Summary
In response to DNA double-strand breaks (DSBs), cells sense the DNA lesions and then activate the protein kinase ATM. Subsequent DSB resection produces RPA-coated ssDNA that is essential for activation of the DNA damage checkpoint and DNA repair by homologous recombination (HR). However, the biochemical mechanism underlying the transition from DSB sensing to resection remains unclear. Using Xenopus egg extracts and human cells we show that the tumor suppressor protein CtIP plays a critical role in this transition. We find that CtIP translocates to DSBs, which is dependent on the DSB sensor complex Mre11-Rad50-NBS1, the kinase activity of ATM and a direct DNA-binding motif in CtIP, and then promotes DSB resection. Thus, CtIP facilitates the transition from DSB sensing to processing: It does so by binding to the DNA at DSBs after DSB sensing and ATM activation, and then promoting DNA resection leading to checkpoint activation and HR.
Introduction
The ability of cells to detect and properly repair DNA DSBs is essential for maintaining genome stability and preventing cancer (Kastan and Bartek, 2004). DSBs are generated by ionizing radiation, perturbations of DNA replication and many cancer therapeutic drugs. Eukaryotic cells have evolved a highly conserved checkpoint mechanism to sense and signal the existence of DSBs (Harper and Elledge, 2007). Central to the DSB checkpoint response are the ATM/ATR protein kinases, which receive signals from DNA damage sensors and then undergo activation (Cimprich and Cortez, 2008; Lee and Paull, 2007). Following activation, ATM and ATR phosphorylate a variety of downstream substrates to initiate both local responses at damage sites, such as chromatin modification and DNA repair, and global responses, such as cell cycle arrest, transcriptional changes and apoptosis (Kurz and Lees-Miller, 2004; Matsuoka et al., 2007). Although ATM and ATR are related kinases and share many substrates, their activation requires different DNA structures at DSBs. While ATM activation occurs on the DNA/chromatin flanking DNA ends and does not require DNA end resection (You et al., 2007), ATR is activated on RPA-coated ssDNA that is generated by 5’ > 3’ DSB resection (Zou and Elledge, 2003).
In addition to activation of ATR, the ssDNA structure generated by DSB resection is also required for formation of the Rad51-ssDNA filament, a key intermediate required for homology probing and D-loop formation during HR repair (Sung and Klein, 2006). Despite its importance, however, the biochemical mechanism and regulation of DSB resection remain poorly understood. In Saccharomyces cerevisiae, genetic studies suggest a 2-step model for DSB resection. In this model, the Sae2 protein and the Mre11-Rad50-Xrs2 (MRX) complex (the counterpart of MRN in metazoans) carry out the initial stage of resection to remove a few hundred nucleotides of the 5’ strand DNA from a DSB end. Subsequently, the Exo1 exonuclease and the Sgs1/DNA2 helicase/exonuclease act redundantly to further resect 5’ strand DNA to generate long 3’ ssDNA tails (Budd and Campbell, 2009; Mimitou and Symington, 2008; Zhu et al., 2008). Whether a similar DSB resection model applies in metazoans remains to be determined.
Recent studies suggest that the transcriptional regulator/tumor suppressor CtIP in mammals is the functional counterpart of Sae2, despite the very limited sequence similarity between these proteins. Human CtIP is recruited to DNA damage sites and is required for formation of RPA foci at DSBs (indicative of ssDNA generation), and subsequent Chk1 phosphorylation by ATR and HR in response to DSB-inducing agents (Chen et al., 2008; Greenberg et al., 2006; Huertas and Jackson, 2009; Sartori et al., 2007; Yu and Chen, 2004; Yuan and Chen, 2009). Studies on the putative CtIP homologs in S. pombe (Ctp1), C. elegans (COM-1), A. thaliana (AtGR1) and chicken support a role for these proteins in DSB resection (Akamatsu et al., 2008; Limbo et al., 2007; Penkner et al., 2007; Uanschou et al., 2007; Yun and Hiom, 2009). Like Sae2 and Ctp1, CtIP apparently also functions in cooperation with the MRN complex in DSB resection (Chen et al., 2008; Lloyd et al., 2009; Sartori et al., 2007; Williams et al., 2009). In addition to its direct involvement in DSB resection, the MRN complex plays a role in DSB sensing and activation of ATM (Difilippantonio and Nussenzweig, 2007). Interestingly, ATM kinase activity is also required for efficient DSB resection and subsequent activation of ATR (Adams et al., 2006; Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006). These findings indicate that MRN and ATM promote the transition from DSB sensing to DNA end processing, although their exact role in this transition is not clear. In this report we have investigated the role of CtIP in the DSB damage response in both Xenopus egg extracts and human cells. Our results reveal that CtIP functions downstream of MRN and ATM and plays a critical role in facilitating the transition from DNA damage sensing to DNA end processing, and linking ATM activation and ATR activation in response to DSBs.
Results
CtIP is required for DSB resection
Depletion of CtIP in mammalian cells using siRNAs abrogates formation of RPA foci, phosphorylation of Chk1 by ATR and DNA repair by HR in response to DSBs, suggesting that CtIP is required for DSB resection (Chen et al., 2008; Greenberg et al., 2006; Huertas and Jackson, 2009; Sartori et al., 2007; Yu and Chen, 2004; Yuan and Chen, 2009). However, we found that CtIP knockdown in U2OS and HeLa cells using a small hairpin RNA (shRNA) caused low levels of chronic ATM activation and reduced cell viability in the absence of exogenous DNA damage (Fig. S1A–C and data not shown). In addition, CtIP is known to regulate gene transcription and Rb-dependent G1/S control in mammalian cells (Liu and Lee, 2006; Wu and Lee, 2006). To investigate the role of CtIP specifically in the DNA damage response, without the interference of cellular transcription or loss of viability, we have characterized CtIP function in the cell-free Xenopus egg extract system that lacks transcription and Rb-dependent G1/S control. By searching EST databases we identified Xenopus CtIP (xCtIP) (NCBI access number: GU207840), which has 45% identity overall with human CtIP, with greater identity in the N-terminal dimerization (65%) and C-terminal “Sae2-like” (81%) domains. Like human CtIP, xCtIP also has central a “PLDLS” CtBP-binding motif (Schaeper et al., 1998) (Fig. 1A).
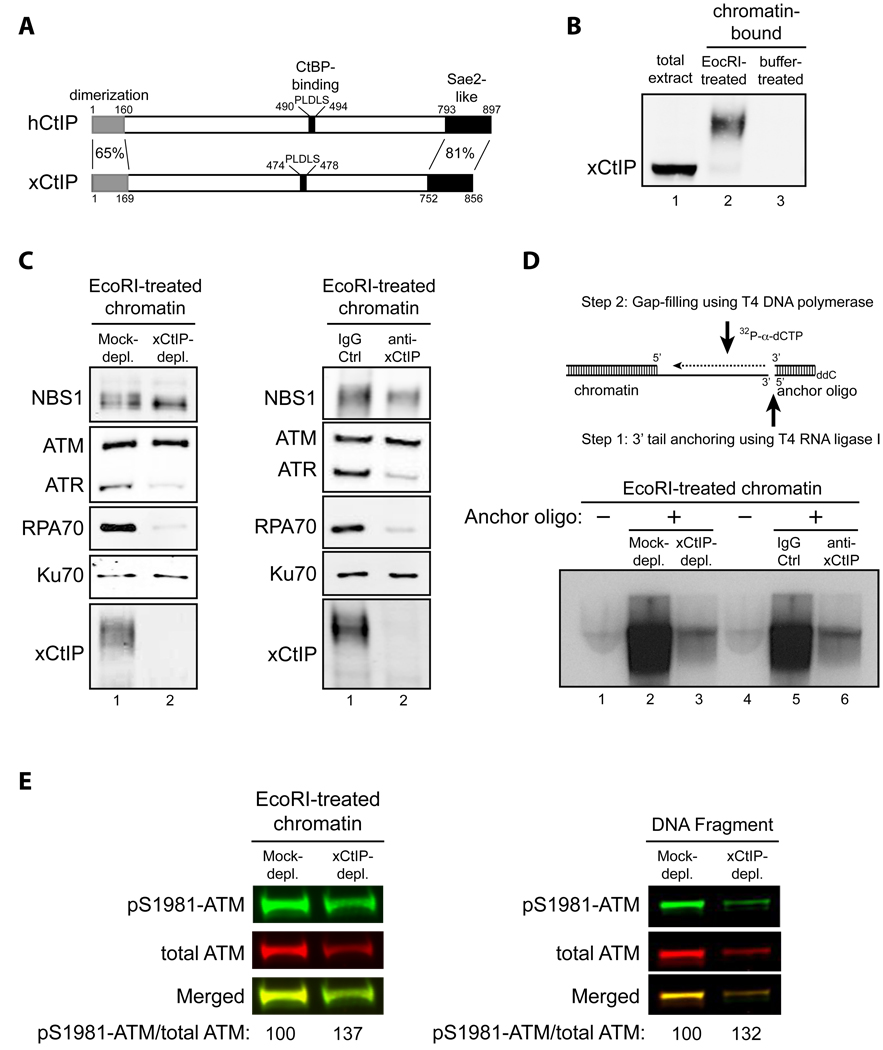
Fig. 1. CtIP is required for DNA end resection, but not ATM activation, in response to DSBs in Xenopus extracts.
