SUMMARY
The N-terminal 17-amino-acids of huntingtin (NT17) can be phosphorylated on serines 13 and 16; however, the significance of these modifications in Huntington’s disease pathogenesis remains unknown. In this study, we developed BAC transgenic mice expressing full-length mutant huntingtin (fl-mhtt) with serines 13 and 16 mutated to either aspartate (phosphomimetic or SD) or alanine (phosphoresistant or SA). Both mutant proteins preserve the essential function of huntingtin in rescuing knockout mouse phenotypes. However, fl-mhtt induced disease pathogenesis, including motor and psychiatric-like behavioral deficits, mhtt aggregation and selective neurodegeneration are abolished in SD but preserved in SA mice. Moreover, modification of these serines in expanded repeat huntingtin peptides modulates aggregation and amyloid fibril formation in vitro. Together, our findings demonstrate that serines 13 and 16 are critical determinants of fl-mhtt-induced disease pathogenesis in vivo, supporting the targeting of huntingtin NT17 domain and its modifications in HD therapy.
INTRODUCTION
Huntington’s disease (HD) is an autosomal dominant and progressive neurodegenerative disorder characterized by movement disorders, cognitive impairment, and psychiatric symptoms (Harper, 1996). HD is caused by a polyglutamine (polyQ) repeat expansion near the N-terminus of huntingtin (Htt), a very large 350 kDa protein (The Huntington’s Disease Research Group, 1993). Huntingtin is thought to function as a scaffolding protein that mediates a variety of protein-protein interactions involved in vesicular trafficking, signaling and transcription (MacDonald, 2003; Li and Li, 2006). HD neuropathology is characterized by the selective loss of predominantly medium-sized spiny neurons (MSNs) in the striatum and to a lesser extent the pyramidal neurons in the deep layers of the cortex (Vonsattel and DiFiglia, 1998). Another pathological hallmark of HD is the accumulation of nuclear and cytoplasmic protein aggregates containing mhtt N-terminal polyQ fragments in the cortex and striatum (DiFiglia et al., 1997; Gutenkast et al., 1999). Currently, it remains unclear how mhtt can cause progressive neuronal dysfunction and selective neurodegeneration in the brain (Orr and Zoghbi, 2007; Li and Li, 2006). Unraveling the underlying pathogenic mechanisms may provide important clues to the development of effective treatments for this lethal neurodegenerative disorder.
Although protein misfolding due to polyQ expansion appears to be a shared pathogenic mechanism in all polyQ disorders (Orr and Zoghbi, 2007; Williams and Paulson, 2008; Bates, 2003; Ross and Poirier, 2004), the critical molecular pathway through which misfolded polyQ proteins elicit disease-specific clinical and pathological outcomes remain unresolved. Recent studies reveal a critical role for the specific protein context beyond the expanded polyQ domain in modifying disease pathogenesis (Gatchel and Zoghbi, 2005; Orr and Zoghbi, 2007; Katsuno et al., 2002). In HD, covalent post-translational modifications of mhtt have been implicated in disease pathogenesis (Schilling et al., 2006; Steffan et al., 2004; Jeong et al., 2009; Humbert et al., 2002; Anne et al., 2007; Yanai et al., 2004). However, in all but one case (i.e. the caspase-6 cleavage site; Graham et al., 2006), the pathogenic significance of htt cis-domain modifications in vivo has been assessed using mhtt N-terminal fragment models. Therefore, it remains to be addressed how these modifications may influence disease pathogenesis in the context of fl-mhtt in a mammalian model of HD.
Emerging data suggest that the N-terminal 1–17 amino-acids of htt (NT17 domain) immediately preceding the polyQ domain may constitute a critical functional domain for htt function and HD pathogenesis (Steffan et al., 2004; Cornett et al., 2005; Rockabrand et al., 2007; Atwal et al., 2007; Aiken et al., 2009). The NT17 domain is highly conserved evolutionarily and can mediate htt binding to peripheral membranous structures (Atwal et al., 2007; Rockabrand et al., 2007). The NT17 domain can function as a cytoplasmic retention signal and deletions or certain point mutations in this domain result in nuclear accumulation of htt in cultured cells (Rockabrand et al., 2007; Cornett et al., 2005; Steffan et al., 2004; Atwal et al., 2007). The NT17 domain can also be covalently modified by ubiquitylation and SUMOylation, which appear to have opposing effects on the toxicity of mhtt fragments in a transgenic Drosophila model (Steffan et al., 2004). Recent biochemical analyses reveal that interaction of the NT17 domain with the adjacent polyQ domain can accelerate mhtt exon 1 peptide aggregation via a novel and complex pathway (Thakur et al., 2009). This converging evidence suggests that the NT17 domain and its modifications may play important roles in the physical and biological properties of wildtype htt, and in the toxicity of fl-mhtt and its toxic fragments.
In neurodegenerative diseases, phosphorylation of disease proteins such as Tau (Ballatore et al., 2007), SCA1 (Emamian et al., 2003), and alpha-Synuclein (Fujiwara et al., 2002) has been shown to play important roles in disease pathogenesis. Recent studies reveal that serines 13 and 16 (S13 and S16) in htt NT17 domain can be phosphorylated in cultured mammalian cells (Aiken et al., 2009; Thompson et al., in press). To address the importance of these modifications in disease pathogenesis elicited by fl-mhtt in a mammalian model of HD, we have introduced either phosphomimetic (SD) or phosphoresistant (SA) mutations into fl-mhtt. Dramatically, SD but not SA fl-mhtt can prevent progressive neuronal dysfunction, mhtt aggregation, and late-onset neurodegenerative pathology in vivo. Moreover, an in vitro aggregation assay reveals that the SD mutations significantly compromise the fibrillization of mhtt-exon 1 peptides, whereas SA mutations do not. Thus, we provide strong evidence that S13 and S16 in htt NT17 domain play a critical role in modulating polyQ-induced misfolding and/or aggregation in vitro and fl-mhtt induced disease pathogenesis in vivo.
RESULTS
Generation and Biochemical Characterization of BAC Transgenic Mice with Mutations at S13 and S16 of Full-length Mutant Huntingtin
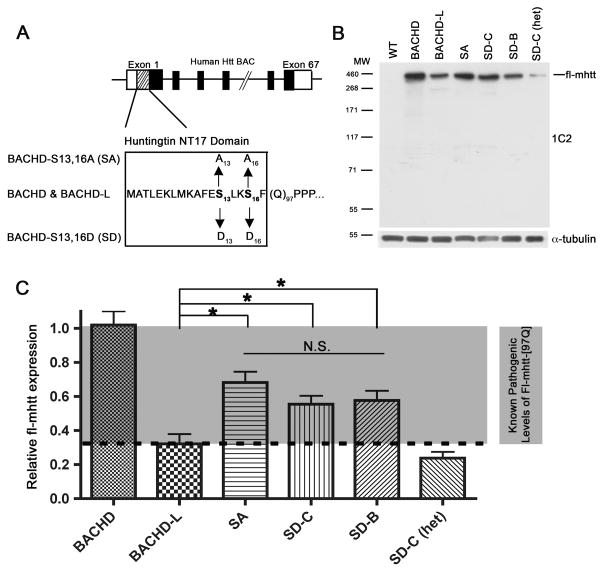
To evaluate the pathogenic relevance of S13 and S16 phosphorylation in mammalian models of HD, we re-engineered the S13 and S16 residues of our previous BACHD construct (fl-mhtt-[97Q]; Gray et al., 2008) to aspartate (SD), a mutation commonly used to mimic serine phosphorylation. We also mutated the two serine residues to alanine (SA), which renders the mhtt NT17 domain incapable of being phosphorylated at these sites (Figure 1A). The structural integrity of the engineered SD and SA BACs were confirmed with pulse field gel electrophoresis (Figure S1A) and the purified DNA was microinjected into FvB/N one-cell embryos to generate transgenic founders (Figures S1B and S1C). Five founders for the BACHD-SD construct and two founders for the BACHD-SA mutant were obtained, and initial expression analyses of the F1 mice from these lines identified two lines with SD mutations (lines B and C) and one line with SA mutations expressing fl-mhtt protein at suitable levels (see below). The mutant htt exon 1 region was PCR amplified and sequenced to confirm the appropriate mutations at S13 and S16, and a polyQ repeat length of 97 (Figures S1D and 1E; Table 1). These mouse lines were expanded by breeding with FvB/NJ wildtype mice, the same inbred mouse background used in the previous phenotypic characterization of BACHD mice (Gray et al., 2008; Spampanato et al., 2008; Menalled et al., 2009).
Figure 1. Generation and biochemical characterization of the phosphomimetic (SD) and phosphoresistant (SA) BACHD transgenic mice.
