Abstract
Interleukin-6 (IL-6) is an important modulator of inflammation and immunity whose dysregulation is associated with a number of disease states. There is evidence of significant heritability in inter-individual variation in IL6 gene expression but the genetic variants responsible for this remain to be defined. We adopted a combined approach of mapping protein and expression quantitative trait loci in peripheral blood mononuclear cells using high-density single-nucleotide polymorphism (SNP) typing for ∼2000 loci implicated in cardiovascular, metabolic and inflammatory syndromes to show that common SNP markers and haplotypes of LEP (encoding leptin) associate with a 1.7- to 2-fold higher level of lipopolysaccharide (LPS)-induced IL-6 expression. We subsequently demonstrate that basal leptin expression significantly correlates with LPS-induced IL-6 expression and that the same variants at LEP which associate with IL-6 expression are also major determinants of leptin expression in these cells. We find that variation involving two other genomic regions, CAPNS1 (encoding calpain small subunit 1) and ALOX15 (encoding arachidonate 15-lipoxygenase), show significant association with IL-6 expression. Although this may be a subset of all such trans-acting effects, we find that the same ALOX15 variants are associated with induced expression of tumour necrosis factor and IL-1beta consistent with a broader role in acute inflammation for ALOX15. This study provides evidence of novel genetic determinants of IL-6 production with implications for understanding susceptibility to inflammatory disease processes and insight into cross talk between metabolic and inflammatory pathways. It also provides proof of concept for use of an integrated expression phenotype mapping approach.
INTRODUCTION
Interleukin-6 (IL-6) is a pleiotropic cytokine that plays critical roles in both innate and adaptive immunity and is vital in the transition from an acute to a sustained inflammatory response. Dysregulation of IL-6 signalling is implicated in many disease processes characterized by chronic inflammation and autoimmunity, including atherosclerosis, sepsis, arthritis, inflammatory bowel disease and oncogenesis together with dysfunctional metabolic states such as obesity and hyperinsulinaemia (1). The importance of IL-6 in autoimmunity has recently been highlighted by the demonstration that, in tandem with TGF-beta, IL-6 is vital to the establishment of pro-autoimmunity T(H)17 lineage T-cells and its continued presence inhibits the emergence of their regulatory counterparts, T(reg) cells (2). Although most cell types are capable of synthesizing IL-6, the majority of circulating IL-6 in the acute inflammatory response is produced by macrophages and monocytes following Toll-like receptor stimulation.
Genetic diversity plays an important role in defining individual levels of expression of IL6 with twin studies demonstrating significant heritability, estimated at between 57 and 61% for ex vivo IL-6 production following endotoxin or mitogen stimulation (3,4). To date attention has focused on DNA sequence variation at the IL6 locus with evidence that specific single-nucleotide polymorphisms (SNPs) in the IL6 promoter region are important determinants of gene expression. This was based on reporter gene analysis and association with IL-6 production in healthy individuals and disease states and remains incompletely understood (5–9). It is probable that determinants of the magnitude of IL-6 response are influenced by diversity at other genomic loci, none of which have yet been resolved.
Mapping gene expression quantitative trait loci (eQTLs) at the RNA level has proved a powerful approach to define putative functional genetic variants (10), while a recent report identified protein QTLs (pQTLs) which may be of more relevance to physiological or disease states (11). Resolution of functionally important regulatory variants requires context-specific effects to be considered. To date, the majority of published studies in human subjects seeking to associate levels of gene expression with SNP diversity have relied upon the use of transformed lymphoblastoid cell lines (12–14) or more recently have analysed basal levels of expression in blood or serum (11,15,16). It is likely that many regulatory polymorphisms determining cytokine responses will only become apparent upon stimulation of an immune response and that accurate assessment of functional studies will depend upon the use of appropriate cell types.
We aimed to define common SNPs associated with IL-6 production in peripheral blood mononuclear cells (PBMCs) from healthy volunteers upon exposure to lipopolysaccharide (LPS) which signals through Toll-like receptor-4. We adopted an integrated expression phenotype mapping approach which analysed both pQTLs and eQTLs using a panel of SNP markers informative for gene loci implicated in immune and inflammatory responses, and with susceptibility to cardiovascular disease (17). This allowed us to resolve specific SNPs and haplotypes in LEP, ALOX15 and CAPNS1 associated with induced IL-6 expression.
RESULTS
Mapping common SNP markers associated with IL-6 expression
We first sought to establish pQTLs for IL-6 expression by analysing PBMCs from 96 healthy volunteers in either the resting state or after induction with LPS. IL-6 production was strongly induced by LPS at 2 ng/ml, from median basal levels of 0.056 ng/ml to a mean of 11.0 ng/ml after 6 h, with significant inter-individual variation observed. Levels of induced IL-6 expression within individuals was reproducible over time for five healthy volunteers investigated on three separate occasions, on average three weeks apart. A linear regression model showed that when considered as categorical variables in a forward stepwise manner using likelihood ratio testing, the day of testing was not significant (P > 0.05); however, the subjects were significantly associated with levels of IL-6 (P = 0.0029), consistent with published data showing reproducibility of individual serum levels of IL-6 over time (18). For the QTL analysis in the full panel of 96 volunteers, three replicate stimulation assays were performed for a given individual on a particular day, and the mean value used in subsequent analyses.