- Schematic comparison of human CtIP and Xenopus CtIP (xCtIP).
- xCtIP associated with damaged, but not undamaged, chromatin in the Xenopus extract, and chromatin-associated xCtIP was highly modified as evidenced by its reduced gel mobility. Xenopus extract (20 µl) was incubated with 10,000 EcoRI-treated or buffer-treated demembranated sperm nuclei/µl for 30 min. After incubation, chromatin was isolated and chromatin-associated xCtIP was detected by immunoblotting (lanes 2 and 3). Lane 1, xCtIP in 1 µl of total extract.
- xCtIP is required for association of RPA and ATR, but not NBS1, ATM and Ku70, with chromatin containing DSBs. Left panel: EcoRI-treated sperm chromatin was incubated with mock-depleted or xCtIP-depleted extract for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting. xCtIP depletion blocked chromatin association of RPA70 and ATR, but not NBS1, ATM and Ku70. Right panel: Extract supplemented with 400 ng/µl of control IgG or affinity-purified, neutralizing xCtIP antibodies was incubated with EcoRI-treated sperm chromatin for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting. Addition of xCtIP antibodies inhibited chromatin association of xCtIP, RPA and ATR, but not NBS1, ATM and Ku70.
- xCtIP is required for efficient DSB end resection in the Xenopus extract. Upper panel: A procedure for detecting DSB end resection in the Xenopus extract. The 3’ tail generated by 5’ strand resection of a DNA DSB in chromatin is ligated to the 5’ phosphorylated end of an anchor oligo using T4 RNA ligase I. After ligation, a second, complementary oligo is annealed to the anchor oligo and T4 DNA polymerase is used to fill the gap between the 3’ end of the second oligo and the 5’ resected end in chromatin in the presence of 32P-α-dCTP (see Experimental Procedures in Supplemental Information for details). The amount of 32P-α-dCTP incorporation represents the extent of DSB resection. Lower panels: EcoRI-treated sperm chromatin was incubated with mock-depleted (lane 2) or xCtIP-depleted extract (lane 3), or with extract supplemented with 400 ng/µl of control IgG (lane 5) or neutralizing xCtIP antibodies (lane 6) for 30 min. After incubation, chromatin was isolated and DSB resection was analyzed on agarose gels after oligo anchoring and gap filling in the presence of 32P-α-dCTP. The anchor oligo was omitted from the control lanes 1 and 4.
- Immunodepletion of xCtIP from the Xenopus extract increased the extent of ATM autophosphorylation at S1981 induced by EcoRI-treated sperm chromatin or by dsDNA fragments, although xCtIP depletion partially removed ATM from the extract. Mock-depleted or xCtIP-depleted extract was incubated with 10,000 sperm nuclei/µl (left panel) or 5 ng/µl of a 2 kbp dsDNA fragment (right panel) for 25 min. After incubation, immunoblotting was performed to detect activated ATM and total ATM in the extracts with rabbit phospho-S1981 specific anti-ATM antibodies and guinea pig anti-Xenopus ATM antibodies, respectively. Primary antibodies were detected by Alexa Fluor 680-labeled anti-rabbit secondary antibodies and IRdye 800-labeled anti-guinea pig second antibodies using Odyssey infrared imaging system. Immunoblotting signals were quantified using Odyssey 3.0 software.
To determine whether xCtIP is involved in DSB resection in the Xenopus extract, we first examined whether xCtIP binds to chromatin in the extract after DSB damage. To this end, we generated antibodies directed against the C-terminal 236 amino acids of xCtIP. Immunoblotting with these antibodies showed that xCtIP is present in the Xenopus extract (Fig. 1B). To assess association of xCtIP with DSBs in the extract, we used EcoRI-treated sperm chromatin, which activates the DSB damage response when added to the extract (Fig. S1D) (You et al., 2005). After incubating damaged chromatin with the extract, we isolated the chromatin fraction through a sucrose cushion and detected chromatin-associated proteins by immunoblotting. xCtIP was indeed associated with damaged chromatin (Fig. 1B), consistent with CtIP recruitment to DSBs in human cells (Chen et al., 2008; Sartori et al., 2007; Yu et al., 2006; Yuan and Chen, 2009). No association of xCtIP with intact chromatin was observed (Fig. 1B). Chromatin-bound xCtIP was highly modified, as evidenced by its reduced gel mobility, which is at least partially due to phosphorylation (Fig. 1B and Fig. S1E).
To determine if xCtIP is required for DSB resection in the Xenopus extract, EcoRI-treated sperm chromatin was incubated with mock- or xCtIP-depleted extract. The chromatin fraction was then isolated from the extract and DSB resection was assessed indirectly by measuring the association of the ssDNA-binding protein RPA with damaged chromatin (Sartori et al., 2007). In the mock-depleted extract, RPA was efficiently recruited to damaged chromatin within 30 min. In contrast, RPA chromatin binding was abolished in the xCtIP-depleted extract (Fig. 1C, left panel), suggesting that xCtIP is indeed required for DSB resection. Consistent with this idea, chromatin association of ATR, which is recruited to and activated on RPA-coated ssDNA (Cimprich and Cortez, 2008; Zou and Elledge, 2003), was also abrogated in the xCtIP-depleted extract (Fig. 1C, left panel). Depletion of xCtIP also removed ~ 50% of ATM protein from the extract due to association of ATM with xCtIP (Fig. S1F). However, the decreased ATM concentration cannot account for the inhibitory effects of xCtIP depletion on chromatin binding of RPA and ATR in the xCtIP-depleted extract, since removal of 50% of ATM protein from the extract using ATM antibodies did not significantly inhibit chromatin binding of RPA and ATR (Fig. S1G). Moreover, addition of affinity-purified xCtIP neutralizing antibodies to extract also inhibited binding of RPA and ATR to damaged chromatin (Fig. 1C, right panel), further indicating that the effects of xCtIP depletion on DSB resection were not caused by removal of an xCtIP-associated protein, such as ATM. In contrast to RPA and ATR, association of ATM and the MRN subunit NBS1 with damaged chromatin was not inhibited by depletion of xCtIP or by addition of xCtIP neutralizing antibodies (Fig. 1C). Chromatin association of Ku70, a protein involved in DSB repair by nonhomologous end-joining that does not require DSB resection, also did not require xCtIP (Fig. 1C).
To further demonstrate the requirement of xCtIP for DSB resection, we developed a gap-filling based assay to directly assess DSB resection in the Xenopus extract. In this method, EcoRI-treated sperm chromatin incubated with the extract was isolated through a sucrose cushion. Subsequently, a 5’ phosphorylated ssDNA oligo (anchor oligo) was ligated to the 3’ ssDNA tails in chromatin produced by DNA resection using T4 RNA ligase I (a single-stranded RNA/DNA ligase). A second oligo complementary to the anchor oligo was added to create a ssDNA gap between the 3’ terminus of the second oligo and the 5’ terminus of a resected DSB, and T4 DNA polymerase was then used to fill the gap in the presence of 32P-α-dCTP (Fig. 1D, see Experimental Procedures in Supplementary Information for details). The level of 32P-α-dCTP incorporation represents the extent of DSB resection. Using this assay we found that disruption of the function of xCtIP in the Xenopus extract by immunodepletion or by addition of neutralizing antibodies greatly inhibited DSB resection (Fig. 1D). Our results in Xenopus egg extracts are consistent with those obtained in mammalian cells and support the notion that CtIP is required for efficient DSB resection.
CtIP is not required for ATM activation
ATM kinase activity is also required for efficient DSB resection in human cells (Adams et al., 2006; Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006), although its exact role in DSB resection is not clear. Interestingly, ATM interacts with CtIP in human cells (Li et al., 2000) and the Xenopus egg extract (Fig. S1F), raising the possibility that the role of CtIP in DSB resection is to promote activation of ATM. To test this, we immunodepleted xCtIP from the extract and examined ATM autophosphorylation at S1981 (a widely used marker for ATM activation (Bakkenist and Kastan, 2003)) in response to damaged chromatin or dsDNA fragments added to the extract. Although the total ATM protein level was reduced in the xCtIP-depleted extract due to the ATM-xCtIP interaction, the extent of ATM autophosphorylation (i.e. the pS1981-ATM/total ATM ratio) induced by DSBs on the unbound ATM was not reduced, but actually slightly increased (Fig. 1E and Fig. S1F). Addition of xCtIP neutralizing antibodies to the extract also had no effect on the extent of ATM autophosphorylation induced by damaged chromatin or DNA fragments (data not shown). Furthermore, disruption of xCtIP function by immunodepletion or adding neutralizing antibodies did not inhibit association of ATM with damaged chromatin, a prerequisite for ATM activation (Fig. 1C) (You et al., 2007). Together these results indicate that xCtIP and DNA end resection are not required for ATM activation in response to DSBs.
