Abstract
Independent transgene insertions differ in expression based on their location in the genome; these position effects are of interest because they reflect the influence of genome organization on gene regulation. Position effects also represent potentially insurmountable obstacles to the rigorous functional comparison of homologous genes from different species because (i) quantitative variation in expression of each gene across genomic positions (generalized position effects, or GPEs) may overwhelm differences between the genes of interest, or (ii) divergent genes may be differentially sensitive to position effects, reflecting unique interactions between each gene and its genomic milieu (lineage-specific position effects, or LSPEs). We have investigated both types of position-effect variation by applying our method of transgene coplacement, which allows comparisons of transgenes in the same position in the genome of Drosophila melanogaster. Here we report an experimental test for LSPE in Drosophila. The alcohol dehydrogenase (Adh) genes of D. melanogaster and Drosophila affinidisjuncta differ in both tissue distribution and amounts of ADH activity. Despite this striking regulatory divergence, we found a very high correlation in overall ADH activity between the genes of the two species when placed in the same genomic position as assayed in otherwise Adh-null adults and larvae. These results argue against the influence of LSPE for these sequences, although the effects of GPE are significant. Our new findings validate the coplacement approach and show that it greatly magnifies the power to detect differences in expression between transgenes. Transgene coplacement thus dramatically extends the range of functional and evolutionary questions that can be addressed by transgenic technology.
Any gene may be expressed at different levels depending on its position in the genome. Such generalized position effect (GPE) on gene expression is well known (1, 2). But let us stand the problem on its head and ask: does the level of expression of different genes, present at the same location in the genome, vary because of unique interactions between each gene and the genomic milieu? We will designate this type of position effect as a lineage-specific position effect (LSPE) to distinguish it from conventional GPE. The term “lineage” is used here because, if it is significant, LSPE could represent a serious impediment to the study of the evolution of gene regulation in different lineages. Any functional comparison of the regulation of homologous but divergent genes present in two species must entail examining them in transgenic organisms in the same genetic background; otherwise, gene effects and background effects would be confounded. Because of GPE, such a comparison also requires an experimental method by which the genes can be inserted at the same position in the genome. But even this may not be sufficient, because the validity and the power of the comparison could be undermined by LSPE.
We recently described a method, transgene coplacement, by which two genes could efficiently be introduced into exactly the same position in the genome of Drosophila melanogaster (3). The feasibility of the method was demonstrated, and we pointed out that this approach provided the opportunity to study LSPE experimentally. How would LSPE be detected? By using this approach, it should be possible to examine a pair of homologous but divergent genes that have been “coplaced” in a variety of genomic positions. Conventional GPE would of course be expected and would be revealed as a large variation in the activity of either gene from one position to the next. LSPE would be revealed differently. It would be reflected in the correlation in gene activity between the pairs of coplaced genes across all of the genomic positions sampled. A low correlation would indicate high LSPE, implying that high or low expression of one gene at a particular location in the genome does not necessarily predict the level of expression of its evolved homolog at the same location. Conversely, a high correlation would argue against LSPE, even in the presence of GPE.
In this paper we present the results of the first of such experiments, comparing the alcohol dehydrogenase (Adh) genes of D. melanogaster and its distant Hawaiian relative Drosophila affinidisjuncta. The genes share a similar structure, with distal and proximal promoters producing distinct transcripts (4). However, the D. affinidisjuncta gene is expressed in strikingly different amounts in different tissues and at different developmental stages from the D. melanogaster gene; these lineage-specific features are largely conserved in D. melanogaster that are transgenic for the D. affinidisjuncta Adh (5). We chose these Adh genes for our study because, in addition to permitting a sensitive quantitative assay for gene expression, they also represent an extreme degree of regulatory divergence between homologous genes. The choice of such divergent genes is deliberate because more divergent genes would have had greater opportunities to evolve allele-specific regulatory differences. This experiment was specifically designed to determine whether LSPE prevents rigorous comparison of homologous genes from different lineages. The evidence we present shows that LSPE is not a serious problem in comparing the activity of Adh genes in these lineages. This finding encourages the hope that the effects of LSPE may often be small enough that rigorous, high-resolution functional comparisons of homologous genes from divergent lineages will be possible.
MATERIALS AND METHODS
Plasmid Construction.
