Abstract
Proteasomes degrade most proteins in mammalian cells and are established targets of anti-cancer drugs. All eukaryotic proteasomes have three types of active sites: chymotrypsin-like, trypsin-like, and caspase-like. Chymotrypsin-like sites are the most important in protein degradation and are the primary target of most proteasome inhibitors. The biological roles of trypsin-like and caspase-like sites and their potential as co-targets of anti-neoplastic agents are not well defined. Here we describe the development of novel, site-specific inhibitors and active-site probes of chymotrypsin-like and caspase-like sites. Using these compounds, we show that cytotoxicity of proteasome inhibitors does not correlate with inhibition of chymotrypsin-like sites and that co-inhibition of either trypsin-like and/or caspase-like sites is needed to achieve maximal cytotoxicity. Thus, caspase-like and trypsin-like sites must be considered as co-targets of anti-cancer drugs.
Introduction
The ubiquitin-proteasome pathway is essential in the maintenance of protein homeostasis in all eukaryotic cells; it is involved in the regulation of numerous biological processes, such as the cell cycle, immune surveillance, inflammatory response, metabolism, circadian rhythms, and others; and in the development of many diseases. Proteasome inhibition causes apoptosis of malignant cells (Adams, 2004; Kisselev and Goldberg, 2001). The proteasome inhibitor bortezomib (Velcade, PS-341) is used for the treatment of multiple myeloma and mantle cell lymphoma. Three other proteasome inhibitors are at different stages clinical trials (Chauhan et al., 2005; Demo et al., 2007; Piva et al., 2008).
The 26S proteasome is a large (1.6–2.4 MDa), hollow cylindrical, multifunctional particle that consists of a 20S proteolytic core and one or two 19S regulatory complexes. Each eukaryotic 20S core particle has three pairs of proteolytic sites with distinct substrate specificities (Arendt and Hochstrasser, 1997; Chen and Hochstrasser, 1996; Dick et al., 1998; Groll et al., 1997; Heinemeyer et al., 1997). The β5 proteolytic sites are “chymotrypsin-like” (Chym-L). The β2 sites are “trypsin-like” (Tr-L). The β1 sites cleave after acidic residues (Glu, Asp) and are referred to as “post-acidic,” PGPH (“post-glutamate peptide hydrolase”), or “caspase-like” (Casp-L).
Tissues of the immune system also express immunoproteasomes, in which β5, β1, and β2 catalytic subunits are replaced by their major histocompatibility complex (MHC)-locus-encoded counterparts, LMP7 (β5i), LMP2 (β1i), and MECL (β2i). Immunoproteasomes have higher Chym-L and Tr-L activities and much lower Casp-L activity, presumably allowing them to generate more peptides for utilization in MHC class I antigen presentation (Cascio et al., 2001).
The biological role of β1, β2, and β5 active sites was first addressed by site-directed mutagenesis of catalytic threonines in the yeast S. cerevisiae. Inactivation of Chym-L (β5) sites caused significant retardation of growth, increase in stress sensitivity, and accumulation of proteasome substrates (Chen and Hochstrasser, 1996; Heinemeyer et al., 1997). Inactivation of Casp-L (β1) sites caused no phenotypic or proteolytic defects (Arendt and Hochstrasser, 1997; Heinemeyer et al., 1997). Inactivation of Tr-L (β2) sites reduced growth rates slightly and reduced the degradation rate of some model substrates (Arendt and Hochstrasser, 1997; Heinemeyer et al., 1997). A strain in which both β1 and β2 sites were inactive had a stronger growth defect than strains in which only the β2 sites were inactivated, but had fewer phenotypic defects than the strain lacking functional β5 sites (Heinemeyer et al., 1997). It should be noted that these mutations also caused defects in the proteasome assembly (Groll et al., 1999) and that some of these phenotypes may have been caused by assembly defects. To distinguish between biological effects caused by inhibition of assembly and inhibition of proteolysis, as well as to study the biological roles of proteasome active sites in mammalian cells, specific inhibitors of active sites are needed.
Because these results from yeast studies showed that Chym-L sites are the most important sites in protein breakdown by the proteasome and because of the ability of hydrophobic peptides to enter cells, various synthetic proteasome inhibitors were optimized to block the β5 sites, which cleave after hydrophobic residues (Kisselev and Goldberg, 2001). Less attention has been paid to the ability of these substances to block the β1 or β2 sites (Kisselev and Goldberg, 2001). Bortezomib was developed as an inhibitor of Chym-L (β5 and β5i) sites (Adams et al., 1999). Only after approval of this agent by the FDA was it discovered that it also inhibits Casp-L (β1 and β1i) sites and Tr-L (β2i) sites in the immunoproteasomes (Altun et al., 2005; Berkers et al., 2005; Kisselev et al., 2006). Similarly, salinosporamide A (NPI-0052) inhibits Chym-L, Tr-L, and, to some extent, Casp-L sites. This agent has a more potent anti-neoplastic activity in mice than bortezomib (Chauhan et al., 2005), further suggesting that co-inhibition of Tr-L and Casp-L sites might be important for the anti-neoplastic activity of proteasome inhibitors. This idea is further supported by two studies in the literature which report that selective inhibition of β5 sites caused moderate inhibition of degradation of model substrates by purified proteasomes and little or no inhibition of protein breakdown inside cells. Significant (more than 50%) inhibition of protein degradation is achieved only when both β5 and either β1 or β2 sites are inhibited (Kisselev et al., 2006; Oberdorf et al., 2001). Thus, β1 and β2 sites play an important role in protein degradation, suggesting that they should be considered as co-targets of anti-cancer drugs.
In this study, we report the development of two novel specific inhibitors of Chym-L and Casp-L sites. Using these compounds, we demonstrate that cytotoxicity of proteasome inhibitors rarely correlates with inhibition of Chym-L (β5) sites alone and that co-inhibition of either β1 or β2 sites is required for β5-specific inhibitors to achieve maximal cytotoxicity.
Results
Novel specific inhibitor of chymotrypsin-like sites
The simplest way to test whether inhibition of β5 sites is sufficient to inhibit cell growth and cause cell death would be to examine the effects of a highly specific inhibitor of these sites on cell growth and viability. For the purpose of this study, “highly specific” would mean that inhibitor does not cause a significant decrease—i.e., more than 20%—in the activity of Casp-L and Tr-L sites under conditions where Chym-L sites are inhibited by at least 95%. We initially intended to use YU-101 (Fig. 1A), developed as specific inhibitor of Chym-L sites (Elofsson et al., 1999), but discovered that it inhibits Tr-L and Casp-L sites before complete inhibition of Chym-L sites can be achieved (Fig. 1B). Hence, we decided to develop a more specific inhibitor.
Figure 1. Characterization of the novel β5-specific proteasome inhibitor NC-005.
A Structures of YU-101 and NC-005. B, C. RPMI-8226 cells were treated with YU-101 and NC-005 for 1h and proteasome activity was measured in cells with ProteasomeGlo assay. Mock-treated cells served as control. Values are averages ± S.E. of two experiments.
YU-101 is a tetrapeptide epoxyketone (Fig. 1A). Contrary to other major groups of proteasome inhibitors, such as peptide vinyl sulfones, peptide boronates, β-lactones and peptide aldehydes, can react with either cysteine or serine proteases or both, epoxyketones are exquisitely proteasome-specific (Groll and Huber, 2004; Kisselev and Goldberg, 2001. One compound of this class, the YU-101 derivative carfilzomib (Demo et al., 2007), is in stage II clinical trials. We have therefore focused our development of site-specific inhibitors on the epoxyketone pharmacophore, varying the peptide portion of the drug to optimize compound specificity.