(A) A schematic representation of the S13,16A and S13,16D mutations within the NT17 domain in the BACHD construct (Gray et al., 2008). (B) A representative Western blot of cortical protein extracts made from one month old BACHD, BACHD-L, SA, SD-B, SD-C, and SD-C heterozygous (het) mice. The blot was probed with polyQ-specific 1C2 antibody followed by anti-tubulin antibody for loading controls. (C) Quantification of the fl-mhtt expression levels in various HD transgenic mouse lines. Densitometry values of individual bands are based on three 1C2 Western blots using independent cortical extracts. The input in each lane is normalized using the anti-α-tubulin control. The results of three independent experiments are expressed as means±SEM. One-way ANOVA analysis reveals that there are significant differences between the fl-mhtt expression levels in BACHD, BACHD-L, SA, SD-C, SD-B, and SD-C (het) mice (F(4,11)=22.090, p=0.000). The Post hoc test (LSD) indicates that the BACHD-L fl-mhtt expression level is significantly lower than the fl-mhtt expression level in the BACHD (p=0.000), SA (p=0.001), SD-C (p=0.017), and SD-B (p=0.010) mice, but does not differ from the SD-C (het) mice (p=0.340).
Table 1.
Phenotype Summary of Different BAC Transgenic Mouse Models of HD
| BAC Lines | PolyQ Repeat | Fl-mhtt Protein (% BACHD) | Motor Dysfunction Rotarod |
Psychiatric-like Anxiety |
Deficits Depressive-like |
Selective Neuropath. | mhtt Aggregates | ||
|---|---|---|---|---|---|---|---|---|---|
| 2 mo | 6 mo | 12 mo | 12 mo | 12 mo | 12 mo | 12 mo | |||
| BACHD | 97 | 100% | + | + | + | + | + | + | + |
| BACHD-L | 97 | 32% | + | + | n.d. | n.d. | n.d. | n.d. | + |
| SA | 97 | 70% | + | + | + | + | + | + | + |
| SD-B | 97 | 54% | − | − | − | − | − | − | − |
| SD-C | 97 | 57% | − | − | − | − | − | − | − |
| SD-B (het) | 97 | 23% | − | − | − | − | − | − | − |
We next used S13- and S16-phosphospecific htt antibodies to confirm the specificity of the S13 and S16 mutations at the protein level in SD and SA mice. The binding of these antibodies should be prevented following either SD or SA mutation in fl-mhtt, and only unmodified, phosphorylated htt will be recognized by phosphospecific antibodies. Consistent with this prediction, we were able to immunoprecipitate fl-mhtt from BACHD brains using S13- and S16-phosphospecific htt antibodies, but not the NT17-mutated fl-mhtt from SD and SA brains (Figure S2). Thus, we were able to show that the phosphorylated species of fl-mhtt can be detected in BACHD brains and confirm the molecular specificity of the S13 and S16 mutations in the context of fl-mhtt-[97Q] protein isolated from SD and SA brains.
We next assessed the fl-mhtt-[97Q] protein levels in BACHD, BACHD-L, BACHD-SD and BACHD-SA mice. BACHD-L mice express fl-mhtt-[97Q] at about one-third the level of the BACHD mice but still demonstrate evidence of disease pathogenesis (Gray et al., 2008; Table 1). We performed quantitative Western blot analysis to ensure that fl-mhtt-[97Q] protein levels in SD and SA mice were at or above the fl-mhtt-[97Q] level in BACHD-L mice and therefore would be expected to elicit at least some disease pathogenesis. We quantified the relative expression of fl-mhtt-[97Q] using two anti-polyQ monoclonal antibodies, 1C2 (Figure 1B) and 4H7H7 (Figure S3A). The mouse monoclonal 4H7H7 antibody can recognize mhtt N-terminal fragment (1–480 amino acid) with 68Q but not the same htt fragment with 17Q (Figure S3B). 4H7H7 can also recognize on Western blots two other expanded polyQ proteins, mutant Ataxin-1 and Ataxin-3, but not their corresponding wild type proteins with non-pathogenic polyQ repeats (Figures S3C and S3D). In our series of BACHD models, both 4H7H7 and 1C2 can specifically recognize human mhtt but not endogenous murine wildtype htt (Figures 1B and S3A). Since these different BACHD mouse lines all have the same length polyQ repeat, Western blot quantification of fl-mhtt with the anti-polyQ antibody 1C2 can accurately determine the relative expression level of soluble, steady state fl-mhtt (Figure 1C and Table 1). We determined the relative levels of fl-mhtt-[97Q] in brain extracts from BACHD and BACHD-L heterozygotes, SA heterozygotes, SD-B line homozygotes, SD-C line homozygotes and heterozygotes [SD-C (het)] (N=3 per genotype). Using one-way ANOVA with Post hoc analysis, we determined that there are no significant differences in fl-mhtt expression levels between the SA, SD-B, and SD-C mice, whereas all of these lines express fl-mhtt-[97Q] protein at significantly higher levels than the BACHD-L line but at lower levels than the BACHD line, suggesting that the SA, SD-B, and SD-C mice express sufficient levels of fl-mhtt-[97Q] to elicit a disease phenotype.
To assess whether the SD or SA mutations affect the subcellular distribution and/or the steady state level of fl-mhtt, we performed a series of Western blot analyses. First, subcellular fractionation of cytosolic, nuclear, microsomal, and mitochondrial fractions of the BACHD, SA, and SD-C mice at 2 months of age (N=3 per genotype) was performed and followed by Western analysis using the 1C2 antibody. At our level of detection in the mouse brain extracts, we do not observe any major shifts of the subcellular localization of fl-mhtt or its detectable fragments in SD or SA mutant mice compared to BACHD mice (Figure S4). Next, we sought to determine the steady state levels of soluble fl-mhtt protein during the aging process to assess whether there is any notable shift in fl-mhtt protein levels or increase in the production of soluble mhtt fragments over time. In each of the three mutant HD mouse lines we have examined, the levels of soluble fl-mhtt are comparable between 2 and 12 months (Figure S5),. Moreover, in SD and SA mice, we do not detect any aberrant increase of soluble mhtt polyQ fragments at 12 months, suggesting that age-dependent differences in the abundance of soluble full-length and fragmented mhtt cannot account for any phenotypic changes.
In summary, our biochemical analyses demonstrate that SA, SD-B and SD-C mice express fl-mhtt-[97Q] at levels well above the range expected to elicit disease pathogenesis in vivo. Furthermore, we cannot detect marked differences in S13 and S16 modified fl-mhtt and its polyQ fragments in their subcellular localization or their steady-state, soluble levels over time.
Full-length Mutant Huntingtin with S13 and S16 Mutations Preserves Normal Htt Function during Development and in Adult Cortical Neurons
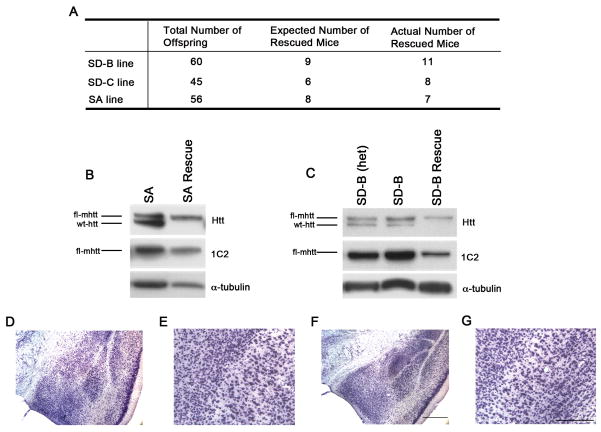
We next asked whether S13 and S16 mutated fl-mhtt could still exhibit the essential functions of htt in vivo. We evaluated whether cis-modified htt can rescue the phenotypes of htt knockout mice, including embryonic lethality in the complete null mutant mice (Zeitlin et al., 1995; White et al., 1997) and late-onset cortical neuron degeneration in the forebrain-specific deletion of the htt gene (Dragatsis et al.., 2005). We previously demonstrated that fl-mhtt-[97Q] in BACHD mice can rescue the embryonic lethality of htt knockout mice, confirming that fl-mhtt retains the essential developmental function of htt (Gray et al., 2008). To assess whether fl-mhtt-[97Q] with SD or SA mutations retain the function of htt, we crossed SA, SD-B and SD-C heterozygous alleles onto the htt knock-out background and scored for the number of the rescued mice born (Gray et al., 2008). In all three lines, the rescued mice were born in a Mendelian ratio (Figure 2A). Overall, we have obtained at least 7 rescued mice per genotype and these rescued mice express only fl-mhtt-[97Q] with S13 and S16 mutations, but not endogenous wild type murine htt (Figures 2B and 2C). Since these rescued mice do not show obvious deterioration with age, we could assess whether S13 and S16 mutated fl-mhtt could also rescue the robust late-onset cortical neurodegeneration phenotype between 6 and 12 month of age in the forebrain-specific htt knockout mice (Dragatsis et al., 2005). We performed neuropathological evaluation of the brain sections from 12 month old SD-B rescue mice and WT control littermates (N=3 per genotype). As shown in Figures 2D to 2G, we did not detect any evidence for neurodegeneration in the cortex of SD-B rescue mice (Figures 2F and 2G), including the piriform cortex near the external capsule where the conditional knockout of htt produced the most notable neurodegeneration (ibid). Together, these studies demonstrate that fl-mhtt with SD or SA mutations retains the essential developmental and neuroprotective function of murine htt.
