Summary
Interaction of hematopoietic progenitors with the thymic stromal microenvironment induces them to proliferate, adopt the T cell fate, and asymmetrically diverge into multiple T lineages. Progenitors at various developmental stages are stratified among different regions of the thymus, implying that the corresponding microenvironments differ from one another, and provide unique sets of signals to progenitors migrating between them. The nature of these differences remains undefined. Here we use novel physical and computational approaches to characterize these stromal subregions, distinguishing gene expression in microdissected tissues from that of their lymphoid constituents. Using this approach, we comprehensively map gene expression in functionally distinct stromal microenvironments, and identify clusters of genes that define each region. Quite unexpectedly, we find that the central cortex lacks distinctive features of its own, and instead appears to function by sequestering unique microenvironments found at the cortical extremities, and modulating the relative proximity of progenitors moving between them.
Introduction
T lymphocytes are constantly lost to a variety of causes (bleeding, cellular senescence, post-activation cell death), and must be replaced to maintain homeostasis. Although homeostatic expansion of peripheral T cells can and does compensate for some of these losses (Surh and Sprent, 2008), new T lymphocytes are produced mainly in the thymus under normal circumstances. Interestingly, the thymus contains no self-renewing stem cells (Wallis et al., 1975; Goldschneider et al., 1986). Instead, intrathymic cells are replenished by recruitment of multipotent hematopoietic progenitors that circulate in the blood (reviewed in Zlotoff et al., 2008). Once inside the thymus, conditions unique to the organ induce new progenitors to undergo a well characterized sequence of developmental events, leading to the generation of several distinct lineages of mature T lymphocytes (reviewed in Zhou et al., 2009; Josefowicz and Rudensky, 2009; Collins et al., 2009; Singer et al., 2008). Progression through this sequence can be tracked by biochemical markers, and many functional characteristics of each stage are known, including the timing of gene recombination and expression of somatically encoded T cell receptors, proliferative status and burst potential, T lineage commitment status and other lineage potentials, etc. (reviewed in Petrie and Zuniga-Pflucker, 2007; Ladi et al., 2006; Shortman et al., 1990). However, the basic nature of the thymic microenvironment remains poorly understood, and only a handful of the signals that specify T lineage developmental processes are known.
Intrathymic differentiation has been closely linked to patterned migration within the organ (reviewed in Petrie and Zuniga-Pflucker, 2007; Ladi et al., 2006). Blood progenitors enter the organ in a narrow region of the peri-medullary cortex (PC), where they are amplified and retained to function as a transient pool of intrathymic “stem cells” (Porritt et al., 2003). Based upon occupancy (or vacancy) of cortical stromal niches (Prockop and Petrie, 2004), cells from this intrathymic progenitor pool are asymmetrically recruited to leave the PC and migrate outwards through the cortex in the direction of the capsule (Porritt et al., 2003). This migration occurs along a matrix composed of reticular epithelial cells (Prockop et al., 2002) that express several of the known signals for T cell differentiation, including Notch ligands, c-kit, and IL-7 (reviewed in Petrie and Zuniga-Pflucker, 2007; Ladi et al., 2006). Outward migration also coincides with progression through two stages of lymphoid development, correlating with loss of non-T lineage potential, accessibility of TCR-γ, -δ, and -β loci to recombination, and several rounds of cell division (reviewed in Petrie and Zuniga-Pflucker, 2007). Arrival in the sub-capsular region (SC) coincides with a major developmental transition, marked by acquisition of the definitive T lineage markers CD4 and CD8 (Lind et al., 2001; Petrie et al., 1990; Nikolic-Zugic et al., 1989) and the immature form of the T cell receptor (preTCR, Groettrup et al., 1993). Arrival in the SC is also characterized by intense proliferative activity (Penit, 1988; Lind et al., 2001), silencing of the TCR-β locus (Dudley et al., 1994), and accessibility of the TCR-α locus to the recombinase (Petrie et al., 1995). Subsequently, the polarity of migration is reversed, and cells move back inwards through the cortex towards the PC (Penit, 1988). Unlike movement outwards, the substrate for inward migration is not clear, and may largely depend on random movement (Li et al., 2007; Witt et al., 2005), although periodic interaction with reticular epithelial cells is still a requirement for proper selection of TCR specificities (Bevan, 1977; Zinkernagel et al., 1978; Teh et al., 1988). Cells that do interact successfully with self-histocompatibility antigens are actively recruited into the medulla (Witt et al., 2005; Ueno et al., 2004), where they undergo additional screening for TCR specificity (Anderson et al., 2002; Kyewski and Derbinski, 2004), as well as final functional maturation (Dyall and Nikolic-Zugic, 1995; Gabor et al., 1997; Boursalian et al., 2004).
The patterned movement of progenitors within the thymus is believed to influence their differentiation by exposing them to an ordered series of inductive microenvironments. This is particularly so in the cortex, where five of the six major stages of lymphoid differentiation occur (reviewed in Petrie and Zuniga-Pflucker, 2007; Ladi et al., 2006). Inherent in this assumption is the concept that different regions of the cortex are, in fact, distinct from one another, yet very few such differences in have been reported. Further, relatively few global gene expression studies have been performed on thymic stroma, and those have focused mainly on medullary epithelial cells and dendritic cells (Derbinski et al., 2005; Popa et al., 2007; Johnnidis et al., 2005; Gotter et al., 2004). Consequently, broad characterization of even gross cortical or medullary microenvironments is lacking. In this study, we use a novel approach to characterize the global landscape of stromal gene expression in thymic tissues, including spatially distinct subregions of the cortex. The approach involves microdissection of intact tissue from anatomically defined tissue regions, followed by characterization of stromal genes either by identifying genes expressed in tissue but not in the corresponding lymphoid cells (stromal-specific genes), or by proportionally subtracting lymphoid gene expression for genes common to both stromal and lymphoid cells (global stromal genes). One of the main benefits of this approach is that changes in stromal gene expression are minimized, because three-dimensional relationships remain intact during tissue isolation, and because lengthy enzymatic incubations are not required. The resulting gene expression signatures for stromal cells from the cortex and medulla show that different thymic regions do, in fact, differ substantially from one another at the gene expression level, including subregions of the cortex. Surprisingly, we find that the central cortex, previously believed to provide a specific inductive microenvironment for key developmental functions, has no unique gene signature of its own, suggesting that its main role may be to modulate the proximity of developing progenitors to inductive microenvironments found in other regions of the thymus.
