis results are provided in Supplementary Material, Table S3.
aMEMN, macrophage-enriched metabolic network (32).
bObesity set: see reference (34).
To assess enrichment for pathways and other functional categories, we employed Expression Analysis Systematic Explorer (EASE) to implement Fisher's exact test-based enrichment analysis of the gene ontology (GO) functional categories and KEGG pathways (33). Categories of metabolism of small molecules and compounds were enriched in females, whereas immune responses and antigen processing were enriched in male data set (Table 1). The set of 56 USF1-induced genes shared between male and female animals showed significant enrichment of type I diabetes pathway, immune responses and antigen processing. Detailed results of the ontology enrichment analyses are provided in Supplementary Material, Table S3.
Characterization of endogenous Usf1 variation in F2 mouse populations
Usf1 has been identified as one of a number of genes supported as having a causal role for relevant metabolic traits using an integrative genetics analysis of an F2 mouse population (the BXH.ApoE F2 cross) (32), in which Usf1 transcript levels are correlated with multiple traits, including plasma lipids, glucose and insulin measures and body fat measures, though not body weight (Supplementary Material, Table S4). In most animals, body fat makes only a modest contribution to body weight, accounting for the apparent discrepancy of Usf1 transcript correlation with the former but not the latter. Hepatic Usf1 transcript levels are under strong cis-regulation in this cross, with transcript levels in the liver ∼1.5-fold higher in mice homozygous for the C3H alleles compared with the B6 alleles, and multiple traits differ according to genotype at the Usf1 locus (Supplementary Material, Fig. S5). This represents a naturally occurring setting in which to examine the impact of genetic variation in Usf1 transcript levels.
The liver global gene expression data from the BXH.ApoE F2 cross were analyzed to identify and functionally characterize the Usf1 molecular signatures, defined as the sets of genes whose expression correlated with Usf1 at P < 0.05. In the BXH.ApoE F2 cross, we identified 2613 and 2802 genes, respectively, in males and females that were significantly correlated with Usf1 transcript levels in the liver (Table 1 and Supplementary Material, Table S5). Approximately one-third of these overlapped between the sexes. There was significant overlap between the Usf1 signature expression profiles of the male F2 mice with the AdUSF1 signatures, but not with the female F2 signatures. Both male and female AdUSF1 signatures significantly overlapped the macrophage-enriched metabolic network (MEMN) and obesity subnetwork-associated gene sets that were previously identified from integrated F2 mouse cross-expression data sets (32,34). Enrichment of GO and KEGG categories in signature gene sets was broadly comparable with that observed in the AdUSF1 signature sets, including the sex-related differences (Table 1).
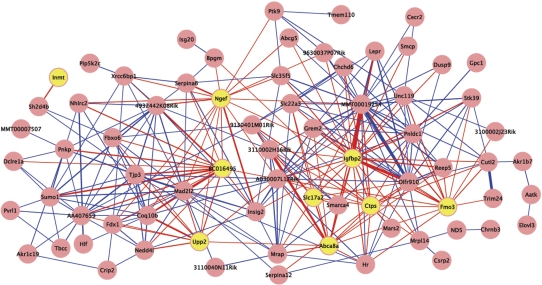
To identify coordinately expressed subsets among genes correlated with Usf1-associated traits, a weighted co-expression network algorithm was applied to partition the molecular signatures of Usf1 from the BXH.ApoE F2 cross-data for each sex (Supplementary Material, Tables S5 and S6). This analysis identified a module (designated light-cyan) of 78 genes from the female signature that was significantly correlated with plasma HDL, insulin, glucose/insulin ratio, adipose mass and leptin (P < 0.0001 for module significance scores). Genes in this module significantly overlapped the MEMN and obesity subnetwork gene sets reported previously, and include several well-characterized genes relevant to metabolic processes such as Insig2 and Lepr (Fig. 5). Nine of the genes were members of the AdUSF1 female signature gene set, and 7 of these were represented among the 20 genes with the highest connectivity (i.e. were ‘hub’ genes), including insulin-like growth factor binding protein 2 (Igfbp2). Approximately 70% of the genes in this module share an edge with one or more of the nine AdUSF1 signature set genes. Several other modules in both the female and male sets had strong module significance scores for metabolic syndrome-related traits, but were not comparably enriched for AdUSF1 signature set genes (Supplementary Material, Tables S5 and S6).
Figure 5.
Light-cyan module from female BXH.ApoE null F2 hepatic co-expression network analysis of Usf1 signature gene set, which is strongly correlated with plasma HDL, insulin, glucose/insulin ratio, adipose mass and leptin. Yellow nodes represent genes that were also part of the female AdUSF1 signature gene set, and red edges indicate a connection to one or more of these AdUSF1 signature gene nodes. Edge width indicates relative strength of connection.