Figure 2. Mutant Huntingtin with S13 and S16 Mutations Preserve Normal Htt Function During Development and in Adult Cortical Neurons.
(A) Heterozygous SD-B, SD-C and SA transgenes were bred in two successive generations with murine Hdh heterozygous knockout mice(Zeitlin et al., 1995) to generate the rescue mice (i.e. fl-mhtt transgene rescue the embryonic lethality of htt null mice). Since the homozygous Hdh null mice are embryonic lethal (ibid), the expected ratio of rescue mice among the live mice born is 1 out of 7. The rescue mice for SD-B, SD-C and SA were born with such Mendelian ratio. (B and C) Western blot analyses with mAb2166 (Htt) and 1C2 to confirm the rescue mice only express fl-mhtt and lack endogenous murine htt. (D–G) SD fl-mhtt retains the essential adult neuronal function of htt in vivo. Nissl stained coronal brain sections of 12-month old SD-B rescue mice (F and G) and WT littermates (D and E) at two different magnifications. No evidence of late-onset cortical neurodegeneration, as previously reported in the forebrain-specific htt null mice (Dragtsis et al, 2000), was detected in these rescue mice. Scale bars = 50 μm.
Phosphomimetic Mutations at S13 and S16 Abolish Motor and Psychiatric-like Deficits Induced by the Expression of Full-length Mutant Huntingtin
BACHD mice exhibit both motor deficits and enhanced psychiatric-like deficits (Gray et al., 2008; Menalled et al., 2009), which can be used to address whether S13 and S16 site mutations affect the neuronal dysfunction elicited by fl-mhtt in vivo. To assess motor dysfunction, we applied a two-way mixed factorial design to repeatedly examine the Rotarod performance in a cohort of SA, SD-B, SD-C mice at 2, 6, and 12 months of age. Similar to BACHD and BACHD-L mice (Gray et al., 2008; Table 1), we found that SA mice also exhibit significant Rotarod deficits starting at 2 months of age (p<0.0001, independent sample Student’s t test), which progressively worsen at 6 and 12 months of age (p=0.0001, p<0.0001 for different ages, Figure 3A). Repeated measure two-way ANOVA further revealed a significant genotype effect (F(1,30) =29.301, p<0.0001). There is also a significant interaction between genotype and age (F(1,30) = 4.965, p=0.034). To further evaluate effects of age on motor performance within each genotype, we used one-way ANOVA to analyze the age contribution to Rotarod performance in separate genotypes. We did not find any significant overall age effect in wild type mice (F(2,45)=0.089, p=0.915), but did find a significant main effect of age in SA mice, (F(2, 45)=3.390, p=0.043), which suggests a progressive impairment of Rotarod performance. Contrary to the findings in SA mice, two independent lines of SD mice (SD-C and SD-B homozygotes) did not exhibit any Rotarod deficits compared to their respective WT littermate controls at 2, 6 and 12 months (Figures 3B and 3C). Repeated measure two-way ANOVA did not detect a significant genotype effect (for SD-B: F(1, 17)=0.695, p=0.416; for SD-C: F(1,22)=0.373, p=0.548). Moreover, there is no significant interaction between genotype and age (for SD-B: F(1, 17)=1.345, p=0.262; for SD-C: F(1, 22)=0.006, p=0.938).
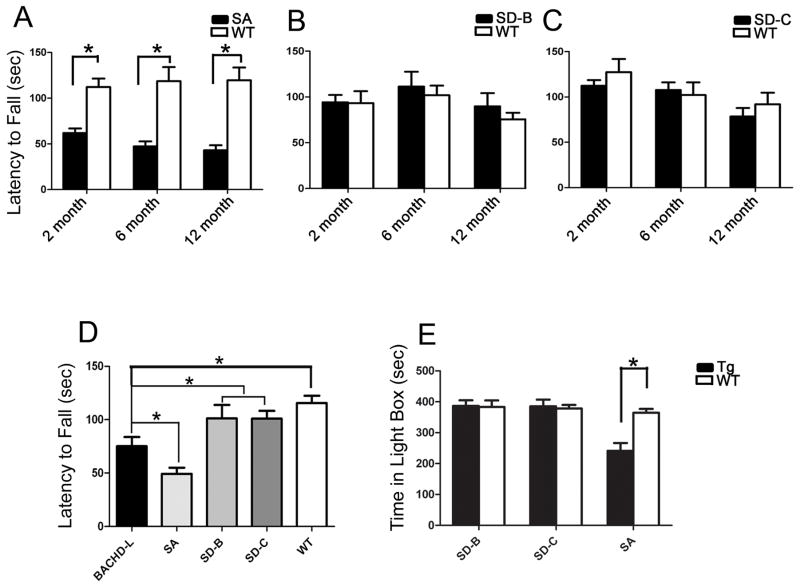
Figure 3. The SA but not the SD mice demonstrate motor and psychiatric-like behavioral deficits.
A mixed model two-way ANOVA design was used to examine the Rotarod performance in a cohort of SA, SD-B, SD-C mice at 2, 6 and 12 months of ages. (A) SA mice exhibit significant Rotarod deficits starting at 2 months of age and persisting at 6 and 12 month ages (* p < 0.0001, Student t test). (B and C) Two independent lines of SD mice (SD-C and SD-B homozygotes) did not exhibit any Rotarod deficits compared to their respective WT littermate controls at 2, 6 and 12 months. (D) Comparison of 6 month age BACHD-L, SA, SD-B and SD-C lines Rotarod performance. SD-B and SD-C mice did not show any significant impairment in their 6-month Rotarod performance compared to wildtype controls. On the other hand, both BACHD-L mice and SA mice exhibited significant impairment compared to WT, SD-B and SD-C mice (one-way ANOVA with Fisher’s LSD post hoc test, p < 0.05, details see main text). (E) The SA mice (p=0.0002, Student’s t test) but not SD mice demonstrate significantly enhanced anxiety in light/dark box exploration test.
We then compared Rotarod performance of SD and SA mice to the BACHD-L line. BACHD L mice have significant Rotarod impairment at 6 months of age (Fig. 3D). We also compared the 6-month Rotarod performance of the WT control mice from different HD mouse lines using one-way ANOVA analysis and found no significant differences among the four groups’ WT controls (F(3, 38) = 1.292, p=0.285 and p>0.05 in pairwise comparisons, Figure S6). Thus, Rotarod performance changes in these lines cannot be attributed to any differences in the WT controls. We then compared the pooled WT controls with SD-B and SD-C mice in their 6-month Rotarod performance (Figure 3D), which did not reveal any significant differences between the genotypes (WT=42, SD-B=10, SD-C=12, one-way ANOVA with Fisher’s LSD post hoc test, p>0.05). On the other hand, both BACHD-L mice and the SA mice exhibited significant motor impairment compared to WT, SD-B and SD-C mice (BACHD-L=12, SA=8, one-way ANOVA with Fisher’s LSD post hoc test, BACHD-L vs SD-B, p=0.032; BACHD-L vs SD-C, p=0.025; SA vs SD-B, p<0.0001; SA vs SD-C, p<0.0001; SA vs BACHD-L, p=0.029). Together, these studies demonstrate that SA mutations still retain the toxicity of fl-mhtt and can elicit motor deficits, while SD mutations abolish this disease phenotype in the BACHD model system.
Although psychiatric symptoms are particularly difficult to model in animals, several tests have been developed to assess psychiatric-like behaviors that have at least some predictive validity in patients (Crawley, 2008). BACHD mice were found to exhibit a higher level of anxiety as measured by two different tasks, the light-dark box and elevated plus maze (Menalled et al., 2009). To assess whether SD or SA mutations in fl-mhtt can modulate these anxiety-like behavioral deficits, we tested 12-month old SD-B, SD-C and SA mice and their respective WT controls. Similar to BACHD, SA mice still exhibit a significant anxiety phenotype compared to the controls (p=0.0002, independent sample Student’s t test), while the SD-B and SD-C mice no longer exhibit the anxiety-like behavior compared to their respective controls (SD-B vs WT p=0.8991; SD-C vs WT, p=0.7606, independent sample Student’s t test; Figure 3E). Thus, consistent with the motor test, the SD but not SA mutations also appear to abolish the psychiatric-like behavioral deficit observed in BACHD mice.