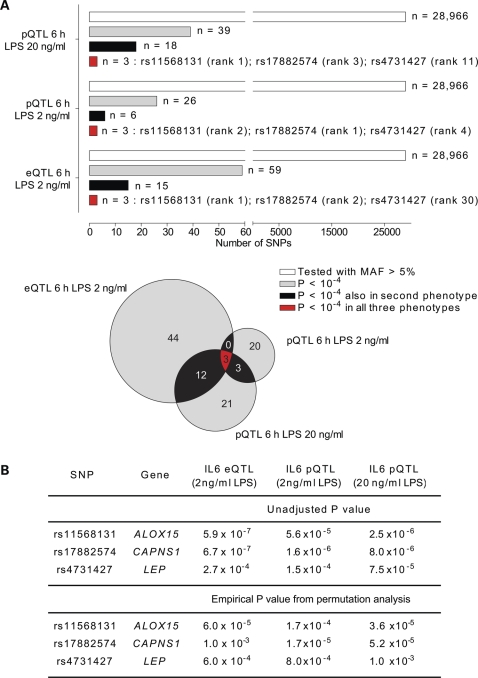
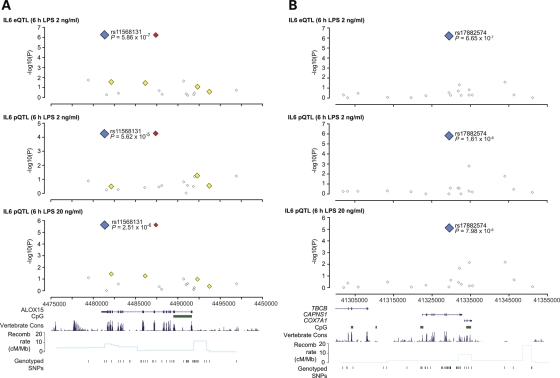
We genotyped the volunteers for a panel of 45 237 SNPs using the Illumina HumanCVD v1 beadchip which includes high-density coverage of ∼2000 genes implicated in immune and inflammatory responses with specific relevance to vascular pathology, metabolic disorders and inflammatory disease states (17). The high density of SNPs on this array increases the probability of capturing functionally relevant polymorphisms within the loci covered on this chip. We found that 26 SNP markers at 17 genomic loci were associated with IL-6 production at the protein level (P < 10−4), notably involving the ALOX15, CAPNS1 and LEP genes at chromosome 17p13.3, 19q13.12 and 7q31.3, respectively (Fig. 1, Supplementary Material, Fig. S1). When we compared these results with IL-6 protein production at 6 h in response to a higher dose of LPS stimulation (20 ng/ml), we saw high concordance between the data sets with SNPs in ALOX15, CAPNS1 and LEP being the most significant hits of association found shared between the two phenotypes (Fig. 1). We then complemented this analysis by mapping eQTL based on IL6 expression at the RNA level in response to LPS stimulation. We found that the same SNP markers at ALOX15, CAPNS1 and LEP were associated with IL6 expression, being the only SNP markers consistently associated across the phenotypes (Fig. 1, Supplementary Material, Fig. S1 and Table S1). For the cohort of volunteers, levels of IL-6 protein and IL6 RNA were seen to be strongly correlated (Supplementary Material, Fig. S2).
Figure 1.
LPS-induced IL-6 transcript and protein expression phenotype mapping. (A) eQTL/pQTL analysis showing concordance of SNP associations for IL-6 expression phenotypes (unadjusted P-values less than 10−4) for rs11568131, rs17882574 and rs4731427 with rank order of association indicated. (B) Unadjusted P-values for SNP associations together with empirical P-values derived from permutation analysis with population correction.
Cis- and trans-associations at LEP
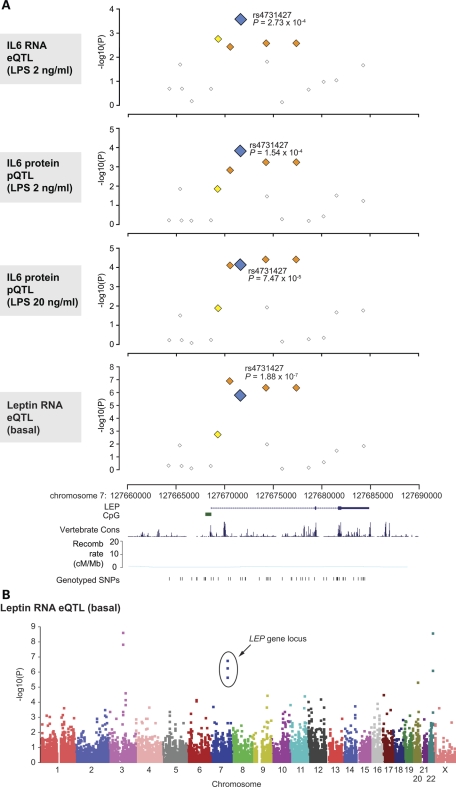
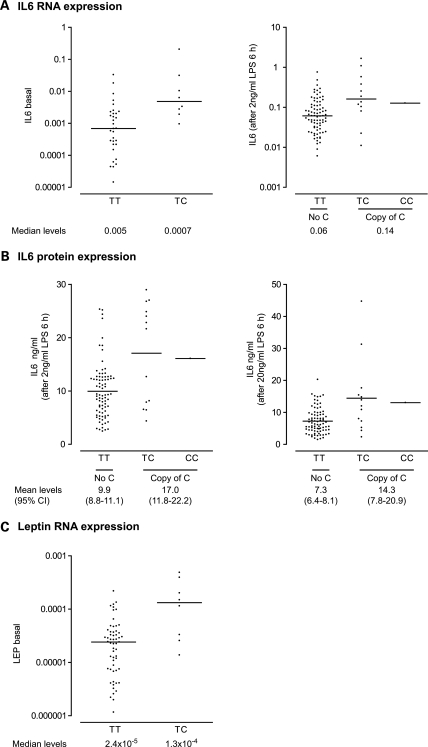
The observed association of IL-6 expression with common SNP markers at or near the LEP gene is of particular biological interest given the evidence that leptin can play an important immunomodulatory role, and specifically published data showing that leptin can both directly activate and augment IL-6 production by PBMCs induced with LPS (19–22). We successfully genotyped 52 SNPs within a 25 kb region spanning LEP of which 16 SNPs were informative with a minor allele frequency (MAF) of greater than 5%. Several common SNP markers showed association with basal and induced IL-6 expression phenotypes at this locus, notably rs4731427 (c.29+3030T>C) (Fig. 2). Individuals possessing a copy of the minor C allele had a 7-fold higher level of basal IL6 expression compared with those without (Fig. 3) (Mann–Whitney test two-tailed P= 0.003), while following LPS induction (2 ng/ml) a 2.3-fold increase in IL6 expression was seen at the RNA level. This was reproduced at the protein level with a 1.7- or 2.0-fold higher level of IL-6 expression (2 or 20 ng/ml LPS, respectively) (P< 0.0001 on unpaired t-test, two-tailed, for either phenotype) (Fig. 3).