Results
Mapping global gene expression in functionally-defined subregions of the thymus
The overall workflow is illustrated in Fig. 1. Regions of thymic tissue that correspond to specific developmental functions (reviewed in Petrie and Zuniga-Pflucker, 2007; Ladi et al., 2006) were selected according to standardized criteria (Fig. 2 and Experimental Procedures). RNA was isolated from microdissected tissues and used to probe Affymetrix MOE430 2.0 arrays (all known mouse genes and ESTs). Initially, MAS5 detection calls (Affymetrix Expression Console) were used to identify genes that were Present (P) in at least one tissue region. For cortical subregions, where 3 independent gene chips were used for each region (peri-medullary cortex, PC; central cortex, CC; sub-capsular cortex, SC), 3/3 P calls were required for designation as present (i.e., highest possible stringency). For the medulla, which was evaluated as a single unit, 7 independent gene chips were used, and genes defined as present were restricted to those receiving at least 6 P calls. A total of 14851 genes (corresponding to 25831 probesets) were identified as being present in at least one thymic tissue region. When genes in this list were subjected to unsupervised hierarchical clustering, replicate (independent) samples clustered together in logical fashion (Fig. 3). Further, cortical subregions were distinct from whole cortex (Fig. 3), showing that there were discernable differences between these subsets. Note that the dynamic range of the heatmap shown in Fig. 3 is dominated by high signals from medullary stromal cells (as discussed later in detail), minimizing visual differences among cortical sub-regions. Nonetheless, visual differences between these regions can be easily distinguished when these regions are clustered alone, without medullary tissue (Suppl. Fig. 1). Since the cellularity of thymic tissues is dominated by lymphoid cells, and since the lymphoid cells themselves are stratified, it would be possible to attribute cortical subregion clustering results to that of the lymphoid cells. However, cortical lymphocytes clustered most closely with medullary lymphocytes, and were less related to either whole cortex or cortical subregions than the latter were to each other, indicating that gene expression by stromal cells strongly influenced the hierarchy of unsupervised clustering, and thus the gene expression results. Even without further analysis, this definitively shows that stromal cells that form functionally-defined subregions of the thymus can clearly be distinguished from one another at the gene expression level.
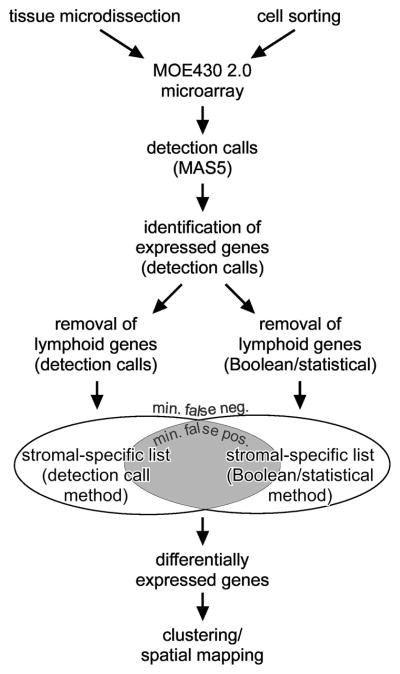
Figure 1.
Workflow for identification of stromal genes and generation of a spatial stromal map of the thymus.
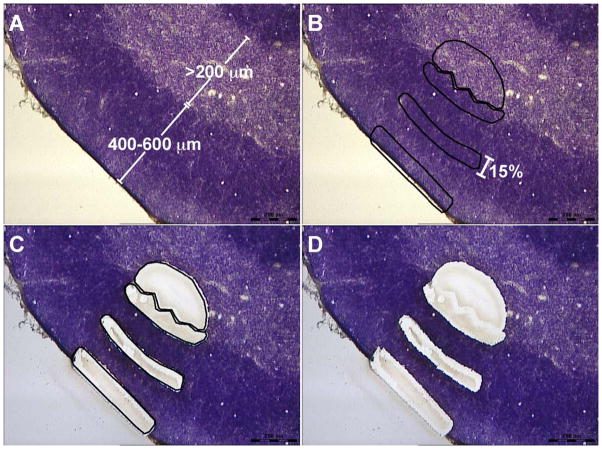
Figure 2. Spatial relationships of subregions used in this study.
Typical medullary and cortical subregions selected for collection are indicated. Panel A shows tissue pre-dissection, including baseline parameters for cortical and medullary tissue depth. Panel B shows the same tissue after defining regions for microdissection; each subregion in the cortex represents approximately 15% of the total cortical depth. Panel C shows the tissue after microdissection; in panel D, the lines defining the areas of interest have been removed to show the final result of the microdissection process. Scale bars on lower right of each panel are 200 μm.
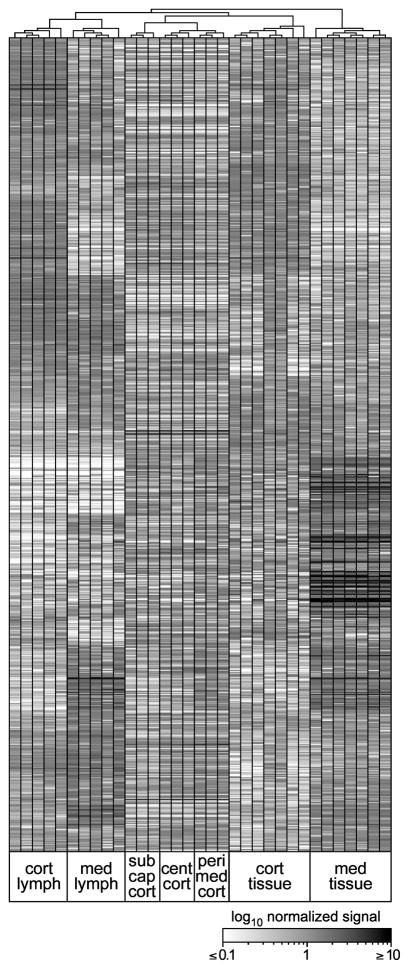
Figure 3. Unsupervised clustering reveals stromal-dominant differences among different subregions of thymic tissue.
RNA from microdissected tissues or purified lymphoid cells (as indicated) was used to probe microarrays, and 25831 probesets present in at least one microdissected tissue region were subjected to two dimensional hierarchical clustering (vertical hierarchy not shown). Black = highest normalized expression. Samples clustered in predictable fashion; notably, cortical subregions were distinct from each other, and were more similar to whole cortex than to other tissue or lymphoid types. Note that, in general, genes with the largest dynamic range are characteristic of medullary stromal cells (i.e., found in whole medulla but not medullary lymphocytes).
Identification of stromal-specific genes from total genes expressed in dissected tissues
Genes expressed by stromal cells can be organized into two different categories: stromal-specific genes, which are expressed by stromal cells but not lymphoid cells, and global stromal genes, some of which may be shared with lymphoid cells (see below). To identify stromal-specific genes, we used two independent approaches. In both cases, the starting list was composed of genes expressed in one or more tissue regions, as described above. The first approach was signal independent, using MAS5 detection calls to identify genes that were present in tissue but Absent (A) in the corresponding lymphoid cells. Since 5 independent lymphoid chips were used for each type, a single wildcard was again allowed (i.e., either 4/5 or 5/5 A calls).