In reviewing the literature, we noticed that peptide aldehyde 1-naptylacetyl(Nac)-4-methyl-tyrosine(mTyr)-phenylalanine-4-methyl-tyrosinal was exceptional in that it did not inhibit Casp-L and Tr-L sites (Momose et al., 2005). We have synthesized an epoxyketone derivative of this compound, Nac-mTyr-Phe-Leu-ek (Fig. 1A), electing to use Leu in the P1 position for simplicity of synthesis and better cell permeability. We have designated this compound NC-005, where NC stays for Norris Cotton Cancer Center and 5 emphasizes that fact that inhibits β5 and β5i subunits. We also decided that for naming all future compounds of this type prepared in our laboratory, the last digit would match the active site inhibited. Using this terminology, the second inhibitor of β5 sites would be NC-015, the first inhibitor of β2 sites NC-002, and the first inhibitor of β1 sites NC-001.
As expected, NC-005 was more specific than YU-101 while maintaining similar potency. Most importantly, NC-005 did not inhibit Casp-L and Tr-L sites at a concentration that caused nearly complete inhibition of Chym-L sites (0.5 µM, Fig. 1C).
Another difference between YU-101 and NC-005 was that NC-005 caused stronger activation of Tr-L and Casp-L activities (50% vs. 20%). This activation is most likely allosteric (Kisselev et al., 1999) and is reduced in YU-101 because inhibition of Casp-L and Tr-L sites occurs at lower concentrations than in NC-005 treated cells. Another possible explanation for this effect— increased proteasome activity because of biosynthesis of new proteasomes—is much less likely because duration of treatment (1h) was too short to activate this transcriptional response.
NC-005 is cytotoxic to multiple myeloma cells, but maximal cytotoxicity is achieved at concentrations where it is no longer chymotrypsin-site specific
To test whether NC-005 is cytotoxic to multiple myeloma cells, cells were treated with NC-005 for 1h, then incubated in a drug-free media for 48h, followed by measurement of cell viability with a mitochondrial dye conversion assay (Fig. 2A). Such short treatment was used because it reflects the clinical situation much better than continuous treatment of cells with proteasome inhibitors as usually used in cell culture experiments. Patients are receiving these drugs as an intravenous bolus injection; within 1 h after such treatment, proteasome inhibition in blood cells reaches its maximum (i.e., 70% inhibition of Chym-L activity), but it recovers within the next 24 h (Hamilton et al., 2005). Similarly, 1 h treatment of RPMI-8226 cells with 0.6 µM NC-005 led to 80% inhibition of Chym-L activity, followed by complete recovery in the next 24 h (Fig. 2B). Recovery was slower in cells treated with higher concentrations of NC-005. An additional reason for using 1 h treatments was that longer incubation led to a slight loss of specificity (data not shown). Recovery was slower at higher concentrations, and this persistent inhibition of proteasomes resulted in the complete or near-complete loss of cell viability.
Figure 2. Cytotoxicity of the novel proteasome inhibitor NC-005 does not correlate with the inhibition of Chym-L sites.
A Overall design of experiments. Cells were treated with inhibitors for 1 h, and then cultured in the absence of the inhibitors for 48 h, when cell viability was assayed with an Alamar Blue. At time points indicated, a fraction of cells was harvested (1, 3, 6, 10, and 23 h) and used for the measurement of apoptosis and of proteasome peptidase activities. B. Recovery of Chym-L activity after 1 h treatment of RPMI-8226 cells with NC-005. Values are averages ± S.E. of two experiments. Recovery of Tr-L and Casp-L activities in RPMI-8226 cells and recovery of all three activities in Dx6 and KMS-18 cells are shown on Fig. S1. C, D. Effect of 1h NC-005 (C) and bortezomib (D) treatment on viability of multiple myeloma cells. Mock-treated cells served as control. Values are averages ± S.E. of 4–6 (C) or 2–3 (D) experiments. E. IC50 (i.e. concentration at which viability was reduced by 50%) was determined by a 4-parameter data fit of dose response curves (C, D) and plotted against specific activity of Chym-L sites. F. Cell viability (from C and D) was plotted against the inhibition of Chym-L (top panel) and Tr-L (middle panel) or Casp-L (bottom panel) sites (values from Table 1). G. Apoptosis of NC-005 treated RPMI-8226 cells. Top panel: caspase-3 activity is expressed in nmole Ac-DEVD-amc cleaved/min/mg protein, and appearance of the cleaved PARP was detected on the Western blot (inset). Bottom panel, percentage of Annexin V-positive apoptotic cells was determined by flow cytometry. Representative results for one out of three different experiments are shown. H. Viability of RPMI-8226, Dox6, and KMS-18 cells (as in C) was plotted against the inhibition of Chym-L (solid lines) and Tr-L (dashed lines) activities at 5 h (top panel) and 10 h (bottom panel) after end of 1 h NC-005-treatment (Fig. S1).
NC-005 was cytotoxic to all myeloma cell lines but sensitivity varied widely, with IC50 ranging from 30 nM to 1.5 µM (Fig. 2C). This was unexpected because these cell lines show little difference in sensitivity to bortezomib (with continuous exposure: (Hideshima et al., 2001). In order to determine whether this difference is a unique feature of NC-005 or a consequence of shortening treatment time to 1 h, we have treated the same cell lines with bortezomib for 1 h. Although the order of sensitivity changed (i.e., RPMI-8226 was the least NC-005-sensitive cell line, but Dox6 and KMS-12-BM the least bortezomib-sensitive lines), similar 50-fold differences in IC50 were observed across the panel (Fig. 2D). Thus, differences in sensitivity in myeloma cells are a general feature of proteasome inhibitors and not a unique feature of NC-005. One possible reason for different sensitivity would be that cell lines that are very sensitive to bortezomib and NC-005 express fewer proteasomes. We determined specific proteasome activity in these cell lines and found little correlation between this parameter and IC50 for either inhibitor (Fig. 2E). The reason for this difference is currently being investigated in the laboratory.
We then asked the question whether inhibiting Chym-L sites alone is sufficient to induce cytotoxicity in multiple myeloma cells. In all cell lines, we measured inhibition of all three activities immediately after the 1 h treatment (Table 1), when inhibition is maximal (Fig. 2B), and noticed that in the majority of them maximal cytotoxicity was achieved only at concentrations where NC-005 co-inhibited Tr-L and sometimes Casp-L sites. In order to test whether cytotoxicity correlates with inhibition of Chym-L sites, we then plotted (Fig. 2F, top panel) cell viability vs. inhibition of these sites (Table 1). Good correlation was observed only for one cell line, NCI-H929, which was the most sensitive to NC-005. Some correlation was observed for three others (MM1.R, MM1.S, and KMS-18). Little or no correlation was observed for the remaining three lines (RPMI-8226, its doxorubicin-resistant derivative Dox 6, and KMS-12-BM). This data is an agreement with the recent report of Parlati et al who found that specific 80% inhibition of the Chym-L sites cause 70% reduction in viability of MM1.S cell but only 20–25% reduction in viability of HS-Sultan and Molt-4 cells (Parlati et al., 2009). For the RPMI-8226 and Dox6 cell lines, lack of viability correlated with inhibition of Tr-L sites (Fig. 2F, middle panel). We have also plotted viability against inhibition of Casp-L sites, but even in these least-NC-005 sensitive sites viability decreased faster than activity (Fig. 2F, bottom panel). Thus, co-inhibition of Tr-L sites appears to be important for NC-005 cytotoxicity.