CtIP is localized to DSB end-proximal regions
In mammalian cells CtIP is recruited to DNA damage sites induced by IR or laser irradiation (Chen et al., 2008; Sartori et al., 2007; Yu et al., 2006; Yuan and Chen, 2009). In the Xenopus extract xCtIP was also associated with DNA fragments or chromatin containing DSBs (Fig. 1B and 1C, and data not shown). Blocking chromatin binding of xCtIP by adding CtIP neutralizing antibodies also inhibited DSB resection (Fig. 1C). These results suggest that CtIP may be directly involved in DNA resection at DSBs. To test this idea we performed chromatin immunoprecipitation (ChIP) experiments on a 7.4 kbp linearized plasmid DNA fragment incubated with the Xenopus extract to determine whether xCtIP is localized to DNA end-proximal regions at a DSB. xCtIP indeed preferentially bound at a DNA end-proximal region (~100 bp away from the DSB end), although a low level of binding was detected ~1.8 kbp away from the DSB end. NBS1 and Ku70 were also enriched at the DNA end-proximal region (Fig. 2A) (You et al., 2007). Like NBS1 and Ku70, xCtIP did not associate with plasmid DNA without DSBs in the extract (Fig. S2A).
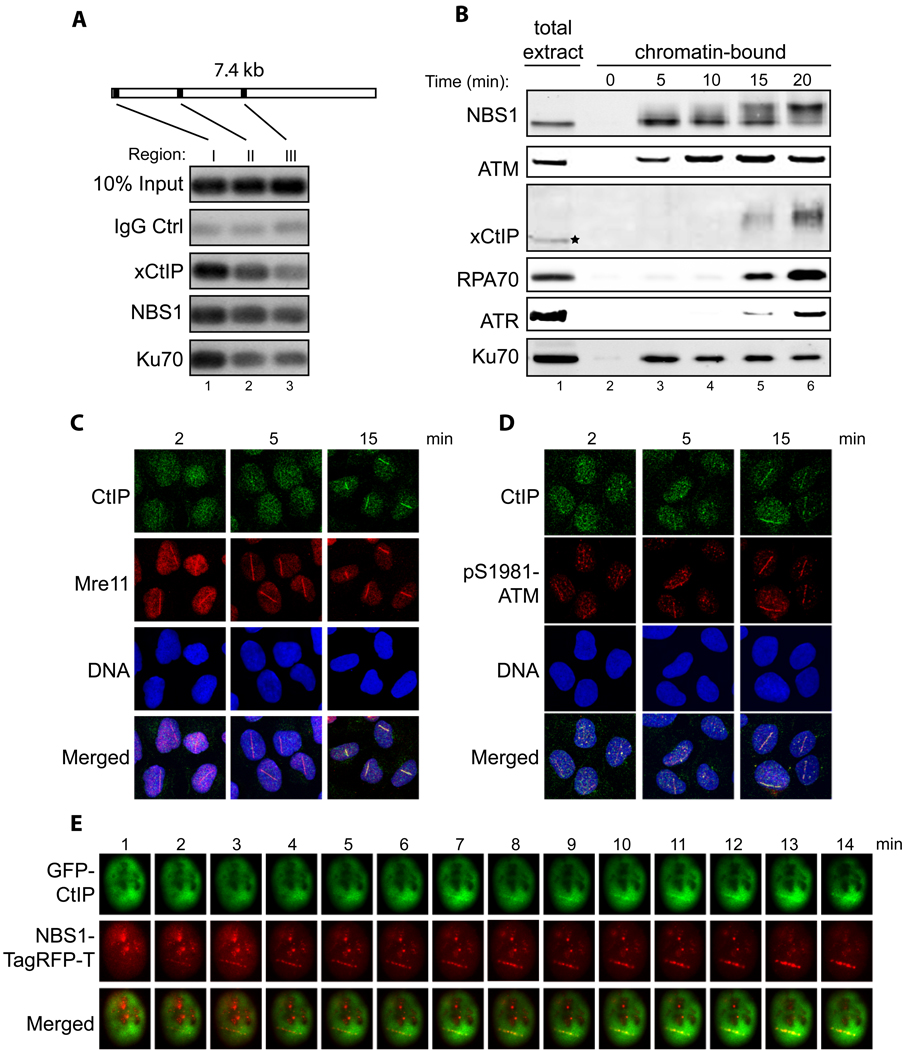
Fig. 2. Spatial and temporal regulation of CtIP recruitment to DNA damage sites.
- Association of xCtIP, NBS1 and Ku70 with different regions of a DNA fragment incubated in the Xenopus extract. A 7.4 kbp linearized plasmid (5 ng/µl) was incubated with the extract for 15 min. After incubation, ChIP experiments were performed to assess the association of proteins at three representative regions of the DNA fragment: 97–298 bp (region I), 1857–2060 (region II) and 3711–3920 (region III).
- Time-course analysis of chromatin association of NBS1, ATM, xCtIP, RPA70, ATR and Ku70 in the Xenopus extract in response to DSBs. EcoRI-treated chromatin incubated in the Xenopus extract was isolated at the indicated times and associated proteins were detected by immunoblotting. Signal in lane 1 represents NBS1, ATM, RPA70, ATR or Ku70 in 0.5 µl of total extract, or xCtIP in 0.1 µl of total extract.
- Accumulation of CtIP and Mre11 at DNA damage sites in human cells at different times after DNA damage. Laser irradiation was performed to generate DSBs in a line pattern in human U2OS cells. Subsequently, cells were fixed at the indicated times followed by indirect immunofluorescence to assess the recruitment of CtIP and Mre11 to DNA damage sites. DNA was stained with DAPI.
- Same as depicted in panel C, except that the accumulation of CtIP and pS1981-ATM at DNA damage sites was assessed at the indicated time points after laser irradiation.
- Laser irradiation was performed in a U2OS cell co-transfected with GFP-CtIP and NBS1-TagRFP-T followed by live cell imaging to assess the kinetics of damage recruitment of CtIP and NBS1 to a damage line.
CtIP is recruited to DSBs after NBS1 and ATM
To determined the timing of CtIP recruitment to DSBs after DNA damage, we isolated EcoRI-treated chromatin incubated in the extract at different times and examined chromatin association of xCtIP and other factors. Chromatin association of NBS1, ATM and Ku70 was readily detectable within 5 min of incubation. In contrast, chromatin association of xCtIP was significantly delayed, being detectable beginning at 15 min. Consistent with the idea that xCtIP directly promotes DNA end resection after its recruitment to DSBs, the timing of chromatin binding of RPA and ATR was similar to that of xCtIP (Fig. 2B).
We also compared the timing of damage recruitment of CtIP, MRN and ATM in human cells. A 532-nm short-pulsed laser microbeam was used to induce DSBs in a line pattern in U2OS cell nuclei (see Experimental Procedures in Supplementary Information), and protein accumulation at laser-induced damage lines was assayed by indirect immunofluorescence (IF) staining. Accumulation of Mre11 and pS1981-ATM at the laser damage lines was readily detectable within 2 min after laser irradiation (Fig. 2C and 2D). In contrast, damage recruitment of CtIP was much delayed, beginning at ~6 min after irradiation (Fig. 2C and 2D, Fig. S2B). In cells co-transfected with GFP-tagged CtIP (GFP-CtIP) and TagRFP-T-tagged NBS1 (NBS1-TagRFP-T) we also observed a substantial delay in the recruitment of GFP-CtIP to laser-induced DSBs, compared with that of NBS1-TagRFP-T (Fig. 2E and Fig. S2C). The above observations in Xenopus extracts and human cells indicate that CtIP is recruited to DSBs after MRN and ATM following DNA damage.
Efficient recruitment of CtIP to DSBs requires NBS1 and ATM kinase activity
This temporal order of damage recruitment suggests that CtIP may function downstream of MRN and ATM in DSB resection. Indeed, addition of neutralizing NBS1 antibodies (You et al., 2005) to the extract abrogated association of xCtIP with EcoRI-treated sperm chromatin (Fig. 3A). Immunodepletion of ATM also inhibited binding of xCtIP to damaged chromatin in the extract (Fig. 3B), although the majority of the xCtIP protein (~80%) was still present in the ATM-depleted extract (Fig. S1F). Moreover, addition of an ATM-specific inhibitor KU-55933 to the extract also inhibited xCtIP association with damaged chromatin (Fig. 3C), indicating that ATM kinase activity is required for damage recruitment of xCtIP. Together, these results strongly suggest that the role of MRN and ATM in DSB resection is, at least in part, to facilitate damage recruitment of CtIP. Consistent with the idea that CtIP damage recruitment is required for DSB resection, disruption of the function of NBS1 or ATM in the extract also inhibited chromatin binding of RPA and ATR (Fig. 3A–C). Chromatin binding of ATM was inhibited by the neutralizing NBS1 antibodies (Fig. 3A), but not by the ATM inhibitor KU-55933 (Fig. 3C), reinforcing the idea that ATM is recruited to damaged chromatin by the MRN complex prior to its activation (You et al., 2005).