In contrast, the second approach was purely signal based. Here, a series of logical (Boolean) and statistical tests were incorporated to identify stromal-specific genes. For accurate estimation of low level gene expression signals, we utilized quantile-normalized signal detection (Probe Logarithmic Intensity Error, PLIER), considered to be more sensitive and accurate for this purpose. The criteria for designation as a stromal-specific gene using this Boolean/statistical method were as follows: [(tissue signal > threshold) AND (lymphoid signal < threshold) AND (tissue signal > lymphoid signal) AND (t-test p-value for differences between tissue and lymphoid signals < 0.05)]. The conventional median value was initially used as a threshold for lymphoid gene expression. However, we observed that many known stromal genes, including several of the Notch ligands that are critical for T cell development (Radtke et al., 1999; Robey et al., 1996; Washburn et al., 1997), were expressed below this threshold (data not shown, but included in the raw microarray results). This is a common problem with signal-based approaches, which depend on an arbitrary cutoff (nominally, the median) to predict which genes are expressed. Since our approach included multiple Boolean and statistical parameters, and since we simultaneously used a parallel, signal-independent approach to determine stromal gene expression (above), we instead utilized the 25th percentile as the threshold for tissue signal, allowing us to include all known stromal genes. A final obstacle to identification of mapped stromal genes using the signal-based method was that the relative contribution of lymphoid cells at different developmental stages will vary widely among different cortical subregions (Lind et al., 2001). Consequently, lymphoid gene expression and tissue:lymphoid ratios would be different in each subregion, making it almost impossible to accurately determine tissue:lymphoid ratios (see discussion of global stromal gene expression, below). To address this, we performed additional experiments where whole cortical regions (spanning all subregions) were microdissected. The Boolean/statistical criteria described above were then used to compare whole cortical tissue to that of total cortical lymphocytes, resulting in a comprehensive list of total cortical stromal-specific genes. This list was then used to mask cortical subregion data, allowing us to regionally map total cortical stromal gene expression.
The detection call (signal-independent) method identified 1989 probesets (mapping to 1775 genes) as being stromal-specific in the cortex, and 2093 probesets (1787 genes) in the medulla. The Boolean/statistical (signal-based) method identified 1420 stromal-specific probesets in cortex (1259 genes), and 2514 stromal-specific probesets in the medulla (2162 genes). 556 genes were common between cortical and medullary compartments. As indicated in Fig. 1, genes appearing at the intersection of the two independent methods constitute a high confidence stromal gene list, with a minimum of false positive (MFP) results. However, as described above, it should be noted that the stringency of each method alone is actually quite high, and thus even the union list for the two approaches (designated here as minimum false negative, MFN) represents a high confidence list. A unified MFP and MFN gene list, indicating the tissue region in which the genes appear, their signals, and their assignment to dynamic spatial map of the thymus (described below) can be found in Suppl. Table 1. When mapped onto functional ontologies or pathways that would be expected to characterize stromal cells, stromal-specific gene lists were highly enriched in classifications that characterize epithelial cells (the dominant component of thymic stroma), antigen presenting cells (Fig. 4), and other similar functions (not shown), and were depleted of genes that characterize lymphoid cells.
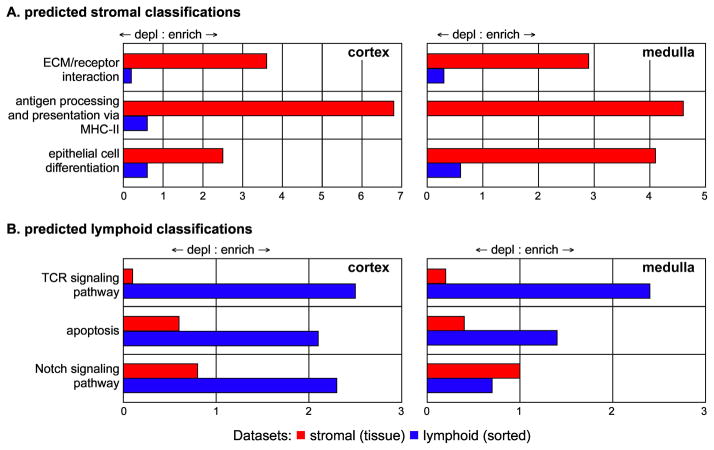
Figure 4. Representation of predicted functional classifications in stromal genes identified by differential mapping.
Genes identified as stromal-specific (Fig. 1) in the cortex or the medulla were mapped onto pathways or functions as described in the text. Mapping onto classifications that would be expected to characterize stromal or lymphoid cells in the thymus are shown. Enrichment or depletion are measured against the number of genes from the whole chip that would be predicted to fall in each category based random chance. Stromal gene lists were enriched for categories that characteristic of stromal cells, and were depleted in categories that characterize lymphoid cells.
Spatial mapping of region-specific gene signatures
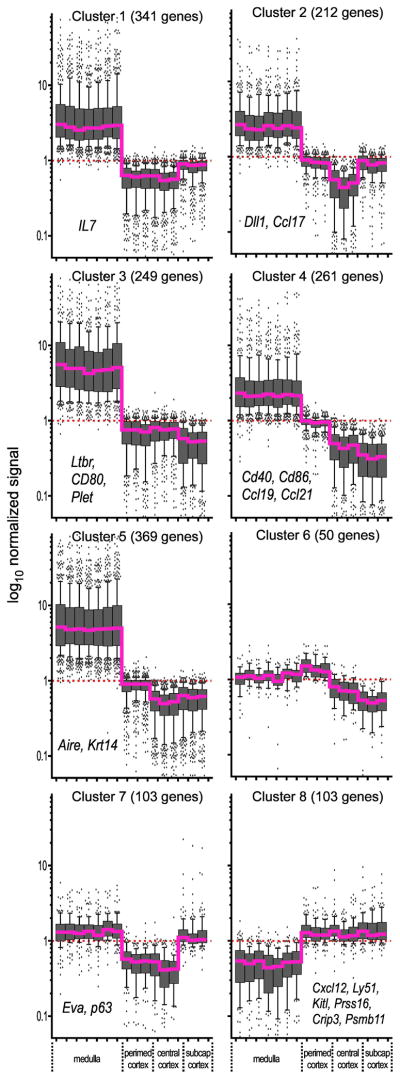
The gene lists presented in Suppl. Table 1 represent a comprehensive map of stromal-specific gene expression in the thymus. In order to identify stromal-specific genes that uniquely characterize each region, we extracted from this list those genes that changed significantly (t test p value < 0.05 AND |fold change| ≥ 2) between different regions. K-means clustering (Gene Spring 7.3.1) was then used to partition these genes into related clusters, which were then mapped in spatial context to the regions from which each sample was derived. Eight dominant spatial patterns, representing all significantly changed stromal-specific genes, are shown in Fig. 5; the genes mapping to each cluster are annotated in Suppl. Table 1.
Figure 5. Spatial mapping of stromal gene regions in the thymus.
Stromal genes that changed significantly between regions (p value <0.05 AND | fold change ≥ 2) were organized into clusters using the k-means algorithm, and 8 clusters representing the dominant patterns are shown. Grey bars indicate the interquartile range (25th–75th percentile) and solid vertical lines indicate the interdecile range (10th–90th percentile) for all genes in the cluster, while outliers are indicated by dots. For the sake of visual clarity, line graphs for individual genes (probesets) are not shown, but assignment to a given cluster mandates that each gene generally follows the trend indicated by the median value (bold magenta line). The per gene normalized value across all experiments (i.e., unity, from which relative change is measured) is indicated by a dashed red line. A panel of genes known to characterize stromal cells in specific compartments are indicated on the corresponding clusters.