Full-length Huntingtin with Phosphomimetic S13 and S16 Mutations Can Prevent the Onset of Neurodegenerative Pathology in vivo
BACHD brains at 12 months of age recapitulate aspects of selective neurodegenerative pathology in HD, particularly the selective atrophy of the cortex and striatum with relative preservation of the cerebellum (Gray et al., 2008). We next addressed whether the SA and SD mutations in fl-mhtt could differentially impact the pathogenesis of selective neurodegeneration in vivo. Similar to BACHD mice, significant forebrain weight loss was detected in SA mice compared to their WT littermate controls (Figure 4A, p=0.0064, Student’s t test). However, both SD-B and SD-C mice do not exhibit the forebrain atrophy phenotype compared to their respective WT controls (SD-B vs WT, p=0.1975; and SD-C vs WT, p=0.7155, Student’s t test). In all three genotypes, no significant cerebellar atrophy was detected (Figure 4B).
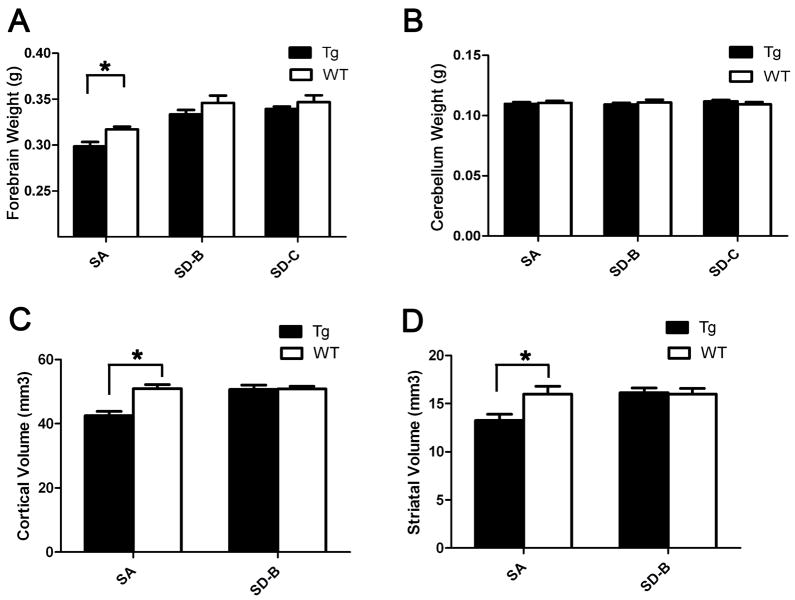
Figure 4. The SA but not the SD mice elicit selective neurodegeneration at 12-month age.
(A) Significant forebrain weight loss was detected in 12-month SA mice compared to their WT littermate controls (p=0.0064, Student’s t test), but not in SD-B or SD-C mice compared to their respective controls. (B) In all three genotypes, SA, SD-B and SD-C, no significant cerebellar atrophy was detected (p>0.05, Student’s t test). (C) Unbiased stereology reveals significant cortical volume loss in SA mice compared with WT littermates (p=0.0030, Student’s t test), but not SD-B mice (p=0.9025, Student’s t test). (D) Unbiased stereology reveals significant striatal volume loss in SA mice compared with their WT littermates (p=0.0336, Student’s t test). SD-B mice do not exhibit significant striatal volume loss compared to their WT controls (p=0.8635, Student’s t test).
To more quantitatively evaluate both the cortical and striatal atrophy in SA and SD mice, we applied the unbiased stereology method to measure both the cortical and striatal volumes in 12-month old SA and SD-B mice. Similar to the BACHD mice (Gray et al., 2008), we found SA mice exhibit 17% reduction of both the striatal and cortical volumes compared with their WT littermates (Figures 4C and 4D; No. of mice: WT=4, SA=5; Striatal volume: WT = 15.97 ± 0.8301 mm3; SA = 13.25 ± 0.6435 mm3; p=0.0336, Student’s t test; Cortical volume: WT = 50.91 ± 1.332 mm3; SA = 42.49 ± 1.318 mm3; p=0.003, Student’s t test). On the contrary, SD-B mice do not exhibit any striatal or cortical volume loss compared to their WT controls (No. of mice: WT=4; SD-B=5; Striatal Volume: WT = 16.00 ± 0.5836 mm3; SD-B=16.13 ± 0.4873 mm3; p=0.8635, Student’s t test; Cortical volume: WT= 50.88 ± 0.7701 mm3; SD-B= 50.67 ± 1.357 mm3; p=0.9025, Student’s t test; Figures 4C and 4D).
Together, these data reveal that the SA and SD mutations have opposing effects on the neurodegeneration phenotypes elicited by fl-mhtt, with the SA fl-mhtt remaining neuropathogenic whereas the SD fl-mhtt no longer induce the selective neurodegenerative pathology in vivo.
Phosphomimetic S13 and S16 Mutations Prevent the Accumulation of Mutant Huntingtin Aggregates in vivo
The ability of mhtt N-terminal fragments to form cytoplasmic aggregates and nuclear inclusions was first discovered in a HD mouse model (Davies et al., 1997), and such aggregates are seen in BACHD mice (Gray et al., 2008). We tested whether the monoclonal antibody 4H7H7 could detect mhtt aggregates containing the expanded polyQ fragments in situ. Using antigen-retrieval followed by immunohistochemistry (Osmand et al., 2006), 4H7H7 can selectively detect at least a subset of mhtt aggregates in the HD patient cortices, but not age-matched control cortices (Figure S7; see Supplementary results). Moreover, we showed the same 4H7H7 immunostaining procedure allows for sensitive detection of mhtt neuropil and nuclear aggregates in the cortex and striatum of four different HD mouse models (Figure S8) (Davies et al., 1997; Gu et al., 2005; Slow et al., 2003; Menalled et al., 2003).
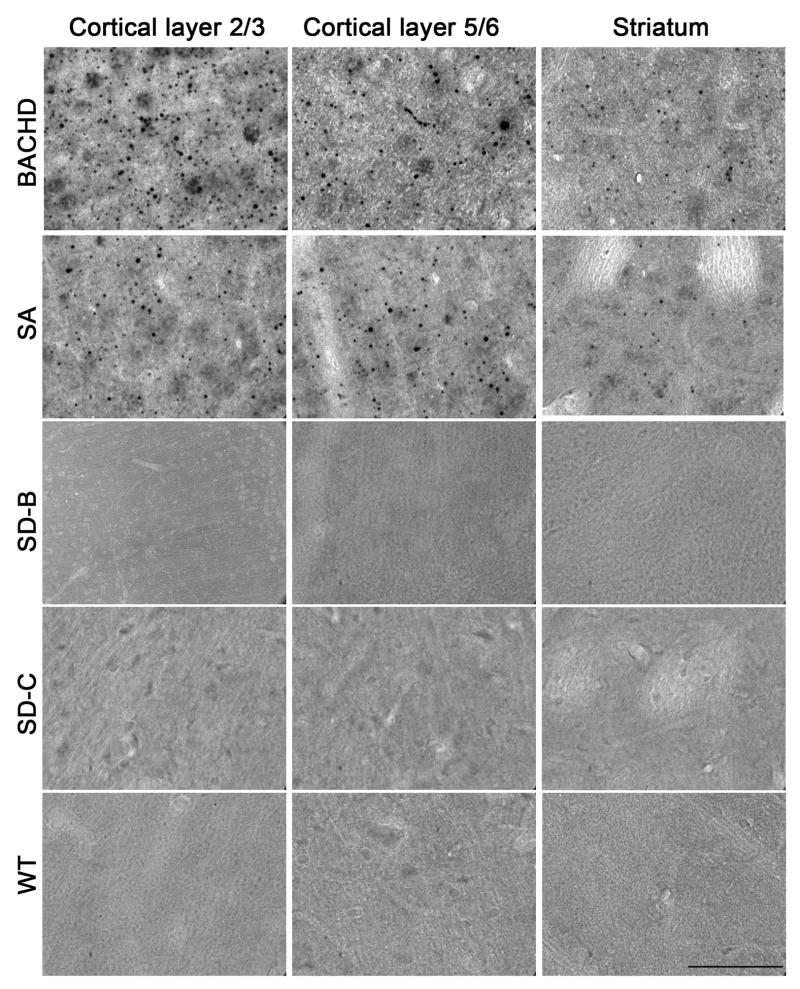
In BACHD mice at 12 months of age, the 4H7H7 antibody can readily detect mhtt aggregates in cortical and striatal sections following the antigen retrieval protocol while the WT controls do not have such staining (Figure 5). We can readily detect mhtt aggregates in the cortex and striatum of 12-month old SA mice, albeit at a somewhat lower level than in BACHD mice consistent with the lower expression of fl-mhtt in SA mice compared to BACHD (Figure 5). The formation of these aggregates in SA mice appears to be progressive, as they were not detected at 2 months of age, and only sparsely detected at 6 months (Figure S9; data not shown). These results are consistent with the progressive accumulation of mhtt aggregation in the brains of HD mice and HD patients (Bates, 2003). Interestingly, we did not detect 4H7H7-immunoreactive mhtt aggregates in brain sections from 12-month old SD mice (SD-B and SD-C) (Figure 5).