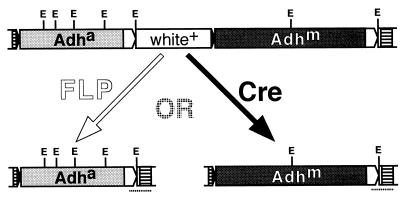
Construction of the Adh transgene coplacement vector, pP[^a>w+^m>], was reported previously (3). Following precedent (3, 6), directly repeated FRT sites are denoted “>>” and directly repeated loxP sites are denoted “^ ^.” As shown in Fig. 1, this vector carries the Wa-s Slow allele of the D. melanogaster Adh locus (“m ” in the constructs, ref. 7) and the D. affinidisjuncta Adh gene (“a ” in the constructs, ref. 5), configured such that the action of Cre excises the D. affinidisjuncta gene, leaving P[^m>], whereas the action of FLP excises the D. melanogaster gene, leaving P[^a>]. The two derivatives are identical except for the cloned Adh genes between the recombinase target sites.
Figure 1.
The transgene coplacement construct, P[^a>w+^m>] (Upper) and its FLP- and Cre-mediated derivatives, P[^a>] and P[^m>] (Lower). Excision by either recombinase is accompanied by loss of white+ phenotype. All genes are transcribed from left to right. loxP sites are shown as solid arrowheads, FRT sites are shown as open arrowheads, and P-element sequences are shown as striped rectangles. The 5′ end of the P element is to the right. EcoRI sites (E) are shown, as is the extent of the probe (dotted lines below the excision derivatives) used to verify identity of the excision derivatives.
Drosophila Transformation and Genetic Manipulations.
Flies were maintained on standard cornmeal-molasses medium at 25°C. Descriptions of mutants and special chromosomes are found in Lindsley and Zimm (8). P element-mediated transformation of Drosophila was carried out using standard means (9, 10) with helper plasmid pπ25.7wc (11) and host strain Df(1)w, y w 67c23. Among 17 independent transformants, 7 homozygous-viable integrations of P[^a>w+^m>] into the third chromosome were isolated and served as the experimental strains in this study. For each insertion of P[^a>w+^m>], females of genotype y w; P[^a>w+^m>] were crossed to males of genotype y w; MKRS, P[ry+, hsFLP]/TM6B, P[w+, cre]. The crosses were brooded once, and one brood was heat-shocked as first-instar larvae for 1 hr in a 38°C water bath to induce FLP expression in those progeny inheriting the MKRS, P[ry+, hsFLP] chromosome; P[w+, cre] does not require heat shock for efficient expression (3). Thus, an “FLP” treatment and a “Cre” treatment were initiated to create D. affinidisjuncta and D. melanogaster derivatives, respectively. Derivative-bearing chromosomes were identified by loss of w+ and extracted into an Adhfn6 cn1 genetic background, such that the X, Y, second, and third chromosomes were common to all experimental strains, with the only differences being the locus of integration of the original transgene coplacement construct and whether the D. affinidisjuncta or D. melanogaster gene remained in that locus.
Cytological Location and Molecular Analysis of Derivatives.
Each of the seven integration sites was mapped cytologically by fluorescent in situ hybridization to polytene chromosome preparations (3). To verify that the experimental strains carried the expected species’ Adh gene and that pairs of strains derived from the same original P[^a>w+^m>] were flanked by the same genomic sequences, genomic DNA was isolated from each of the P[^a>] and P[^m>] strains, digested with EcoRI, separated on a 0.8% agarose gel, and transferred to a nylon filter. The filter was probed with a subcloned sequence corresponding to the 5′ P-element sequence and part of the neighboring FRT in P[^a>w+^m>] (Fig. 1); all expected species-specific and integration-specific bands were present.
ADH Assays.