DISCUSSION
USF1 has been implicated as a genetic determinate of FCHL and the common complex metabolic disorders of metabolic syndrome and its related traits. Mouse models can provide supportive evidence for the relevance and potential mechanisms of candidate genes identified from human studies. In this work, we utilized three such models. Tg mice constructed utilizing the human gene and its flanking regions are informative because the inclusion of regulatory regions allows for more physiologic expression patterns by tissue and over time. Adenoviral transfection models provide short-term over-expression of the human gene. As it is primarily expressed in the liver, it allows assessment of the significance of hepatic expression apart from other tissues and other genes. The F2 intercross model allows examination of the endogenous gene across a segregating genetic background. When combined with global gene expression analyses, it is a powerful approach for assessing possible causal associations with physiologic traits and for inferring potential pathways involved. Application of these three independent mouse model systems validates the role of USF1 as a determinate of plasma lipid, insulin/glucose and body mass-related traits. They also highlight the significant sex differences that are found with variation in USF1. Analysis of microarray data from the liver suggests that USF1 influences sets of genes involving metabolism and inflammation.
Significant evidence of association of FCHL with USF1 was first observed in 60 extended Finnish families in males affected with high serum triglyceride levels (10). The association and linkage disequlibrium extended over a 46 kb region in affected men. In this and other studies (9,22), the region encompasses a portion of the adjacent F11R gene, making F11R a potential candidate as well. In the present study, only the first two exons of the F11R gene were included in the BAC used to construct the tg mice. The BAC included two other genes, ARHGAP30 (Rho GTPase activating protein 30) and PVRL4 (poliovirus receptor-related 4), which were not candidates from the human studies, but might be confounding in the mouse transgenics. Although these genes are similarly positioned near Usf1 in the mouse, in the F2 data set, they do not show evidence of having genetic variation in or near the gene that would influence transcript levels. They also have not been implicated as being causal for any relevant traits using the causality test, or evidence from the literature that they play a role in metabolic diseases. In the adenoviral vector mediated over-expression study, only USF1 was expressed, excluding any possible direct effect of F11R, ARHGAP30 or PVRL4. Thus, we were able to assess the effects of USF1 separate from F11R, and other flanking genes, and provide strong support that USF1 is the responsible gene at this locus for the positive associations found in the human studies.
Besides the FCHL phenotype, significant associations of USF1 for one or more traits involving measures of plasma lipids, obesity and body composition and glucose metabolism have been observed in multiple studies (9,13–15,17,22). Similar to these clinical observations, we found effects on this same set of traits, though to varying degrees in the different models studied. Modifying conditions among the models used here included genetic background, gene dosage, organs affected and duration of effect for the transgenic versus the adenoviral vector experiments. In the F2 population, where there is significant linkage, interaction with co-inherited genes likely played a role, along with possible species-related differences relative to the other two models that could affect activity. Additionally, the strong hyperlipidemia-inducing setting of ApoE null genotype and 16 weeks of a Western diet likely contributed to differences with the other models as well. It was of interest to observe in this setting that the genotype associated with greater Usf1 levels showed increased rather than decreased total cholesterol and other lipid fractions that were seen in the transgenic and transfection models. The apoE null genotype creates a hyperlipidemic state by impairing clearance of low and very low density lipoproteins as well as chylomicrons. When combined with a Western diet, plasma total and LDL/VLDL cholesterol levels are typically 10 times or more than those observed in chow-fed wt mice.
Multiple, though not all, clinical studies have noted distinct sex effects, affecting plasma lipids, obesity and body mass index, cardiovascular disease risk and mortality (14,17,21). Also, different USF1 alleles have been mapped for males (common allele) and females (minor allele) in FCHL populations for the same traits (13). Our findings in the mice were consistent with these clinical observations, as there were clear sex-related differences in trait expression in each of the three models studied. As discussed subsequently, gene expression patterns were also notably different between the sexes.
There are little direct data from in vivo studies as to how genetic variation in USF1 may influence trait expression. To address this, we analyzed global gene expression data from liver tissues of the adenovirus vector-treated mice over-expressing human USF1 and controls, and F2 mice from a population with an endogenous variation in relative Usf1 mRNA expression. We elected not to study the tg mice because of the presence of the adjacent genes in the BAC used and the differing inbred strain background. The signature gene sets derived from these studies showed obvious similarity between the two models when analyzed for pathway and gene set enrichment, with a very apparent sex effect. In both adenoviral vector and F2 mice, males showed enrichment primarily for inflammation and immune-related GO categories, whereas females showed enrichment for metabolism-related categories, as well as some immune response categories. Both showed significant overlap with two gene sets that we identified previously as being associated with obesity, metabolic and cardiovascular traits. One of these is the ‘macrophage enriched metabolic network’ set that involves inflammation and immune response-related genes among others (32), which was overrepresented in all groups, including the 54 genes that were differentially expressed in common between the male and female adenoviral vector-treated mice, with P-values after Bonferroni correction of 10−5 to 10−31. GO biological process categories ‘immune response’, ‘defense response’, and ‘response to biotic stimulus’ were also very significantly over-represented in all the adenoviral transfected gene sets and the F2 ApoE null male signature set (Supplementary Material, Table S3).