Figure 5. SD mutations but not SA mutations prevent the accumulation of mhtt polyQ aggregate in the brain.
Mutant huntingtin aggregates in HD patient brains and existing HD mouse brains can be readily detected by immunostaining with 4H7H7, a monoclonal antibody selectively recognizes the expanded polyQ epitope (see Figures S3, S6, S7, and Supplementary Results). Using the same antibody, we can readily detect mhtt aggregates accumulated in the cortical layers 2–3, layers 5–6 and striata of 12 month old BACHD and SA mice but not WT littermate controls brains. Such staining are absent in the 2-month old SA mice (Figure S8). However, in the 12-month old SD-B and SD-C brains, 4H7H7 stained mhtt aggregates are absent in all brain regions including cortical layers 2–3, layers 5–6 and striata. Scale bar = 50 μm.
Taken together, the in vivo results suggest that the SD mutation prevents the onset of behavioral deficits and neuropathology normally arising from polyQ repeat expansion in the context of full-length human mhtt in HD mice, including motor and psychiatric-like behavioral deficits, selective neurodegenerative pathology, and mhtt aggregation.
Phosphomimetic mHtt Exon1 Peptides Retard Aggregation in vitro
Since the SD or SA mutations appear to have differential effects on disease pathogenesis, we hypothesize that they may confer distinct biophysical properties that alter the folding, association, and/or aggregation properties of mhtt. In htt exon-1 related peptide study in vitro, the N17 sequence engages in a reciprocal crosstalk with expanded polyQ sequences to radically alter the aggregation pathway while greatly enhancing aggregation rate and producing an array of prefibrillar intermediates and mature amyloid fibrils (Thakur et al., 2009). In this study, reduction of net hydrophobicity via multiple mutations within NT17 clearly suppressed the aggregation rate (Thakur et al., 2009). Based on this result, we expected that the SA mutation (increased hydrophobicity) in NT17 might enhance aggregation, while the SD mutation (decreased hydrophobicity) might decrease aggregation.
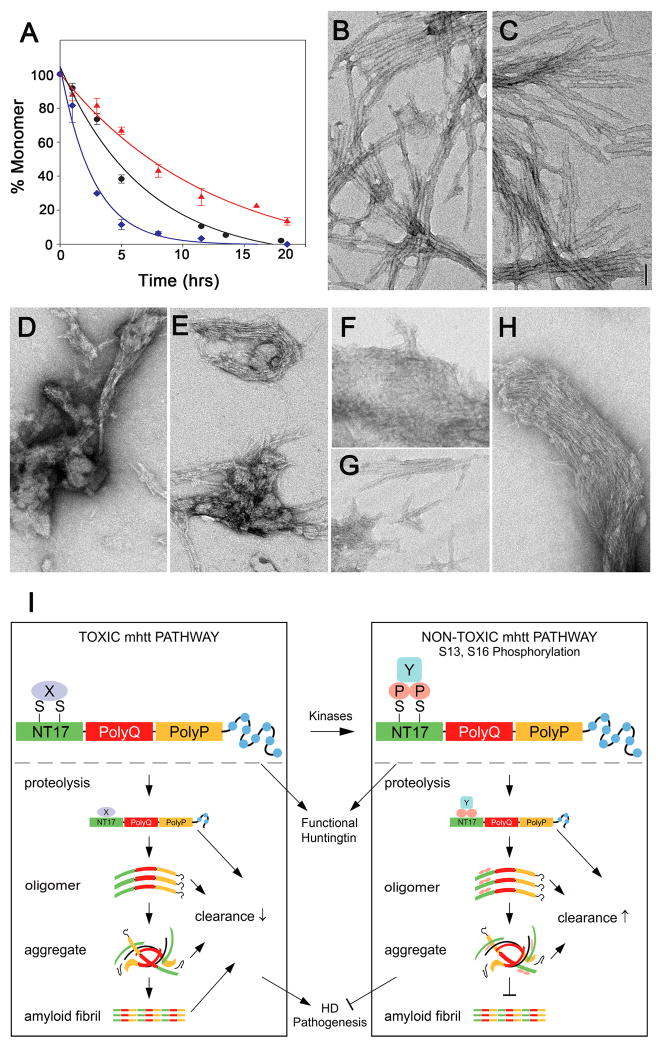
We used synthetic exon-1 peptides containing the WT, SD, and SA NT17 domains to test these predictions. For synthetic feasibility and convenience in the kinetics, we studied peptides containing a Q37 repeat, a polyQ length at the threshold for HD disease risk. Using an HPLC sedimentation assay, we found that the SA double mutant of the exon-1 peptide aggregates more rapidly than the same peptide with the WT NT17 sequence, while the SD double mutant peptide aggregates more slowly (Figure 6A; p < 0.005 for both, Student’s t-test). More qualitative differences are seen when the aggregation products are analyzed by electron microscopy (EM). We harvested aggregates at 96 hours, when aggregation was essentially complete. The aggregates of the SA double mutant exon-1 peptide (Figure 6C) are morphologically indistinguishable from the aggregates of the WT peptide (Figure 6B), both being highly homogeneous preparations of long, straight, relatively thick fibrils. In contrast, the aggregates from the SD double mutant exon-1 peptide are quite heterogeneous, consisting of an assortment of short, protofibril-like aggregates and bundles of very thin filaments (Figures 6D to 6H). Based on the EM data, the SD aggregates appear to be a mixture of stabilized, but morphologically normal, exon1 aggregation intermediates, and products of an alternative aggregation pathway (A. K. Thakur, M. Jayaraman, R. Kodali, R. Mishra and R. Wetzel, unpublished).
Figure 6. In vitro aggregation of exon-1 peptides and a schematic model on serines 13 and 16 as a critical molecular switch of HD pathogenesis.
(A). HPLC based sedimentation assays indicate differences in the aggregation of WT:10.8uM (●, R2 = 0.9832, S.E. = 6.7), SA:10.9uM (◆, R2 = 0.9843, S.E. = 6.3) and SD:10uM (▲, R2 = 0.9863, S.E. = 3.2) NT17Q37P10K2 peptides. The experiments were done in duplicate (for WT) and triplicate (SA and SD mutants) and the data was fit using Sigma Plot 10.0 software to a single exponential three-parameter decay function. (B–H). Electron microscope images on aggregates harvested after 96 hrs aggregation time. WT (B) and SA mutant (C) peptide aggregates are morphologically equivalent. SD mutant peptide aggregates (D–H) include none of these mature amyloids, but rather are composed of a mixture of alternative morphologies associated with aborted and/or alternative aggregation pathways.
(I) A schematic model of how serines 13 and 16 act as a molecular switch to dichotomize fl-mhtt induced disease pathogenesis. Depending on the molecular state of S13 and S16, fl-mhtt may be involved in two distinct molecular pathways. In the toxic mhtt pathway, when S13 and S16 are not phosphorylated, a state mimicked by SA mutations, fl-mhtt may mediate its toxicity via interaction with yet undefined protein X, or it may be cleaved into toxic mhtt polyQ fragments which may misfold and form toxic monomers, oligomers, aggregates and mature amyloid fibrils. Moreover, these mhtt species may be difficult to be cleared by the lysosome or proteosome (Thompson et al., in press). Overall, the unphosphorylated or SA form of fl-mhtt can elicit HD pathogenesis in vivo. In contrast, phosphorylation of S13 and S16, modeled by SD mutations, may induce biophysical changes to alter its polyQ-induced protein misfolding and/or protein-protein interactions (e.g. interacting with protein Y instead of protein X), and slow the kinetics of htt aggregation and impair the formation of mature amyloid fibrils, thereby enhancing the clearance of mhtt species. Overall, our study provides the proof-of-principle that molecular targeting of S13 and S16 (i.e. SD mutations) may convert fl-mhtt into a functional but non-toxic form in vivo, and hence represents an attractive novel target for HD therapy.
We next asked whether a single phosphomimetic mutation at S13 or S16 can recapitulate the effects of SD double mutations on mhtt-exon 1 peptide aggregation. Interestingly, the S16D single mutant can mimic the reduced aggregation rate of the SD double mutant in the HPLC sedimentation assay, while the S13D mutant aggregates at a comparable rate to the WT peptide (Figure S10). However, when we examined the morphologies of the completed aggregation product at 96 hours, both S13D and S16D single mutant peptides resulted in mixtures of mature amyloid fibrils similar to those from WT NT17 and short protofibril-like aggregates similar to those from SD double mutants (Figure S10). Since the SD double mutant exhibits negligible mature fibrils, it appears that both the S13D and S16D mutations contribute, in a cumulative fashion, to altering the exon1 aggregation pathway.