As expected, based on the known functions of these medullary and cortical compartments, significant differences were observed in their stromal gene expression patterns. Five of the 8 clusters (designated 1, 2, 3, 4, and 5 in Fig. 5) were dominated by gene expression in medullary stromal cells. This finding is obvious even prior to identification of stromal-specific genes, since it can be clearly seen in unsupervised clustering of tissue and lymphoid results (Fig. 3), where the most differentially regulated genes map to whole medulla (but not medullary lymphoid cells or any cortical cells or tissues). The significance of this high-level gene expression in medullary stromal cells is not known. In any case, these five clusters are further distinguished by patterns of stromal gene expression in cortical subregions. For instance, genes in cluster 1 are more highly expressed in the SC than in other cortical regions, while those in cluster 5 distinguish the PC region. Genes in cluster 2 characterize both SC and PC, while those in cluster 3 represent both PC and CC. Cluster 4 represents a set of genes whose expression constantly decreases when moving outward from the medulla. Clusters 6 and 8 represent genes that are dominant in cortex over medulla, with cluster 8 being essentially unchanged in all cortical regions, and cluster 6 being highest in the PC region. Cluster 7 includes genes that are expressed at similar levels in both the medulla and SC region, but are decreased in the PC and CC. Conspicuous by its absence, however, is a cluster that uniquely defines gene expression in the CC; the implications and potential significance of this observation are discussed later in this manuscript.
Identification of global stromal gene expression
The processes described above accurately identifies stromal-specific genes in the context of a physical map of the thymus. Stromal-specific gene lists characterize many important stromal genes, including all known stromal signals for developing lymphocytes (see Fig. 5, Suppl. Table 1, and Discussion). However, they are limited in what they can reveal about stromal cell biology, since important genes that are common to lymphocytes (e.g., metabolic enzymes, signal transduction molecules, etc.) are removed from these lists. Theoretically, it is possible to calculate the proportion of gene expression contributed by stromal cells to a given tissue region if the proportion of lymphoid signal is known. This can be represented by the following formula (Gosink et al., 2007):
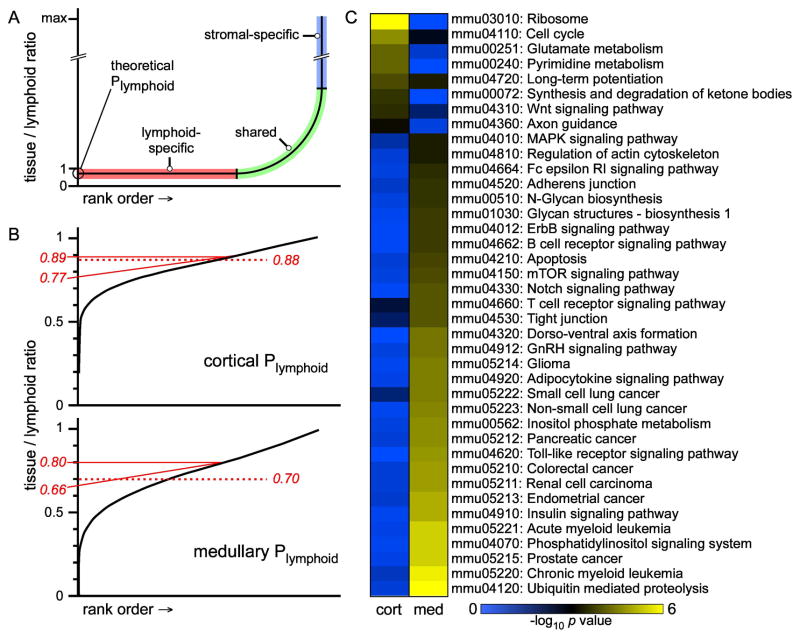
Stissue and Slymphoid are experimentally measured values (microarray), and since the tissue is represented by only two cell types (stromal or lymphoid; see Introduction), Pstromal is equal to 1 − Plymphoid. Thus, calculation of stromal signals can be carried out simply by determining the proportion of total tissue signal that is contributed by lymphoid cells (Gosink et al., 2007). In theory, this can be determined in a straightforward fashion by plotting rank ordered tissue:lymphoid signal ratios for all genes on the chip, and determining the y-axis intercept; since stromal cells do not add any signal to lymphoid-specific genes, the ratio of normalized signals from purified lymphocytes to that of the whole tissue will accurately estimate Plymphoid, and allow the above equation to be solved for Sstromal (Fig. 6A).
Figure 6. Calculation of global stromal gene signatures by proportional subtraction of lymphoid signals from tissues.
Panel A shows hypothetical behavior for noiseless data. Stromal-specific genes are infinitely higher in tissue than in lymphoid cells, and thus form a portion of the curve with infinite slope. Lymphoid-specific genes (no additional signal provided by stromal cells) likewise form the section of the curve with a slope of zero. The point at which the lymphoid-specific line intersects the y-axis defines the proportion of lymphoid signal in total tissue (per chip normalized data). In real (noisy) data, the hypothetical curve is deformed into a sigmoidal function in which the y-intercept does not exist (panel B). Note that only the region of the curve with tissue:lymphoid values <1 is shown, since this is the theoretical maximum contribution of lymphoid signal to tissue). Three independent methods were used to predict the y-intercept; the tangent to the inflection point (diagonal red line), a line perpendicular to the inflection point (horizontal red line), or the average ratio for a panel of known lymphoid-specific genes (dashed red line). Panel C shows enrichment in KEGG pathways for the most-changed genes in each compartment (medulla or cortex; yellow = highest statistical significance, black = threshold of significance, and blue = lowest significance). Note that enriched pathways in cortical stroma are strongly biased towards metabolic functions, while those in medulla are heavily biased towards signaling, particularly epithelial growth factor signaling pathways.
Unfortunately, several factors inherent to microarray experiments, including biological variation, experimental variation, digital signal processing noise (including large signal deviations at the low end of the range) distort the rank-ordered tissue:lymphoid curve such that it does not intersect the y-axis in a predictable fashion (Fig. 6B and Gosink et al., 2007). Consequently, we used several independent approaches to estimate the y-axis intercept (Plymphoid). These included the y-intercept of a line tangent to the inflection point at the low end of tissue:lymphoid ratios (Fig. 6B), the y-intercept of a line perpendicular to the inflection point, and the mean tissue:lymphoid signal ratios for a series of known lymphoid-specific genes (e.g., Rag1, CD8, etc.; see Experimental Procedures and Suppl. Table 2). Each of the three methods generated Plymphoid values in a relatively narrow range; 0.77–0.89 for the cortex, and 0.66–0.80 for the medulla. These values are consistent with relative cortical and medullary stromal cellularity as indicated by conventional staining approaches (e.g., keratins, CD11b, CD11c, etc). Depending on the specific goals, more inclusive (lower Plymphoid values) or more exclusive (higher Plymphoid values) global stromal signatures can be calculated from the raw data associated with this study, by applying Plymphoid values in the indicated range (Fig. 6B) to the equation described above. It is also possible to exceed the predicted range to include fewer or more genes, depending on the specific objectives.