ADH specific-activity assays were performed using the spectrophotometric method of Sofer and Ursprung (12) on preparations of five third-instar larvae or five adults collected 6–8 days post-eclosion, except as noted. Live larvae or adults were homogenized in 300 μl of 0.05 M sodium phosphate (pH 7.5). An additional 200 μl of this buffer was added, the sample was centrifuged for 1 min at maximum speed in a microcentrifuge, and 400 μl of supernatant was transferred to a Millipore Ultrafree-MC 0.45-μm low-binding filter unit, which was centrifuged for 1 min at 10,000 × g. This extra filtration step considerably reduced within-sample variability in spectrophotometric readings. Activity was measured as the change in 340-nm absorbance at 30°C in 2-ml reactions using 100 μl of homogenate. The reaction buffer was as described (12), but with 2-propanol as the substrate. ADH units are expressed as nanomoles of NAD+ reduced per minute. Specific activity is units per microgram of total protein, which was determined by using the Bradford procedure (ref. 13; Bio-Rad). Each strain was assayed in at least two independent preparations per developmental stage and sex. Each preparation was assayed twice for activity and twice for total protein. ADH histochemical analysis was performed on individuals aged as for the activity measurements using standard means (14). For activity measurements on 5-, 7-, and 9-day-old adults, cohorts of males from each strain were collected over an 8-hr period and allowed to age 5, 7, or 9 days longer, at which time they were transferred to 1.5-ml microcentrifuge tubes, immediately frozen in a slurry of dry ice in ethanol, and transferred to a −80°C freezer until all of the flies could be assayed together. Each strain was assayed in two independent preparations per age group.
RESULTS
Transgene Coplacement Efficiently Produces Pairs of Strains in Which the Adh Genes of D. affinidisjuncta and D. melanogaster Are Present in the Same Location in the Genome.
Of seven third-chromosome homozygous-viable insertions of P[^a>w+^m>], all were successfully altered by FLP and Cre to produce seven pairs of P[^a>] and P[^m>] lines. The excision reactions of FLP and Cre approach 100% efficiency, making routine the genetic manipulations outlined in Materials and Methods. Southern hybridization (not shown) confirmed that each derivative contained the expected species’ gene and that pairs of derivatives from the same original P[^a>w+^m>] integration shared the same flanking genomic sequences. The strains are referred to by the cytological location of the transgene insertion, as determined by in situ hybridization to polytene chromosomes.
ADH Specific Activity for D. affinidisjuncta and D. melanogaster Genes Is Highly Correlated with Genomic Position.
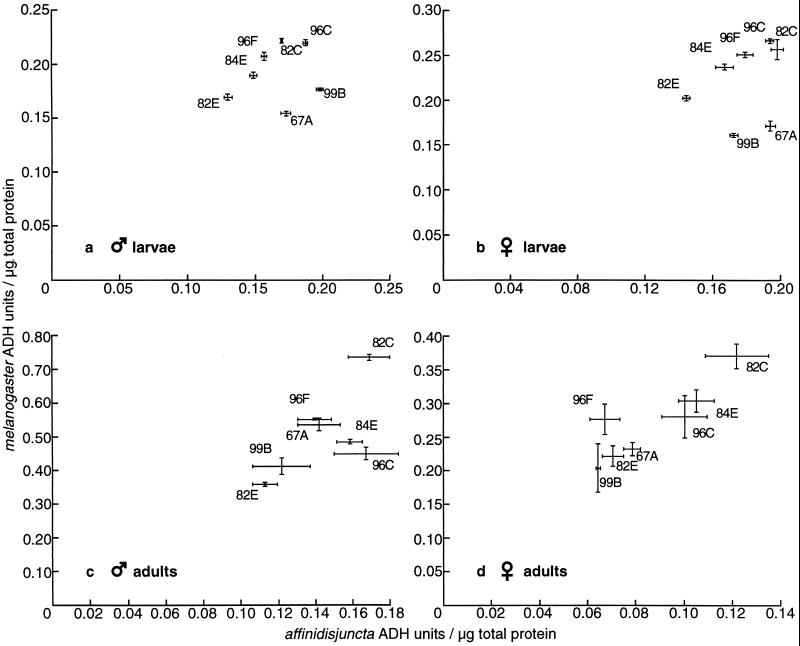
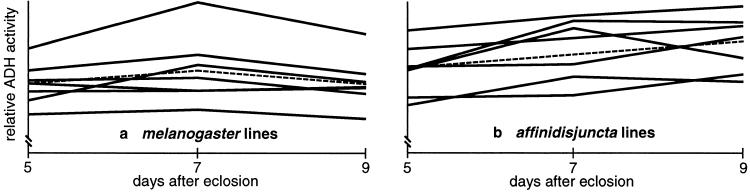
To determine the degree to which genomic position similarly affects the expression of the divergent Adh genes of D. affinidisjuncta and D. melanogaster, we assayed ADH specific activity in male and female larvae and adults of the seven P[^a>] and seven P[^m>] strains described above. Fig. 2 a and b show the results for male and female larvae. Omitting the two striking outliers (99B and 67A) yields correlation coefficients of 0.93 for males and 0.91 for females. As described below, these outliers are justifiably omitted from the analysis because they represent misexpression of the transgenes owing to a tissue-specific silencing effect. Fig. 2 c and d show the results for male and female adults. Standard errors for adult specific activities are greater than for larvae, presumably because of individual variability in enzyme level during days 6–8 of adult life. Indeed, the situation is even more complicated because the time course of ADH activity, as assayed in 5-, 7-, and 9-day-old adults, manifests a significantly