While more work is required to understand the mechanism of the SD mutations’ effect on aggregation, structure, and physical properties of the htt aggregation products, our in vitro results suggest that SD mutations somehow modulate the misfolding and aggregation of mhtt fragment induced by polyQ expansion. The SD mutations appear to act by either redirecting aggregation down a different assembly pathway or by retarding the normal assembly pathway, in either case resulting in reduced aggregation kinetics and altered aggregate morphologies.
DISCUSSION
Our study provides direct in vivo mammalian genetic evidence for a critical role for htt S13 and S16 residues in preventing fl-mhtt-induced disease pathogenesis in HD mouse models. Both phosphomimetic mutations (SD) and phosphoresistant mutations (SA) can rescue phenotypes of htt null mice. The most important finding of this study is that SD but not SA mutations in the context of fl-mhtt can prevent behavioral deficits, selective neurodegeneration, and accumulation of mhtt aggregates in the BAC transgenic mouse models of HD (Table 1). In vitro kinetic studies with purified mhtt exon 1 peptides reveal that modifications of serines 13 and 16 alter the course and kinetics of aggregation, with SD reducing fibrillization while accumulating alternative aggregate products, and SA maintaining the normal aggregation patterns of the expanded repeat length (Thakur et al., 2009). These results suggest that modification of the two serine residues is capable of altering the structural properties of this domain. Taken together, the results demonstrate that subtle molecular changes of only two amino acids in the NT17 domain in the context of the large full-length human mutant htt protein can dramatically reduce the pathogenic potential of the mutant protein in vivo. Hence, our results provide strong support for the hypothesis that S13 and S16 in the htt NT17 domain may act as a molecular switch to regulate disease pathogenesis elicited by fl-mhtt, and that targeting such a critical molecular pathogenic mechanism may have a dramatic therapeutic impact.
Significance of S13 and S16 Residues in HD Pathogenesis in vivo
A critical challenge in HD research is to identify which subset of molecular mechanisms could have a significant role in determining disease pathogenesis and in developing therapeutics (Orr and Zoghbi, 2007; Beal and Ferrante, 2004; Li and Li, 2006). One fruitful approach to study polyQ disease pathogenesis is to assess how the distinct protein context of the polyQ repeat (i.e. other domains or covalent modifications) may be involved in controlling disease pathogenesis (Orr and Zoghbi, 2007). In HD, multiple htt protein domains and covalent htt modifications have been implicated in disease pathogenesis (e.g. Steffan et al., 2004; Duennwald et al., 2006; Jeong et al., 2009). However, only the caspase cleavage sites have been tested thus far for pathogenic significance in the context of fl-mhtt in transgenic mice (Graham et al., 2006). The in vivo significance of the modification of S13 and S16 in the context of fl-mhtt-induced disease pathogenesis in a mammalian model of HD is directly addressed by this study.
An important goal in studying HD pathogenesis is to identify critical molecular pathogenic mechanisms that can prevent the early-onset neuronal dysfunction and late-onset neurodegeneration induced by mhtt (Gusella and MacDonald, 2000). Our study clearly demonstrates that the S13 and S16 sites may act as a molecular switch to prevent these pathogenic events. We determined that SD mice do not display early neuronal dysfunction as demonstrated by motor and psychiatric-like behavioral deficits, which are readily detectable in mice with SA mutations or with unmodified fl-mhtt (Gray et al, 2008; Menalled et al., 2009). In addition, we were able to demonstrate that SD, but not SA mutants can rescue late-onset neurodegeneration as assayed by selective forebrain weight loss and cortical and striatal atrophy. Our study suggests that the NT17 domain and its modifications are critical regulators of very early pathogenic initiation events that lead to the prevention of the cascade of downstream pathogenic events.
S13 and S16 Modification May Regulate Mutant Huntingtin Misfolding and Aggregation
Protein misfolding leading to amyloid fibril formation is a widely investigated molecular pathogenic mechanism for a large group of age-dependent neurodegenerative disorders (Ross and Poirier, 2004; Williams and Paulson, 2008; Bossy-Wetzel et al., 2004). A key mechanistic finding of this study is the effect of S13 and S16 mutations on mhtt misfolding and aggregation. Accumulation of nuclear and cytoplasmic aggregates is a pathological hallmark of HD (DiFiglia et al., 1997; Gutekunst et al., 1999). Prior studies have demonstrated that mutations in the NT17 domain may affect the aggregation and/or nuclear accumulation of mhtt N-terminal fragments in transfected cells (Rockabrand et al., 2007; Atwal et al., 2007). We have found that modification of the NT17 Ser residues can dramatically alter the course of aggregation. In particular, the SD double mutant slows the aggregation rate and prevents the normal course of the aggregation reaction. In contrast, the SA double mutant aggregates more rapidly than the WT NT17 peptide to generate products with morphology indistinguishable from that of the WT. Moreover, the single S13D or S16D mutants appear to offer partial protection against formation of mature amyloid fibrils, suggesting a cumulative effect of Ser to Asp mutations underlies the ability of the SD double mutant to counteract the propensities of mhtt exon1 peptides for misfolding and aggregation in vitro. These results are consistent with previous in vitro results showing that mutations within the NT17 domain that reduce hydrophobicity reduce aggregation rates (Thakur et al., 2009).
Importantly, the aggregation behavior of these exon-1 model peptides correlates well with the differential mhtt aggregate accumulation in the SD and SA brains in vivo, suggesting that the SD mutations may also have a significant impact on the structure and physical properties of fl-mhtt or its fragments in vivo. We have found that 12-month old SA mutants, which have phenotypes similar to BACHD mice, exhibit mhtt aggregates in the cortex and striatum, while 12-month old SD mutants exhibit no detectible aggregates (Figure 5). Consistent with the in vitro aggregation results (Figure 6), the lack of detectible aggregates in the SD mice may be due to a slow and incomplete aggregation process, or prolonged lifetimes of aggregation intermediates. These aggregation intermediates might be less readily detected by the 4H7H7 antibody or may be more easily cleared in affected neurons compared to mature amyloid fibrils from SA and WT mhtt. The latter possibility is supported by the findings that SD but not SA mutations in the context of mhtt exon 1 fragments or fl-mhtt can enhance its clearance via lysosome or proteosome in cultured mammalian cells (Thompson et al., in press). In summary, our study provides convergent in vitro and in vivo evidence that SD but not SA mutations alter the biophysical properties of NT17 domain, counteracting the molecular consequences of mhtt polyQ expansion such as protein misfolding and aggregation.
Molecular Pathways Implicated in S13 and S16 Phosphorylation in Huntingtin
Our study suggests the phosphorylation status of the two serine residues in the NT17 domain can influence the pathogenic consequences of expression of fl-mhtt protein in the mammalian brain (Figure 6I). Our study is consistent with the converging evidence supporting the important role of NT17 domain and its modifications in mhtt-induced neuronal toxicity in cellular and invertebrate models of HD (Steffan et al., 2004; Rockabrand et al, 2007; Atwal et al, 2007; Subramaniam et al., 2009; Thompson et al., in press). Our result highlights the importance of elucidating the molecular pathways, particularly the kinases and phosphatases, which modulate these phosphorylation events in the HD brain. Also, it will be important to uncover the downstream molecular cascades resulting from S13 and S16 phosphorylation of fl-mhtt that mediate the suppression of fl-mhtt induced disease pathogenesis (Figure 6I).
Currently, very little is known about the potential partners that can interact with htt via the N17 domain. However, two recent studies may shed some light onto this issue. In both a yeast and mammalian cell line model (Omi et al., 2008; Wang et al., 2009), the htt NT17 domain is shown to bind to 14-3-3 proteins, which are known to bind to phosphorylated proteins. Since the NT17 domain has two serine and one threonine residue, we speculate that 14-3-3 proteins may be able to bind to S13 or S16 upon its phosphorylation and such binding may modulate mhtt toxicity in vivo. This hypothetical model for HD is reminiscent of SCA1 pathogenesis in which 14-3-3 proteins can aggravate polyQ expanded ataxin-1 mediated disease via binding to phosphorylated serine 776 (Chen et al., 2003). Thus, future molecular studies should focus on establishing the signaling pathways that lead to alteration of htt S13 and S16 phosphorylation, and in identifying phosphorylation dependant protein-protein interaction changes that may be responsible for the suppression of fl-mhtt toxicities in vivo.