Figure 2.
Common SNP markers in LEP are associated with IL-6 and leptin expression. (A) Single marker allelic association results for LEP gene locus with IL-6 or leptin expression plotted as −log10(P) values by genomic coordinate. With reference to rs4731427, SNPs with MAF greater than 5% and r2 < 0.2 are shown as white squares, 0.2–0.5 (yellow) and 0.5–0.8 (orange). LEP gene structure and genotyped SNPs are shown below. Estimated recombination rates are shown from HapMap (using Build 35 coordinates). Gene structure, vertebrate multiz alignment and conservation track (17 species) (49) and genotyped SNP locations adapted from screenshot of the UCSC Genome Browser (Human March 2006 Assembly). (B) Manhattan plot showing strength of association from PLINK analysis plotted as −log10(P) values by chromosome for leptin expression.
Figure 3.
Association of IL-6 and leptin expression with rs4731427. Scatter plots are shown for IL-6 expression at either the (A) RNA (relative to actin and plotted on log scale with median value indicated) or (B) protein level (with mean value and 95% confidence intervals shown) together with data for (C) LEP RNA.
These results are consistent with the hypothesis that differences in levels of expression of leptin modulate IL-6 responsiveness. We proceeded to analyse basal expression of leptin in our cohort of healthy volunteers and found that this was significantly correlated with induced levels of IL-6 (Supplementary Material, Fig. S2). Strikingly, when we analysed leptin expression as a quantitative trait using the panel of 45 237 SNPs, we found strong evidence of association involving the same common SNP markers at the LEP locus which had been associated with higher induced IL-6 expression (Fig. 2). Possession of a copy of the rarer C allele of rs4731427 associated with IL-6 expression phenotypes was associated with a 5.5-fold higher LEP expression (Mann–Whitney test two-tailed P= 0.0045) (Fig. 3).
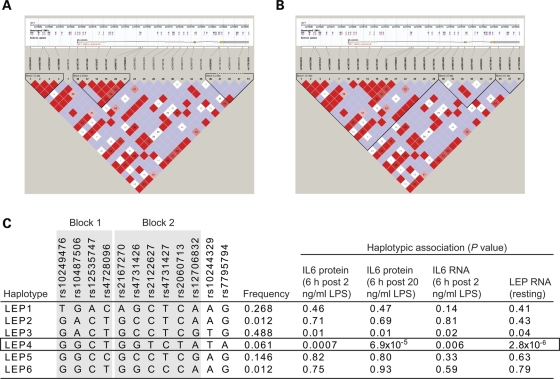
We found evidence of substantial linkage disequilibrium (LD) across the LEP gene with a very low recombination rate (Fig. 2) and we proceeded to analyse gene expression in terms of the haplotypic structure at this locus for individuals of Caucasian ancestry (Fig. 4). This defined six haplotypes and showed that for IL-6 and leptin expression phenotypes, haplotype LEP4 was most strongly associated (Fig. 4). This haplotype refines the observed association with rs4731427 and suggests the functional variant may be localized within this haplotype.
Figure 4.
Haplotypic analysis of LEP. For volunteers of Caucasian ancestry, genotyped SNPs of greater than 5% MAF were used to infer the underlying haplotypic structure using Haploview. Haplotype blocks shown defined based on (A) confidence intervals (47) or (B) solid spine of LD. The Haploview plot for D′ shows the following: white and blue squares have a LOD score of less than 2 (indicating lower confidence) with the white having D′ < 1 and blue having a D′ = 1. Those squares that are shades of pink to red have a LOD score greater than 2 and have D′ < 1, while the red squares represent D′ = 1. (C) Haplotypic association for expression phenotypes shown based on six inferred haplotypes at LEP locus.
Our data are supported by the previous independent reports of associations with leptin expression and obesity for a common non-coding SNP 2.5 kb upstream of LEP, rs7799039 (23–26). We observed a significant association with rs10487506, an SNP marker in complete LD with rs7799039 (Supplementary Material, Fig. S3); this SNP also shows a weak association with IL-6 expression at 6 h after LPS induction (2 ng/ml) (unadjusted P-value 0.03). However, our dense SNP genotyping at the LEP gene locus resolves the likely underlying functional haplotype with much greater precision and suggests that the functional cis-acting variant is not rs7799039, but carried on the LEP4 haplotype.
Signals of association with IL-6 expression at ALOX15 and CAPNS1
We found two SNP markers in complete LD (rs11568131 and rs11078527) (c.*287C>T and c.808-94C>T) located in the 3′-UTR and sixth intron of ALOX15 which showed a robust signal of association with IL-6 expression at both the RNA and protein level, and with different doses of LPS (Fig. 5, Supplementary Material, Figs S1 and S3). LD was relatively low and haplotypic analysis was not more informative than single SNP association for IL-6 expression. In our population sample, possession of one or more copies of the haplotype carried by the T alleles of rs11568131 and rs11078527 was associated with a 3-fold higher level of IL6 RNA expression following 2 ng/ml LPS induction for 6 h (Mann–Whitney test two-tailed P= 0.0002) and a 1.5- or 1.7-fold higher level of IL-6 protein expression (following 2 or 20 ng/ml LPS induction, respectively) (unpaired t-test two-tailed P= 0.0007 and P= 0.0008) (Supplementary Material, Fig. S3). The SNPs are present in non-conserved regions of DNA not associated with regulatory potential based on seven species alignments (27) or any predicted transcription factor binding site (28). We note that the associated SNPs occur twice as frequently among individuals of Caucasian geographic ancestry (MAF in CEU 0.183) compared with individuals of African or Asian ancestry (29).