different profile for the D. affinidisjuncta and the D. melanogaster genes. As shown in Fig. 3, ADH specific activity reaches a peak at around day 7 for flies transformed with the D. melanogaster gene, whereas ADH activity tends still to be rising at day 9 for flies transformed with the D. affinidisjuncta gene. ANOVA with linear regression (Table 1) confirms the observed pattern. As expected, both the P[^a>] and P[^m>] lines show a highly significant position effect (within-strain variance; P < 0.001 and P < 10−8, respectively). In addition, the P[^a>] lines show a highly significant linear term (P = 0.002) and no significant deviations from linear (P = 0.928). On the other hand, the P[^m>] lines show no significant linear trend (P = 0.778) but show significant deviations from this zero slope (P = 0.040), consistent with the peaked profile. Indeed, when the linear regression model is changed for the P[^m>] lines to incorporate a single linear slope term that is of opposite sign for the transition from day 5 to day 7 and for the transition from day 7 to day 9, this slope term is significant (P = 0.014), whereas deviations from this peaked linear model are not significant (P = 0.212). In neither the P[^m>] nor the P[^a>] lines was there found a significant day-by-strain interaction (two-way ANOVA, not shown), although the power to detect such an interaction may be low. It is tempting to speculate that the D. affinidisjuncta gene retains expression-timing properties of the species from which it derives, which has a 12-week generation time and reaches sexual maturity at around 25 days post-eclosion. More detailed analysis of the time course of expression in particular tissues, both in these transgenic animals and in the native species, would be necessary to address directly the fascinating issue of regulatory divergence in the timing of gene expression. In any event, despite the dramatic regulatory divergence in adult expression between the D. affinidisjuncta and D. melanogaster genes and the higher SEs of activity measurements, the adult correlations are relatively high: 0.66 for males and 0.88 for females.
Figure 2.
Specific activity measurements for preparations of male (a) and female (b) third-instar larvae and male (c) and female (d) adults (6–8 days post-eclosion). In each graph, ADH specific activity of P[^m>] derivatives is plotted against ADH specific activity of P[^a>] derivatives for each genomic location (identified by cytological position in polytene chromosomes). SE bars are shown.
Figure 3.
Time course of ADH specific activity in male adults on days 5, 7, and 9 post-eclosion. P[^m>] strains are shown in (a) and P[^a>] strains are shown in (b). The vertical axis is scaled differently in the two graphs to allow easier comparison of the shapes of the activity profiles. Regression lines are shown dashed. Statistical analyses by ANOVA with linear regression are shown in Table 1.
Table 1.
ANOVA with linear regression analysis of time course of ADH expression
| Source of variation | df | P[^a>] strains P value | P[^m>] strains P value |
|---|---|---|---|
| Strains | 6 | <0.001 | <10−8 |
| Linear slope | 1 | ||
| monotonic | 0.002 | 0.778 | |
| peaked at day 7 | na | 0.014 | |
| Deviations from linear | 13 | ||
| monotonic | 0.928 | 0.040 | |
| peaked at day 7 | na | 0.212 | |
| Error | 21 | ||
| Total | 41 |
ANOVA with linear regression was performed on the P[^a>] and P[^m>] strains separately. In each analysis, the total sum of squares was partitioned into a term for variation among strains, a single linear slope term, a term for deviations from linear regression, and an error term associated with replicates for each measurement. The linear slope term was either “monotonic” (standard linear regression) or “peaked” at day 7 (same slope magnitude with opposite sign for the transitions from day 5 to day 7 and day 7 to day 9).