To evaluate the major differences between cortical and medullary pathways, global stromal gene expression was calculated for each compartment using the middle (lymphoid-specific gene method) Plymphoid value (0.88 for cortex, 0.70 for medulla). Signal ratios were then determined for each probeset, and genes corresponding to the top 10% of ratios for cortex or medulla were mapped onto KEGG pathways (www.genome.jp/kegg) using the DAVID Bioinformatic Resource (/david.abcc.ncifcrf.gov, Huang da et al., 2009; Dennis et al., 2003). Somewhat unexpectedly, we found that genes highly expressed in cortical stromal cells were strongly biased towards metabolic processes, while those in medullary stromal cells were strongly biased towards signaling functions, especially those related to epithelial cell growth/survival factors (Fig. 6C). The significance of this finding is not known, but underscores the utility of our global approach to identify unique properties of stromal cells in different tissue regions. Notably, the pathways that characterize lymphoid cells in each compartment (using the same method) differ substantially from that of the corresponding (computationally identified) stromal lists (data not shown), indicating that the proportional removal of lymphoid signals from tissues does indeed facilitate the identification of global stromal signals.
Discussion
The thymus is unique among tissues that undergo steady-state differentiation for several reasons. A major distinction is that it contains no long-term self-renewing stem/progenitor cells (Wallis et al., 1975; Goldschneider et al., 1986), and instead relies on semi-continuous recruitment (Foss et al., 2001) of stem/progenitor cells that circulate in the bloodstream. Upon entry into the thymus, microenvironmental conditions specific to the thymus induce these multi-potent progenitors to adopt the T lineage fate, undergo multiple asynchronous rounds of proliferation, and diverge into at least five major functionally distinct sub-lineages (helper, cytotoxic, regulatory, NK-T, and γ/δ). Stromal elements forming the thymic microenvironment are also responsible for screening the repertoire of non-germline encoded T cell receptors (TCR) generated during somatic recombination, eliminating cells with self-reactive specificities and selecting for those that are self-tolerant (Kisielow et al., 1988; Bevan, 1977; Kappler et al., 1987; Zinkernagel et al., 1978). While the role of the thymic stromal microenvironment in TCR screening has been relatively well characterized, only a few of the stromal signals that regulate the other processes have been identified, and are mainly limited to Notch ligands (Robey et al., 1996; Radtke et al., 1999), kit ligand (Rodewald et al., 1995), and interleukin-7 (Peschon et al., 1994). A major obstacle has been that stromal cells, especially the epithelial cells that form the majority of thymic stroma (van Ewijk et al., 1994; Boyd et al., 1993), are difficult to isolate efficiently, especially in spatial context, which is a critical influence in all embryonic and steady-state developmental systems, including the thymus.
The generation of a functional map of the thymus (reviewed in Petrie and Zuniga-Pflucker, 2007; Ladi et al., 2006), together with the relative ease of isolating the corresponding lymphoid cells in a pristine state, prompted us to devise a computational approach to identify stromal gene expression in tissues from microdissected thymic regions. The use of laser microdissection to isolate intact tissue regions allows spatial context to be defined, while simultaneously ensuring a panoramic, unbiased representation of all stromal cells within any region of interest. Further, since tissues are frozen intact within minutes of harvest, this approach can also be expected to maintain stromal gene expression in a nearly unadulterated state. Together with data from isolated lymphoid cells, this computational approach thus provides an unprecedented opportunity to globally assess stromal:parenchymal relationships, in spatial context, and in an essentially unperturbed tissue.
Our stromal gene expression data can be divided into two partially overlapping sets, namely, stromal-specific genes (genes expressed by stromal but not lymphoid cells) or global stromal genes (all stromal genes, including some that may be shared with lymphoid cells). The former is particularly useful for validation, since known markers that distinguish stromal versus lymphoid cells can be used to assess the outcomes. Even using very high stringency criteria in each of two completely independent computational approaches (illustrated in Fig. 1), almost all known stromal-specific genes were captured (Suppl. Table 1), including the dominant signals mediating lymphoid differentiation (Notch ligands, kit ligand, IL-7, or MHC), signals controlling intrathymic migration (Cxcl12, Ccl19, Ccl21, Ccl25, or Vcam1), or canonical markers of epithelial, mesenchymal, and hematopoietic stroma (all reviewed in Petrie and Zuniga-Pflucker, 2007). The only notable exception was the Notch ligand Dll4, which was absent from stromal-specific lists because it was also found (using high-stringency criteria) at low levels in lymphoid cells, consistent with previously published findings (Yan et al., 2001), and informatic analysis of multiple thymus or T lymphoid datasets in public databases (http://www.ncbi.nlm.nih.gov/geo). Known, differentially expressed stromal genes (from RNA in situ or immunohistochemistry) were also completely recapitulated by our spatial mapping approach, as indicated on Fig. 5. The novel genes included in our results thus provide a validated resource of genes that may control lymphoid differentiation, and further elucidate the biology of the stromal cells themselves. Many novel stromal-specific genes (Suppl. Table 1) are predictive of the biology for the region in which they are found. For instance, the gene signature defining the PC (Fig. 5, cluster 6), where multipotent progenitors first enter the thymus from the bloodstream, includes multiple genes known to regulate chemotaxis, extravasation, or cell migration (for examples, see Kanda et al., 2008; Yang et al., 2003; Rauch et al., 2005), while the signature that discriminates the SC from other cortical regions (cluster 7) includes genes that regulate classical signals for differentiation, including regulators of Wnt (Tremblay et al., 2009), Shh (Allen et al., 2007), or Nodal signaling (Beck et al., 2002), as well as chemoattractive and chemorepulsive (Toyofuku et al., 2008; Mertsch et al., 2008) signals. Such genes, for which known function in other tissues correlates with regional processes in the thymus, represent high-priority candidates for probing the biology of these stromal regions.
While stromal cells from the central cortex (CC) shared expression of many genes with other cortical subregions and/or the medulla, we find that they did not express any unique genes of their own. This result was unexpected, since this subregion is generally thought to induce several specific developmental processes, including migration out from the PC, lymphoid lineage specification, proliferation, and accessibility of TCR γ, δ, and β loci to enzymes mediating somatic recombination (reviewed in Petrie and Zuniga-Pflucker, 2007). It is important to note that the CC does share with the PC, SC, and/or medulla several key regulators of these processes, including Kitl, IL7, Dll1, Cxcl12, and Vcam1 (Suppl. Table 1), even though it does not appear to have additional genes. This suggests that one of the main functions of the CC may be to ensure that unique microenvironments found in the PC (and/or medulla) remain insulated from those in the SC, and further suggests that the predominant function of the CC may be to modulate the proximity of immature progenitors to signals from distal regions, rather than by direct induction of its own specific processes. Such proximal:distal relationships are important influences in virtually all systems of metazoan development. We cannot rule out the possibility that quantitative, rather than qualitative, differences in gene expression and protein levels may play an important role in establishing different microenvironments, as has been suggested by a similar approach to study the developing kidney (Brunskill et al., 2008). Nonetheless, our findings substantially challenge many of the assumptions that have been made regarding the roles of stratified tissue regions in the control of lymphoid development, and simultaneously facilitate an in depth understanding of the biology of these regions.
Finally, it is worth noting that unlike most solid tissues, the structure of the thymus is somewhat amorphous, with cortical regions sometimes flanked by two or more medullary regions, as well as cortical and/or medullary regions of varying depths and ratios. Of necessity, our approach idealized the structure of the thymus, restricting tissues harvested to those with uniform borders and concentric cortical/medullary regions of defined depths (see Methods). It is possible, if not likely, that regions of the thymus that do not conform to these idealized criteria may be distinct in function as well as structure, and may have different gene expression patterns than those found in more structured regions. While we can only speculate on the function of these atypical regions, it is tempting to consider that they may distort the developmental process in a way that could add to the diversity of distinct T lineages and/or their relative functions.