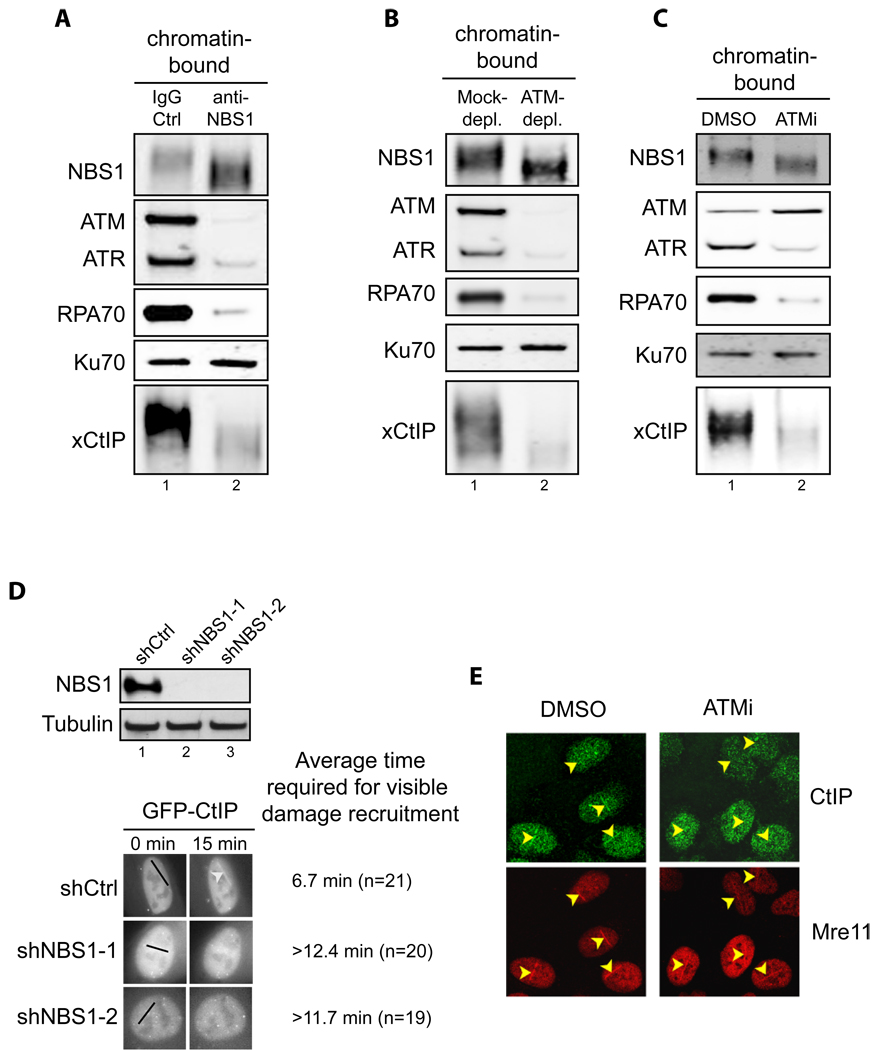
Fig. 3. Efficient recruitment of CtIP to DSBs requires NBS1 and ATM kinase activity.
- Addition of NBS1 neutralizing antibodies inhibited association of ATM, ATR, RPA70, Ku70 and xCtIP, but not NBS1 itself, with damaged chromatin in the Xenopus extract. Extract preincubated with 300 ng/µl of control IgG or affinity-purified NBS1 antibodies were incubated with EcoRI-treated sperm chromatin for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting.
- ATM depletion abrogated association of xCtIP, RPA70 and ATR, but not NBS1 and Ku70, with damaged chromatin in the Xenopus extract. EcoRI-treated sperm chromatin was incubated with mock-depleted or ATM-depleted extract for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting.
- ATM inhibition blocked the association of xCtIP, RPA70 and ATR, but not NBS1, Ku70 and ATM, with damaged chromatin in the Xenopus extract. EcoRI-treated sperm chromatin was incubated with the extract supplemented with DMSO or the ATM inhibitor KU-55933 (100 µM) for 30 min (ATMi). After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting.
- Knockdown of NBS1 in human cells delayed damage recruitment of GFP-CtIP. Upper panel: Lentiviral vectors encoding a control non-targeting shRNA or NBS1-targeting shRNAs were transfected into cells and NBS1 protein levels were examined two days after transfection. Lower panel: GFP-CtIP and the shRNA-encoding vectors were co-transfected into U2OS cells. Two days after transfection, laser irradiation was performed on cells that were transfected with both GFP-CtIP and shRNA-encoding vectors (mCherry positive). Damage recruitment of GFP-CtIP to DNA damage sites was examined by live cell imaging for 20 min after laser irradiation. The average time required for detectable accumulation of GFP-CtIP at laser-induced DNA damage sites in cells treated with shCtrl, shNBS1-1 and shNBS1-2 is 6.7 min, >12.4 min and >11.7 min, respectively. Whereas GFP-CtIP exhibited damage recruitment in all shCtrl treated cells, no GFP-CtIP damage recruitment was observed in 25% of shNBS1-1 treated cells and 20% of shNBS1-2 treated cells 20 min after laser irradiation.
- ATM inhibition resulted in delay in damage recruitment of CtIP, but not Mre11, in human cells. U2OS cells were treated with DMSO or the ATM inhibitor KU-55933 (10 µM) for 30 min. Subsequently, laser irradiation was performed to induce DSBs in cells. Ten min after irradiation cells were fixed followed by indirect immunofluorescence to assess the association of CtIP and Mre11 with DNA damage sites.
Consistent with the observations in the Xenopus extract, we found that shRNA-mediated knockdown of NBS1 delayed recruitment of GFP-CtIP to DSBs in human U2OS cells (Fig. 3D). Treating cells with the ATM inhibitor KU-55933 also inhibited recruitment of CtIP, but not Mre11, to DNA breaks induced by laser irradiation (Fig. 3E, also see Fig. 6B). Taken together, these data strongly suggest that CtIP is recruited to DSBs after MRN-dependent DSB sensing and ATM activation. After recruitment, CtIP then promotes DNA resection, leading to production of RPA-bound ssDNA and subsequent recruitment and activation of ATR. In agreement with this model, a recent study by Shiotani and Zou (Shiotani and Zou, 2009) shows that overexpression of CtIP (together with the exonuclease Exo1) speeds up the transition from ATM activation to ATR activation in response to DSBs.
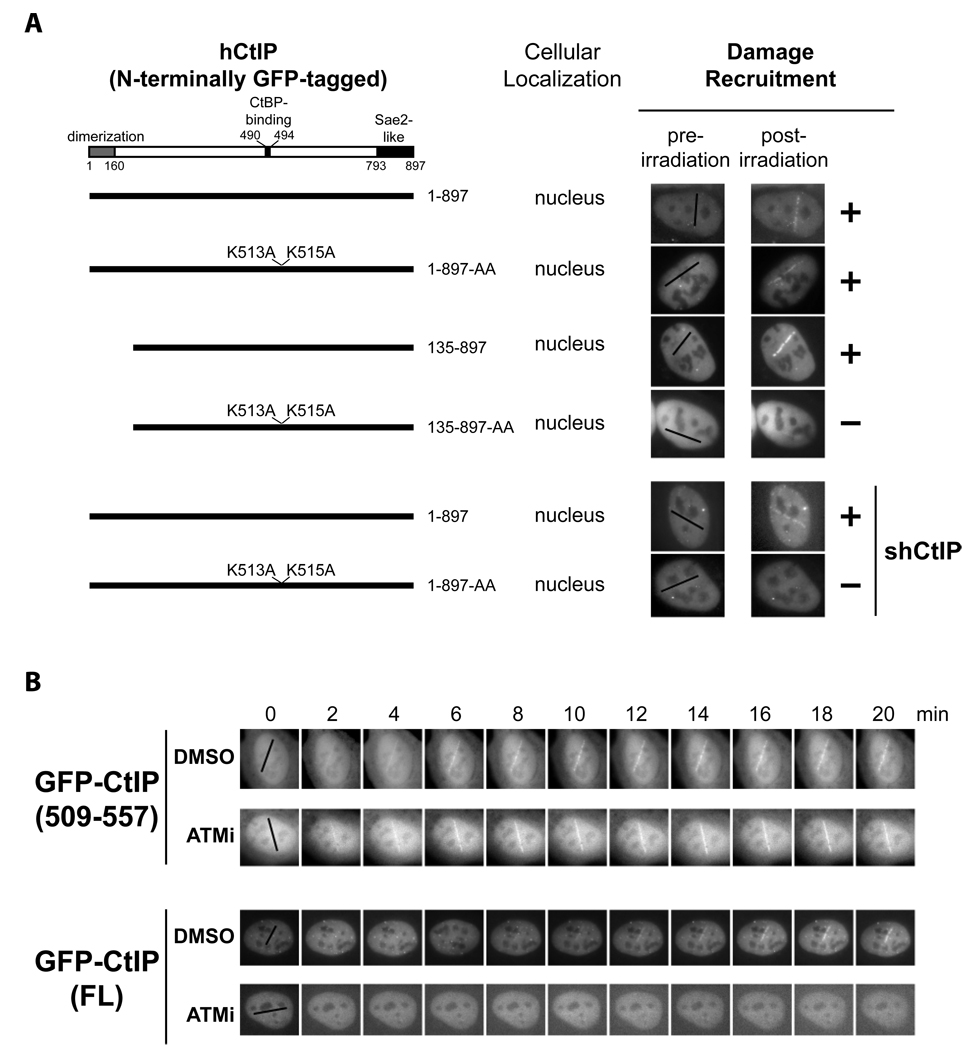
Fig. 6. The CtIP DR motif and ATM kinase activity cooperate in promoting damage recruitment of CtIP.