Transgene Coplacement Reveals a Tissue-Specific Silencing Effect in Larvae.
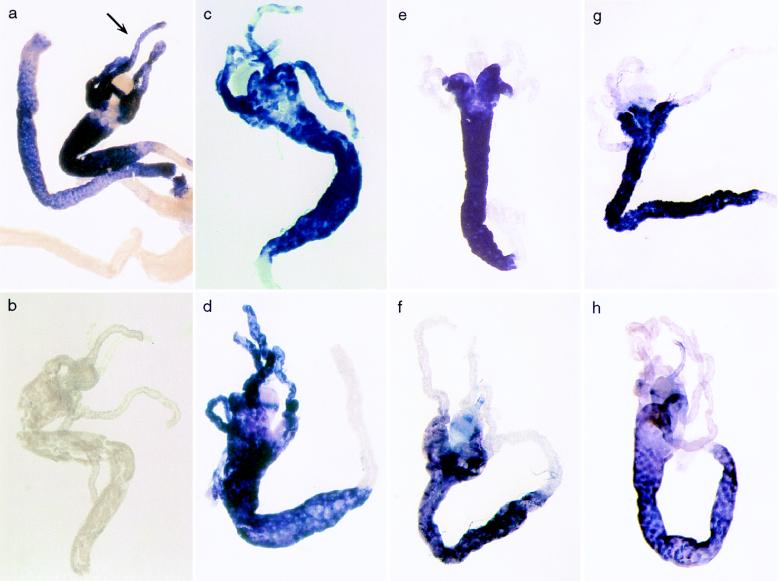
In both male and female larvae, the 99B and 67A lines do not obey the strong correlation evident among the other genomic positions (Fig. 2 a and b). Histochemical analysis (Fig. 4) reveals that expression of ADH in the gastric ceca is abolished in both the P[^a>] and P[^m>] derivatives of the 99B and 67A lines, whereas other tissues have normal expression. Because the gastric ceca are thought to secrete digestive enzymes into the alimentary system, it is not possible to assess from static histochemical data the degree to which this silencing effect reduces total ADH output for P[^a>] and P[^m>] larvae. However, the whole-larva specific activity data suggest that the output of the D. melanogaster gene suffers a greater reduction. Note that the silencing effect itself is not gene-specific; instead, its effect on overall specific activity depends on which species’ gene is the source of ADH.
Figure 4.
Histochemical staining of third-instar larvae. The strains represented are Canton-S (wildtype AdhS allele; a), Adhfn6 (null Adh allele; b), P[^m>] 82E (c), P[^a>] 82E (d), P[^m>] 99B (e), P[^a>] 99B (f), P[^m>] 67A (g), and P[^a>] 67A (h). In a, the gastric ceca are marked by an arrow. Strain 82E is consistently among the lowest-expressing strains for both P[^m>] and P[^a>] derivatives in male and female larvae and adults. Nevertheless, expression of both species’ genes is readily apparent in the gastric ceca, as it is in strains 82C, 84E, 96C, and 96F (not shown). In contrast, expression in the gastric ceca is abolished in the 99B and 67A lines.
The High Correlation with Position Translates into much Greater Power to Detect Quantitative Expression Differences Between Transgenes.
When transgene classes are not matched by genomic position, a standard t test (or one-way ANOVA) is used to test the null hypothesis that there is no difference in expression between the transgenes (15, 16). In this case, the power of the two-tailed test is equal to the value of the cumulative t distribution for (n − 1) degrees of freedom, evaluated from −∞ to
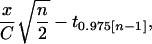 |
where n is the sample size for each transgene class, x is the true difference between transgene classes (expressed as a proportion of the mean of one class), C is the coefficient of variation across genomic positions, and t0.975[n−1] is the critical value of the t distribution with (n − 1) degrees of freedom for a type I error rate of α = 0.05. More power is possible when transgene classes can be matched by genomic position, as with transgene coplacement. In this case, the same null hypothesis is tested by a paired-comparisons t test (or two-way ANOVA), which will have greater power than the standard t test when there is a correlation between members of a pair. Specifically, the power is equal to the value of the cumulative t distribution for (n − 1) degrees of freedom, evaluated from −∞ to
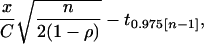 |
where ρ is the correlation between pairs.