Therapeutic Implications of Huntingtin S13 and S16 Phosphorylation
Our study suggests that targeting the mhtt NT17 domain may be therapeutic for HD, particularly prior to the onset of disease. Consistent with this, a single-chain Fv intrabody against htt NT17 domain reduced neurotoxicity and aggregation of mhtt N-terminal fragments in vivo (Wolfgang et al, 2005; Southwell et al., 2008). Another important finding of our study is that S13 and S16 phosphomimetic mutations do not elicit disease pathogenesis but do maintain normal htt function in embryonic development and the adult brain (Figures 2 and 6I). This result suggests an exciting therapeutic possibility of converting a toxic form of fl-mhtt into a functional but non-toxic form. Molecular analyses of the differences between SD mhtt and other forms of mhtt may help to design high-throughput screening assays for identification of compounds that modify fl-mhtt elicited disease pathogenesis
In conclusion, we have demonstrated the significance of S13 and S16 residues in disease pathogenesis elicited by HD transgenic mouse models. Our findings suggest that specific S13 and S16 modifications in fl-mhtt may act as a molecular switch to suppress HD pathogenesis in the mammalian brain, thus providing an important avenue for the development of new therapeutics for this currently incurable neurodegenerative brain disorder.
EXPERIMENTAL PROCEDURES
Generation of BAC Transgenic Mice
We used site-directed mutagenesis to modify the shuttle vector plasmid, pLD53-SCAB-HDexon1-97Q (Gray et al., 2008). Serines 13 and 16 were mutated to either aspartic acid (SD) or alanine (SA) using QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene). The following oligonucleotide primers were used to introduce the SD mutation (5′-AAG CTG ATG AAG GCC TTC GAG GAC CTC AAA GAC TTC CAA CAG CAG CAA CAG CAA C-3′) or SA mutation (5′-AAG CTG ATG AAG GCC TTC GAG GCC CTC AAA GCC TTC CAA CAG CAG CAA CAG CAA C-3′). PCR products from mutagenesis reactions were used to transfect Pir2 competent cells (Invitrogen). Correct RecA-based shuttle vector plasmids were used to engineer a human htt BAC (Gray et al, 2008) to introduce the SD and SA mutations (Yang et al., 1997; Gong et al, 2002; Yang and Gong, 2005). The final modified BAC were confirmed by DNA sequencing to contain the SD or SA mutations and to have 97 mixed CAA-CAG codons encoding the expanded polyQ repeat (see Supplementary Methods). The modified BAC DNA was prepared according to our published protocols and microinjected into FvB fertilized oocytes at the UCLA Transgenic Core. BACHD, BACHD-L, SD and SA mice were maintained in the FvB/NJ background and bred and maintained under standard conditions consistent with National Institutes of Health guidelines and approved by the UCLA Institutional Animal Care and Use Committees.
Preparation of Brain Extracts and Western Blotting
We used our published protocols for the preparation of brain protein extracts and Western blot analyses (Gray et al., 2008; see Supplementary Methods). Immunoblots were probed with anti-Htt 2166 (Abcam, mAb2166, 1:3000), 1C2 (Millipore, MAB1574, 1:3000), S13-P snd S16-P phosphor-specific antibodies (1:1000; Thompson et al., in press), and -tubulin (Sigma, 1:10,000); biotinylated monoclonal 4H7H7 antibody (Brooks et al, 2004).
Genetic Rescue of Murine Huntingtin Null Phenotypes by SD and SA BAC Transgenes
The genetic rescue of murine htt knockout phenotypes during embryonic development is similar to our published study (Gray et al., 2008). Briefly, heterozygous BACHD-SD and BACHD-SA transgene alleles were bred with murine Hdh heterozygous mice (Zeitlin et al., 1995) to generate double heterozygous mice. These double heterozygous mice were then crossed to Hdh heterozygotes to create homozygous Hdh mice either with or without the BAC transgenes, and the number of mice with these genotypes were scored among the offsprings born. The rescue mice were defined as mice without two endogenous Hdh alleles but with the heterozygous BACHD-SD or BACHD-SA transgenes. No mice with both Hdh null alleles but without any BACHD-SD or SA transgenes were among the live littermates in our study (Zeitlin et al., 1995).
To assess the ability of BACHD-SD transgene in rescuing the adult neurodegeneration phenotype of Hdh forebrain-specific knockout mice, three BACHD-SD rescue mice and WT littermate controls were sacrificed at 12 months of age. Coronal brain sections were prepared (Gray et al., 2008) and used to examine for marked cortical neurodegeneration as described before (Dragatsis et al., 2000).
Behavioral Tests of Mutant HD mice
Mice were tested for motor impairment using our established Rotarod test (Gray et al., 2008). We tested the following group of mice at 2, 6 and 12 months: SA mice (WT=12, Tg=13), SD-B (WT=10, Tg=13), SD-C (WT=10, Tg=13), and BACHD-L (WT=17; Tg=20). For each genotype, we used approximately equal number of males and females. The mice were trained on an Ugo Basile 7650 Accelerating Rotarod (0 to 40 RPM in 5 minutes) for 3 trials per day for 2 days at 2 months of age (Gray et al., 2008). The same cohort of mice was tested for 3 trials per day for 3 consecutive days at 2, 6, and 12 months of age. All Rotarod tests were performed during the light phase of the light cycle.
Anxiety-like behavior was assessed in the light/dark box exploration test (File, 1997). The light/dark exploration box is made of plexiglass with a clear (light) side (27 × 27 × 30 cm) and a smaller, fully opaque (dark) side (18 × 27 × 30 cm) separated by a partition with a small opening. The dark side is completely closed to light. The lighted side is illuminated with a 60-watt light bulb or light with an illuminance of about 400 lux. Light/dark exploration was performed during the early phase of the light/dark cycle. Mice were placed in testing room at least 1 h before testing. The mice are tested for 10 min in the light/dark box and assessed for movements from one side to another. The same cohort of SD and SA mice and respective wild type controls from the above Rotarod tests were tested at 12 months of age in this light-dark exploration test.
Antigen Retrieval and Immunohistochemical Staining of polyQ Mutant Htt Aggregates with Monoclonal 4H7H7 Antibody
Mice were perfused transcardially with 4% paraformaldehyde in 0.1 M PBS. After 6 hours postfix at 4 °C, brains were cryoprotected with 30% sucrose in 0.1 M PBS at 4 °C, and frozen coronal brain sections (40 μm) were cut on a cryostat (Gu et al., 2005; Gray et al., 2008). The antigen retrieval procedure is described in more detail elsewhere (Osmand et al., 2006). Briefly, the brain sections were antigen treated with formic acid and followed by 1% sodium borohydride. Biotinylated monoclonal antibody 4H7H7 was then used at a concentration of 50 ng/ml and incubated with the brain sections overnight at 4°C. Subsequently, the immunostaining signal was amplified with Tyramide Signal Amplification kit (Perkin Elmer) and detected with SD-AG chromogen (Vector Lab.).
Brain Weight and Stereological Brain Volume Measurements
The quantitative analyses of brain weights and stereological measurements of cortical and striatal volumes using the optical fractionator method using Stereo Investigator software (MicroBrightField) were identical to our published protocols for BACHD mice (Gray et al., 2008).
Analysis of in vitro Htt-exon 1 Peptide Aggregate Formation
Chemically synthesized peptides were obtained from the Keck Biotechnology Center at the Yale University (http://keck.med.yale.edu/ssps/), purified by reverse phase HPLC, and the purity determined and identities confirmed by mass spectrometry using an Agilent 1100 Electrospray MSD. The structures of the peptides used were NT17-Q37P10K2, in which the sequence of wild type NT17 is MATLEKLMKAFESLKSW and the underscored Ser residues are the ones replaced by Ala or Asp in the SA, SD, and single S13D and S16D mutants. Phe17 was replaced by Trp in these peptides to be used as a fluorescent probe; we have observed no effects on aggregation or other properties in Phe17->Trp mutants (Thakur et al., 2009). Purified peptides were lyophilized and freshly disaggregated before use as described (O’Nuallain et al., 2006), then dissolved in phosphate buffered saline and incubated at 37 °C. The aggregation propensity of each peptide was determined by measuring the amount of monomer remaining in solution at different times using the sedimentation assay (O’Nuallain et al., 2006). Electron micrographs of the aggregates were obtained by directly analyzing aliquots of the reaction mixture collected at different times. For each determination, 5 μl of the sample was placed on a freshly glow-discharged Formvar carbon coated grid and adsorbed for 2 min, after which the grid was washed with deionized water before staining the protein with 2 μl of 1% (w/v) uranyl acetate and removing excess stain by blotting. Grids were imaged on a Tecnai T12 microscope (FEI) operating at 120 kV and 30,000X magnification and equipped with an UltraScan 1000 CCD camera (Gatan) with post-column magnification of 1.4X.