Therefore, one common mechanism by which genetic variation in USF1 may be influencing the lipid and metabolic traits is by altering inflammation-related processes in the liver and likely other tissues. There has been substantial research linking inflammation with these traits in humans and in animal models, particularly obesity and insulin resistance, where both adipose tissue and the liver are critical sites (35–38). The mechanisms are complex and involve both changes in inflammatory cell populations in each organ and altered function in the respective parenchymal cells. Multiple inflammatory mediators can lead to impairment of insulin signaling in hepatocytes and other cells, which in turn affects diverse metabolic processes including lipid metabolism (35,39). Evidence that this is occurring in humans in association with USF1 variation exists from expression analyses of fat biopsies of individuals with FCHL, comparing a set with a protective USF1 allele versus those having the risk allele. Many of the same immune/inflammatory response GO categories we observed were significantly overrepresented in the differentially expressed gene set in that study (10). Additional support for the relevance to humans is provided by a report of USF1 associations with traits measured in the Cardiovascular Health Study, which involved older individuals, which described significant association of USF1 alleles with C-reactive protein and interleukin-6 levels in plasma (22). Systemic inflammatory markers were not measured in our studies, except for plasma MCP-1 in the F2 mice, which showed no correlation with Usf1 transcript levels (P > 0.4, males and females). However, we did observe a correlation of hepatic Usf1 transcript levels with the extent of atherosclerosis in the aortic root in males (r = −0.21; P = 0.01), and with hepatic Il6 transcript levels in females (r = 0.22; P = 0.006). All together, these results suggest that it is involved in a complex network of genes that regulate metabolic trait expression and inflammation.
The most clearcut findings from our co-expression network analyses that suggest how USF1 might influence such networks were observed in the female F2 mice. One module of tightly correlated genes was significantly correlated with insulin/glucose traits, and a third of the most highly connected genes (hubs) were ones differentially expressed in the short-term AdUSF1 study. Among the most interesting of these was Igfbp2. IGFBP2 tg mice have greater insulin sensitivity and lower body fat, presumably acting through its modulating effects on IGF-I (40), and low plasma levels of IGFBP2 have been associated with the metabolic syndrome traits in a cohort of diabetic patients (41). In networks, variation in hub gene activity has a disproportionate impact. Thus, by having a direct impact on one or more hub genes, the effect of Usf1 would be propagated to many more genes in the module. Several genes in this module have been associated with metabolic related traits in some genome-wide association studies, including Igfbp2, Insig2, Ngef and Lepr (42–45). Thus, although such analyses do not provide specific biochemical explanations for the mechanisms of Usf1 action, they do provide evidence that Usf1 can influence gene networks that in turn influence trait expression and suggest that specific genes may be more significantly involved. It is important to note that these analyses were restricted to the liver, and Usf1 expression in adipose and other relevant tissues is likely important in determining the impact on traits. Also, modulation of activity by non-transcriptional regulation, such as post-translational modifications (46), would not be reflected in these analyses.
The mechanisms by which genetic variation of USF1 influences medically relevant phenotypes are undoubtedly complex, and it is unlikely that the apparent association with inflammatory processes accounts for all of the findings observed in the different models. There was enrichment of metabolism-related GO categories, particularly in female mice, that suggests a role for pathways independent of inflammation, though these were not the ones that obviously could explain particular phenotypes. In both the F2 and the adenoviral transfected mice, we did not observe a significant relationship between USF1 and Apoa1 or Apob transcripts in the liver, suggesting that changes in the clearance of lipoproteins rather than altered hepatic synthesis were involved in the USF1-induced changes in plasma lipid fractions in these models. USF1 is ubiquitously expressed, and recent genome-wide ChIP-seq studies have shown that it is associated with thousands of genes (29). Human studies have suggested that there are significant influences of other genes, sex, age and physiologic state on the phenotypic effect of genetic variation of USF1 (13,17,21,22). Our work supports the importance of genetic variation in USF1 as a determinant of traits relevant to hyperlipidemia, obesity and metabolic syndrome and highlights the significance of sex in this regard. It also suggests that USF1 may act in part through influencing gene networks in the liver closely linked to metabolism and inflammation, such as that described earlier containing Igfbp2 and closely correlated genes.
MATERIALS AND METHODS
Tg animal models
BAC clone CTD3003012 including the human USF1 gene sequence was purchased from Invitrogen (Supplementary Material, Fig. S1A). The sequence was confirmed using PCR primers specific to the two BAC ends and human USF1 gene sequence. The clones were cultured in LB medium containing chloramphenicol. BAC DNA was extracted and purified with a large-construct kit (Qiagen) and pulse-field electrophoresis. The purified circular BAC DNA was injected into pronuclei of fertilized FVB eggs, and surviving eggs were transferred to pseudopregnant FVB female mice. Transgenic founders were identified using DNA isolated from tail biopsies by PCR using BAC-end-specific and USF1-specific primers. The integration of the entire BAC sequence was confirmed in all founders.