The experimenter generally has no ability to change x, ρ, or C, so it is instructive to compute sample sizes, n, necessary to detect a given difference, x, with a given power (1 − β) for various values of ρ and C as shown in Table 2. In the table, the desired power (acceptance rate of true hypotheses) is set at 1 − β = 0.8, the type I error rate is set at α = 0.05, and the true mean difference between transgene classes is either 10% or 20%. Representative values of C are those computed by Laurie-Ahlberg and Stam (16) for transgenes expressing AdhS, AdhF, or xanthine dehydrogenase (Xdh) as well as that computed by Parsch et al. (17) for an AdhF transgene flanked by binding sites for the su(Hw) protein (18). It is clear from Table 2 that high values of ρ, as measured in the present study, dramatically reduce the sample size needed to detect differences between transgenes. Indeed, in those situations when the unpaired experiment would require impracticably large sample sizes—as with 291 strains of each transgene class for detecting a 10% mean difference when the coefficient of variation across genomic positions is equal to that of AdhS—paired comparisons provide a disproportionally great reduction in sample size, returning feasibility to the experiment. Note that although AdhF transgenes flanked by su(Hw)-binding sites still experience a highly significant position effect (17) and insulation does not reduce sample sizes as much as does transgene coplacement when there is even a moderately high correlation between transgenes, combining the two appoaches promises to increase power synergistically (Table 2).
Table 2.
Sample sizes needed to detect differences between transgene classes
| t test | AdhS | Xdh | AdhF | {{AdhF}} |
|---|---|---|---|---|
| Difference in means = 10%, α = 0.05, 1 − β ≥ 0.8, n ≥ | ||||
| Unpaired | ||||
| ρ = 0 | 291 | 133 | 35 | 14 |
| Paired | ||||
| ρ = 0.7 | 89 | 42 | 12 | 6 |
| ρ = 0.8 | 60 | 29 | 9 | 5 |
| ρ = 0.9 | 31 | 16 | 6 | 4 |
| Difference in means = 20%, α = 0.05, 1 − β ≥ 0.8, n ≥ | ||||
| Unpaired | ||||
| ρ = 0 | 75 | 35 | 11 | 6 |
| Paired | ||||
| ρ = 0.7 | 24 | 12 | 5 | 4 |
| ρ = 0.8 | 13 | 9 | 4 | 3 |
| ρ = 0.9 | 10 | 6 | 4 | 3 |
Sample sizes (n per class) required to detect (by t test) mean expression differences between transgene classes are calculated based on given true difference of 10% or 20%; type I error rate (α) of 0.05; power (1 − β) of at least 0.8; correlation with position (ρ) of 0, 0.7, 0.8, or 0.9; and coefficient of variation (SD/mean) among genomic positions. A range of typical coefficients of variation is represented by AdhS (0.429), Xdh (0.288), AdhF (0.145), and AdhF flanked by binding sites for the su(Hw) protein (0.086), denoted as {{AdhF}}.
DISCUSSION
As expected, GPE exerts a profound influence over the expression of transgenes of each of the highly divergent Adh homologues. In spite of the large GPE, the correlation in specific activity between D. melanogaster and D. affinidisjuncta Adh genes that occupy the same genomic position is very high in both larvae (0.93 for males and 0.91 for females) and adults (0.66 for males and 0.88 for females). This result is the key finding because it argues against LSPEs. Furthermore, the lack of significant LSPE validates the transgenic approach for comparing divergent homologues: any system that matches transgenes precisely for genomic position will provide a powerful and sensitive approach to detecting differences between the transgenes. At present, transgene coplacement may be the only practical means of detecting very subtle differences because the method is more efficient than alternative systems such as template-directed gap repair of P-element excisions (19) and FLP-mediated site-specific transposition (20).
In the course of this study we have also demonstrated a tissue-specific, but not a lineage-specific, silencing effect in the larval gastric ceca, which is associated with two genomic positions. It is intriguing that two independent positions yield the same misexpression phenotype. Further study could elucidate the nature of the silencing effect in each case, to assess whether the same mechanism is at work. We also have demonstrated that the time course of ADH expression in adults is different for the D. melanogaster and D. affinidisjuncta transgenes. Further study could address the issue of whether the D. affinidisjuncta gene actually retains the expression-timing properties of the native species, as well as how this information is encoded and how it evolves.