Figure 5.
Association with IL-6 and SNP markers at ALOX15 and CAPNS1. Single marker allelic association results for IL-6 eQTL and pQTLs are shown plotted as −log10(P) versus genomic coordinate for (A) ALOX15 gene region and (B) CAPNS1 gene region. For each of the top associated SNPs in the two gene loci (plotted with blue squares), SNPs with MAF greater than 5% and r2 < 0.2 are shown as white squares, 0.2–0.5 (yellow) and >0.8 (red). In total, 45 SNPs over a 20 kb region spanning ALOX15 and 47 SNPs over a 50 kb region at CAPNS1 were genotyped.
We also observed a strong signal of association with IL-6 expression for a common SNP marker at CAPNS1, rs17882574 (c.721 + 391A>G) (Fig. 5, Supplementary Material, Figs S1 and S3). This locus also has low LD within individuals of Caucasian ancestry. Individuals with one or more copies of the minor G allele had a 2.8-fold increased IL6 RNA expression following 2 ng/ml LPS induction for 6 h (Mann–Whitney test two-tailed P= 0.03); and a 1.8- or 2.0-fold increased IL-6 protein expression (2 or 20 ng/ml LPS induction, respectively) (unpaired t-test P < 0.0001 for either phenotype) (Supplementary Material, Fig. S3). The SNP marker rs17882574 lies within the middle of the ninth intron of CAPNS1 in a non-conserved region of DNA not associated with regulatory potential, or predicted transcription factor binding. No data is publically available from HapMap or other resources comparing allele frequencies for this SNP between populations.
Finally, among all the IL-6 phenotypes studied, no strong signal of association was found with IL-6 production at the IL6 locus itself, including previously reported promoter SNPs IL6-174C/G (rs1800795), IL6-572C/G (rs1800796) and IL6-6331T/C (rs10499563), although the literature is controversial and a number of other studies find no evidence of association (5–7,30–33) (Supplementary Material, Fig. S4). We successfully genotyped a total of 40 SNPs across a 20 kb region spanning IL6 at chromosome 7p15.3, of which 22 were informative with an MAF greater than 5%.
Several genetic factors contribute to variation in IL-6 expression
Our data set from healthy volunteers leads to the hypothesis that genetic variation at LEP, ALOX15 and CAPNS1 is significantly associated with observed variation in induced IL-6 expression between individuals. We used a linear regression model to assess this relationship further. We found that the associated common SNP markers at each of these genes (rs4731427, rs11568131 and rs17882574, respectively) were each significant and independent predictors of IL-6 protein expression, together explaining approximately one-third of observed variation in induced IL-6 protein levels in this data set (Supplementary Material, Table S2). For IL6 RNA, rs17882574 was not a significant predictor and the final model only included rs4731427 and rs11568131, explaining 23% of observed variance in IL6 RNA expression. The LEP SNP rs4731427 was significant for all the IL-6 phenotypes and when analysed in a linear regression model for leptin RNA explained 17% of observed variance in leptin expression (Supplementary Material, Table S2).
Evidence of association with other cytokines involved in the inflammatory response
These data identify a number of candidate variants associated with differential expression of IL-6 at the transcript and protein level. For LEP, we noted evidence of cis-acting regulatory variants modulating leptin expression and in turn IL-6 levels, while for genetic variation at ALOX15 and CAPNS1 the basis of our observed association with IL-6 expression is unknown. The evidence of a pro-inflammatory role for the products of 12/15-lipoxygenase (12/15 LOX) encoded by ALOX15 in stimulating cytokines such as IL-6 (34–36), and of calpain (encoded in part by CAPNS1) in regulating NF-κB signalling through cleavage of IκB (37), would suggest that the associated polymorphisms in these genes might modulate the expression of cytokines other than IL-6. To investigate this, we assayed LPS induced expression of a panel of cytokines whose expression remained elevated at 6 h following LPS stimulation (the time RNA was harvested) and looked for evidence of association with the IL-6-associated SNPs rs11568131, rs17882574 and rs4731427. Transcript abundance was assayed for IL-1beta (IL-1B), IL-10, IL-12 subunit alpha (IL-12A) and beta (IL12-B) and IL-23. Tumour necrosis factor (TNF) was also assayed but as transcript levels virtually normalise by 6 h (data not shown), ELISAs of supernatant were performed to quantify TNF protein abundance. This analysis showed a significant association between the rs11568131 polymorphism in ALOX15, previously associated with increased IL-6 expression, and IL1B and TNF expression. Namely, individuals with a copy of the T allele had 1.4-fold higher levels of IL1B transcript compared with those without (Mann–Witney test two-tailed P= 0.009) and 1.5-fold higher levels of TNF protein (P= 0.005 unpaired t-test, two-tailed) (Supplementary Material, Fig. S5). No significant association was noted for the other cytokines assayed with this SNP or the two IL-6-associated SNPs at CAPNS1 and LEP, respectively.
DISCUSSION
IL-6 is a potent pro-inflammatory cytokine and tight regulation of its synthesis and release is critical to its role in modulating acute and chronic inflammation. For cytokines such as IL-6, the resolution of functionally important genetic variation requires analysis in a relevant cell type following induction of gene expression with a physiologically appropriate stimulus. Here we report a significant association between levels of IL-6 production on LPS stimulation of primary human PBMCs and possession of common SNP markers at the ALOX15, CAPNS1 and LEP gene loci which together explained up to one-third of the variance in induced IL-6 expression in our cohort. Given the sample size, it is possible that the observed effect sizes may be an overestimate, as previously noted in QTL mapping studies in which the proportion of phenotypic variance explained for correctly identified loci was dependent on the number of progeny examined (the so-called Beavis effect) (38). However, our data identifies complex cis- and trans-acting variants modulating IL-6 expression which are of high biological plausibility, and for ALOX15 provides evidence of association with the expression of other cytokines involved in the acute inflammatory response.