All mice were bred at UCLA and were fed a 4% chow diet (Harlan Teklad7017; 4% fat, 0% cholesterol) ad libium and maintained on a 12 h light/dark cycle until euthanasia at the age indicated. Genomic DNA was isolated from tail samples using a DNeasy kit (Qiagen) and genotyped using PCR. All reactions were carried out using initial denaturing at 98°C for 30 s, followed by 35 cycles at 98°C for 10 s, 55°C for 30 s, 72°C for 30 s and completed with an extension at 72°C for 2 min. All mice were weaned at 3 weeks of age, when started with a 4% chow diet.
Phenotypic characterization of the tg mouse model
All tg mice were studied as heterozygotes, and controls (wt) were sex-matched non-tg littermates. Starting at 8 weeks of age, each mouse was monitored for body weight, and body composition was evaluated by NMR (Brucker Minispec). Lean mass, fat mass and water content were monitored over the course of chow diet for 15 weeks. Mice were fasted and anesthetized via exposure to isoflurane before blood was collected through the retro-orbital sinus. All procedures were performed in accordance with the current National Research Council Guide for the Care and Use of Laboratory Animals and were approved by the UCLA Animal Research Committee.
Analysis of phenotypic data
Student's t-test was used to analyze the differences in the clinical traits between tg animals and their wt littermate controls. The significance level was set to P < 0.05. The significance of the difference in the growth curves of total body weight, fat mass, lean mass and adiposity between tg and wt controls was determined using an auto-regressive method described previously (34).
Adenovirus-mediated over-expression of USF1
Recombinant adenovirus serotype 5 (DE1/E3) expressing human USF1 and/or enhanced GFP (AdUSF1 and AdGFP) was produced by Vectors Biolab (Philadelphia, PA, USA) (Supplementary Material, Fig. S1B). Both 293 cells and mouse primary hepatocytes were cultured in monolayer and infected with 10 MOI (multiplicity of infection) of AdUSF1 or AdGFP. Quantitative PCR was employed to detect expression of human USF1 transcript, and western blot analysis was performed to detect human USF1 using the mouse monoclonal anti-human USF1 antibody AB58100 (Abcam). To transiently over-express USF1 in the liver, mice received ∼3 × 109 plaque-forming units of recombinant adenovirus vector particles in 0.2 ml saline diluent by tail vein injection. Five days post-injection, mice were fasted for 6 h prior to euthanasia. All animals were exposed to isoflurane for blood collection via retro-orbital sinus, and tissues were snap-frozen using liquid nitrogen. For western blot analysis, 40 µg of total protein extracted from each liver was applied to a 4–12% Bis–Tris NuPAGE gel in MOPS/SDS running buffer (Invitrogen, Carlsband, CA, USA). After transblotting, PVDF membrane was incubated in 5% non-fat milk-TBS/T blotto at room temperature for 1 h on shaker, followed by incubation with primary antibody (diluted 1:1000 in blotto) overnight at 4°C on a shaker. After three TBS/T washings, the membrane was incubated with secondary antibody (1:6000, ECL Mouse IgG, HRP-linked whole antibody, GE Health, Piscataway, NJ, USA) for 1 h at room temperature on a shaker and washed again three times in TBS/S. ECL-plus (GE Health) was employed to visualize protein bands on film.
RNA analysis
Total RNA isolation of tissues and cells was performed with TRIzol reagent (Invitrogen), followed by an RNeasy Mini Kit (Qiagen) cleanup step. Complementary DNA was synthesized with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) for RT–PCR. Quantitative PCR was carried out by using QuantiTect SYBR Green PCR Kit (Qiagen) on an Opticon machine (MJ Research). Each qPCR was carried out at least in duplicate. The primer sets used for real-time RT–PCR were: human USF1 (AATGGAACGGGGGTAGAAAG; AGATAACACCTGCAGCCACC) and mouse Usf1 (GGGAGTTTGGCAAGTGTGTT; CAGTGTCCAGCAAGGAGACA), and sets from Qiagen for murine TATA box-binding protein (Tbp; QT00198443), beta-actin (Actb; QT01136772), apolipoprotein A-I (Apoa1; QT00110663) and apolipoprotein B (Apob; QT01061494). The PCR conditions were 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, 72°C for 1 min. All qPCR readouts were normalized to either β-actin or TATA box-binding protein transcript levels.
Global gene expression analysis
Illumina Mouse whole-genome expression BeadChip (MouseRef-6-v2 Expression BeadChip) was used to profile gene expression of adenovirus-infected animals. All amplifications and hybridizations were performed according to the Illumina protocol by the Southern California Genome Consortium Microarray Core Laboratory at UCLA. Briefly, 200 ng of total RNA was reverse-transcribed to cDNA using an Ambion cDNA synthesis kit (AMIL 1791) and then converted to cRNA and labeled with biotin. Biotinylated cRNA product (800 ng) was hybridized to arrays prepared and allowed to incubate overnight (16–20 h) at 55°C. Arrays were washed and then incubated with Cy3-labeled streptavidin. After washing, arrays were dried and scanned on an Illumina BeadScan confocal laser scanner. Robust spline normalization and cubic root transformation methods with LumiR package were employed to process raw data of gene expression. The microarray data for these analyses have been deposited in GEO under accession number GSE17442.