Table 1. Effects of different concentrations of NC-005 on proteasome activity and cell viability.
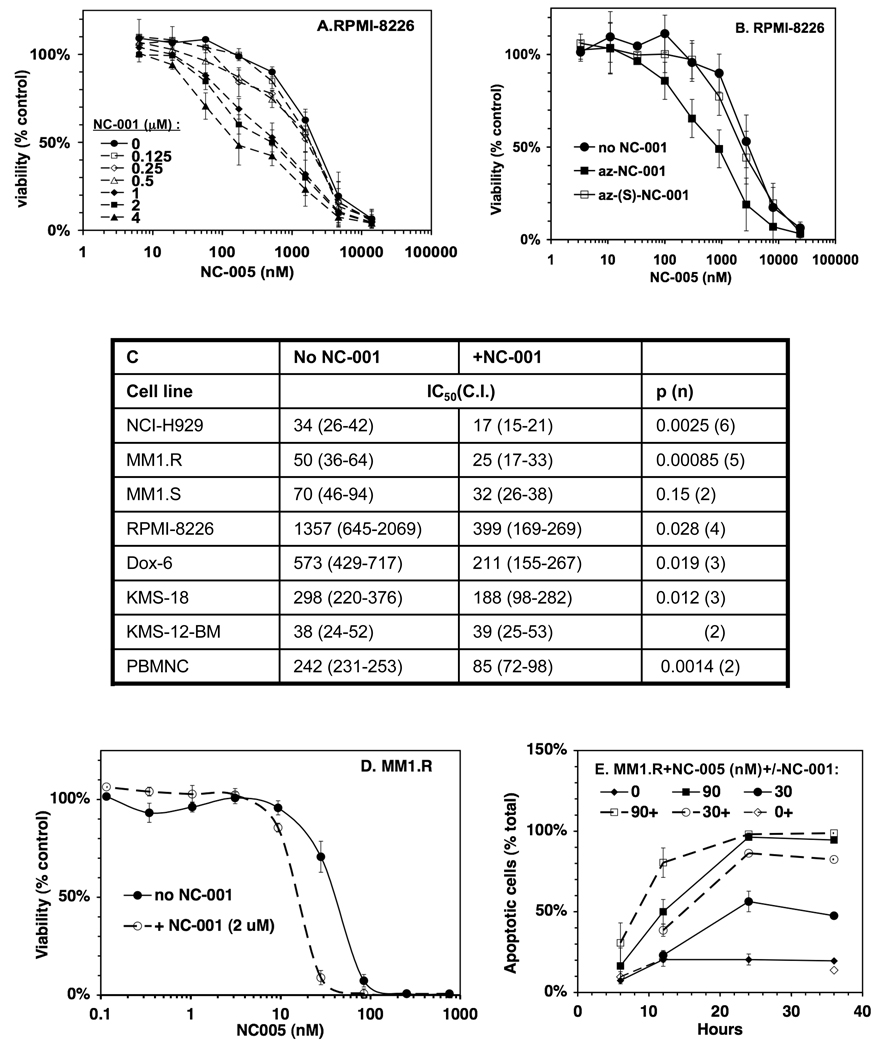
Cells were treated with NC-005 for 1h, followed by the measurement of proteasome peptidase activities; another faction of NC-005-treated cells was cultured for 48 hours in the presence or absence of 2 µM NC-001 (10 µM in case of Dox-6 cells), after which cell viability was measured with Alamar Blue assay. Viability data is the same as on Fig. S3. MS-maximal sensitization by NC-001; MC-maximal cytotoxicity (NC-005 alone); n.t.-not tested. Value are averages±S.E. of 2–3 (inhibition) or 2–5 (viability) independent measurements.
| NC-005 (µM) |
Inhibition of active sites (by NC-005 alone, % control) |
Viability (% control) |
|||||
|---|---|---|---|---|---|---|---|
| Cell line | β5 | β2 | β1 | −NC-001 | +NC-001 | ||
| RPMI-8226 | 0.058 | 87±2.8 | −12±9 | 2±9 | 103±2 | 92±3 | |
| 0.175 | 98±0.5 | 7±7 | 5±2 | 92±3 | 67±6 | ||
| 0.52 | 98±0.3 | 25±8 | 20±1 | MS | 84±7 | 40±15 | |
| 1.6 | 99±0.2 | 63±5 | 37±4 | 38±12 | 14±4 | ||
| 4.8 | 99±0.1 | 88±3 | 48±2 | MC | 14±5 | 7±2 | |
| Dox6 | 0.058 | 94±0.3 | −12±4 | 9±12 | 101±2 | 92.5±5 | |
| 0.175 | 98±0.0 | 12±2 | 17±4 | 98±5 | 58±6 | ||
| 0.52 | 99±0.1 | 46±2 | 32±3 | MS | 55±8 | 13±6 | |
| 1.6 | 99±0.1 | 78±1 | 42±4 | MC | 6.0±3.5 | 2.5±1.3 | |
| KMS-18 | 0.019 | 46±8 | −47±40 | −4±8 | 111±9 | 104±3 | |
| 0.058 | 88±3.5 | 12±10 | 20±3.5 | 110±6 | 80±8 | ||
| 0.175 | 98±0.2 | 17±1 | 17±6 | MS | 80±6 | 52±10 | |
| 0.52 | 99±0.1 | 36±6 | 37±1 | 20±6 | 16±8 | ||
| 1.6 | 99±0.2 | 64±5 | 49±5 | MC | 3.4±0.6 | 1.6±0.7 | |
| MM1.R | 0.028 | 40±2 | −44±10 | −80±24 | MS | 71±8 | 9±2 |
| 0.084 | 89±1 | −14±12 | −45±24 | 7±3 | 0.7±0.1 | ||
| 0.252 | 91±2 | −4±11 | −22±13 | MC | 0.8±0.1 | 0.7±0.1 | |
| MM1.S | 0.0064 | 31±2 | 0±10 | 9±3 | 106±2 | 99±3 | |
| 0.019 | 54±8 | −13±4 | 1±8 | 91±2 | 73±2 | ||
| 0.058 | 92±1.5 | 2±12 | 14±1 | MS | 56±6 | 21±6 | |
| 0.175 | 98±0.1 | 11±9 | 20±3 | 15±12 | 2.2±2 | ||
| 0.52 | 99±0.1 | 38±9 | 36±5 | MC | 0.65±0.07 | 0.2±0.0 | |
| H929 | 0.0064 | 17±2 | −16±0.5 | −4±3 | 97±11 | 82±9 | |
| 0.019 | 45±5 | −15±20 | −3±10 | MS | 79±6 | 45±10 | |
| 0.058 | 87±1 | 1±21 | 9±10 | 22±7 | 8±5 | ||
| 0.175 | 97 | 7 | 15 | MC | 8±7 | 5±4 | |
| KMS-12-BM | 0.019 | 83 | 0 | 8 | 112±9 | n.t. | |
| 0.058 | 97±0 | 11±2 | 17±2 | 72±21 | n.t. | ||
| 0.17 | 98±0 | 21±8 | 19±9 | 31±5 | n.t. | ||
| 0.52 | 99±0 | 50±5 | 35±0.4 | 14±4 | n.t. | ||
| 1.6 | 100±0 | 83±3 | 48±4 | MC | 2.4±0.1 | n.t. | |
| 4.8 | 100±0 | 92±0.4 | 58±4 | 1.2±0.3 | n.t. | ||
| PBMNC | 0.019 | 63±3 | 0±3 | 3±5 | 96±7 | 90±10 | |
| 0.058 | 92±3 | 0±7 | −15±1 | MS | 77±2 | 39±1 | |
| 0.175 | 98±1 | 21±12 | −9±3 | 43±1 | 22±1 | ||
| 0.52 | 99±1 | 59±18 | 9±7 | 15±1 | 7±0 | ||
| 1.6 | 100±2 | 97±6 | 42±13 | MC | 4±0.3 | 3.5±0.4 | |
| MDA-MB-231 | 0.009 | 39±14 | −20±32 | −25±30 | 99±4 | 100±9 | |
| 0.028 | 81±8 | −33±48 | −43±48 | 101±0 | 84±3 | ||
| 0.084 | 95±6 | −12±18 | −26±14 | MS | 85±4 | 33±6 | |
| 0.252 | 96±3 | 8±18 | −42±23 | 39±6 | 7±1 | ||
| 0.756 | 99±3 | 59±2 | −9±4 | MC | 6±1 | 5±1 | |
A caveat of this analysis is that proteasome activity might recover, wholly or partially, before apoptosis is induced (Fig. 2B). In that case, average proteasome inhibition between the end of NC-005 treatment and commitment to apoptosis would be less than inhibition at 1h, which was used for the correlation analysis presented in Fig. 2F. To test whether this is the case, we have measured apoptosis (Fig. 2G) and proteasome activity during the first 24 h after treatment (Fig. 2B and S1). In RPMI-8226 cells, caspase activation and PARP-cleavage were observed 5 h after the treatment (Fig. 2G, top panel) and annexin V-positive apoptotic cells were detected at 10 h (bottom panel). Throughout this period, recovery of proteasome activity at cytotoxic concentrations did not exceed 30% (Fig. 2B, S1). When we plotted cell viability versus inhibition of Chym-L and Tr-L sites at 5 and 10 h after the treatment, viability of RPMI-8226 and Dox6 cell lines again did not correlate with inhibition of Chym-L sites; much better correlation was observed between viability and inhibition of Tr-L sites (Fig. 2H). Thus, co-inhibition of Tr-L sites contributes to the cytotoxic effects of NC-005.