When LSPE is negligible, as seems to be the case with the Adh homologues studied, the high correlation in expression between genes occupying the same genomic position can be exploited in the detection of subtle expression differences between related sequences, thereby dramatically extending the range of functional and evolutionary questions that can be addressed by transgenic technology. Although our results pertain only to the divergent Adh alleles studied, it seems likely that LSPE will be minimized when comparing genes that share much of their regulatory sequences—for example, when comparing different cDNA sequences driven by the same regulatory sequences. Therefore, it may be possible to detect differences between alleles differing by single amino acid substitutions or even between alleles sharing primary sequence but differing in codon usage. Furthermore, because coplaced genes occupy allelic positions in homologous chromosomes, pairing- or location-dependent phenomena—such as transvection (21), position-effect variegation (22), and meiotic recombination (23)—become more amenable to transgenic-based investigation. Allelism of coplaced genes also dictates that coplaced genes will segregate from one another in meiosis, making it possible to construct large experimental populations segregating for any two coplaced alleles. Changes in allele frequency in these populations may be analyzed to yield inferences about relative reproductive success of the different genotypes without the confounding effect of initial linkage disequilibrium that impedes similar studies of standing intraspecific variation (24). In these ways, the coplacement approach opens a whole range of functional and evolutionary questions to empirical analysis.
Acknowledgments
We thank C. Laurie and M. Brennan for the Adh genes, D. Sullivan for advice on ADH assays, I. Beerman for assistance with ADH assays, W. J. Dickinson for helpful comments, J. Parsch and W. Stephan for sharing raw data, P. Goss and D. Petrov for helpful discussions, and the members of the Hartl laboratory for advice and support. M.L.S. is a Howard Hughes Medical Institute Predoctoral Fellow. This work was supported by National Institutes of Health Grants GM-33741 and HG-01250 to D.L.H.
ABBREVIATIONS
- ADH
alcohol dehydrogenase
- su(Hw)
suppressor of Hairy-wing
- GPE
generalized position effect
- LSPE
lineage-specific position effect
References
- 1.Spradling A C, Rubin G M. Cell. 1983;34:47–57. doi: 10.1016/0092-8674(83)90135-6. [DOI] [PubMed] [Google Scholar]
- 2.Wilson C, Bellen H J, Gehring W J. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- 3.Siegal M L, Hartl D L. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowan R D, Dickinson W J. J Mol Evol. 1988;28:43–54. doi: 10.1007/BF02143496. [DOI] [PubMed] [Google Scholar]
- 5.Brennan M D, Dickinson W J. Dev Biol. 1988;125:64–74. doi: 10.1016/0012-1606(88)90059-0. [DOI] [PubMed] [Google Scholar]
- 6.Golic K G, Lindquist S. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 7.Kreitman M. Nature (London) 1983;304:412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- 8.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 9.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 10.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 11.Karess R E, Rubin G M. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 12.Sofer W, Ursprung H. J Biol Chem. 1968;243:3110–3115. [PubMed] [Google Scholar]
- 13.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 14.Ashburner M. Drosophila: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Sokal R R, Rohlf F J. Biometry. 3rd Ed. New York: Freeman; 1995. [Google Scholar]
- 16.Laurie-Ahlberg C C, Stam L F. Genetics. 1987;115:129–140. doi: 10.1093/genetics/115.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsch J, Tanda S, Stephan W. Proc Natl Acad Sci USA. 1997;94:928–933. doi: 10.1073/pnas.94.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patton J S, Gomes X V, Geyer P K. Nucleic Acids Res. 1992;20:5859–5860. doi: 10.1093/nar/20.21.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gloor G B, Nassif N A, Johnson-Schlitz D M, Preston C R, Engels W R. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- 20.Golic M M, Rong Y S, Petersen R B, Lindquist S L, Golic K G. Nucleic Acids Res. 1997;25:3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C-T. J Cell Biol. 1993;120:587–590. doi: 10.1083/jcb.120.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henikoff S. Genetics. 1994;138:1–5. doi: 10.1093/genetics/138.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawley R S, McKim K S, Arbel T. Annu Rev Genet. 1993;27:281–317. doi: 10.1146/annurev.ge.27.120193.001433. [DOI] [PubMed] [Google Scholar]
- 24.Árnason E. Genetics. 1991;129:145–168. doi: 10.1093/genetics/129.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]