For LEP, we note that irrespective of genotype, the basal expression of leptin strongly correlates with subsequent LPS-induced IL-6 levels. Furthermore, we find that LEP SNPs showing trans association with IL-6 expression are themselves highly associated with increased basal expression of leptin, supporting a mechanism whereby relative expression of leptin can influence subsequent induced IL-6 responses. Genetic factors play a significant role in determining circulating levels of leptin, accounting for 34% of the observed variance in women and 45% in men among adult Finnish twins (39). Our dense SNP typing at LEP fine maps a previously reported SNP (rs7799039, ‘−2548G/A’) (23,24) association with leptin expression to a specific haplotype and has important potential implications for our understanding of the pathophysiology associated with IL-6, notably in inflammation-related disease. The observed differences at a protein level in IL-6 expression of 1.7- to 2-fold associating with LEP genotype may be clinically highly significant, emphasising the potential disease relevance of these data.
Although leptin was initially identified for its role in regulating appetite and metabolism, it also plays an important immunomodulatory role in both infectious and inflammatory states with increased leptin expression in response to LPS described (40,41). Leptin pre-treatment has been shown to augment LPS-induced IL-6 production, while obese/obese mice with deficient leptin receptor signalling have significantly reduced LPS-induced IL-6 production (20,21). Similarly, individuals with severe lipodystrophy exhibit reduced blood leptin levels and subnormal LPS responses with altered lymphocyte subsets; both of which can be corrected by exogenous leptin replacement (42). In microglia leptin leads to dose- and time-dependent increases in IL-6 production with enhanced recruitment of NF-κB, the coactivator p300 and histone acetylation at the IL6 promoter (22).
ALOX15 is implicated in multiple pathologies including atherosclerosis, diabetes and myeloproliferative disease (43). The gene is highly expressed in monocytes and macrophages and encodes 12/15 LOX, an enzyme regulating fatty acid and lipoprotein metabolism and leukotriene synthesis. Recent biological evidence supports a link with expression of inflammatory cytokines including IL-6. Activity of 12/15 LOX leads to the generation of unstable lipid products from arachidonate that stimulate expression of IL-6 and TNF in mouse adipocytes (35) and macrophages (34), while targeting 12/15 LOX with siRNAs results in reduced expression of inflammatory genes including IL-6 (44). Moreover, 12/15 LOX knock out mice show reduced expression of cytokines including TNF by peritoneal macrophages in response to bacterial infection, suggesting a key role for this enzyme in the acute inflammatory response (36). These mice also fail to show high fat diet induced IL-1B, IL-6 and TNF expression in adipocytes (45), implicating a role for 12/15 LOX in the chronic inflammatory metabolic syndrome.
Our data demonstrates that specific genetic variants in ALOX15 not only affect IL-6 expression, but also influence the expression of IL-1B and TNF, cytokines both intimately associated with the acute inflammatory response. The mechanism for this remains unclear but does not appear to be through a non-specific pro-inflammatory effect as evidenced by the absence of association with expression of the other cytokines assayed including IL-10 and IL-12. Specificity is also indicated by the absence of association of TNF and IL-1B expression with the LEP and CAPNS1 SNPs, which suggests that the effect of ALOX15 activity on the induction of these cytokines is not through an effect on IL-6. Likewise, the association of leptin and IL-6 adds another tier to the wealth of data implicating a tight relationship between these cytokines, in particular highlighting how basal leptin may modulate subsequent IL-6 levels.
Additional work is required to confirm and fine map our common SNP marker associations with IL-6, to define the causative functional variants and their role in the broader inflammatory cascade. Our data set provides proof of concept for a combined eQTL/pQTL approach utilizing a carefully phenotyped cohort which includes different dose effects on induction of gene expression, allowing for replication of association within the cohort and resolution of SNP markers and haplotypes consistently associated with differential gene expression.
MATERIALS AND METHODS
Ethics statement
This study was approved by the Oxfordshire Research Ethics Committee (REC reference 06/Q1605/55). All volunteers provided written informed consent for the collection of samples and subsequent analysis.
Study volunteers
Healthy volunteers from the Oxfordshire area were recruited and 50 ml of blood was venesected into EDTA-containing vacuutainer tubes. Sex, age and self-reported ethnicity were recorded and the samples anonymized. The median age of the 96 volunteers was 30 years (range 21–64 years); 59 were female; 82 were of European Caucasian origin while the remainder were of Asian descent (10 Indian and 4 Chinese). Self-reported ethnicity was confirmed with reference to SNP data using principal components analysis from the HapMap. A distance matrix was generated from the whole genome autosomal genotyping data using an agglomerative clustering algorithm. The subjects were observed to cluster in three groups corresponding to the reported ethnicities. Although a distinct structure was observed with the entire data set, it did not influence the results. Ethnicity was not significant in both regression models and in within-cluster permutations.