Experimental Procedures
Mice
Male C57BL6 mice at 4–8 weeks of age were used; procedures were approved by an institutional animal committee.
Isolation of microdissected tissues
Intact thymuses were removed, immediately placed in ice cold mounting medium, and frozen. Transverse 20 μm sections were cut from the middle third of the organ (A/P axis) and mounted on PEN membrane slides (Leica). Sections selected for microdissection were chosen to have cortical regions 400–600 μm deep, medullary regions at least 200 μm deep, and symmetrical cortical/medullary borders. Tissue was fixed in cold acetone/ethanol (3:1 v/v), rehydrated through graded ethanol, stained with cresyl violet (LCM Staining Kit, Ambion), and dehydrated through graded ethanol. Slides were dried at room temperature and immediately used for microdissection on a Leica AS LMD system. Sections adjacent to those used for microdissection were mounted on glass slides for archival purposes. Additional sections 100 μm above and below microdissected sections were also mounted on glass slides and examined to rule out tissue irregularities outside microdissected plane. Cortical subregions were prepared from tissues as described above, with each subregion representing 15% of the total cortical depth. RNA was prepared using RNAqueous Micro kits (Ambion). Each independent RNA pool included tissue from multiple sections and mice.
Isolation of purified lymphoid cells
Thymuses were removed and placed immediately in ice cold medium (Hanks’ balanced salt solution, 5% FBS, 10 g/ml DNAse); all subsequent steps were at 4°C. Single cell suspensions were stained with labeled antibodies (below), followed by cell sorting. Sort gates for medullary lymphoid cells were CD3hi AND CD45+, and for cortical lymphoid cells were CD3−/lo AND CD45+ AND (CD90+ OR CD117+). Dead cells were excluded using DAPI. Cells for microarray analysis were selected to be >99% pure, and were generally >99.8% pure. RNA was prepared using RNeasy Mini Kits (Qiagen) according to recommended protocols.
In vitro transcription, RNA labeling, and array hybridization
These were carried out by the Genomics Core Facility at The Scripps-Florida Research Institute. Briefly, 0.2–1.0 μg of total RNA was used for 3′ in vitro one- (whole tissues or sorted cells) or two- (cortical subregions) cycle target labeling (Affymetrix). Biotin-labeled cRNA probes were prepared as recommended by the manufacturer (Affymetrix), using oligo-dT primers, SuperScript, and BioArray High Yield RNA Transcript kits (Affymetrix). Fragmentation/hybridization to MOE430v2.0 arrays (Affymetrix) were performed according to Affymetrix protocols. Imaging was performed on a Hewlett Packard GeneArray Scanner. MIAME-compliant raw data has been deposited into the NCBI Gene Expression Omnibus database (accession number pending).
Unsupervised clustering of microarray results
A list of all genes defined as being Present in at least one tissue region (see Results) was generated. The Pearson correlation similarity measure and average linkage algorithm in GeneSpring 7.3.1 (Agilent) was used to identify relative similarities in gene expression profiles.
Signal-independent identification of stromal-specific genes
Detection calls (Present, Marginal, Absent) were calculated using the MAS5 algorithm in Affymetrix Expression Console software (v.1.0). This algorithm calculates the statistical significance (Wilcoxon’s Signed Rank test) of differences between matched/mismatched probes in each probeset, and assigns a detection call of P to those where p < 0.05, M where 0.05 ≤ p < 0.065, and A where p ≥ 0.065. For medulla, stromal-specific genes received P calls for at least 6/7 independent tissue results and A calls at least 4/5 lymphoid results. For cortical sub-regions, where 3 chips were used for each region, stromal specific genes received 3/3 P calls in tissue and 4/5 A calls in the corresponding lymphocytes.
Signal-based identification of stromal-specific genes
Mean quantile-normalized (PLIER) signals for each group of biological replicates were calculated for each probeset on the chip. Default parameters in the PM-MM PLIER algorithm in the Affymetrix Expression Console (v1.0) were used. In the case of total cortical or medullary tissues or cells, where 5 to 7 gene chip experiments were performed, the single-most outlying value (largest Z score) was removed for each probeset. As described in Results, the 25th percentile was used as the threshold for genes present in microdissected tissue; noise induced by this low threshold was offset by the implementation of other processing criteria, including a requirement that the lymphoid signal be below the median value (i.e., absent), that the differences in normalized signals between tissue and lymphoid samples be statistically significant (Student’s two-tailed t test p value < 0.05), and that the normalized tissue signal be greater than that of lymphoid cells.
Assessment of enrichment for predicted functions
Four lists were analyzed using the NIAID’s web-based tool DAVID (david.abcc.ncicrf.gov) (Dennis et al., 2003; Huang da et al., 2009): cortical stromal MFP, medullary stromal MFP (both as defined above), and similar lists for cortical or medullary lymphoid cells, representing all probesets called Present in 4/5 lymphoid experiments AND lymphoid signal > corresponding tissue signal AND Student’s t-test p value for tissue:lymphoid differences < 0.05). Statistical p values were calculated in GeneSpring 7.3.1. Multiple testing correction was set to Benjamini and Hochberg. Each list of probesets was uploaded into DAVID and analyzed using the Functional Annotation Tool. The background was set to the MOE430 2.0 chip and species was set to Mus musculus. Within the “chart” function for Gene Ontologies (All Biological Processes) and Kegg Pathways, the “options” feature was used to include fold enrichments. Also within the “options” feature the EASE score (modified Fisher’s exact p value) threshold was set to 1 and gene count threshold was set to 0, in order to obtain results for all terms in all four lists. Gene Ontology and Kegg pathway results, including enrichment and EASE scores, were downloaded as tab-delimited text files for each of the four lists using the “download file” feature. The text files were imported into Excel in order to compare results for terms or pathways across each each of the four lists. Results for terms or pathways expected to be enriched in either stromal or lymphoid lists were evaluated and enrichment values for some representative terms and pathways are charted for each of the lists in figure 4. All terms charted were also statistically over-represented (EASE score < 0.05) in at least one compartment (cortex or medulla) in the expected tissue type (stroma or lymphoid).
Identification of differentially expressed stromal-specific genes
Per gene normalized values (median = 1) for each probeset in the minimum false positive list were generated using the MAS5 algorithm (Affymetrix Expression Console). Signals that changed at least 2-fold between any two regions and were statistically different (Student’s t-test p value < 0.05) were identified. The k-means clustering algorithm in GeneSpring GX 7.3.1 (100 iterations, Pearson correlation similarity measure) was used to assign patterns of expression in the context of thymic space.