Statistical Analysis
Statistical analyses of the behavioral phenotypes and neuropathological phentoypes of BACHD-SD and SA mice and BACHD-L mice were similar to our published protocols (Gray et al., 2008). All data is shown as the mean ± standard error of the mean (SEM). For Rotarod analysis, SPSS 14.0 statistics software (Chicago, IL) was used. The significance level was set at 0.05. A general linear model ANOVA with repeated measures was performed to assess the main effects of age, genotype and their interaction. In some datasets, one-way ANOVA was used to analyze the time effect on Rotarod performance of BACHD mice and wild type mice from 2, 6, and 12 months of age. Fisher’s LSD post hoc analysis was performed for pairwise comparisons among ages. For neuropathology and stereology studies, all comparisons between BACHD and wildtype mice are shown as mean ± standard error of the mean (SEM). Comparisons were made with an independent sample Student’s t test with a significance level of 0.05. For electrophysiological studies, data in the text and figures are presented as mean ± standard error of the mean (SEM). Differences between groups were assessed with Student’s t-tests or appropriate designed ANOVAs. The significance level was set at 0.05.
Supplementary Material
Acknowledgments
We would like to thank Nancy Wexler, Carl Johnson, and the Hereditary Disease Foundation (HDF) for their tremendous support of this project. HD research in X. William Yang lab at UCLA is supported by a contract from HDF, and by grants from NINDS/NIH (R01NS049501; 3R01NS049501-05S1) and CHDI Foundation. R. Wetzel was supported by a research grant from the Huntington Society of Canada and the Huntington’s Disease Society of America (HDSA), and by NIH RO1 grant AG019322. The work of J.S. Stefan and L.M. Thompson was supported by HDF, the Fox Family Foundation (J.S.S.), the CHDI Foundation, the HDSA, and NIH awards NS52789 and NS045283 (L.M.T.). S. Finkbeiner was supported by 2R01NS39074, 2R01NS45491, 2P01AG022074, CHDI, and the Taube-Koret Center for HD Research. A.P. Osmand was supported by HDF. We also would like to thank Scott Zeitlin for the Hdh Knockout mice; M. A. Hickey, M.F. Chesselet and M.S. Levine for providing R6/2 and CAG140 brain sections, R. Truant for sharing unpublished studies, and members of the Yang Lab, particularly X.H. Lu, M. Gray, J.P. Cantle, Y. Cui and T.K. Murphy for discussions and critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aiken CT, Steffan JS, Guerrero CM, Khashwji H, Lukacsovich T, Simmons D, Purcell JM, Menhaji K, Zhu YZ, Green K, et al. Phosphorylation of threonine-3: Implications for huntingtin aggregation and neurotoxicity. J Biol Chem. 2009 doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne SL, Saudou F, Humbert S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J Neurosci. 2007;27:7318–7328. doi: 10.1523/JNEUROSCI.1831-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- Bates G. Huntingtin aggregation and toxicity in Huntington’s disease. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- Beal MF, Ferrante RJ. Experimental therapeutics in transgenic mouse models of Huntington’s disease. Nat Rev Neurosci. 2004;5:373–384. doi: 10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. Molecular pathways to neurodegeneration. Nat Med . 2004;10(Suppl):S2–9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- Brooks E, Arrasate M, Cheung K, Finkbeiner SM. Using antibodies to analyze polyglutamine stretches. Methods Mol Biol. 2004;277:103–128. doi: 10.1385/1-59259-804-8:103. [DOI] [PubMed] [Google Scholar]
- Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, Skoulakis EM, Orr HT, Botas J, Zoghbi HY. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- Cornett J, Cao F, Wang CE, Ross CA, Bates GP, Li SH, Li XJ. Polyglutamine expansion of huntingtin impairs its nuclear export. Nat Genet. 2005;37:198–204. doi: 10.1038/ng1503. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57:809–818. doi: 10.1016/j.neuron.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian ES, Kaytor MD, Duvick LA, Zu T, Tousey SK, Zoghbi HY, Clark HB, Orr HT. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–387. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- File SE. Animal tests of anxiety. Current Protocols in Neuroscience . 1997;8:3.1–3.15. doi: 10.1002/0471142301.ns0803s26. [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Hasegawa M, Dohmae N, Kawashima A, Masliah E, Goldberg MS, Shen J, Takio K, Iwatsubo T. alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat Cell Biol. 2002;4:160–164. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006a;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Li C, Wei W, Lo V, Gong S, Li SH, Iwasato T, Itohara S, Li XJ, Mody I, et al. Pathological cell-cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;46:433–444. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li XJ. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper PS. New genes for old diseases: the molecular basis of myotonic dystrophy and Huntington’s disease. The Lumleian Lecture 1995. J R Coll Physicians Lond. 1996;30:221–231. [PMC free article] [PubMed] [Google Scholar]
- Humbert S, Bryson EA, Cordelieres FP, Connors NC, Datta SR, Finkbeiner S, Greenberg ME, Saudou F. The IGF-1/Akt pathway is neuroprotective in Huntington’s disease and involves Huntingtin phosphorylation by Akt. Dev Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia TJ, Jr, Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobayashi Y, Doyu M, Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- Li S, Li XJ. Multiple pathways contribute to the pathogenesis of Huntington disease. Mol Neurodegener. 2006;1:19. doi: 10.1186/1750-1326-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ME. Huntingtin: alive and well and working in middle management. Sci STKE 2003. 2003:pe48. doi: 10.1126/stke.2003.207.pe48. [DOI] [PubMed] [Google Scholar]
- Menalled L, El-Khodor B, Patry M, Suarez-Farinas M. Systematic behavioral evaluation of transgenic and knock-in mouse models of Huntington’s Disease. Neurobiology of Disease . 2009 doi: 10.1016/j.nbd.2009.05.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- Omi K, Hachiya NS, Tanaka M, Tokunaga K, Kaneko K. 14-3-3zeta is indispensable for aggregate formation of polyglutamine-expanded huntingtin protein. Neurosci Lett. 2008;431:45–50. doi: 10.1016/j.neulet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- O’Nuallain B, Thakur AK, Williams AD, Bhattacharyya AM, Chen S, Thiagarajan G, Wetzel R. Kinetics and thermodynamics of amyloid assembly using a high-performance liquid chromatography-based sedimentation assay. Methods Enzymol. 2006;413:34–74. doi: 10.1016/S0076-6879(06)13003-7. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Osmand AP, Berthelier V, Wetzel R. Imaging polyglutamine deposits in brain tissue. Methods Enzymol. 2006;412:106–122. doi: 10.1016/S0076-6879(06)12008-X. [DOI] [PubMed] [Google Scholar]
- Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, Sullivan PG, Steffan JS, Sensi SL, Thompson LM. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med . 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Schilling B, Gafni J, Torcassi C, Cong X, Row RH, LaFevre-Bernt MA, Cusack MP, Ratovitski T, Hirschhorn R, Ross CA, et al. Huntingtin phosphorylation sites mapped by mass spectrometry. Modulation of cleavage and toxicity. J Biol Chem. 2006;281:23686–23697. doi: 10.1074/jbc.M513507200. [DOI] [PubMed] [Google Scholar]
- Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, Oh R, Bissada N, Hossain SM, Yang YZ, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- Southwell AL, Khoshnan A, Dunn DE, Bugg CW, Lo DC, Patterson PH. Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J Neurosci. 2008;28:9013–9020. doi: 10.1523/JNEUROSCI.2747-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato J, Gu X, Yang XW, Mody I. Progressive synaptic pathology of motor cortical neurons in a BAC transgenic mouse model of Huntington’s disease. Neuroscience. 2008;157:606–620. doi: 10.1016/j.neuroscience.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–30. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon IJ, Anjum DH, Kodali R, Creamer TP, Conway JF, et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington’s Disease Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, Martinez-Vincente M, Arrasate M, O’Rourke JG, Khashwji H, Lukacsovich T, Zhu YZ, Lau AL, Massey A, Hayden MR, Zeitlin SO, Finkbeiner S, Green K, LaFerla F, Bates G, Huang L, Patterson PH, Lo DC, Cuervo AM, Marsh JL, Steffan JS. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. Journal Cell Biology. 2009 doi: 10.1083/jcb.200909067. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, Costello CE, Sherman MY. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J. 2009;23:451–63. doi: 10.1096/fj.08-117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Auerbach W, Duyao MP, Vonsattel JP, Gusella JF, Joyner AL, MacDonald ME. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang WJ, Miller TW, Webseter JM, Thompson LM, Marsh JL, Messer A. Suppression of Huntington’s disease pathology in Drosophila by human single-chain Fv antibodies. Proc Natl Acad Sci U S A. 2005;102:11563–8. doi: 10.1073/pnas.0505321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai A, et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat Neurosci. 2006;9:824–31. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XW, Gong S. An overview on the generation of BAC transgenic mice for neuroscience research. Curr Protoc Neurosci . 2005;Chapter 5(Unit 5):20. doi: 10.1002/0471142301.ns0520s31. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.