PBMC purification and ex vivo stimulation assays
Volunteers were venesected in the morning and PBMC purified using a Ficoll (Sigma-Aldrich) gradient within 2 h of sample collection. In order to minimize differential expression patterns due to circadian expression oscillations all blood samples were collected at the same time of day and cultured and processed in batches. Using a pasteur pipette, the PBMC layer was gently aspirated and washed twice in excess Hanks buffered saline solution buffer (without Ca2+ or Mg2+) and then with culture media. Cells were counted with a haemocytometer using trypan blue staining and resuspended at 2.5 × 106 cells per ml in RPMI 1640 growth medium (Sigma) supplemented with l-glutamine (2 mm), penicillin/streptomycin (100 U/ml penicillin, 0.1 mg/ml streptomycin) and 10% v/v heat-inactivated Gold standard fetal calf serum prior to being aliquoted into 14 ml polystyrene tubes (Becton Dickinson) tubes at 1 ml per tube. Three biological replicate assays per condition were set up for each volunteer. After incubation overnight at 37°C in 5% humidified CO2, cells were washed once with fresh media and then resuspended in fresh media alone (untreated) or fresh media with LPS at either 2 or 20 ng/ml (final concentration). LPS was purchased from Sigma (L-4391) lot 114K4133. Cells were incubated for a further 6 h prior to the separation of cells from supernatant by centrifugation. Supernatants for IL-6 ELISA were stored at −80°C prior to quantification and cell pellets were resuspended in RLT buffer (Qiagen) supplemented with beta-mercaptoethanol and stored at −80°C for subsequent RNA extraction.
ELISAs
ELISAs were performed in 96-well plates following the manufacturer's instructions using IL-6 and TNF Duoset kits (DY 206, DY210 R&D Systems), with protein concentrations estimated at 450 nm from a standard calibration curve included on each plate. All samples were analysed in duplicate.
RNA extraction, cDNA synthesis and quantitative PCR
Total RNA was prepared using the RNAeasy Mini kit (Qiagen) following the manufacturer's instructions with inclusion of a DNase I digestion step on the RNAeasy minicolumn. Following quantification, cDNA was prepared using SuperScript III (Invitrogen) using random primers and following the manufacturer's instructions which included digestion with RNase H (Invitrogen) following cDNA synthesis. Control RT reactions omitting the reverse transcriptase were included for each sample. Quantitative PCR was performed using SYBR Green Supermix (BioRad) on an iQ Cycler (BioRad). PCR efficiency was determined and melt curve analysis performed for gene specific primer sets (ACTB: fwd CCAGCCTTCCTTCCTGGGC rev TGTGTTGGCGTACAGGTCTTTGC; IL6: fwd GCTATGAACTCCTTCTCCACAAGC, rev CCCTACATCTTTGGAATCTTCTCC; IL10: fwd GGTGAAGAATGCCTTTAATAAGCTCC, rev TTTCGTATCTTCATTGTCATGTAGGC; IL12A: fwd CAGCAACATGCTCCAGAAGGC, rev TAAATACTACTAAGGCACAGGGCCATC; IL12B: fwd AGTGTCAAAAGCAGCAGAGGCTC, rev TTGGGTGGGTCAGGTTTGATG; IL1B: fwd ATGGCCCTAAACAGATGAAGTGC, rev GAAGGGAAAGAAGGTGCTCAGG; IL23: fwd CACTAGTGGGACACATGGATCTAAG, rev GATCCTTTGCAAGCAGAACTGAC). Relative gene transcript levels were determined by the ΔΔCt method. Leptin was quantified using the ABI ‘assay on demand’ Hs00174877m1 probe set (Applied Biosystems).
Genechip hybridization platform (Illumina BeadArray)
Genomic DNA was prepared for each of the healthy volunteers and quantified using the Nanodrop technology. Genotyping was performed using the humanCVD bead array chip (Illumina) following manufacturers recommendations as previously described (17).
LD and haplotype analysis
LD estimates and haplotypes were computed using the HaploView 3.3.2 program (46). For volunteers of Caucasian ancestry, genotyped SNPs of greater than 5% MAF were used to infer the underlying haplotypic structure with LD blocks predicted by the Confidence Intervals algorithm as previously described (47). An accelerated expectation-maximization algorithm was used to create highly accurate population frequency estimates of the phased haplotypes based on maximum likelihood as determined from unphased input (46).
Statistical analysis
PLINK was the primary software utilized for analysis of the data sets within this study (48). Statistics were verified using SPSS, R package and SNPTest, and concordant statistical results were obtained. Standard QC measures included exclusion criteria of maximum per-person missing (MIND > 0.1), maximum per-SNP missing (GENO > 0.1) and MAF < 0.01. After frequency and genotyping pruning, and removing individuals with low genotyping success rates, the total genotyping rate in the remaining individuals was more than 98.8% for the Illumina humanCVD beadchip. PLINK was used to perform quantitative trait analysis which generates a P-value using the Wald test. For each SNP, PLINK generates a phenotypic mean for the three genotypic states and compares these means using the Wald test statistic to generate a P-value. The Wald test is useful especially in this instance, since it does not require that the data fit a normal distribution. Covariates age, ethnicity and sex were included as additional terms in PLINK analysis to further interrogate observed associations. Non-parametric statistics and log transformation were applied where data were not normally distributed, otherwise unpaired t-tests were used for analysis of expression data for specific SNPs, data passing tests of normality. Permutation analysis was performed using a label-swapping procedure which swaps phenotypes while retaining the genotype linkage structure. An adaptive algorithm was used which speeds computations by dropping SNPs less likely to be significant in each iteration. Analysis was performed with one million permutations using a within-cluster algorithm which swaps labels within each population cluster. The analysis was repeated without any restrictions. The empirical P-value is robust with respect to normality of phenotype and multiple testing issues. Haplotypic association analysis was also performed using the haplotypes defined as described earlier. A linear regression model was used to estimate the contribution of the most strongly associated SNPs to IL-6 expression. Potential confounding factors (age, sex and ethnicity) were then added to the core model. Factors which did not contribute significantly to the fit of the model on likelihood ratio testing (P< 0.05) were excluded. Linear regression analysis was carried out using STATA v10 software.