Calculation of global stromal gene expression
As described in Results, global stromal gene expression can be calculated by proportionally removing lymphoid gene expression from tissue results. However, several factors (discussed in Results and Gosink et al., 2007) induce distortion that makes precise calculation of the proportion of lymphoid signal (Plymphoid) impractical. Consequently, three independent methods were used to estimate Plymphoid. The first two relied on identification of the inflection point of the deformed (sigmoid) curve generated by plotting rank-ordered tissue:lymphoid ratios (see Fig. 6B). The first of these assumes that the y-axis value of the inflection point represents the true Plymphoid, while the second assumes that the y-intercept of a line tangent to the inflection point is the actual Plymphoid. To find the inflection point, a third-order polynomial equation (cubic spline, least-squares) was fitted to the data using Prism 5.0 (GraphPad, La Jolla, CA). To most accurately define the curve in the area of the inflection point, probesets in the top 5th percentile of lymphoid signals, or those with tissue:lymphoid ratios ≥ 2, were removed prior to modeling the equation. The x-axis value of the inflection point (xi) was calculated by setting the second derivative of the polynomial regression equation to zero, and the y-axis value of the inflection point (yi, an estimate of Plymphoid) was determined by substituting xi into the regression equation and solving for y. To determine the y-intercept (b) of a line tangent to the inflection point of the regression curve, the slope of the tangent (m) was calculated by solving the first derivative of the regression equation described above, and then using the standard equation for slope and intercept: yi = mxi + b. The third method for estimating Plymphoid involved using the average tissue:lymphoid ratio for a set of genes generally accepted to be lymphoid-specific (see Suppl. Table 2).
To evaluate the major differences between cortical and medullary pathways, global stromal gene expression was calculated for each compartment using the middle (lymphoid-specific gene ratio) Plymphoid value (0.88 for cortex, 0.70 for medulla). Signal ratios were then determined for each probeset, and genes corresponding to the top 10% of ratios for cortex or medulla were mapped onto KEGG pathways (www.genome.jp/kegg) using the DAVID Bioinformatic Resource (/david.abcc.ncifcrf.gov, Dennis et al., 2003; Huang da et al., 2009). Enrichment was considered to be significant with an EASE score (modified Fisher’s exact p value, Hosack et al., 2003) of <0.05. The entire gene chip was used as the background. Heat mapping was carried out using GeneSpring GX 7.3.1 and displays enrichment scores (negative log10 of the enrichment p value) using a blue-yellow scale, where blue corresponds to least significant and yellow to most significant.
Supplementary Material
RNA from microdissected cortical subregions (as indicated) was used to probe microarrays, and probesets defined as being Present in at least one microdissected tissue region (see Methods) were subjected to two dimensional hierarchical clustering (vertical hierarchy not shown). Black = highest normalized expression. Whereas the normalized range for all tissues was dominated by medullary stromal cells (see Fig. 3), in the absence of medullary results, clusters of region-specific gene expression in the cortex can be easily visualized.
Acknowledgments
This work was supported by PHS grants AI033940, AI067453, and AG031576 (to HTP). The authors declare no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen BL, Tenzen T, McMahon AP. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007;21:1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Beck S, Le Good JA, Guzman M, Ben Haim N, Roy K, Beermann F, Constam DB. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat Cell Biol. 2002;4:981–985. doi: 10.1038/ncb890. [DOI] [PubMed] [Google Scholar]
- Bevan MJ. In a radiation chimaera, host H-2 antigens determine immune responsiveness of donor cytotoxic cells. Nature. 1977;269:417–418. doi: 10.1038/269417a0. [DOI] [PubMed] [Google Scholar]
- Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ. Continued maturation of thymic emigrants in the periphery. Nat Immunol. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- Boyd RL, Tucek CL, Godfrey DI, Izon DJ, Wilson TJ, Davidson NJ, Bean AG, Ladyman HM, Ritter MA, Hugo P. The thymic microenvironment. Immunol Today. 1993;14:445–459. doi: 10.1016/0167-5699(93)90248-J. [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Yu J, Grimmond S, McMahon AP, Patterson LT, Little MH, Potter SS. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, Peltonen L, Walter J, Kyewski B. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley EC, Petrie HT, Shah LM, Owen MJ, Hayday AC. T cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity. 1994;1:83–93. doi: 10.1016/1074-7613(94)90102-3. [DOI] [PubMed] [Google Scholar]
- Dyall R, Nikolic-Zugic J. The majority of postselection CD4+ single-positive thymocytes requires the thymus to produce long-lived, functional T cells. J Exp Med. 1995;181:235–245. doi: 10.1084/jem.181.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss DL, Donskoy E, Goldschneider I. The importation of hematogenous precursors by the thymus is a gated phenomenon in normal adult mice. J Exp Med. 2001;193:365–374. doi: 10.1084/jem.193.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor MJ, Godfrey DI, Scollay R. Recent thymic emigrants are distinct from most medullary thymocytes. Eur J Immunol. 1997;27:2010–2015. doi: 10.1002/eji.1830270827. [DOI] [PubMed] [Google Scholar]
- Goldschneider I, Komschlies KL, Greiner DL. Studies of thymocytopoiesis in rats and mice. I. Kinetics of appearance of thymocytes using a direct intrathymic adoptive transfer assay for thymocyte precursors. J Exp Med. 1986;163:1–17. doi: 10.1084/jem.163.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink MM, Petrie HT, Tsinoremas NF. Electronically subtracting expression patterns from a mixed cell population. Bioinformatics. 2007;23:3328–3334. doi: 10.1093/bioinformatics/btm508. [DOI] [PubMed] [Google Scholar]
- Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J Exp Med. 2004;199:155–166. doi: 10.1084/jem.20031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groettrup M, Ungewiss K, Azogui O, Palacios R, Owen MJ, Hayday AC, von Boehmer H. A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor beta chain and a 33 kd glycoprotein. Cell. 1993;75:283–294. doi: 10.1016/0092-8674(93)80070-u. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Johnnidis JB, Venanzi ES, Taxman DJ, Ting JP, Benoist CO, Mathis DJ. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci U S A. 2005;102:7233–7238. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda H, Newton R, Klein R, Morita Y, Gunn MD, Rosen SD. Autotaxin, an ectoenzyme that produces lysophosphatidic acid, promotes the entry of lymphocytes into secondary lymphoid organs. Nat Immunol. 2008;9:415–423. doi: 10.1038/ni1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- Kyewski B, Derbinski J. Self-representation in the thymus: an extended view. Nat Rev Immunol. 2004;4:688–698. doi: 10.1038/nri1436. [DOI] [PubMed] [Google Scholar]
- Ladi E, Yin X, Chtanova T, Robey EA. Thymic microenvironments for T cell differentiation and selection. Nat Immunol. 2006;7:338–343. doi: 10.1038/ni1323. [DOI] [PubMed] [Google Scholar]