Identification of molecular signatures of Usf1 in segregating mouse populations
Data were analyzed from a mouse intercross described in previous studies (32,47–50). These were constructed from C57BL/6J (B6) and C3H/HeJ (C3H) strains on an ApoE null background (BXH.ApoE). RNA hybridization and transcript quantification on custom Agilent gene expression microarrays have been described previously (48). Individual transcript intensities were corrected for experimental variation and normalized and were reported as the mean log 10 ratio (mlratio) of an individual experiment relative to a pool reference made from the same F2 population. The F2 mouse cross-microarray data for this study have been deposited in GEO under accession number GSE2814. Other studies using data from this cross have been published (32,34,48–51). Correlations between transcripts and Usf1 message level were calculated using Pearson's correlation, with data from male and female mice analyzed independently. The significance level was set at P < 0.05. Among 23 574 probes (60mer oligonucleotides) for mouse genes and ESTs and 2186 control sequences (Agilent Technologies), 2898 and 3082 probe readouts were identified to significantly correlate with Usf1 mRNA levels at a nominal P-value of <0.05, which represented transcripts from 2613 and 2802 genes in male and female animals, respectively.
Construction of weighted co-expression networks with Usf1 molecular signature sets
Weighted co-expression gene networks were constructed with Usf1 molecular signature genes identified in each sex independently as described (52). Briefly, Pearson's correlation between each pair of signature genes was computed to yield a similarity matrix that is subsequently transformed into adjacency matrix by applying power functions to generate a scale-free weighted co-expression gene network. The connectivity between any two genes was determined by taking the sum of their connection strengths with all other genes in the network. The adjacency matrix was employed to measure node dissimilarity, based on the topological overlap matrix. Hierarchical clustering on the topological overlap matrix was performed to define gene modules. Modules are identified by an assigned color designation; there is no relationship between modules from different sexes that have the same assigned color designation, as the co-expression networks were constructed independently. The first principal component of a given module was defined as module eigengene, which is considered the most representative gene expression in a module. For each module, Pearson's correlation coefficient between its eigengene and each clinical trait was determined and defined as module significance. Detailed information of the algorithm and R packages can be downloaded at http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/ CytoScape 1 was employed to visualize the co-expression network for a given module, where node and edge correspond to transcript and pair-wise connectivity (53).
Pathway and gene set enrichment analysis
To assess whether gene signature sets and modules were significantly enriched for GO function categories and KEGG pathways, one-tailed Fisher exact test implemented in the EASE analysis tool was employed (33). Overlap between gene sets was also determined using the one-tailed Fisher exact test as implemented in EASE.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the US National Institutes of Health (DK072260 to T.A.D., HL28481 to A.J.L., HL082762 and HL095056 to P.P., and T32 HG02536 for R.M.-H.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Judy S. Wang for assistance in mouse husbandry and assessment of physiologic phenotypes; Diana Shih for advice and assistance with transgenic and transfection vector construction and analysis; Sharda Charugundla for performance of plasma lipid analyses; Leslie Ingram-Drake for assistance with data management and analysis; Susanna Wang for BXH.ApoE null F2 cross-construction and characterization, and Eric Schadt for extensive collaboration, including microarray performance and analysis concerning this data set; Xia Yang for advice concerning with data analysis and interpretation; and Anatole Ghazalpour for advice concerning co-expression analyses.