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Wellcome Trust (074318 to J.C.K. and 088891 to B.P.F.) and a Principal Research Fellowship to A.V.S.H. Funding to pay the open access charge was provided by the Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to Marian Knight for help with statistical analysis and volunteer recruitment. We would like to thank all members of the Knight Laboratory for helpful discussion and suggestions relating to this study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Naugler W.E., Karin M. The wolf in sheep's clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends Mol. Med. 2008;14:109–119. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., Kuchroo V.K. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 3.de Craen A.J., Posthuma D., Remarque E.J., van den Biggelaar A.H., Westendorp R.G., Boomsma D.I. Heritability estimates of innate immunity: an extended twin study. Genes Immun. 2005;6:167–170. doi: 10.1038/sj.gene.6364162. [DOI] [PubMed] [Google Scholar]
- 4.Worns M.A., Victor A., Galle P.R., Hohler T. Genetic and environmental contributions to plasma C-reactive protein and interleukin-6 levels—a study in twins. Genes Immun. 2006;7:600–605. doi: 10.1038/sj.gene.6364330. [DOI] [PubMed] [Google Scholar]
- 5.Muller-Steinhardt M., Ebel B., Hartel C. The impact of interleukin-6 promoter -597/-572/-174 genotype on interleukin-6 production after lipopolysaccharide stimulation. Clin. Exp. Immunol. 2007;147:339–345. doi: 10.1111/j.1365-2249.2006.03273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith A.J., D'Aiuto F., Palmen J., Cooper J.A., Samuel J., Thompson S., Sanders J., Donos N., Nibali L., Brull D., et al. Association of serum interleukin-6 concentration with a functional IL6 -6331T>C polymorphism. Clin. Chem. 2008;54:841–850. doi: 10.1373/clinchem.2007.098608. [DOI] [PubMed] [Google Scholar]
- 7.Tischendorf J.J., Yagmur E., Scholten D., Vidacek D., Koch A., Winograd R., Gressner A.M., Trautwein C., Wasmuth H.E., Lammert F. The interleukin-6 (IL6)-174 G/C promoter genotype is associated with the presence of septic shock and the ex vivo secretion of IL6. Int. J. Immunogenet. 2007;34:413–418. doi: 10.1111/j.1744-313X.2007.00712.x. [DOI] [PubMed] [Google Scholar]
- 8.Hegedus C.M., Skibola C.F., Bracci P., Holly E.A., Smith M.T. Screening the human serum proteome for genotype–phenotype associations: an analysis of the IL6 -174G>C polymorphism. Proteomics. 2007;7:548–557. doi: 10.1002/pmic.200600366. [DOI] [PubMed] [Google Scholar]
- 9.Walston J., Arking D.E., Fallin D., Li T., Beamer B., Xue Q., Ferrucci L., Fried L.P., Chakravarti A. IL-6 gene variation is not associated with increased serum levels of IL-6, muscle, weakness, or frailty in older women. Exp. Gerontol. 2005;40:344–352. doi: 10.1016/j.exger.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melzer D., Perry J.R., Hernandez D., Corsi A.M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G., et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 14.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 16.Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 17.Keating B.J., Tischfield S., Murray S.S., Bhangale T., Price T.S., Glessner J.T., Galver L., Barrett J.C., Grant S.F., Farlow D.N., et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dugue B., Leppanen E. Short-term variability in the concentration of serum interleukin-6 and its soluble receptor in subjectively healthy persons. Clin. Chem. Lab. Med. 1998;36:323–325. doi: 10.1515/CCLM.1998.054. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Margalet V., Martin-Romero C., Santos-Alvarez J., Goberna R., Najib S., Gonzalez-Yanes C. Role of leptin as an immunomodulator of blood mononuclear cells: mechanisms of action. Clin. Exp. Immunol. 2003;133:11–19. doi: 10.1046/j.1365-2249.2003.02190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zarkesh-Esfahani H., Pockley G., Metcalfe R.A., Bidlingmaier M., Wu Z., Ajami A., Weetman A.P., Strasburger C.J., Ross R.J. High-dose leptin activates human leukocytes via receptor expression on monocytes. J. Immunol. 2001;167:4593–4599. doi: 10.4049/jimmunol.167.8.4593. [DOI] [PubMed] [Google Scholar]
- 21.Loffreda S., Yang S.Q., Lin H.Z., Karp C.L., Brengman M.L., Wang D.J., Klein A.S., Bulkley G.B., Bao C., Noble P.W., et al. Leptin regulates proinflammatory immune responses. FASEB J. 1998;12:57–65. [PubMed] [Google Scholar]
- 22.Tang C.H., Lu D.Y., Yang R.S., Tsai H.Y., Kao M.C., Fu W.M., Chen Y.F. Leptin-induced IL-6 production is mediated by leptin receptor, insulin receptor substrate-1, phosphatidylinositol 3-kinase, Akt, NF-kappaB, and p300 pathway in microglia. J. Immunol. 2007;179:1292–1302. doi: 10.4049/jimmunol.179.2.1292. [DOI] [PubMed] [Google Scholar]
- 23.Mammes O., Betoulle D., Aubert R., Giraud V., Tuzet S., Petiet A., Colas-Linhart N., Fumeron F. Novel polymorphisms in the 5' region of the LEP gene: association with leptin levels and response to low-calorie diet in human obesity. Diabetes. 1998;47:487–489. doi: 10.2337/diabetes.47.3.487. [DOI] [PubMed] [Google Scholar]