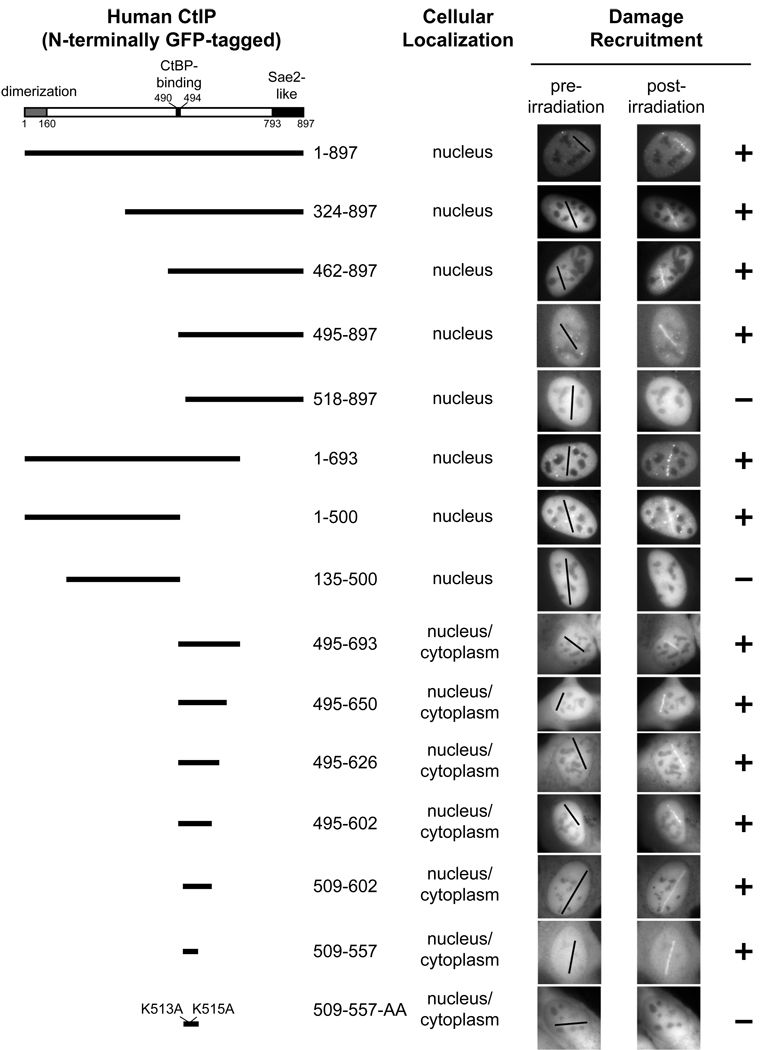
- The DR motif is essential for damage recruitment of CtIP. N-terminally GFP-tagged, wild type or mutant CtIPs were transfected into U2OS cells or U2OS cells expressing an shRNA that depletes endogenous CtIP. Cells were then subjected to laser irradiation to induce DSBs. Damage recruitment of GFP-CtIP proteins was examined 20 min after laser irradiation.
- Unlike GFP-CtIP, damage recruitment of GFP-CtIP(509–557) does not require ATM kinase activity. GFP-CtIP (containing full length CtIP) and GFP-CtIP(509–557) (containing the DR motif) were transfected into U2OS cells. Subsequently, cells were treated with DMSO or the ATM inhibitor KU-55933 (10 µM) for 30 min before laser irradiation. The damage recruitment kinetics of GFP-CtIP or GFP-CtIP(509–557) were assessed by live cell imaging.
Identification of a minimal “Damage Recruitment (DR)” motif in CtIP
To further investigate CtIP recruitment to DSBs and its importance for DNA resection, we mapped the domain(s) in human CtIP that mediate its damage recruitment by examining the recruitment of a series of N-terminally GFP-tagged human CtIP mutants to laser-induced DSBs in transfected U2OS cells. GFP-tagged, wild type CtIP was efficiently recruited to DNA damage sites (Fig. 2E and Fig. 4). Several domains/motifs, including an N-terminal homo-dimerization domain (1–160), a C-terminal Sae2-like domain (793–897) and a CtBP-binding motif PLDLS (490–494) have been identified in CtIP (Dubin et al., 2004; Limbo et al., 2007; Sartori et al., 2007; Schaeper et al., 1998). Interestingly, both GFP-CtIP (495–897) and GFP-CtIP (1–693) were recruited to DNA damage sites (Fig. 4), indicating that these known domains/motifs are dispensable for CtIP damage recruitment. Through further mutagenesis studies, we identified a minimal “damage recruitment (DR)” motif (509–557) containing 49 amino acids in a central region of CtIP. When fused to GFP, this DR motif efficiently translocated to laser-induced DNA damage sites (Fig. 4). Interestingly, two lysines (K513 and K515), which are conserved from chicken to humans in the N-terminus of this DR motif (Fig. 5A), were required for the recruitment of this motif to DNA damage sites. The GFP-CtIP(509–557)-AA mutant with K513A and K515A mutations lacked damage recruitment capacity (Fig. 4). We also observed low levels of damage recruitment of GFP-CtIP(1–500), which likely resulted from association of the mutant with endogenous CtIP that translocated to DNA damage sites. In support of this idea, a partial deletion in the dimerization domain in GFP-CtIP(1–500) completely abolished its damage recruitment (see the result for GFP-CtIP(135–500) in Fig. 4). Importantly, all the deletion and point mutations in CtIP described above caused an all-or-none effect on the damage recruitment of GFP-CtIP.
Fig. 4. Identification of a minimal “Damage Recruitment (DR)” motif in CtIP.
N-terminally GFP-tagged, wild type CtIP or various deletion mutant CtIPs were transfected into human U2OS cells. After laser irradiation, recruitment of these GFP-CtIP proteins to DNA damage sites was monitored by live cell imaging. “Post-irradiation” images were taken 20 min after laser irradiation.
Fig. 5. The conserved DR motif exhibits direct DNA-binding activity.
- Sequence comparison of the DR motif and its surrounding regions in CtIP homologs in human, mouse, rat, chicken and Xenopus. The highly conserved CtBP-binding motif (“PLDLS”) is located immediately upstream of the DR motif (509–557) in human CtIP. Numbers above and below the sequences denote the amino acid positions in human CtIP and Xenopus CtIP, respectively.
- Purified recombinant GST-His, GST-His tagged hCtIP(509–557) and GST-His tagged hCtIP(509–557)-AA proteins were resolved in a denaturing polyacrylamide gel and detected with Coomassie blue dye.
- Recombinant proteins shown in panel B were incubated with a 5’ 32P-labled 83 nt ssDNA oligo for 10 min at room temperature prior to gel shift analysis. hCtIP(509–557), but not the GST-His tag or hCtIP(509–557)-AA, associated with the ssDNA oligo, resulting in a mobility shift of the DNA substrate on a native polyacrylamide gel.
- Same as depicted for panel C, except that a 5’ 32P-labeled 100 bp dsDNA fragment was incubated with the recombinant proteins. hCtIP(509–557), but not the GST-His tag or hCtIP(509–557)-AA, associated with the dsDNA fragment.
- Purified recombinant GST, GST-tagged xCtIP(479–584) and GST-tagged xCtIP(479–584)-AA proteins resolved in a denaturing polyacrylamide gel were detected with Coomassie blue dye.
- Same as depicted for panel C except that recombinant proteins shown in panel E were used for gel mobility shift analysis. xCtIP(479–584), but not the GST tag or xCtIP(479–584)-AA, associated with the ssDNA oligo.
- Same as depicted for panel D except that recombinant proteins shown in panel E were used for gel mobility shift analysis. xCtIP(479–584), but not the GST tag or xCtIP(479–584)-AA, associated with the dsDNA fragment.
The DR motif of CtIP exhibits direct DNA-binding activity
Since we did not detect the interaction of the DR motif with other known DSB-associated proteins, such as MRN and ATM both in the presence or absence of DNA damage (data not shown), we performed gel mobility shift assays to test if the DR motif has direct DNA-binding activity. Incubation of a recombinant GST-His fusion protein containing the DR motif (CtIP(509–557)) with a 32P-labeled ssDNA oligo or dsDNA fragment reduced the mobility of the DNA substrates on native polyacrylamide gels (Fig. 5B–D). In contrast, the GST-His tag alone did not cause a mobility shift of the ssDNA oligo or the dsDNA fragment. Alanine substitution of K513 and K515, which are essential for the damage recruitment of the DR motif in cells (Fig. 4), completely abolished the DNA-binding activity of this motif in vitro (Fig. 5C and 5D). Together these results strongly suggest that the CtIP DR motif is a direct DNA-binding motif and that its DNA-binding activity is important for its ability to translocate to DNA damage sites in cells. We have not observed obvious DNA sequence specificity for the DNA-binding activity of the DR motif (data not shown). In addition to linear ssDNA and dsDNA fragments, the DR motif apparently could bind closed circular ssDNA or dsDNA without DNA ends (Fig. S3A).
Although the DR motif in CtIP is well conserved from chicken to humans, Xenopus CtIP has sequence homology only in the N-terminal part of the DR motif (Fig. 5A). Interestingly, we found that a GST-fusion protein containing xCtIP(479–584), but not GST, could also bind to the 32P-labeled ssDNA oligo and dsDNA fragment in vitro (Fig. 5E–G). Moreover, GST-xCtIP(479–584), but not GST, could bind to damaged (but not undamaged) chromatin incubated with the extract (Fig. S3B). Alanine substitution of R496 and K498 (which correspond to K513 and K515, respectively, in human CtIP) in xCtIP(479–584) abolished its DNA-binding activity in vitro, and chromatin association activity in the extract (Fig. 5E–G and Fig. S3B). Taken together, these results suggest that the presence of a direct DNA-binding motif is a conserved feature of CtIP in vertebrates.