Another explanation for the lack of correlation between inhibition of Chym-L sites and cytotoxicity would be an off-target effect of NC-005. Although we considered such scenario as unlikely because of specificity of epoxyketones, we decided to use the chemical tools at our disposal to demonstrate that NC-005 does not, in fact, interact with other cellular proteins. We therefore converted it into an active site probe (i.e., irreversible inhibitor with a reporter group attached to it). We have used a two-step labeling strategy, which we successfully applied in our previous studies (van Swieten et al., 2007), in which a small azido group is added to the molecule of interest. After the probe irreversibly attaches to its targets inside cells, cell extracts are prepared and treated with azido-reactive biotinylated phosphane. The phosphane reacts selectively with azide in a Staudinger-Bertozzi ligation, resulting in the biotinylation of the inhibitor targets. Biotinylated polypeptides can be then visualized on Western blots or isolated by affinity chromatography and identified by mass-spectrometry. The advantage of this two-step strategy is that an azide normally does not alter cell permeability or active-site specificity of the compound. Contrary to this, direct synthetic incorporation of biotin or a fluorescent moiety, which is required for a one-step labeling procedure, often alters active site specificity and decreases cell permeability.
When phosphane-treated extracts of az-NC-005-treated RPMI-8226 cells were separated on SDS-PAGE, a well-characterized pattern of bands of proteasomes active subunits (Altun et al., 2005) was detected. Labeling was concentration-dependent, with the β5/β5i band appearing first, followed by β2i, β2, β1, and β1i bands. Other bands were background bands from non-specific reaction of phosphane with cellular proteins and a band of endogeneously biotinylated protein of ~70 kDa (which can serve as a loading control). Appearance of proteasome subunit bands generally correlated with the inhibition of the corresponding activities (Fig. 3A). One notable exception was that the β2i band was detected on the gel at 0.3 µM az-NC-005, when Tr-L activity was still at 100%. However, it should be noted that maximal Tr-L activity in this experiment is 140%. Thus, az-NC-005 and, presumably, NC-005 itself do not have an off-target effect due to irreversible modification of non-proteasomal targets.
Figure 3. NC-005 derived active site probe and its inactive diastereomer demonstrate that NC-005 is proteasome specific.
A RPMI-8226 cells were treated with az-NC-005 active site probe. Activities were measured (values are averages ± S.E. of two experiments) and, in parallel, extracts of these cells were treated with biotinylated phosphane (Fig. 4G) and analyzed by Western blotting. Az-NC-005 reacting polypeptides were visualized by staining with fluorescently labeled streptavidin and identified based on the well-characterized pattern of migration of proteasome’s catalytic subunits (Altun et al., 2005). Endogeneously biotinylated protein of ~70 kDa served as a loading control. B. RPMI-8226 cells were treated with 1.6 µM NC-005 and (S)-NC-005 for 6 h. Proteasome activities were measured immediately after the treatment using peptides substrates (Table); cell viability was measured after subsequent 42 h cultivation in inhibitor-free media.
These experiments with az-NC-005 did not exclude the possibility that some of NC-005’s effects are due to non-covalent binding to other cellular proteins. In order to rule out this possibility, we inverted the stereochemistry of the Cα atom of the epoxy-ring from (R) to (S) configuration, producing a compound with dramatically reduced inhibitor potency (Fig. 3B). When used at the same concentration as the concentration of NC-005 that caused 93% reduction of cell viability, (S)-NC-005 was not cytotoxic to RPMI-8226 cells (Fig. 3B). Thus, cytotoxicity of NC-005 is dependent on its ability to covalently modify proteasome active sites. This lack of off-target effects of NC-005 supports the view that a need to co-inhibit Tr-L sites is a major reason for the lack of correlation between inhibition of Chym-L sites and cytotoxicity.
Development of potent, specific, cell-permeable inhibitor and active site probe of caspase-like sites
That inhibition of Chym-L sites is often insufficient to achieve maximal cytotoxicity suggests that inhibitors of Casp-L and Tr-L sites should enhance the cytotoxic effect of the inhibitor of Chym-L sites. Building on our expertise in the development of highly specific peptide aldehydes and peptide vinyl sulfone inhibitors of Casp-L (β1i and β1) sites (Kisselev et al., 2003; van Swieten et al., 2007), we have synthesized an epoxyketone analogue of this compound, Ac-APnLL-ek, which we designate NC-001 (Fig. 4A).
Figure 4. NC-001 is a highly specific, cell-permeable inhibitor of Casp-L sites.