- 24.Hoffstedt J., Eriksson P., Mottagui-Tabar S., Arner P. A polymorphism in the leptin promoter region (-2548 G/A) influences gene expression and adipose tissue secretion of leptin. Horm. Metab. Res. 2002;34:355–359. doi: 10.1055/s-2002-33466. [DOI] [PubMed] [Google Scholar]
- 25.Ben Ali S., Kallel A., Ftouhi B., Sediri Y., Feki M., Slimane H., Jemaa R., Kaabachi N. Association of G-2548A LEP polymorphism with plasma leptin levels in Tunisian obese patients. Clin. Biochem. 2009;42:584–588. doi: 10.1016/j.clinbiochem.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Le Stunff C., Le Bihan C., Schork N.J., Bougneres P. A common promoter variant of the leptin gene is associated with changes in the relationship between serum leptin and fat mass in obese girls. Diabetes. 2000;49:2196–2200. doi: 10.2337/diabetes.49.12.2196. [DOI] [PubMed] [Google Scholar]
- 27.Taylor J., Tyekucheva S., King D.C., Hardison R.C., Miller W., Chiaromonte F. ESPERR: learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wingender E., Dietze P., Karas H., Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boiardi L., Casali B., Farnetti E., Pipitone N., Nicoli D., Cantini F., Macchioni P., Bajocchi G., Catanoso M.G., Pulsatelli L., et al. Relationship between interleukin 6 promoter polymorphism at position -174, IL-6 serum levels, and the risk of relapse/recurrence in polymyalgia rheumatica. J. Rheumatol. 2006;33:703–708. [PubMed] [Google Scholar]
- 31.Brull D.J., Montgomery H.E., Sanders J., Dhamrait S., Luong L., Rumley A., Lowe G.D., Humphries S.E. Interleukin-6 gene -174g>c and -572g>c promoter polymorphisms are strong predictors of plasma interleukin-6 levels after coronary artery bypass surgery. Arterioscler. Thromb. Vasc. Biol. 2001;21:1458–1463. doi: 10.1161/hq0901.094280. [DOI] [PubMed] [Google Scholar]
- 32.Malarstig A., Wallentin L., Siegbahn A. Genetic variation in the interleukin-6 gene in relation to risk and outcomes in acute coronary syndrome. Thromb. Res. 2007;119:467–473. doi: 10.1016/j.thromres.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Ravaglia G., Forti P., Maioli F., Chiappelli M., Dolzani P., Martelli M., Bianchin M., Mariani E., Bolondi L., Licastro F. Associations of the -174 G/C interleukin-6 gene promoter polymorphism with serum interleukin 6 and mortality in the elderly. Biogerontology. 2005;6:415–423. doi: 10.1007/s10522-005-4908-x. [DOI] [PubMed] [Google Scholar]
- 34.Wen Y., Gu J., Chakrabarti S.K., Aylor K., Marshall J., Takahashi Y., Yoshimoto T., Nadler J.L. The role of 12/15-lipoxygenase in the expression of interleukin-6 and tumor necrosis factor-alpha in macrophages. Endocrinology. 2007;148:1313–1322. doi: 10.1210/en.2006-0665. [DOI] [PubMed] [Google Scholar]
- 35.Chakrabarti S.K., Cole B.K., Wen Y., Keller S.R., Nadler J.L. 12/15-Lipoxygenase products induce inflammation and impair insulin signaling in 3T3-L1 adipocytes. Obesity. 2009;17:1657–1663. doi: 10.1038/oby.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dioszeghy V., Rosas M., Maskrey B.H., Colmont C., Topley N., Chaitidis P., Kuhn H., Jones S.A., Taylor P.R., O'Donnell V.B. 12/15-Lipoxygenase regulates the inflammatory response to bacterial products in vivo. J. Immunol. 2008;181:6514–6524. doi: 10.4049/jimmunol.181.9.6514. [DOI] [PubMed] [Google Scholar]
- 37.Schaecher K., Goust J.M., Banik N.L. The effects of calpain inhibition on IkB alpha degradation after activation of PBMCs: identification of the calpain cleavage sites. Neurochem. Res. 2004;29:1443–1451. doi: 10.1023/b:nere.0000026410.56000.dd. [DOI] [PubMed] [Google Scholar]
- 38.Xu S. Theoretical basis of the Beavis effect. Genetics. 2003;165:2259–2268. doi: 10.1093/genetics/165.4.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaprio J., Eriksson J., Lehtovirta M., Koskenvuo M., Tuomilehto J. Heritability of leptin levels and the shared genetic effects on body mass index and leptin in adult Finnish twins. Int. J. Obes. Relat. Metab. Disord. 2001;25:132–137. doi: 10.1038/sj.ijo.0801526. [DOI] [PubMed] [Google Scholar]
- 40.Grunfeld C., Zhao C., Fuller J., Pollack A., Moser A., Friedman J., Feingold K.R. Endotoxin and cytokines induce expression of leptin, the ob gene product, in hamsters. J. Clin. Invest. 1996;97:2152–2157. doi: 10.1172/JCI118653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lord G.M., Matarese G., Howard J.K., Baker R.J., Bloom S.R., Lechler R.I. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature. 1998;394:897–901. doi: 10.1038/29795. [DOI] [PubMed] [Google Scholar]
- 42.Oral E.A., Javor E.D., Ding L., Uzel G., Cochran E.K., Young J.R., DePaoli A.M., Holland S.M., Gorden P. Leptin replacement therapy modulates circulating lymphocyte subsets and cytokine responsiveness in severe lipodystrophy. Js. Clin. Endocrinol. Metab. 2006;91:621–628. doi: 10.1210/jc.2005-1220. [DOI] [PubMed] [Google Scholar]
- 43.Kuhn H., O'Donnell V.B. Inflammation and immune regulation by 12/15-lipoxygenases. Prog. Lipid Res. 2006;45:334–356. doi: 10.1016/j.plipres.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Li S.L., Dwarakanath R.S., Cai Q., Lanting L., Natarajan R. Effects of silencing leukocyte-type 12/15-lipoxygenase using short interfering RNAs. J. Lipid Res. 2005;46:220–229. doi: 10.1194/jlr.M400328-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Sears D.D., Miles P.D., Chapman J., Ofrecio J.M., Almazan F., Thapar D., Miller Y.I. 12/15-lipoxygenase is required for the early onset of high fat diet-induced adipose tissue inflammation and insulin resistance in mice. PLoS ONE. 2009;4:e7250. doi: 10.1371/journal.pone.0007250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 47.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 48.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.