- Li J, Iwanami N, Hoa VQ, Furutani-Seiki M, Takahama Y. Noninvasive intravital imaging of thymocyte dynamics in medaka. J Immunol. 2007;179:1605–1615. doi: 10.4049/jimmunol.179.3.1605. [DOI] [PubMed] [Google Scholar]
- Lind EF, Prockop SE, Porritt HE, Petrie HT. Mapping precursor movement through the postnatal thymus reveals specific microenvironments supporting defined stages of early lymphoid development. J Exp Med. 2001;194:127–134. doi: 10.1084/jem.194.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertsch S, Schmitz N, Jeibmann A, Geng JG, Paulus W, Senner V. Slit2 involvement in glioma cell migration is mediated by Robo1 receptor. J Neurooncol. 2008;87:1–7. doi: 10.1007/s11060-007-9484-2. [DOI] [PubMed] [Google Scholar]
- Nikolic-Zugic J, Moore MW, Bevan MJ. Characterization of the subset of immature thymocytes which can undergo rapid in vitro differentiation. Eur J Immunol. 1989;19:649–653. doi: 10.1002/eji.1830190412. [DOI] [PubMed] [Google Scholar]
- Penit C. Localization and phenotype of cycling and post-cycling murine thymocytes studied by simultaneous detection of bromodeoxyuridine and surface antigens. J Histochem Cytochem. 1988;36:473–478. doi: 10.1177/36.5.2895787. [DOI] [PubMed] [Google Scholar]
- Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, Meyer JD, Davison BL. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie HT, Livak F, Burtrum D, Mazel S. T cell receptor gene recombination patterns and mechanisms: cell death, rescue, and T cell production. J Exp Med. 1995;182:121–127. doi: 10.1084/jem.182.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie HT, Pearse M, Scollay R, Shortman K. Development of immature thymocytes: initiation of CD3, CD4, and CD8 acquisition parallels down-regulation of the interleukin 2 receptor alpha chain. Eur J Immunol. 1990;20:2813–2815. doi: 10.1002/eji.1830201243. [DOI] [PubMed] [Google Scholar]
- Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- Popa I, Zubkova I, Medvedovic M, Romantseva T, Mostowski H, Boyd R, Zaitseva M. Regeneration of the adult thymus is preceded by the expansion of K5+K8+ epithelial cell progenitors and by increased expression of Trp63, cMyc and Tcf3 transcription factors in the thymic stroma. Int Immunol. 2007;19:1249–1260. doi: 10.1093/intimm/dxm092. [DOI] [PubMed] [Google Scholar]
- Porritt HE, Gordon K, Petrie HT. Kinetics of steady-state differentiation and mapping of intrathymic-signaling environments by stem cell transplantation in nonirradiated mice. J Exp Med. 2003;198:957–962. doi: 10.1084/jem.20030837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prockop SE, Palencia S, Ryan CM, Gordon K, Gray D, Petrie HT. Stromal cells provide the matrix for migration of early lymphoid progenitors through the thymic cortex. J Immunol. 2002;169:4354–4361. doi: 10.4049/jimmunol.169.8.4354. [DOI] [PubMed] [Google Scholar]
- Prockop SE, Petrie HT. Regulation of thymus size by competition for stromal niches among early T cell progenitors. J Immunol. 2004;173:1604–1611. doi: 10.4049/jimmunol.173.3.1604. [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Rauch BH, Millette E, Kenagy RD, Daum G, Fischer JW, Clowes AW. Syndecan-4 is required for thrombin-induced migration and proliferation in human vascular smooth muscle cells. J Biol Chem. 2005;280:17507–17511. doi: 10.1074/jbc.M410848200. [DOI] [PubMed] [Google Scholar]
- Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, Weinmaster G, Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- Rodewald HR, Kretzschmar K, Swat W, Takeda S. Intrathymically expressed c-kit ligand (stem cell factor) is a major factor driving expansion of very immature thymocytes in vivo. Immunity. 1995;3:313–319. doi: 10.1016/1074-7613(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Shortman K, Egerton M, Spangrude GJ, Scollay R. The generation and fate of thymocytes. Semin Immunol. 1990;2:3–12. [PubMed] [Google Scholar]
- Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4− versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Teh HS, Kisielow P, Scott B, Kishi H, Uematsu Y, Bluthmann H, von Boehmer H. Thymic major histocompatibility complex antigens and the alpha beta T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature. 1988;335:229–233. doi: 10.1038/335229a0. [DOI] [PubMed] [Google Scholar]
- Toyofuku T, Yoshida J, Sugimoto T, Yamamoto M, Makino N, Takamatsu H, Takegahara N, Suto F, Hori M, Fujisawa H, Kumanogoh A, Kikutani H. Repulsive and attractive semaphorins cooperate to direct the navigation of cardiac neural crest cells. Dev Biol. 2008;321:251–262. doi: 10.1016/j.ydbio.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Tremblay F, Revett T, Huard C, Zhang Y, Tobin JF, Martinez RV, Gimeno RE. Bidirectional modulation of adipogenesis by the secreted protein Ccdc80/DRO1/URB. J Biol Chem. 2009;284:8136–8147. doi: 10.1074/jbc.M809535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Saito F, Gray DH, Kuse S, Hieshima K, Nakano H, Kakiuchi T, Lipp M, Boyd RL, Takahama Y. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J Exp Med. 2004;200:493–505. doi: 10.1084/jem.20040643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ewijk W, Shores EW, Singer A. Crosstalk in the mouse thymus. Immunol Today. 1994;15:214–217. doi: 10.1016/0167-5699(94)90246-1. [DOI] [PubMed] [Google Scholar]
- Wallis VJ, Leuchars E, Chwalinski S, Davies AJ. On the sparse seeding of bone marrow and thymus in radiation chimaeras. Transplantation. 1975;19:2–11. doi: 10.1097/00007890-197501000-00002. [DOI] [PubMed] [Google Scholar]
- Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- Witt CM, Raychaudhuri S, Schaefer B, Chakraborty AK, Robey EA. Directed migration of positively selected thymocytes visualized in real time. PLoS Biol. 2005;3:e160. doi: 10.1371/journal.pbio.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan XQ, Sarmiento U, Sun Y, Huang G, Guo J, Juan T, Van G, Qi MY, Scully S, Senaldi G, Fletcher FA. A novel Notch ligand, Dll4, induces T-cell leukemia/lymphoma when overexpressed in mice by retroviral-mediated gene transfer. Blood. 2001;98:3793–3799. doi: 10.1182/blood.v98.13.3793. [DOI] [PubMed] [Google Scholar]
- Yang D, Rosenberg HF, Chen Q, Dyer KD, Kurosaka K, Oppenheim JJ. Eosinophil-derived neurotoxin (EDN), an antimicrobial protein with chemotactic activities for dendritic cells. Blood. 2003;102:3396–3403. doi: 10.1182/blood-2003-01-0151. [DOI] [PubMed] [Google Scholar]
- Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Zinkernagel RM, Callahan GN, Althage A, Cooper S, Klein PA, Klein J. On the thymus in the differentiation of “H-2 self-recognition” by T cells: evidence for dual recognition? J Exp Med. 1978;147:882–896. doi: 10.1084/jem.147.3.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotoff DA, Schwarz BA, Bhandoola A. The long road to the thymus: the generation, mobilization, and circulation of T-cell progenitors in mouse and man. Semin Immunopathol. 2008;30:371–382. doi: 10.1007/s00281-008-0133-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA from microdissected cortical subregions (as indicated) was used to probe microarrays, and probesets defined as being Present in at least one microdissected tissue region (see Methods) were subjected to two dimensional hierarchical clustering (vertical hierarchy not shown). Black = highest normalized expression. Whereas the normalized range for all tissues was dominated by medullary stromal cells (see Fig. 3), in the absence of medullary results, clusters of region-specific gene expression in the cortex can be easily visualized.