A Structures of NC-001, NC-001- derived probe and it’s inactive diastereomers. B– D. Casp-L (B) and Chym-L and Tr-L activities (C) of the proteasome during continuous treatment of cells with NC-001. Panel D shows all three activities 6 hours after treatment. E. Proteasome peptidase activities in cells treated with az-NC-001 for 6 h. F. Casp-L activity of the proteasome in cells treated for 6 h with az-NC-005 diastereomers. All activity measurements were performed in RPMI-8226 cells and are averages ± S.E. of two independent experiments, in which mock-treated cells served as control. G. Extracts of these cells (i.e., from experiment shown in E and F) were treated with biotinylated phosphane (BioP), separated on 12% gel, transferred on PVDF membrane and probed with fluorescently labeled streptavidin. Polypeptides modified by the probes were identified by incubating the membrane with fluorescently labeled streptavidin. Extracts treated with biotinylated non-site-specific inhibitor Ada-Bio-Ahx3-Leu3-VS were used to identify the migration pattern of proteasome bands (Altun et al., 2005). Endogeneously biotinylated protein of ~70 kDa can be used as a loading control. H. Isolation of az-NC-001-modified subunits by Streptavidin affinity chromatography. Extracts of cells treated overnight with 5 µM az-NC-001 or mock-treated were treated with BioP, passed through a Streptavidin-Sepharose HP column (to remove excess BioP), treated with SDS (to dissociate proteasome subunits), and re-incubated with Sterptavidin-Sepharose. After extensive washing of the beads, specifically bound proteins were eluted with 1 mM biotin in LDS gel loading buffer at 95°C, fractionated on SDS-PAGE, and analyzed by Western blotting with antibodies to proteasome subunits indicated. Percentage of total sample loaded on the gels are indicated. Fig. S2 demonstrates that modification with az-NC-001 and BioP results in the mobility shift of the β1 subunit.
Treatment of cells with NC-001 results in a specific, time- and concentration-dependent inhibition of β1 sites (Fig. 4B). Maximal (close to 100%) inhibition was achieved upon 5 h treatment with 2 µM inhibitor. The IC50 of the inhibitor after 6 h treatment was ~0.5 µM. Longer treatment (up to 24 h) with NC-001 slightly improved inhibition at lower concentration (Fig. 4B) without any loss of specificity, even at 4 µM (Fig. 4C). Thus, NC-001 is a potent, cell-permeable and highly specific inhibitor of Casp-L sites. NC-001 specifically inhibited Casp-L sites in all cell lines tested (data not shown).
To confirm that NC-001 does not have any off-target effects and to compare it’s specificity towards Casp-L sites of constitutive and immunoproteasomes (β1 and β1i sites), we converted it into the active-site probe and synthesized its inactive analogs. Using the same strategy as for synthesis of NC-005 derivatives, we have generated an NC-001 derivative carrying an azido-group (az-NC-001) and an az-(S)-NC-001 diastereomer with the inverted configuration of the Cα atom of the epoxygroup (Fig. 4A). In addition, we have purified and isolated az-D-NC-001, a compound with D-Nle in the P2 position, which is generated as a by-product at the last step of the synthesis.
Az-NC-001 specifically inhibited Casp-L sites in RPMI-8226 cells (Fig. 4D, E). Treatment of extracts of az-NC-001–treated cells with biotinylated phosphane revealed dose-dependent labeling of β1 and β1i subunits (Fig. 4G). We could not detect any other modified polypeptide. Proteasome-specific labeling was substantially reduced in az-(S)-NC-001 and az-D-NC-001 (Fig. 4G), which were also much less potent in inhibiting Casp-L activity (Fig. 4F). In order to confirm that all signal in the β1 and β1i bands indeed comes from β1 and β1i subunits and not from non-resolved β5 and β5i subunits, we denatured proteasomes in extracts of cells treated with high concentrations (5 µM) of az-NC-001and isolated individual subunits on Streptavidin-Sepharose beads (Fig. 4H). β1 and β1i subunit were abundantly detected in the eluates; no β5 and only trace amounts β5i were detected eluted from these columns. This analysis also revealed that β1 and β1i band shifts upward slightly upon modification of the probe (see also Fig. S2). Thus, az-NC-001 is a specific probe for Casp-L sites of constitutive proteasomes and immunoproteasomes.
NC-001 sensitizes cells to NC-005
Treatment of cells with NC-001 alone did not cause any growth inhibition or cytotoxicity. This is an agreement with yeast data, where inactivation of this site by mutation caused no phenotypic defect (Arendt and Hochstrasser, 1997; Heinemeyer et al., 1997). We next set out to determine whether inhibiting Casp-L sites increases the cytotoxic effects of Chym-L sites inhibitors. In the initial experiment, we treated RPMI-8226 cell lines with different concentrations of NC-005 for 1 h and then with different concentrations of NC-001 for 48 h, whereupon cell viability was measured with the Alamar Blue mitochondrial dye conversion assay (Fig. 5A). High concentration of NC-001 (1–4 µM) sensitized cells to NC-005 leading to up to 5-fold decrease in IC50 (Fig. 5A). These concentrations inhibit Casp-L sites by more than 90% (Fig. 4D). Lower concentration of NC-001 (<0.5 µM), which caused less than 80% inhibition of Casp-L sites, did not sensitize RPMI-8226 cells to NC-005. Inactive NC-001 analogue, az-(S)-NC-001, did not sensitize RPMI-8226 cells to NC-005 (Fig. 5B). Thus, sensitization of cells to inhibitors of Chym-L sites is due to the inhibition of Casp-L sites.
Figure 5. NC-001 sensitizes cells to NC-005.
A, B RPMI-8226 cells were treated with NC-005 for 1 h and then split into two equal portions. One was treated with NC-001 (at concentrations indicated, -----A), or az-NC-001, or az-(S)-NC-001 (B) and the other was mock-treated for 48 h, when cell viability was measured with Alamar Blue. Values are averages ± S.E. of two independent experiments. C. NC-001 reduces IC50 of NC-005 (i.e. concentration at which cell viability is reduced by ½ compared to mock-treated cells) in multiple cell lines. IC50 for each individual experiment (performed as in A) were determined by a four-parameter fit of dose-response curves (Figs. S3A (RPMI-8226 cells), 5D (MM1.R cells), and S3B (all other cell lines)). Values in the table are averages (confidence intervals). Results of the t-test are provided in the last column (p-value, n-number of times experiment was repeated). NC-001 was used at 2 µM except in Dox 6 cells where it was at 10 µM. D, E. NC-001 (2 µM) sensitizes MM1.R cells to NC-005 by reducing IC50 (D) and accelerating cell death. Experiment in (E) was performed as in (A), except that at different time points aliquots of cells were collected and analyzed for apoptosis using Annexin V staining (E). Fig. S3A demonstrates that sensitization of cells to NC-005 by NC-001 does not depend on the order of treatment with these reagents. Fig. S3B demonstrates sensitization of other myeloma cells lines to NC-005 by NC-001.
We then tested whether sensitization is affected by the order of inhibitors in treatment (Fig. S2). In the first experiment, cells were treated with NC-005 for 1 h and then by 2 µM NC-001 for 48 h. In the second experiment, cells were co-treated with NC-005 and 18 µM NC-001 for 1 h. (1 h incubation with 18 µM NC-001 caused similar inhibition of Casp-L sites as 6–24 h treatment with 2 µM compound). In the third experiment, RPMI-8226 cells were pretreated with 2 µM NC-001 for 6 h, then treated with NC-005 for 1 h. Similar sensitization was observed under these different conditions. We then decided to use 1 h-treatment with NC-005 followed by continuous treatment with NC-001, as this allowed a simpler experimental set-up than 1 h co-treatment or pre-treatment with NC-001 and allowed us to keep NC-001 concentrations as low as possible. Duration of NC-005 was limited to 1 h for the same reasons as in initial experiments (Fig. 2).