Author contributions: Study design: T.A.D., A.J.L., P.P., S.W. and R.M.-H; performance of experiments: S.W., R.M.-H., E.Z.D., H.Q. and L.W.C.; data analysis: S.W., R.M.-H., A.J.L. and T.A.D.; manuscript preparation: S.W., R.M.-H., A.J.L. and T.A.D.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Genest J.J., Jr, Martin-Munley S.S., McNamara J.R., Ordovas J.M., Jenner J., Myers R.H., Silberman S.R., Wilson P.W., Salem D.N., Schaefer E.J. Familial lipoprotein disorders in patients with premature coronary artery disease. Circulation. 1992;85:2025–2033. doi: 10.1161/01.cir.85.6.2025. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein J.L., Schrott H.G., Hazzard W.R., Bierman E.L., Motulsky A.G. Hyperlipidemia in coronary heart disease. II. Genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J. Clin. Invest. 1973;52:1544–1568. doi: 10.1172/JCI107332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopkins P.N., Heiss G., Ellison R.C., Province M.A., Pankow J.S., Eckfeldt J.H., Hunt S.C. Coronary artery disease risk in familial combined hyperlipidemia and familial hypertriglyceridemia: a case–control comparison from the National Heart, Lung, and Blood Institute Family Heart Study. Circulation. 2003;108:519–523. doi: 10.1161/01.CIR.0000081777.17879.85. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.C., Lusis A.J., Pajukanta P. Familial combined hyperlipidemia: upstream transcription factor 1 and beyond. Curr. Opin. Lipidol. 2006;17:101–109. doi: 10.1097/01.mol.0000217890.54875.13. [DOI] [PubMed] [Google Scholar]
- 5.Pajukanta P., Nuotio I., Terwilliger J.D., Porkka K.V., Ylitalo K., Pihlajamaki J., Suomalainen A.J., Syvanen A.C., Lehtimaki T., Viikari J.S., et al. Linkage of familial combined hyperlipidaemia to chromosome 1q21–q23. Nat. Genet. 1998;18:369–373. doi: 10.1038/ng0498-369. [DOI] [PubMed] [Google Scholar]
- 6.Coon H., Eckfeldt J.H., Leppert M.F., Myers R.H., Arnett D.K., Heiss G., Province M.A., Hunt S.C. A genome-wide screen reveals evidence for a locus on chromosome 11 influencing variation in LDL cholesterol in the NHLBI Family Heart Study. Hum. Genet. 2002;111:263–269. doi: 10.1007/s00439-002-0773-8. [DOI] [PubMed] [Google Scholar]
- 7.Allayee H., Krass K.L., Pajukanta P., Cantor R.M., van der Kallen C.J., Mar R., Rotter J.I., de Bruin T.W., Peltonen L., Lusis A.J. Locus for elevated apolipoprotein B levels on chromosome 1p31 in families with familial combined hyperlipidemia. Circ. Res. 2002;90:926–931. doi: 10.1161/01.res.0000015885.27134.f0. [DOI] [PubMed] [Google Scholar]
- 8.Pei W., Baron H., Muller-Myhsok B. Linkage of familial combined hyperlipidemia to chromosome 1q21–23 in Chinese and German families. Zhonghua Yi Xue Za Zhi. 2000;80:25–27. [PubMed] [Google Scholar]
- 9.Huertas-Vazquez A., Aguilar-Salinas C., Lusis A.J., Cantor R.M., Canizales-Quinteros S., Lee J.C., Mariana-Nunez L., Riba-Ramirez R.M., Jokiaho A., Tusie-Luna T., et al. Familial combined hyperlipidemia in Mexicans: association with upstream transcription factor 1 and linkage on chromosome 16q24.1. Arterioscler. Thromb. Vasc. Biol. 2005;25:1985–1991. doi: 10.1161/01.ATV.0000175297.37214.a0. [DOI] [PubMed] [Google Scholar]
- 10.Pajukanta P., Lilja H.E., Sinsheimer J.S., Cantor R.M., Lusis A.J., Gentile M., Duan X.J., Soro-Paavonen A., Naukkarinen J., Saarela J., et al. Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1) Nat. Genet. 2004;36:371–376. doi: 10.1038/ng1320. [DOI] [PubMed] [Google Scholar]
- 11.Ng M.C., So W.Y., Lam V.K., Cockram C.S., Bell G.I., Cox N.J., Chan J.C. Genome-wide scan for metabolic syndrome and related quantitative traits in Hong Kong Chinese and confirmation of a susceptibility locus on chromosome 1q21–q25. Diabetes. 2004;53:2676–2683. doi: 10.2337/diabetes.53.10.2676. [DOI] [PubMed] [Google Scholar]
- 12.Langefeld C.D., Wagenknecht L.E., Rotter J.I., Williams A.H., Hokanson J.E., Saad M.F., Bowden D.W., Haffner S., Norris J.M., Rich S.S., et al. Linkage of the metabolic syndrome to 1q23–q31 in Hispanic families: the Insulin Resistance Atherosclerosis Study Family Study. Diabetes. 2004;53:1170–1174. doi: 10.2337/diabetes.53.4.1170. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.C., Weissglas-Volkov D., Kyttala M., Sinsheimer J.S., Jokiaho A., de Bruin T.W., Lusis A.J., Brennan M.L., van Greevenbroek M.M., van der Kallen C.J., et al. USF1 contributes to high serum lipid levels in Dutch FCHL families and U.S. whites with coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2007;27:2222–2227. doi: 10.1161/ATVBAHA.107.151530. [DOI] [PubMed] [Google Scholar]
- 14.Choquette A.C., Bouchard L., Houde A., Bouchard C., Perusse L., Vohl M.C. Associations between USF1 gene variants and cardiovascular risk factors in the Quebec Family Study. Clin. Genet. 2007;71:245–253. doi: 10.1111/j.1399-0004.2007.00755.x. [DOI] [PubMed] [Google Scholar]
- 15.Auro K., Kristiansson K., Zethelius B., Berne C., Lannfelt L., Taskinen M.R., Jauhiainen M., Perola M., Peltonen L., Syvanen A.C. USF1 gene variants contribute to metabolic traits in men in a longitudinal 32-year follow-up study. Diabetologia. 2008;51:464–472. doi: 10.1007/s00125-007-0892-9. [DOI] [PubMed] [Google Scholar]