The DR motif is required for CtIP recruitment to DSBs
To determine if the DR motif is important for damage recruitment of CtIP in cells, a GFP-CtIP-AA mutant containing K513A and K515A substitutions in full length CtIP was transfected into U2OS cells. The GFP-CtIP-AA mutant was still recruited to laser-induced DNA damage sites (Fig. 6A, upper panel), but removal of the N-terminal 134 amino acids in the CtIP homo-dimerization domain in GFP-CtIP-AA completely abrogated damage recruitment (see the result for GFP-CtIP(135–897)-AA in Fig. 6A, upper panel). In contrast, removal of the same region from GFP-CtIP did not inhibit its damage recruitment (see the result for GFP-CtIP(135–897) in Fig. 6A, upper panel). These data suggest that the observed damage recruitment of GFP-CtIP-AA was due to its association with endogenous CtIP that was recruited to DSBs. To further demonstrate the importance of the DR motif and its DNA-binding activity in CtIP damage recruitment, we generated U2OS cells depleted of endogenous CtIP by infection with lentiviruses expressing a CtIP-targeting shRNA (shCtIP) (Fig. S4). Subsequently, shCtIP-resistant GFP-CtIP and GFP-CtIP-AA expression vectors were transfected into these cells. Laser irradiation and live cell imaging were then performed to monitor the damage recruitment of GFP-CtIP and GFP-CtIP-AA. In contrast to GFP-CtIP, which exhibited efficient damage recruitment, GFP-CtIP-AA was not recruited to DNA damage sites in the absence of endogenous CtIP (Fig. 6A, lower two panels). Together, these results indicate that the DR motif and its DNA-binding activity are essential for CtIP recruitment to DSBs.
Functional relationship between the DR motif and ATM kinase activity in damage recruitment of CtIP
We next asked whether the damage recruitment of the DR motif of CtIP is also controlled by ATM kinase activity. Damage recruitment of full length GFP-CtIP in transfected U2OS cells was severely inhibited by the ATM inhibitor KU-55933 (Fig. 6B), consistent with the results for endogenous CtIP (Fig. 3). In contrast, no obvious delay in damage recruitment of GFP-CtIP(509–557) was observed in the presence of the ATM inhibitor (Fig. 6B), indicating that the DR motif and ATM kinase activity play non-redundant roles in the CtIP recruitment to DSBs. Phosphorylation of CtIP by ATM (directly or indirectly) after DNA damage may unmask the DR motif, which then mediates CtIP damage recruitment through its direct binding to the DNA at DSBs. Consistent with this idea, damage recruitment of GFP-CtIP was much delayed in untreated cells compared with that of GFP-CtIP(509–557) (Fig. 6B and data not shown).
CtIP damage recruitment is required for DSB resection, checkpoint activation and cell survival after DNA damage
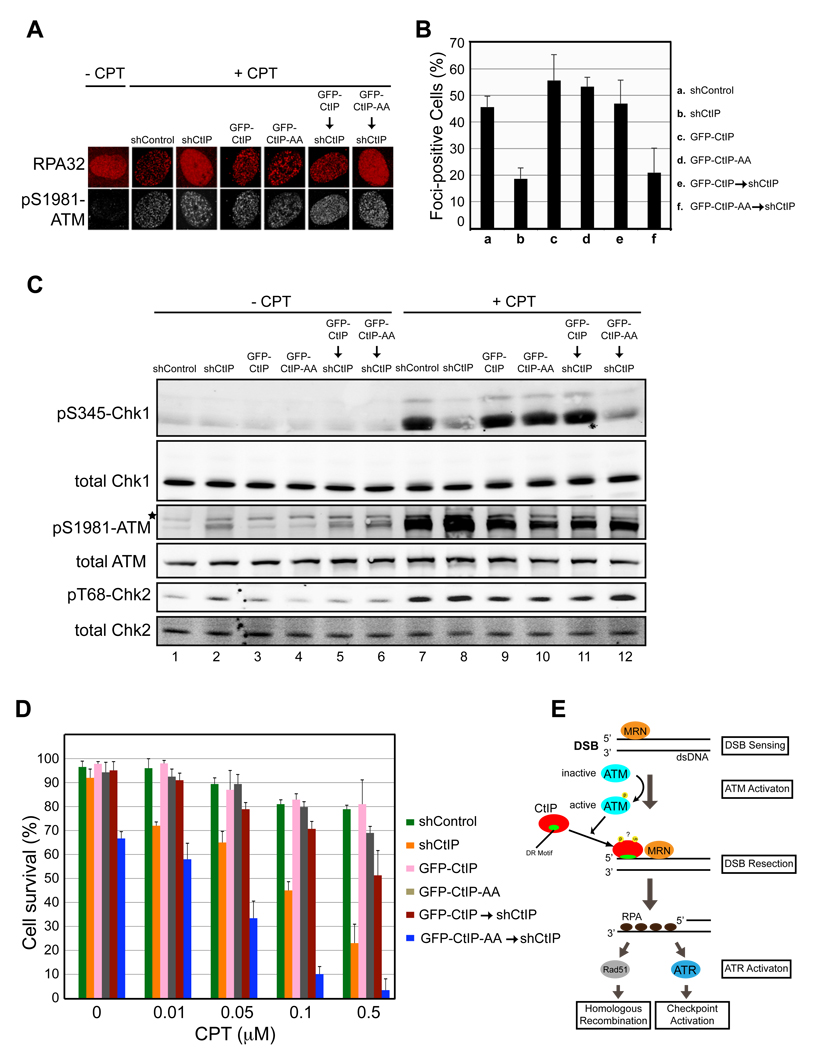
To determine its functional significance in the transition from DSB sensing to resection, we asked whether CtIP damage recruitment is required for DNA end resection, checkpoint activation and cell survival after DSB damage. To address this issue, U2OS cells were first infected with lentiviruses expressing shCtIP-resistant GFP-CtIP or GFP-CtIP-AA. GFP-positive cells were then infected with lentiviruses expressing shCtIP to deplete endogenous CtIP (Fig. S4). To assess the importance of CtIP damage recruitment for DSB resection, CtIP replacement cells were treated with camptothecin for 2 h to induce DSBs (Sartori et al., 2007) and then fixed, and formation of RPA and pS1981 ATM foci at DNA damage sites was analyzed by IF. In agreement with published data (Sartori et al., 2007), CtIP knockdown partially inhibited formation of RPA foci at DNA damage sites, resulting in a more diffuse RPA IF staining pattern (Fig. 7A and 7B). CtIP knockdown did not inhibit formation of pS1981-ATM foci at DNA damage sites (Fig. 7A and 7B), consistent with the observation that ATM functions upstream of CtIP in the DSB damage response (Fig. 3). The defects in RPA focus formation in CtIP-depleted cells were reversed by expression of GFP-CtIP, but not GFP-CtIP-AA (Fig. 7A and 7B). Expression of GFP-CtIP or GFP-CtIP-AA in U2OS cells in the presence of endogenous CtIP did not cause obvious effects on RPA focus formation after DNA damage (Fig. 7A and 7B). Together these results indicate that CtIP damage recruitment is required for efficient DSB resection.
Fig. 7. Damage recruitment of CtIP is required for efficient DSB resection, checkpoint activation and cell survival after DSB damage.
-
Aand B. CtIP damage recruitment is required for efficient formation of RPA foci at DNA damage sites. Proliferating U2OS cells expressing a control shRNA (shControl), an shRNA targeting endogenous CtIP only (shCtIP), GFP-CtIP, GFP-CtIP-AA, GFP-CtIP and shCtIP or GFP-CtIP-AA and shCtIP were treated with 1 µM camptothecin (CPT) for 2 hr. Subsequently, cells were fixed and indirect immunofluorescence was performed using antibodies against RPA and pS1981-ATM. Primary antibodies were detected using donkey anti-mouse Alexa 568-labeled and donkey anti-rabbit Cy5-labled secondary antibodies. pS1981-ATM signal was used as a marker for CPT-induced DSB damage. Representative images were shown in A. The percentage of RPA foci-positive cells among the pS1981-ATM-positive cells for each sample is shown in B. Data represent the mean ± sd. Defects in RPA focus formation in CtIP-depleted cells were reversed by expression of GFP-CtIP, but not GFP-CtIP-AA.
-
CCtIP damage recruitment is required for efficient Chk1 phosphorylation after DSB damage. U2OS cells described in panel A were treated with 1 µM camptothecin for 2 hr followed by immunoblotting to assess Chk1 phosphorylation at S345, Chk2 phosphorylation at T68 and ATM autophosphorylation at S1981. Defects in Chk1 phosphorylation in CtIP-depleted cells were reversed by expression of GFP-CtIP, but not GFP-CtIP-AA.
-
DMutations in the DR motif of CtIP caused hypersensitivity of cells to DSB damage. U2OS cells described in panel A were treated with various concentrations of camptothecin for 2 hr. Subsequently, 1000 treated cells were re-plated in triplicate and clonogenic analysis was performed to assess the sensitivity of the cells to DSB damage. Data represent mean ± sd. DNA damage hypersensitivity caused by CtIP depletion was reversed by expression of GFP-CtIP, but not GFP-CtIP-AA. Asterisk, nonspecific band.