We then tested the effect of NC-005 and NC-001 on other multiple myeloma cell lines (Fig. S3B, 5C and Table 1). In these experiments, we used only one concentration of NC-001, namely which caused 90–99% inhibition of Casp-L activity (10 µM in Dox-6 cells, 2 µM in all others). NC-001 sensitized other multiple myeloma cell lines to NC-005, causing a 2–3.5-fold decrease in IC50 (Fig. 5C). This group contained the majority of cell lines where inhibition of Chym-L sites was insufficient to achieve maximal cytotoxicity (Table 1) but also included the MM1.R (Fig. 5D) and NCI-H929 and cell lines, where inhibition of Chym-L sites alone was strongly cytotoxic (Table 1). NC-001 also sensitized MDA-MB-231 breast cancer cell lines and peripheral blood mononuclear cells (PBMNCs, Fig. S3B). Although this sensitization effect was not observed at high and low concentrations of NC-005, at concentrations close to IC50 for viability, NC-001 caused a 30–50% decrease in cell viability in all cell lines (Table 1). Thus, the inhibitor of Casp-L sites NC-001 sensitizes cells to inhibitors of Chym-L sites.
In addition to the end-point increase of cytotoxicity, NC-001 increased the rate of NC-005– induced cell death. For example, in MM1.R cells treated with 30 nM NC-005, the rate of apoptosis was doubled in the presence of NC-001 (Fig. 5E). Similar results were obtained in RPMI-8226 cells (data not shown). Thus, the inhibition of Casp-L sites not only increases the number of cells that undergo cell death in response to the inhibition of Chym-L sites but also enhances the rate of this process.
Is sensitization of cells to the inhibitor of Chym-L sites by NC-001 clinically significant? To be so, sensitization should be observed upon inhibition of β5 sites at levels that are clinically achievable. At the maximal tolerated dose, bortezomib causes 80% inhibition of Chym-L activity of the proteasome in blood (Hamilton et al., 2005); newer agents can achieve 90% inhibition (Demo et al., 2007). We have used measurements of inhibition of all three active sites in NC-005–treated cells (Table 1) to determine the extent of Chym-L sites inhibition needed for sensitization by NC-001. In NCI-H929 and MM1.R cells, sensitization was observed upon 40– 60% inhibition of Chym-L activity, and thus is clinically relevant (Table 1). In other myeloma cells, maximal sensitization occurred upon 90–99% inhibition of Chym-L sites (Table 1). This exceeds in vivo inhibition achievable by bortezomib, but can be achieved by three new agents, carfilzomib, salinosporamide A, and CEP-18770 (Chauhan et al., 2005; Demo et al., 2007; Piva et al., 2008), which are undergoing clinical trials. Thus, sensitization of MM1.R cells by NC-001 is of potential clinical significance.
Another interesting question is whether NC-001 treatment alters recovery of Chym-L and Tr-L activities in NC-005–treated cells. In MM1.R (Fig. 6A) and NCI-H929 cells (Fig. S4A), NC-001 treatment did not change inhibition of the chymotrypsin- and Tr-L sites. In RPMI-8226 (Fig. 6B) and Dox 6 (Fig. S4A) cells, NC-001 reduced the recovery of Chym-L activity. However, the effect was small during first 11 h and became significant only at 24 h, long after apoptosis has been triggered (Fig. 2G). It was most pronounced at 175 nM, was smaller at 520 nM, and at 1.6 µM there was no effect. Thus, it occurred only at concentrations that cause partial loss of viability (Fig. 5A), suggesting that recovery occurs only in the cells that do not undergo apoptosis; these still have functional protein biosynthesis machinery and can synthesize new proteasomes. NC-001 reduces this fraction and thus decreases recovery. NC-005–treated MM1.R and H929 cells die at faster rates (compare rate of RPMI-8226 cell death on Fig. 2G and of MM1.R cell death on Fig. 5E), and activity does not get a chance to recover.
Figure 6. Effect of NC-001 on recovery of proteasome activity in NC-005-treated cells.
A. MM1.R cells were treated with NC-001 in the presence or absence of 2 µM NC-001 for 1h, followed by incubation in media in the presence or absence of NC-001 respectively. At times indicated, some cells were harvested and proteasome activity was measured in extracts with fluorogenic peptide substrates and normalized per mg total protein (determined by Bradford). No inhibition of Tr-L activity was observed. B. RPMI-8226 cells were treated with NC-005 for 1h, followed by the NC-001 or mock-treatment. At times indicated proteasome activity was measured with ProteasomeGlo assay and normalized per number cell at time zero. Time-dependent decrease in Tr-L and Casp-L activity in RPMI-8226 cells treated with higher concentrations of NC-005 reflects time-dependent decrease in the number of cells due to inhibition of cell proliferation or cell death. Solid lines, no NC-001; dashed lines, with NC-001. Values are averages ± S.E. of two independent experiments. Fig. S4A demonstrates effects of NC-001 on recovery of proteasome activity in NC-005 treated Dox6 and KMS-18 cells. Fig. S4B demonstrates that ProteasomeGlo and fluorogenic substrate assays give identical results.
Discussion
Earlier studies have firmly established Chym-L sites of proteasomes as targets of anti-neoplastic agents (Adams, 2004). The Casp-L and Tr-L sites were not initially considered as such, but recent studies have suggested that the ability to co-target them can be important for the anti-neoplastic activity of proteasome inhibitors (Altun et al., 2005; Berkers et al., 2005; Chauhan et al., 2005) and for their ability to inhibit protein breakdown (Kisselev et al., 2006). Lack of highly specific, cell-permeable active site inhibitors has prevented investigators from directly testing this hypothesis. In this study, we describe the development of such inhibitors and provide direct evidence that Casp-L sites have to be considered co-targets of proteasome inhibitors alongside with Chym-L sites. These data also strongly suggest that co-targeting Tr-L sites could be at least as important as co-targeting Casp-L sites.
First, cytotoxicity of NC-005 to several multiple myeloma cell lines correlates poorly with the inhibition of Chym-L sites (Fig. 2F,H). Second, in the majority of cell lines tested, maximal cytotoxicity is achieved only when Tr-L sites are co-inhibited (Table 1). Third, the specific inhibitor of Casp-L sites, although non-cytotoxic to these cell lines when used as a single agent, sensitizes cells to NC-005 (Fig. 5, S3).
The conclusion that Chym-L sites are the primary targets of anti-neoplastic agents was based on earlier reports in which panels of different peptide boronates (Adams et al., 1999) or peptide epoxyketones (Elofsson et al., 1999) were tested for ability to inhibit cell growth. This ability correlated with their capacity to inhibit Chym-L sites in vitro assays of the purified proteasomes. The extent of inhibition of these sites inside cells and whether Casp-L and Tr-L site were also inhibited at cytotoxic and growth-inhibitory concentrations was not tested. These differences in the experimental design between those studies and our work are the most likely reasons for our differing conclusions. An opposite result, in that cell death can be achieved without inhibition of Chym-L sites, was reported by two studies claiming that inhibition of β1i sites (i.e. Casp-L sites of immunoproteasomes) is sufficient to induce apoptosis in cells that express high amounts of immunoproteasomes (Ho et al., 2007; Kuhn et al., 2008). This is in a disagreement with the present work, as we demonstrate that complete inhibition of both β1 and β1i sites did not cause any growth inhibition or cytotoxicity in any of the cell lines tested. Possible reasons for this difference are (a) that effects of β1i-specific inhibitors are cell-line specific or (b) that β1i inhibitors are not as specific as NC-001 and co-inhibit Chym-L activity (β5i) at cytotoxic concentrations.