- 16.Zeggini E., Damcott C.M., Hanson R.L., Karim M.A., Rayner N.W., Groves C.J., Baier L.J., Hale T.C., Hattersley A.T., Hitman G.A., et al. Variation within the gene encoding the upstream stimulatory factor 1 does not influence susceptibility to type 2 diabetes in samples from populations with replicated evidence of linkage to chromosome 1q. Diabetes. 2006;55:2541–2548. doi: 10.2337/db06-0088. [DOI] [PubMed] [Google Scholar]
- 17.Holzapfel C., Baumert J., Grallert H., Muller A.M., Thorand B., Khuseyinova N., Herder C., Meisinger C., Hauner H., Wichmann H.E., et al. Genetic variants in the USF1 gene are associated with low-density lipoprotein cholesterol levels and incident type 2 diabetes mellitus in women: results from the MONICA/KORA Augsburg case-cohort study, 1984–2002. Eur. J. Endocrinol. 2008;159:407–416. doi: 10.1530/EJE-08-0356. [DOI] [PubMed] [Google Scholar]
- 18.Meex S.J., van Vliet-Ostaptchouk J.V., van der Kallen C.J., van Greevenbroek M.M., Schalkwijk C.G., Feskens E.J., Blaak E.E., Wijmenga C., Hofker M.H., Stehouwer C.D., et al. Upstream transcription factor 1 (USF1) in risk of type 2 diabetes: association study in 2000 Dutch Caucasians. Mol. Genet. Metab. 2008;94:352–355. doi: 10.1016/j.ymgme.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 19.Collings A., Hoyssa S., Fan M., Kahonen M., Hutri-Kahonen N., Marniemi J., Juonala M., Viikari J.S., Raitakari O.T., Lehtimaki T.J. Allelic variants of upstream transcription factor 1 associate with carotid artery intima-media thickness: the Cardiovascular Risk in Young Finns study. Circ. J. 2008;72:1158–1164. doi: 10.1253/circj.72.1158. [DOI] [PubMed] [Google Scholar]
- 20.Kristiansson K., Ilveskoski E., Lehtimaki T., Peltonen L., Perola M., Karhunen P.J. Association analysis of allelic variants of USF1 in coronary atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2008;28:983–989. doi: 10.1161/ATVBAHA.107.156463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komulainen K., Alanne M., Auro K., Kilpikari R., Pajukanta P., Saarela J., Ellonen P., Salminen K., Kulathinal S., Kuulasmaa K., et al. Risk alleles of USF1 gene predict cardiovascular disease of women in two prospective studies. PLoS Genet. 2006;2:e69. doi: 10.1371/journal.pgen.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reiner A.P., Carlson C.S., Jenny N.S., Durda J.P., Siscovick D.S., Nickerson D.A., Tracy R.P. USF1 gene variants, cardiovascular risk, and mortality in European Americans: analysis of two US cohort studies. Arterioscler. Thromb. Vasc. Biol. 2007;27:2736–2742. doi: 10.1161/ATVBAHA.107.154559. [DOI] [PubMed] [Google Scholar]
- 23.Lusis A.J., Attie A.D., Reue K. Metabolic syndrome: from epidemiology to systems biology. Nat. Rev. Genet. 2008;9:819–830. doi: 10.1038/nrg2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson F., Hercberg S., Froguel P. Common polymorphisms in the USF1 gene are not associated with type 2 diabetes in French Caucasians. Diabetes. 2005;54:3040–3042. doi: 10.2337/diabetes.54.10.3040. [DOI] [PubMed] [Google Scholar]
- 25.Ng M.C., Miyake K., So W.Y., Poon E.W., Lam V.K., Li J.K., Cox N.J., Bell G.I., Chan J.C. The linkage and association of the gene encoding upstream stimulatory factor 1 with type 2 diabetes and metabolic syndrome in the Chinese population. Diabetologia. 2005;48:2018–2024. doi: 10.1007/s00125-005-1914-0. [DOI] [PubMed] [Google Scholar]
- 26.Plaisier C.L., Horvath S., Huertas-Vazquez A., Cruz-Bautista I., Herrera M.F., Tusie-Luna T., Aguilar-Salinas C., Pajukanta P. A systems genetics approach implicates USF1, FADS3, and other causal candidate genes for familial combined hyperlipidemia. PLoS Genet. 2009;5:e1000642. doi: 10.1371/journal.pgen.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sirito M., Walker S., Lin Q., Kozlowski M.T., Klein W.H., Sawadogo M. Members of the USF family of helix–loop–helix proteins bind DNA as homo- as well as heterodimers. Gene Expr. 1992;2:231–240. [PMC free article] [PubMed] [Google Scholar]
- 28.Naukkarinen J., Gentile M., Soro-Paavonen A., Saarela J., Koistinen H.A., Pajukanta P., Taskinen M.R., Peltonen L. USF1 and dyslipidemias: converging evidence for a functional intronic variant. Hum. Mol. Genet. 2005;14:2595–2605. doi: 10.1093/hmg/ddi294. [DOI] [PubMed] [Google Scholar]
- 29.Rada-Iglesias A., Ameur A., Kapranov P., Enroth S., Komorowski J., Gingeras T.R., Wadelius C. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 2008;18:380–392. doi: 10.1101/gr.6880908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirito M., Lin Q., Deng J.M., Behringer R.R., Sawadogo M. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc. Natl Acad. Sci. USA. 1998;95:3758–3763. doi: 10.1073/pnas.95.7.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallet V.S., Casado M., Henrion A.A., Bucchini D., Raymondjean M., Kahn A., Vaulont S. Differential roles of upstream stimulatory factors 1 and 2 in the transcriptional response of liver genes to glucose. J. Biol. Chem. 1998;273:20175–20179. doi: 10.1074/jbc.273.32.20175. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y., Zhu J., Lum P.Y., Yang X., Pinto S., MacNeil D.J., Zhang C., Lamb J., Edwards S., Sieberts S.K., et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 34.Yang X., Deignan J.L., Qi H., Zhu J., Qian S., Zhong J., Torosyan G., Majid S., Falkard B., Kleinhanz R.R., et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat. Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bensinger S.J., Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454:470–477. doi: 10.1038/nature07202. [DOI] [PubMed] [Google Scholar]