-
EA model for damage recruitment of CtIP and its functional relationships with MRN and ATM in DSB resection. In response to DSBs, the DSB sensor complex MRN recruits ATM to the DNA/chromatin flanking a DNA end where ATM activation occurs. Following activation, ATM promotes recruitment of CtIP to DSBs, possibly by phosphorylating CtIP directly or indirectly. MRN may also directly facilitate CtIP damage recruitment. In addition to MRN and ATM, damage recruitment of CtIP requires its DR motif with direct DNA-binding activity. It is likely that conformational changes in CtIP after modifications (e.g. ATM-dependent phosphorylation and BRCA1-dependent ubiquitination) unmask the DR motif in CtIP, which then directly binds to the DNA at DSBs, leading to damage recruitment of CtIP. After damage recruitment, CtIP functions in cooperation with MRN to promote resection of 5’ strand DNA, resulting in generation of 3’ ssDNA tails that are bound by RPA. RPA-coated ssDNA then serves as a signal for activation of ATR and the DNA damage checkpoint. It also serves as a substrate for formation of the Rad51-ssDNA filament that is required for HR-mediated DSB repair.
DSB resection is required for Chk1 phosphorylation by ATR, which in turn is required for full activation of the DNA damage checkpoint (Cimprich and Cortez, 2008). We found that CtIP knockdown significantly reduced Chk1 phosphorylation in U2OS cells treated with camptothecin for 2 hr (compare lane 7 with 8 in Fig. 7C), as reported (Greenberg et al., 2006; Sartori et al., 2007; Yu and Chen, 2004). Chk1 phosphorylation in CtIP-depleted cells was restored by expression of GFP-CtIP, but not GFP-CtIP-AA (compare lane 11 with 12 in Fig. 7C), indicating that CtIP damage recruitment is critical for activation of the DNA damage checkpoint. These results also reinforce the notion that CtIP damage recruitment is required for DSB resection.
Consistent with its role in DSB resection, which is required for checkpoint activation and DSB repair by HR, depletion of CtIP caused hypersensitivity of cells to camptothecin treatment (Fig. 7D) (Sartori et al., 2007; Yun and Hiom, 2009). The hypersensitivity of CtIP-depleted cells to camptothecin could be reversed by expression of GFP-CtIP, but not GFP-CtIP-AA, indicating that CtIP recruitment to DSBs is required for cell viability after DNA damage. Taken together, these data strongly suggest that in response to DSBs CtIP is recruited to DNA damage sites through its DNA-binding motif after ATM activation. Following damage recruitment CtIP then promotes DNA end resection and subsequent checkpoint activation and DNA repair, thereby enhancing cell survival after DNA damage.
Discussion
Requirement of CtIP and its damage recruitment for DSB resection
Resection of the 5’ strand DNA at a DSB is a key process required for both checkpoint activation and DNA repair by HR after DNA damage (Bernstein and Rothstein, 2009). Our in vitro studies in the Xenopus egg extract and in vivo studies in human cells have revealed that CtIP plays a critical role in promoting DNA end resection after DSB sensing (Fig. 1 and Fig. 7), corroborating the published observations in mammalian cells (Chen et al., 2008; Greenberg et al., 2006; Huertas and Jackson, 2009; Sartori et al., 2007; Yu and Chen, 2004). Moreover, we have found that in Xenopus extracts xCtIP associates with damaged chromatin and with DNA fragments, and blocking xCtIP chromatin association by addition of xCtIP neutralizing antibodies inhibits DSB resection (Fig. 1 and Fig. 2). In human cells, CtIP also translocates to DSBs (Fig. 2–Fig. 4 and Fig. 6), and CtIP mutations that disrupt its damage recruitment cause reduced DSB resection, checkpoint signaling and cell survival after DSB damage (Fig. 6 and Fig. 7). Together, these data indicate that CtIP recruitment to DSBs is crucial for DNA end resection.
Spatio-temporal regulation of CtIP recruitment to DSBs
We found that CtIP is localized to the DSB end-proximal regions in the Xenopus extract (Fig. 2A), further reinforcing the idea that CtIP acts directly at DSBs to promote DNA end resection. Previously, we and others have shown that the MRN subunit NBS1 directly interacts with ATM and is required for damage recruitment and activation of ATM (Difilippantonio et al., 2005; Falck et al., 2005; You et al., 2005). Here we show that MRN and ATM are required for damage recruitment of CtIP, which in turn is required for the association of RPA and ATR with damaged DNA/chromatin (Fig. 1 and Fig. 3). Consistent with this functional order, the damage recruitment timing of CtIP coincides with that of RPA and ATR, and is delayed compared with that of MRN and ATM (Fig. 2). This delay may reflect the time required for MRN-dependent DSB sensing, local chromatin structure remodeling and ATM activation that are prerequisites for CtIP relocation to DSBs.
Functional relationships between CtIP, MRN and ATM
The nuclease activity of Mre11 in the MRN/MRX complex is believed to be directly involved in resection of the 5’ strand DNA at a DSB (Bernstein and Rothstein, 2009). Our study indicates that the MRN complex also plays an indirect role in DSB resection by promoting the damage recruitment of CtIP (Fig. 3). Consistent with this observation, fission yeast Ctp1 also requires Mre11 and NBS1 for its damage localization (Limbo et al., 2007; Williams et al., 2009), and MRN interacts with CtIP in human cells (Chen et al., 2008; Sartori et al., 2007; Yuan and Chen, 2009) and Ctp1 in fission yeast (Lloyd et al., 2009; Williams et al., 2009). However, CtIP apparently is not brought to DSBs passively through its interaction with MRN because the timing of CtIP damage recruitment is much delayed compared with that of MRN (Fig. 2). The role of MRN in the initial damage recruitment of CtIP is likely to be indirect; it facilitates activation of ATM which, in turn, promotes CtIP relocation to DSBs. However, this model does not rule out the possibility that the CtIP-MRN interaction helps retain CtIP at DSBs.
After damage recruitment, CtIP apparently promotes a transition in MRN function from DSB sensing to DSB resection. Sartori et al. (Sartori et al., 2007) have proposed that CtIP acts to regulate MRN’s nuclease activity in DSB resection, since CtIP directly interacts with MRN in cells and can stimulate the endonuclease activity of a Mre11-Rad50 complex on a circular ssDNA in vitro. Lengsfeld et al. (Lengsfeld et al., 2007) reported that the CtIP homolog Sae2 in budding yeast also has intrinsic ssDNA endonuclease activity, like Mre11. MRX can stimulate Sae2-mediated cleavage of ssDNA in the presence of an adjacent hairpin structure, suggesting that MRX plays an accessory role in the Sae2-dependent processing of at least a subset of DSBs (Lengsfeld et al., 2007). Precisely how CtIP/Sae2 and MRN/MRX function together at DSBs to promote DNA end resection awaits further study. Given the very limited sequence similarities shared by the CtIP homologs in different organisms, differences may exist with regard to the mechanism of their cooperation with MRN/MRX in DSB resection.
It is unclear at this point how ATM kinase activity promotes CtIP recruitment to DSBs. One possibility is that phosphorylation of CtIP by ATM promotes CtIP damage recruitment. In an attempt to test this, we generated a GFP-CtIP-8A mutant in which all 8 S/TQ sites (ATM-preferred phosphorylation sites), including the two known ATM phosphorylation sites S664 and S745, in human CtIP were mutated into alanine. However, we have not observed obvious delay in recruitment of the GFP-CtIP-8A mutant to laser-induced DNA damage sites, compared with that of GFP-CtIP containing wild type CtIP (Fig. S5). CtIP phosphorylation at other sites by ATM or by an ATM downstream kinase such as Chk2 may facilitate CtIP damage recruitment. Work is underway to test this possibility.
A DNA-binding motif in CtIP mediates its damage recruitment
By monitoring the recruitment of GFP-tagged CtIP mutant proteins to laser-induced DSBs, we have identified a 49 amino acid DR motif in human CtIP that can recruit fused GFP to DNA damage sites (Fig. 4). Interestingly, this motif can bind to both ssDNA and dsDNA with or without DNA ends (Fig. 5 and Fig. S3). Two conserved, positively charged residues in the DR motif, K513 and K515, are essential for both its DNA binding in vitro and its damage recruitment in cells (Fig. 4 and Fig. 5). Importantly, the DR motif is required for damage recruitment of full length CtIP and its function in DSB resection, checkpoint activation and cell survival after DNA damage (Fig. 6 and Fig. 7). These results suggest that CtIP directly binds to the DNA at DSBs and then promotes DNA end resection. It is not clear at present how the DR motif mediates the recruitment of CtIP specifically to DSBs, but not to other chromatin regions without DNA damage. One possibility is that the binding affinity of the DR motif to DNA with ends is regulated by other regions of CtIP. Alternatively (but not mutually exclusively), association of CtIP with DSBs may be enhanced by other protein factors (e.g. MRN) after damage recruitment.