What are the implications of these results for the development of therapeutic proteasome inhibitors? The first significant observation of this study is that at clinically achievable 70% inhibition of Chym-L sites (Hamilton et al., 2005), cytotoxicity is achieved only in a fraction of cell lines tested, and even in these, stronger inhibition is needed to achieve maximal cytotoxicity (Fig. 2F, Table 1). Similar results were observed with bortezomib (Fig. 2D and manuscript in preparation) and carfilzomib (Parlati et al., 2009). These results suggest that insufficient proteasome inhibition is the main reason why only a fraction of patients respond to bortezomib as a single agent (Richardson et al., 2003). Based on this data, we predict that the next generation of proteasome inhibitors (e.g., carfilzomib, NPI-0052, and CEP-18770) that can achieve stronger (90–95%) in vivo inhibition of Chym-L activity (Chauhan et al., 2005; Demo et al., 2007; Piva et al., 2008), will be more potent anti-neoplastic agents. Early results of phase II trials of carfilzomib confirm this prediction, as this agent is able to achieve responses in bortezomib-refractory myeloma patients (Jagannath et al., 2008).
Our data clearly indicate that co-inhibiting Chym-L and Casp-L sites should lead to a more potent anti-neoplastic agent and strongly suggests that co-inhibiting Tr-L sites should have similar effect. Of particular importance is that, in at least two cell lines, sensitization by NC-001 occurs upon clinically achievable (<70%) inhibition of Chym-L sites. In a number of other cell lines, stronger inhibition was needed to achieve this sensitization. Such stronger inhibition can be achieved in vivo by the second generation of proteasome inhibitors. However, if this leads to a higher toxicity to normal cells (as our data in PBMNCs indicates) the clinical usefulness of this phenomenon may be limited, and in vivo experiments are needed to address this issue. Poor solubility of NC-001 and NC-005 has prevented us from performing these experiments, which will be done after analogues of these compounds with improved pharmacological properties are developed.
Allosteric interactions between active sites observed in these and earlier studies (i.e., activation of Casp-L and Tr-L activities upon occupancy of Chym-L sites by inhibitors, activation of Tr-L sites by inhibitors of Casp-L sites, cooperativity between Chym-L sites (Kisselev et al., 1999)) may further contribute to inhibitor resistance, as inactivation of one site by an inhibitors would automatically lead to other sites cleaving proteins at faster rates. Thus, allostericity may also be one of the mechanisms behind sensitizing effects, as inhibiting of the second site may take away the benefits of the allosteric activation upon inhibition of the first site.
The importance of our work goes beyond treating cancer. The proteasome is involved in the regulation of a variety of cellular processes, and this work will inevitably raise questions about the roles of individual active sites in these processes. The most interesting application of compounds developed in this study would be to investigate the role of individual active sites in antigen presentation (Groettrup and Schmidtke, 1999). The fact that all three types of site are different in immunoproteasomes strongly suggests that they are important in this process. Even if an active site (e.g., Casp-L) is of little importance to overall protein degradation, its activity may be critical for the precise excision of certain epitopes. Specific inhibitors of this site would block presentation of this epitope. Conversely, some epitopes may be destroyed by the specific action of an active site, and their presentation would be enhanced by its specific inhibitor. Site-specific inhibitors, active site probes, and their inactive analogues developed in these studies, and specific cell-permeable inhibitors of Tr-L sites we are currently developing, will allow us to test this hypothesis in future work.
Significance
This study provides the first direct evidence that Casp-L proteasome sites must be considered co-targets of anti-neoplastic drugs (along with Chym-L sites) and strongly suggest that co-targeting of Tr-L sites would increase anti-neoplastic activity of proteasome inhibitors. The highly-specific, potent, and cell-permeable inhibitors of Casp-L and Chym-L sites described here would be excellent tools to study the role of these active sites in a variety of biological processes (e.g., antigen presentation). Used together, these inhibitors would enable study of the effect of combined inhibition in which inhibition of each site is varied to any desired extent (e.g., 50% inhibition of Chym-L sites and 95% inhibition of Casp-L sites), which cannot be achieved by any of the currently available compounds. The active-site probes and inactive analogues described here would provide a unique capability to infer that observed biological effects are indeed due to inhibition of the proteasome’s active sites.
Experimental Procedures
Materials
26S proteasome was purified from rabbit muscle as described (Kisselev et al., 2002), except that 1 h centrifugation of extract at 100,000 g was replaced by a 30 min centrifugation at 40,000g. YU-101 was purchased from Calbiochem, bortezomib was purchased from DHMC pharmacy. MV-151, AdaBio-Axh3-Leu3-VS and biotinylated phosphane were synthesized as described (van Swieten et al., 2007). Synthesis of NC-001, NC-005, az-NC-001, az-D-NC-001, and az-(S)-NC-001, az-NC-005, and (S)-NC-005 is described in the Supplementary Material section, which also contains analytical data for these inhibitors. Stock solutions of inhibitors were prepared in DMSO and their concentrations were determined by amino acid analysis.
Measurements of proteasome activity in cells extracts were performed as described by (Kisselev and Goldberg, 2005). Suc-LLVY-7-amido-4-methylcoumarine (amc, Bachem), Ac-nLPnLD-amc, and Ac-RQR-amc (both custom synthesized by MP Biomedicals) were used for the measurements of Chym-L, Casp-L, and Tr-L activities respectively. Proteasome activity in cells was measured using Promega ProteasomeGlo™ Cell-Based Assay (Promega) (Moravec et al., 2009). Results of cell- and extracts-based assays were indistinguishable (Fig. S4B). See Supplementary materials for details of both procedures.
Experimental Procedures section of the Supplement contains detailed information on following procedures: visualization of polypeptides modified by the subunit-specific active site probes in a 2-step procedure, isolation of modified subunits on Streptavidin Sepharose, apoptosis and cell viability assay.