- 36.de Luca C., Olefsky J.M. Inflammation and insulin resistance. FEBS Lett. 2008;582:97–105. doi: 10.1016/j.febslet.2007.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karalis K.P., Giannogonas P., Kodela E., Koutmani Y., Zoumakis M., Teli T. Mechanisms of obesity and related pathology: linking immune responses to metabolic stress. FEBS J. 2009;276:5747–5754. doi: 10.1111/j.1742-4658.2009.07304.x. [DOI] [PubMed] [Google Scholar]
- 38.Tilg H., Moschen A.R. Inflammatory mechanisms in the regulation of insulin resistance. Mol. Med. 2008;14:222–231. doi: 10.2119/2007-00119.Tilg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tilg H., Moschen A.R. Insulin resistance, inflammation, and non-alcoholic fatty liver disease. Trends Endocrinol. Metab. 2008;19:371–379. doi: 10.1016/j.tem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Wheatcroft S.B., Kearney M.T., Shah A.M., Ezzat V.A., Miell J.R., Modo M., Williams S.C., Cawthorn W.P., Medina-Gomez G., Vidal-Puig A., et al. IGF-binding protein-2 protects against the development of obesity and insulin resistance. Diabetes. 2007;56:285–294. doi: 10.2337/db06-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heald A.H., Kaushal K., Siddals K.W., Rudenski A.S., Anderson S.G., Gibson J.M. Insulin-like growth factor binding protein-2 (IGFBP-2) is a marker for the metabolic syndrome. Exp. Clin. Endocrinol. Diabetes. 2006;114:371–376. doi: 10.1055/s-2006-924320. [DOI] [PubMed] [Google Scholar]
- 42.Grarup N., Rose C.S., Andersson E.A., Andersen G., Nielsen A.L., Albrechtsen A., Clausen J.O., Rasmussen S.S., Jorgensen T., Sandbaek A., et al. Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: validation and extension of genome-wide association studies. Diabetes. 2007;56:3105–3111. doi: 10.2337/db07-0856. [DOI] [PubMed] [Google Scholar]
- 43.Lyon H.N., Emilsson V., Hinney A., Heid I.M., Lasky-Su J., Zhu X., Thorleifsson G., Gunnarsdottir S., Walters G.B., Thorsteinsdottir U., et al. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet. 2007;3:e61. doi: 10.1371/journal.pgen.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norris J.M., Langefeld C.D., Talbert M.E., Wing M.R., Haritunians T., Fingerlin T.E., Hanley A.J., Ziegler J.T., Taylor K.D., Haffner S.M., et al. Genome-wide association study and follow-up analysis of adiposity traits in hispanic Americans: The IRAS Family Study. Obesity (Silver Spring) 2009;17:1932–1941. doi: 10.1038/oby.2009.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salopuro T., Pulkkinen L., Lindstrom J., Eriksson J.G., Valle T.T., Hamalainen H., Ilanne-Parikka P., Keinanen-Kiukaanniemi S., Tuomilehto J., Laakso M., et al. Genetic variation in leptin receptor gene is associated with type 2 diabetes and body weight: The Finnish Diabetes Prevention Study. Int. J. Obes. (Lond.) 2005;29:1245–1251. doi: 10.1038/sj.ijo.0803024. [DOI] [PubMed] [Google Scholar]
- 46.Wong R.H., Chang I., Hudak C.S., Hyun S., Kwan H.Y., Sul H.S. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136:1056–1072. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S., Yehya N., Schadt E.E., Wang H., Drake T.A., Lusis A.J. Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet. 2006;2:e15. doi: 10.1371/journal.pgen.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Nas A., Guhathakurta D., Wang S.S., Yehya N., Horvath S., Zhang B., Ingram-Drake L., Chaudhuri G., Schadt E.E., Drake T.A., et al. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology. 2009;150:1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S.S., Schadt E.E., Wang H., Wang X., Ingram-Drake L., Shi W., Drake T.A., Lusis A.J. Identification of pathways for atherosclerosis in mice: integration of quantitative trait locus analysis and global gene expression data. Circ. Res. 2007;101:e11–e30. doi: 10.1161/CIRCRESAHA.107.152975. [DOI] [PubMed] [Google Scholar]
- 51.Ghazalpour A., Doss S., Zhang B., Wang S., Plaisier C., Castellanos R., Brozell A., Schadt E.E., Drake T.A., Lusis A.J., et al. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4:Article 17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 53.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.