While this paper was under review, Gu and Chen (Gu and Chen, 2009) reported that a region (amino acids 515–537) within the DR motif (amino acids 509–557) of human CtIP interacts with PCNA at replication forks, raising the possibility that PCNA may also play a role in facilitating CtIP recruitment to DSBs and/or its retention at DSBs. However, the role of PCNA in CtIP damage recruitment/retention, if any, apparently is not conserved in Xenopus since xCtIP lacks a PCNA-interacting protein box (PIP box) in the PIP box region identified in human CtIP. Interestingly, unlike full length CtIP, recruitment of the CtIP DR motif to DSBs does not require ATM kinase activity (Fig. 6B). This indicates that the DR motif and ATM kinase activity play non-redundant roles in facilitating CtIP damage recruitment. ATM-dependent modification of CtIP (or a CtIP-interacting protein) may unmask the DR motif in CtIP, which then binds to the DNA at DSBs, resulting in CtIP recruitment to DSBs. We are currently pursuing this possibility. Interestingly, Lengsfeld et al. (Lengsfeld et al., 2007) have shown that Sae2 also has DNA-binding activity in vitro, though it is not clear whether the DNA-binding activity is required for Sae2 damage recruitment in cells. Although we have not identified a region in Sae2 that is similar to the CtIP DR motif in sequence, it is tempting to speculate that CtIP and Sae2 are recruited to DSBs via a similar mechanism, i.e. through their direct binding to the DNA at DSBs.
Taken together, our study suggests that in response to DSBs CtIP receives a signal from ATM after MRN-dependent DSB sensing and ATM activation, and then binds to damaged DNA through its DNA-binding motif. Subsequently, CtIP promotes ssDNA resection (in collaboration with MRN) at DSBs, generating ssDNA that is bound by RPA. The RPA-bound ssDNA then serves as the signal for activation of the checkpoint kinase ATR and formation of the HR intermediate Rad51 nucleofilament (Fig. 7E). Thus, CtIP acts as a molecular switch that facilitates the transition from DSB sensing to processing and repair, and from ATM activation to ATR activation. Further studies on the role of CtIP in DSB resection and its functional relationships with other damage checkpoint and repair proteins such as MRN, ATM and BRCA1 will add to our understanding of how cells sense and respond to DNA damage in order to maintain genome integrity and stability.
Experimental Procedures
Cell culture, transfection, lentivirus production and generation of CtIP-replacement cells
Plasmid constructs, antibodies and chemical inhibitors used in this study are described in the Supplemental Experimental Procedures. Human U2OS and 293T cells were cultured at 37 °C in DMEM with 10% FBS in the presence of antibiotics (penicillin and streptomycin) and 10% CO2. Plasmid DNA was transfected into U2OS cells using Effectene transfection reagent (Qiagen) according to the manufacturer’s protocol. Lentiviruses were generated in 293T cells as described in Supplemental Experimental Procedures. To generate the CtIP-replacement cells, U2OS cells were first infected with lentiviruses expressing GFP-CtIP or GFP-CtIP-AA. Subsequently, GFP-positive cells were enriched by cell sorting (Becton-Dickinson FACS Vantage SE DiVa). Finally, cells were infected with shCtIP-expressing lentiviruses to deplete endogenous CtIP.
Xenopus egg extracts, immunodepletion, immunoblotting, chromatin immunoprecipitation, chromatin binding and DNA end resection assays
Interphase Xenopus egg extract was prepared as previously described (Smythe and Newport, 1991). Immunodepletion of ATM was as described (You et al., 2007). To deplete xCtIP from the Xenopus extract, 30 µl of protein A agarose beads coupled with 90 µl of xCtIP anti-serum were incubated with 150 µl of extract for 45 min at 4 °C. Beads were then removed from the extract by low-speed centrifugation. The extract supernatant was then subjected to two additional rounds of depletion under the same conditions. Immunoblotting was performed using IRDye 800- and Alexa 680-conjugated secondary antibodies and an Odyssey infrared imaging system (LI-COR Biosciences) as described previously (You et al., 2007; You et al., 2005). Chromatin immunoprecipitation (ChIP) and Chromatin binding assays in the Xenopus extract were performed as described (You et al., 2007; You et al., 2005). The direct DNA resection assay is described in Supplemental Experimental Procedures.
Expression and purification of recombinant proteins, probe labeling and gel mobility shift assay
Expression and purification of recombinant proteins and labeling of ssDNA and dsDNA probes are described in Supplemental Experimental Procedure. To perform gel shift assays, 1×105 cpm of labeled DNA probes and 1 µg of recombinant proteins were incubated with the binding buffer (10 mM HEPES (pH 7.8), 75 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 3% Ficoll and 1 µg BSA) for 10 min at room temperature. Samples were then directly loaded onto and resolved in 5% native polyacrylamide gels containing 5% glycerol. Gels were then dried and exposed to Kodak Biomax XAR films.
Laser irradiation, live cell imaging and immunofluorescence staining
Laser irradiation of U2OS cells using a customized system is described in Supplemental Experimental Procedures. To monitor the recruitment of EGFP- or TagRFP-T-tagged proteins to DNA damage site, live cell imaging was performed with appropriate filter sets (Chroma) after laser irradiation. Fluorescence images were gathered every 60 seconds after laser irradiation. Recruitment of endogenous proteins to laser-induced DNA damage sites was detected by immunofluorescence staining and confocal microscopy as described in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Matthew Weitzman, Paul Russell, Walter Eckhart, Douglass Forbes, Helen Piwnica-Worms, Julie Bailis, Nicole Orazio and Charly Chahwan for critical discussions, Jill Meisenhelder, Suzy Simon, Justin Zimmermann and David Chambers for technical support. We thank Jeremy Copp and Gerald Pao for providing Xenopus and human CtIP cDNA, Graeme Smith and Mark O’Connor for providing the ATM inhibitor KU-55933. T.H. is a Frank and Else Schilling American Cancer Society Research Professor. M.B. is the Arnold and Mabel Beckman Professor at UC Irvine and Adjunct Professor of Bioengineering at UC San Diego. I.V. is an American Cancer Society Professor of Molecular Biology. This work was supported by a grant from the American Cancer Society IRG-58-010-52 (Z.Y.), US Public Health Service Grants CA14195 and CA80100 from the NCI (T.H.), grants from the Air Force Office of Scientific Research (F9620-00-1-0371) and the Beckman Laser Institute Foundation (M.B.), and grants from the NIH, Leducq Foundation, Lustgarten Foundation, Ellison Medical Foundation, and the H. N. and Frances C. Berger Foundation (I.V.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006;25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu Y, Murayama Y, Yamada T, Nakazaki T, Tsutsui Y, Ohta K, Iwasaki H. Molecular characterization of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol Cell Biol. 2008;28:3639–3651. doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Rothstein R. At loose ends: resecting a double-strand break. Cell. 2009;137:807–810. doi: 10.1016/j.cell.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS ONE. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, Yang YP, Petukhova GV, Eckhaus M, Feigenbaum L, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Nussenzweig A. The NBS1-ATM connection revisited. Cell Cycle. 2007;6:2366–2370. doi: 10.4161/cc.6.19.4758. [DOI] [PubMed] [Google Scholar]
- Dubin MJ, Stokes PH, Sum EY, Williams RS, Valova VA, Robinson PJ, Lindeman GJ, Glover JN, Visvader JE, Matthews JM. Dimerization of CtIP, a BRCA1- and CtBP-interacting protein, is mediated by an N-terminal coiled-coil motif. J Biol Chem. 2004;279:26932–26938. doi: 10.1074/jbc.M313974200. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Chen PL. Expression of PCNA-binding domain of CtIP, a motif required for CtIP localization at DNA replication foci, causes DNA damage and activation of DNA damage checkpoint. Cell Cycle. 2009;8:1409–1420. doi: 10.4161/cc.8.9.8322. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Huertas P, Jackson SP. Human CtIP mediates cell-cycle control of DNA-end resection and double-strand-break repair. J Biol Chem. 2009 doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kurz EU, Lees-Miller SP. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst) 2004;3:889–900. doi: 10.1016/j.dnarep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ting NS, Zheng L, Chen PL, Ziv Y, Shiloh Y, Lee EY, Lee WH. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature. 2000;406:210–215. doi: 10.1038/35018134. [DOI] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Lee WH. CtIP activates its own and cyclin D1 promoters via the E2F/RB pathway during G1/S progression. Mol Cell Biol. 2006;26:3124–3134. doi: 10.1128/MCB.26.8.3124-3134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A, Portik-Dobos Z, Tang L, Schnabel R, Novatchkova M, Jantsch V, Loidl J. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 2007;26:5071–5082. doi: 10.1038/sj.emboj.7601916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeper U, Subramanian T, Lim L, Boyd JM, Chinnadurai G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe C, Newport JW. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Uanschou C, Siwiec T, Pedrosa-Harand A, Kerzendorfer C, Sanchez-Moran E, Novatchkova M, Akimcheva S, Woglar A, Klein F, Schlogelhofer P. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lee WH. CtIP, a multivalent adaptor connecting transcriptional regulation, checkpoint control and tumor suppression. Cell Cycle. 2006;5:1592–1596. doi: 10.4161/cc.5.15.3127. [DOI] [PubMed] [Google Scholar]
- You Z, Bailis JM, Johnson SA, Dilworth SM, Hunter T. Rapid activation of ATM on DNA flanking double-strand breaks. Nat Cell Biol. 2007;9:1311–1318. doi: 10.1038/ncb1651. [DOI] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chen J. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J Biol Chem. 2009;284:31746–31752. doi: 10.1074/jbc.M109.023424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases dna2 and exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.