Supplementary Material
Acknowledgements
We are grateful to Hans van der Elst (Leiden Institute of Chemistry) for the high-resolution mass-spectrometry analysis; to Cassie Nghiem (Dartmouth College) for help with solid phase peptide synthesis; to Elizabeth McCoy and Laurent Perreard (the Kisselev laboratory) for assistance with Streptavidin pull-down experiments; to Jaqueline Channon-Smith and John DeLeong (Dartmouth Immune Monitoring Laboratory) for providing PBMNCs; to Wayne Casey (Department of Chemistry, Dartmouth College) for help with NMR analysis; and to Alan Eastman (Norris Cotton Cancer Center) for advice on cell viability and apoptosis assays. These studies were supported by the Research Scholar Grant and the Institutional Research Grant from the American Cancer Society, by grants from NCI (5RO1-CA124634) and International Myeloma Foundation to AFK, by the Prouty Pilot Grant from the Friends of the Norris Cotton Cancer Center to AFK and DLW, by a grant from the American Cancer Society to DLW, and by grants from the Netherlands Organization for Scientific Research (NWO) and the Netherlands Genomics Initiative (NGI) to HSO.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- Adams J. The proteasome: a suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, Maas J, Pien CS, Prakash S, Elliott PJ. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 1999;59:2615–2622. [PubMed] [Google Scholar]
- Altun M, Galardy PJ, Shringarpure R, Hideshima T, LeBlanc R, Anderson KC, Ploegh HL, Kessler BM. Effects of PS-341 on the activity and composition of proteasomes in multiple myeloma cells. Cancer Res. 2005;65:7896–7901. doi: 10.1158/0008-5472.CAN-05-0506. [DOI] [PubMed] [Google Scholar]
- Arendt CS, Hochstrasser M. Identification of the yeast 20S proteasome catalytic centers and subunit interactions required for active-site formation. Proc Natl Acad Sci. 1997;94:7156–7161. doi: 10.1073/pnas.94.14.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkers CR, Verdoes M, Lichtman E, Fiebiger E, Kessler BM, Anderson KC, Ploegh HL, Ovaa H, Galardy PJ. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nat Methods. 2005;2:357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- Cascio P, Hilton C, Kisselev A, Rock K, Goldberg A. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D, Catley L, Li G, Podar K, Hideshima T, Velankar M, Mitsiades C, Mitsiades N, Yasui H, Letai A, et al. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Can Cell. 2005;8:407–419. doi: 10.1016/j.ccr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- Demo SD, Kirk CJ, Aujay MA, Buchholz TJ, Dajee M, Ho MN, Jiang J, Laidig GJ, Lewis ER, Parlati F, et al. Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Res. 2007;67:6383–6391. doi: 10.1158/0008-5472.CAN-06-4086. [DOI] [PubMed] [Google Scholar]
- Dick TP, Nussbaum AK, Deeg M, Heinemeyer W, Groll M, Schirle M, Keilholz W, Stevanovic S, Wolf DH, Huber R, et al. Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J Biol Chem. 1998;273:25637–25646. doi: 10.1074/jbc.273.40.25637. [DOI] [PubMed] [Google Scholar]
- Elofsson M, Splittgerber U, Myung J, Mohan R, Crews CM. Towards subunit-specific proteasome inhibitors: synthesis and evaluation of peptide alpha ',beta '-epoxyketones. Chem Biol. 1999;6:811–822. doi: 10.1016/s1074-5521(99)80128-8. [DOI] [PubMed] [Google Scholar]
- Groettrup M, Schmidtke G. Selective proteasome inhibitors: modulators of antigen presentation? Drug Discov Today. 1999;4:63–71. doi: 10.1016/s1359-6446(98)01292-6. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik H, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Groll M, Heinemeyer W, Jager S, Ullrich T, Bochtler M, Wolf DH, Huber R. The catalytic sites of 20S proteasomes and their role in subunit maturation: a mutational and crystallographic study. Proc Natl Acad Sci. 1999;96:10976–10983. doi: 10.1073/pnas.96.20.10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AL, Eder JP, Pavlick AC, Clark JW, Liebes L, Garcia-Carbonero R, Chachoua A, Ryan DP, Soma V, Farrell K, et al. Proteasome inhibition with bortezomib (PS-341): a phase I study with pharmacodynamic end points using a day 1 and day 4 schedule in a 14-day cycle. J Clin Oncol. 2005;23:6107–6116. doi: 10.1200/JCO.2005.01.136. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J Biol Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliot PJ, Adams J, Anderson KC. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–3076. [PubMed] [Google Scholar]
- Ho YK, Bargagna-Mohan P, Wehenkel M, Mohan R, Kim KB. LMP2-specific inhibitors: chemical genetic tools for proteasome biology. Chem Biol. 2007;14:419–430. doi: 10.1016/j.chembiol.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath S, Vij R, Stewart AK, Somlo G, Jakubowiak A, Reiman T, Trudel S, Taylor J, Fuhrman D, Cruickshank S, et al. Initial Results of PX-171-003, An Open-Label, Single-Arm, Phase II Studyof Carfilzomib (CFZ) in Patients with Relapsed and Refractory Multiple Myeloma (MM) Blood. 2008;112:A. 864. [Google Scholar]
- Kisselev AF, Akopian TN, Castillo V, Goldberg AL. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol Cell. 1999;4:395–402. doi: 10.1016/s1097-2765(00)80341-x. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Callard A, Goldberg AL. Importance of different active sites in protein breakdown by 26S proteasomes and the efficacy of proteasome inhibitors varies with the protein substrate. J Biol Chem. 2006;281:8583–8590. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Garcia-Calvo M, Overkleeft HS, Peterson E, Pennington MW, Ploegh HL, Thornberry NA, Goldberg AL. The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J Biol Chem. 2003;278:35869–35877. doi: 10.1074/jbc.M303725200. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Measuring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods in Enzymology. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Kaganovich D, Goldberg AL. Binding of hydrophobic peptides to several non-catalytic sites promotes peptide hydrolysis by all active sites of 20S proteasomes. Evidence for peptide-induced channel opening in the alpha-rings. J Biol Chem. 2002;277:22260–22270. doi: 10.1074/jbc.M112360200. [DOI] [PubMed] [Google Scholar]
- Kuhn DJ, Hunsucker SA, Chen Q, Voorhees PM, Orlowski M, Orlowski RZ. Targeted inhibition of the immunoproteasome is a potent strategy against models of multiple myeloma that overcomes resistance to conventional drugs and non-specific proteasome inhibitors. Blood. 2008 doi: 10.1182/blood-2008-07-171637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose I, Umezawa Y, Hirosawa S, Iinuma H, Ikeda D. Structure-based design of derivatives of tyropeptin A as the potent and selective inhibitors of mammalian 20S proteasome. Bioorg Med Chem Lett. 2005;15:1867–1871. doi: 10.1016/j.bmcl.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Moravec RA, O'Brien MA, Daily WJ, Scurria MA, Bernad L, Riss TL. Cell-based bioluminescent assays for all three proteasome activities in a homogeneous format. Anal Biochem. 2009;387:294–302. doi: 10.1016/j.ab.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Oberdorf J, Carlson EJ, Skach WR. Redundancy of mammalian proteasome beta subunit function during endoplasmic reticulum associated degradation. Biochemistry. 2001;40:13397–13405. doi: 10.1021/bi011322y. [DOI] [PubMed] [Google Scholar]
- Parlati F, Lee SJ, Aujay M, Suzuki E, Levitsky K, Lorens JB, Micklem DR, Ruurs P, Sylvain C, Lu Y, et al. Carfilzomib can induce tumor cell death through selective inhibition of the chymotrypsin-like activity of the proteasome. Blood. 2009;114:3439–3447. doi: 10.1182/blood-2009-05-223677. [DOI] [PubMed] [Google Scholar]
- Piva R, Ruggeri B, Williams M, Costa G, Tamagno I, Ferrero D, Giai V, Coscia M, Peola S, Massaia M, et al. CEP-18770: A novel, orally active proteasome inhibitor with a tumor-selective pharmacologic profile competitive with bortezomib. Blood. 2008;111:2765–2775. doi: 10.1182/blood-2007-07-100651. [DOI] [PubMed] [Google Scholar]
- Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, Rajkumar SV, Srkalovic G, Alsina M, Alexanian R, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348:2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- van Swieten PF, Samuel E, van Nieuwendijk AMCH, Leeuwenburgh MA, van der Marel GA, Kessler BM, Overkleeft HS, Kisselev AF. A cell-permeable inhibitor and activity-based probe for the caspase-like activity of the proteasome. Bioorg Med Chem Lett. 2007;17:3402–3405. doi: 10.1016/j.bmcl.2007.03.092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.